Abstract
Presenilin 1 (PS1) is required for the proteolytic processing of Notch and the β-amyloid precursor protein (APP), molecules that play pivotal roles in cell-fate determination during development and Alzheimer's disease pathogenesis, respectively. In addition, PS1 interacts with β-catenin and promotes its turnover through independent mechanisms. Consistent with this activity, we report here that PS1 is important in controlling epidermal cell proliferation in vivo. PS1 knockout mice that are rescued through neuronal expression of human PS1 transgene develop spontaneous skin cancers. PS1-null keratinocytes exhibit higher cytosolic β-catenin and β-catenin/lymphoid enhancer factor-1/T cell factor (β-catenin/LEF)-mediated signaling. This effect can be reversed by reintroducing wild-type PS1, but not a PS1 mutant active in Notch processing but defective in β-catenin binding. Nuclear β-catenin protein can be detected in tumors. Elevated β-catenin/LEF signaling is correlated with activation of its downstream target cyclin D1 and accelerated entry from G1 into S phase of the cell cycle. This report demonstrates a function of PS1 in adult tissues, and our analysis suggests that deregulation of β-catenin pathway contributes to the skin tumor phenotype.
Mutations in presenilin 1 (PS1) are linked to early onset of familial Alzheimer's disease (FAD) (1). The pathogenic mechanisms of these mutations are likely mediated by their enhanced activities in the proteolytic cleavage of the β-amyloid precursor protein (APP) to generate the longer form of β-amyloid peptide, Aβ42 (2). PS1 is a ubiquitously expressed multipass transmembrane protein. Although localized mainly to the endoplasmic reticulum and Golgi membranes, it has also been detected on nuclear envelopes and plasma membrane, where it complexes with cadherin/catenin molecules (3–5). Full-length PS1 undergoes proteolytic processing to generate an amino-terminal and a carboxyl-terminal fragment of 28 and 18 kDa, respectively, that stay as a stable and biologically active complex (6).
Gene knockout studies revealed that PS1 is required for the proteolytic processing of two distinct molecules, Notch and APP (7–9). PS1 null mice die perinatally as a result of impaired Notch pathway, and this activity is highly conserved (8, 10–13). The lethal phenotype of the PS1-deficient mice prevents the postnatal evaluation of any additional physiological roles of PS1 whose expression persists throughout adulthood. We reported earlier that transgenic mice expressing either the wild-type (WT) human PS1 (hPS1) or hPS1 containing the A246E FAD mutation, under the control of neuronal-specific human Thy-1 promoter, could rescue the PS1 null mice from embryonic lethality (14). These mPS1−/−, Thy-hPS1 “rescue” (abbreviated herein as hPS1 rescue) mice thus provide a unique model of PS1 deficiency in most adult peripheral tissues.
In addition to its role in Notch and APP processing, PS1 has been shown to interact with β-catenin (15–17), an Armadillo (arm) repeats-containing protein involved in cell adhesion and Wingless/Wnt signaling (18). The Wnt signaling pathway is tightly controlled by, among others, cytosolic β-catenin levels. In the absence of Wnt ligands, cytosolic β-catenin is rapidly phosphorylated by glycogen synthase kinase-3β and degraded through the proteasome-mediated pathway (19). Binding of Wnt ligands to cell surface receptors leads to the inactivation of glycogen synthase kinase-3β and accumulation of cytosolic β-catenin, which translocates to the nucleus. In nucleus, β-catenin binds to the lymphoid enhancer factor-1/T-cell factor (LEF/TCF, abbreviated herein as LEF) family of transcription factors and activates its downstream target genes (20), among which are c-myc and cyclin D1 (21–23). Deregulation of β-catenin signaling either by mutations in β-catenin or its modulators such as axin and APC is associated with various malignancies, both in humans and transgenic mice (24–27).
The interaction of PS1 with β-catenin has been implicated in regulating β-catenin stability and its downstream signaling (16, 28, 29). Recently, we generated key data to show that PS1 normally acts as a negative regulator of β-catenin and that this activity is independent from its mechanism in Notch processing (30). Here, we report a novel function of PS1 in controlling cell proliferation and skin tumorigenesis and provide data to suggest that activated β-catenin/LEF signaling contribute to the phenotype.
Materials and Methods
Cell Culture.
For preparation of primary keratinocytes, skin tissue samples were taken from 1-day-old pups immediately after euthanasia. After washing in calcium-free PBS, each skin was floated in 2 ml of 0.25% trypsin at 4°C for 15 h. The epidermis was then peeled off from the skin and placed in a collagen-coated dish containing complete keratinocyte medium (GIBCO). The cells were then released mechanically by gentle stirring, and cells of the same genotype were mixed and plated (106 for each 35-mm dish). Extraction and immunoblotting of PS1 and soluble β-catenin was performed as described (30).
Luciferase Reporter Assays.
The widely used TOP-flash reporter gene construct was used to assay β-catenin-mediated downstream signaling. It contains four consensus LEF binding sites, a minimal Fos promoter, and luciferase reporter (ref. 34, gift of M. van de Wetering, Univesiteit Utrecht, The Netherlands). The FOP-flash vector with mutated LEF sites was used as negative control. WT and hPS1 rescue primary keratinocytes were plated onto 6-well plates. Transfections were performed on day 3 when cells reached 80% confluence. The cells were transfected by Lipofectamine 2000 reagent (GIBCO) using 2 μg of the TOP-flash reporter or the FOP-flash control construct according to the manufacturer's recommended conditions. To suppress β-catenin/LEF signaling effect, 2 μg of empty vector, cytomegalovirus (CMV)-hPS1 or CMV-PS1Δcat DNA, was cotransfected in hPS1 rescue cells. A plasmid expressing enhanced green fluorescent protein was introduced in each case, and transfection efficiency was normalized to green fluorescent protein-positive cells. Transfected cells were harvested 24 h later. Reporter assays were performed by using the luciferase reporter system (Promega). Results are expressed as mean ± SD of three independent experiments performed in triplicates.
Fluorescence-Activated Cell Sorting (FACS).
WT and hPS1 rescue primary keratinocytes were cultured for 3 days. The cells were then washed, trypsinized, spun down, and fixed in 1% paraformaldehyde at a final concentration of 106 cells/ml followed by centrifugation. The cells were stained with propidium iodide (Sigma) and counted on a FACSCalibur cell sorter using CELLQUEST software (Becton Dickinson). The percentages of cells in the G1, S, and G2/M phases of the cell cycle were determined.
Histology, Immunofluorescence Staining, and Microscopy.
Tissues for histology were fixed in 10% neutral buffered formalin, embedded in paraffin blocks, cut into 5-μm sections, and stained with hematoxylin/eosin. Slides were examined microscopically.
For immunofluorescence staining of PS1, fresh newborn skin tissues were fixed. The sections were pretreated with a microwave oven for antigen recovery and incubated with a blocking solution containing 0.2% goat serum. A polyclonal antibody against PS1 (PS1-NT) was applied followed by incubation with an FITC-anti-rabbit IgG (Sigma) secondary antibody.
Confocal images were captured on an Axiovert 100 microscope using LSM software (Zeiss). The primary antibody was mouse anti-β-catenin (Sigma). The secondary antibody was Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes). The sections were counterstained with 0.5 μg/ml propidium iodide (Sigma) in PBS to visualize cell nuclei.
BrdUrd Labeling.
For BrdUrd labeling in live animals, 1-day-old hPS1 rescue and control littermates were injected (i.p.) with BrdUrd (Roche Molecular Biochemicals) resuspended in 0.9% NaCl at 100 mg/kg. The mice were killed after 1 h. Skin sections were fixed, processed, and stained with FITC-conjugated mAb to BrdUrd (Becton Dickinson). The labeling index was expressed as the mean of BrdUrd-positive cells/mm epidermis ± SD.
Results
Epidermal Hyperplasia and Tumorigenesis in hPS1 Rescue Mice.
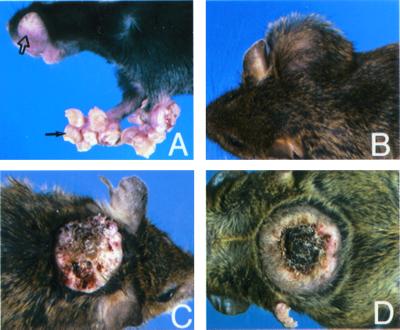
We have previously described the generation of viable hPS1 rescue mice expressing either WT or A246E FAD mutant hPS1 on mPS1 null background (14). Although these hPS1 rescue mice were generally healthy and exhibited normal lifespan, as the animals aged, skin abnormalities were readily observed in multiple lines of both WT and A246E hPS1 rescue mice (three lines each). The most common phenotypes were excessive growth of the perioral skin, paws, and feet and the formation of large cutaneous masses indicative of malignancy (Fig. 1). These skin lesions could be identified as early as 3 months of age and were obvious in 90% of the animals 9 months and older.
Figure 1.
Representative skin lesions in hPS1 rescue mice. (A) Formation of cystic dermal nodules in perioral skin and front paws (open and solid arrows). (B–D) Non-ulcerated (B) and ulcerated (C and D) cutaneous masses present in hPS1 rescue mice.
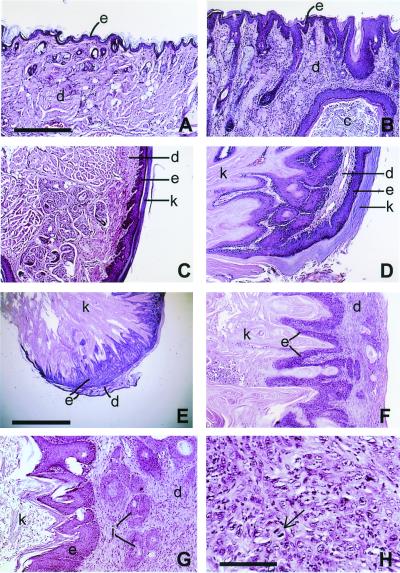
Although the skin lesions in hPS1 rescue mice occurred in various areas of the body with dramatic differences in their gross appearance, histological examination of the skin revealed that they shared common pathological features. These include epidermal hyperplasia and hyperkeratosis (Fig. 2 B and D), formation of epithelioid cysts (Fig. 2B), and neoplasm ranging from keratoacanthoma-like appearance (Fig. 2 D and E) to the more severe squamous cell carcinomas (Fig. 2 G and H) (31). Overall, 25 mice with skin tumors were diagnosed from a total of 45 mice analyzed (4–24 months with average age of 16 months). A summary of the histological analysis is shown in Table 1.
Figure 2.
Histopathology of hPS1 rescue mice. (A and C) Sections of perioral skin and paws of WT mice, respectively. (B and D) The same areas of hPS1 rescue mice, showing epidermal hyperplasia and hyperkeratosis. The epidermal layers (e, darker stain) were thickened and expanded with numerous projections of stratified squamous epithelium that were covered by massive amounts of dense, layered keratin (k in D). A large keratin-filled epidermal cyst (c) is embedded in dermal layer in B. D also shows that the two layers of hyperplastic epithelium were folded on one another, sharing a common dermal layer and resulting in a keratoacanthoma-like appearance. The same pathology is also detected in a cutaneous tumor (E and F, lower and higher power view, respectively). (G and H) The expanded dermis with infiltrating clusters of neoplastic squamous epithelial cells (i in G), which are characteristic of locally invasive squamous cell carcinoma. The arrow in H indicates a dividing neoplastic cell in a disorganized cluster of invading epithelial cells. e, epidermis; d, dermis; c, epidermal cyst; k, keratin; i, islands of infiltrating epidermal cells. A–D, F, and G are of the same magnification; scale bar in A = 275 μm. Scale bars in E and H represent 1.5 mm and 70 μm, respectively.
Table 1.
Summary of skin phenotype in hPS1 rescue mice
| Line | 10-8 | 17-3 | 16-4 |
|---|---|---|---|
| hPS1 genotype | WT | WT | A246E |
| No. with skin lesion/total* | 15/15 | 15/15 | 12/15 |
| No. with tumor/total† | 10/15 | 7/15 | 8/15 |
Mice from two WT (10-8 and 17-3) and one A246E FAD mutant (16-4) hPS1 rescue lines were subject to detailed analysis.
Number of mice that developed skin lesion/total number of mice evaluated.
Number of mice with tumor/number of mice analyzed.
The skin tumor phenotypes were specific to hPS1 rescue animals, as they were never seen in littermate controls containing endogenous mouse PS1. Absence of skin tumor in PS1 heterozygous mice indicates that 50% of the normal mPS1 level is sufficient at maintaining normal proliferative cycles in epidermis.
Lack of PS1 Expression in Skin Tissues of hPS1 Rescue Mice.
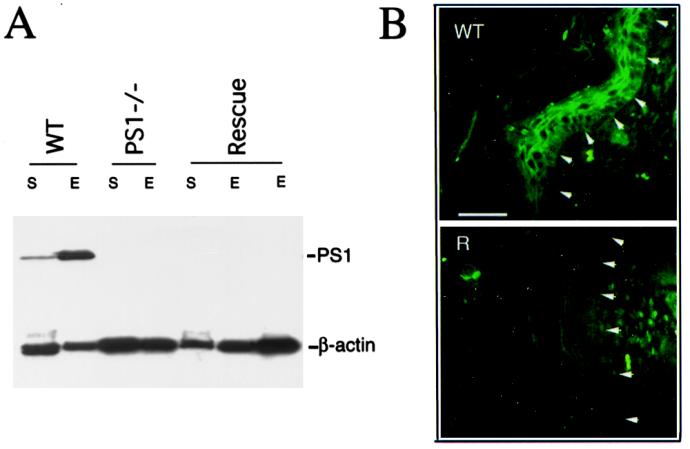
In view of the central nervous system-restricted expression pattern of the human Thy-1 promoter (32), we suspected that the phenotype of the hPS1 rescue mice was because of the PS1 deficiency in skin. We thus determined PS1 levels in both the whole skin (S) and the epidermis (E) of newborn hPS1 rescue mice (Fig. 3). Using the PS1NT antibody (33), we showed that PS1 protein was below the level of detection in hPS1 rescue neonates and, on Western blots, was indistinguishable from that of PS1−/− neonates (Fig. 3A). Endogenous mPS1 was readily detected in the same tissues from WT control animals (Fig. 3A). Absence of PS1 protein was also evident when adult skin of hPS1 rescue mice was analyzed (data not shown). To further define the expression pattern of PS1, we performed immunofluorescence staining of newborn skin using the PS1NT antibody (Fig. 3B). Consistent with the Western blot result, endogenous PS1 was predominantly expressed in the epidermis in WT mouse skin (Fig. 3B Upper). PS1 protein could be detected in both the cytoplasm and along the cell membrane in the basal cell layer of the epidermis (layer highlighted by arrows). In the more differentiated cells in the suprabasal layers, PS1 was concentrated at the cell periphery resembling the intercellular contacts described in other epithelial tissues (5). Staining for PS1 was absent in the skin of hPS1 rescue mice (Fig. 3B Lower).
Figure 3.
PS1 deficiency in newborn hPS1 rescue mouse skin. (A) Western blot analysis of PS1 expression. PS1 protein can be detected from both whole skin (S) and epidermis (E) of WT mouse but was found below the level of detection in either PS1 null (PS1−/−) or mPS−/−, hPS1 rescue (Rescue) newborns. PS1NT antibody was used, which detected a 28-kDa PS1-NTF fragment (top bands). β-Actin staining serves as loading control. (B) Immunofluorescence staining of newborn mouse skin using PS1NT antibody. In WT skin (Upper), PS1 expression is enriched in the epidermis (epidermis and dermis separated by arrows). PS1 protein can be detected in both the cytoplasm and plasma membrane in the basal cell layer of the epidermis (highlighted by arrows) but is mostly concentrated at the intercellular contacts toward more differentiated suprabasal layers (layers distal to arrows). No specific PS1 staining can be detected in hPS1 rescue skin (R, Lower). (Scale bar = 100 μm.)
Elevated Cytosolic β-Catenin and Its Downstream Signaling in the Absence of PS1.
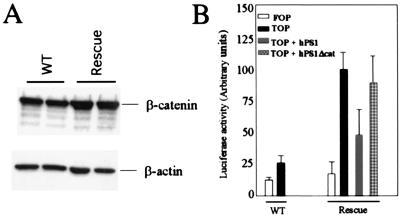
Abnormal stabilization and accumulation of β-catenin has been implicated in various neoplasms. As we have shown previously that PS1 is a negative regulator of β-catenin stability and signaling in cultured fibroblasts (30), we examined whether there was β-catenin up-regulation in cells of epidermal origin that lacked PS1 expression. Primary keratinocytes were isolated and cultured from WT and hPS1 rescue neonates on the day of birth, and soluble β-catenin protein was extracted 2 days later. The result showed that there were higher steady-state levels of cytosolic, hence signaling pool of β-catenin protein in hPS1 rescue keratinocytes as compared with the controls (Fig. 4A). Neither the total detergent-extractable β-catenin nor its distribution along the epidermal layers as judged by immunohistostaining using β-catenin-specific antibody was changed, suggesting that the membrane-associated β-catenin, which consists of the majority of β-catenin protein in cells, was not grossly altered (data not shown). To quantify β-catenin/LEF-mediated transcriptional activation, we transfected either WT or hPS1 rescue primary keratinocytes with the TOP-flash reporter containing the LEF binding motifs and measured luciferase activities (34). FOP-flash vector with mutated LEF sites was used as control. As shown in Fig. 4B, the luciferase activity was significantly higher in PS1-null keratinocytes as compared with control cells when both were transfected with the TOP-flash vector (filled bars in rescue vs. WT). Importantly, accelerated β-catenin/LEF-mediated transcriptional activation in PS1 null cells could be suppressed by reintroducing WT PS1 but not by PS1Δcat, a PS1 mutant deleting the essential β-catenin binding site but retaining full Notch activity (P < 0.001 between TOP + PS1 vs. TOP + PS1Δcat; P > 0.05 between TOP vs. TOP + PS1Δcat, Student's t test) (30, 33). These results demonstrate that the down-regulation of PS1 in β-catenin signaling is mediated through direct interaction of the two molecules and that abnormal β-catenin signaling in the absence of PS1 cannot be suppressed by restoring the PS1 Notch activity.
Figure 4.
Accumulation of soluble β-catenin and elevation of β-catenin/LEF signaling in the absence of PS1. (A) Higher levels of soluble β-catenin in hPS1 rescue primary keratinocytes (lanes labeled as Rescue) as compared with controls (WT). β-Actin staining was used as loading control. Shown are representative results from six independent experiments, and the average increase is ≈2-fold. (B) Effects of PS1 on β-catenin/LEF signaling. Primary keratinocytes isolated from WT or hPS1 rescue neonates were transfected with either the FOP-flash control construct containing the mutated LEF binding sites (FOP) or the TOP-flash reporter construct containing the LEF-responsive elements (TOP). In addition, the rescue culture was cotransfected with the TOP-flash vector plus wild-type hPS1 (TOP + hPS1) or the TOP-flash vector plus β-catenin binding-defective hPS1 (TOP + hPS1Δcat). The cells were assayed for luciferase activity 24 h posttransfection.
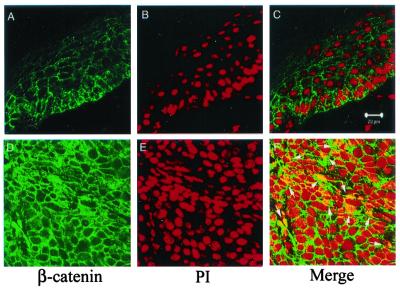
Accumulation of the signaling β-catenin was also evident by immunofluorescence staining and confocal imaging analysis of β-catenin protein in tumor samples (Fig. 5). In contrast to control epidermis, where β-catenin staining was mostly localized in the membrane (Fig. 5A), there was significant amount of cytosolic β-catenin accumulation in the tumor cells (Fig. 5D). Furthermore, nuclear β-catenin protein could be detected in some tumor cells (Fig. 5F).
Figure 5.
Nuclear localization of β-catenin in tumors. Immunostaining and confocal image analysis shows that in WT epidermis (Upper), β-catenin is predominately localized on the plasma membranes (A), and there is no overlapping staining between β-catenin (green) and nucleus (visualized by staining with propidium iodode, red) (C). In a representative tumor sample (Lower), there is a dramatic increase of β-catenin protein in the cytoplasm (D). In addition, nuclear β-catenin can be readily detected as shown by overlapping staining (highlighted by arrows) between β-catenin (green) and nucleus (red) (F). (Scale bar = 20 μM.)
Enhanced Cyclin D1 Protein Levels and Accelerated G1 into S Phase Transition in the Absence of PS1.
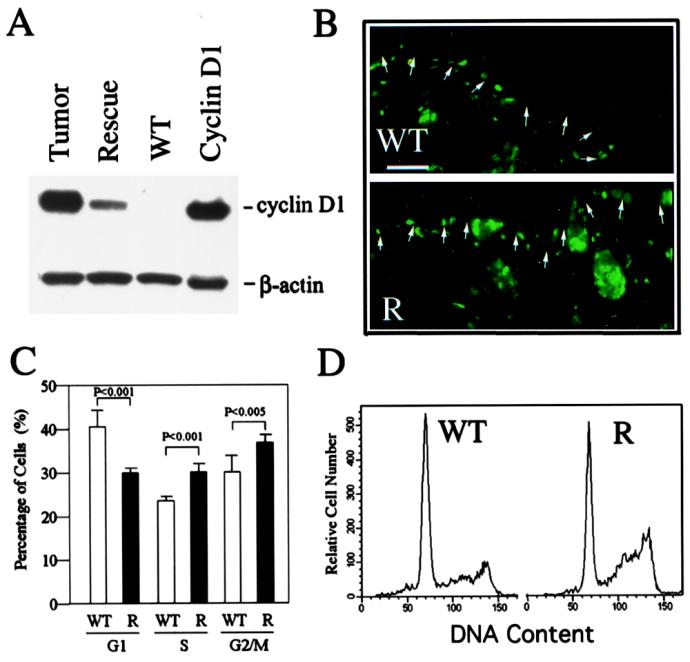
It has been shown that cyclin D1 acts as a direct target gene for β-catenin/LEF signaling (22, 23). Consistent with this finding, Western blot analysis showed that cyclin D1 protein was indeed activated in the epidermis of hPS1 rescue neonates when there was no frank hyperplasia (Fig. 6A). This level was further elevated in skin tumor samples and likely represents other events during tumorigenic process. Cyclin D1 has been shown to be a rate-limiting factor in cell proliferation by regulating the entry through G1 and into the S phase of the cell cycle (35, 36). To test the effect of cyclin D1 up-regulation on cell-cycle progression, we measured in vivo incorporation of BrdUrd in basal cells of the newborn epidermis. As seen in Fig. 6B, BrdUrd incorporation (cells entering S phase) in the basal cell layer of hPS1 rescue mice (Lower) was higher than the control littermates (Upper). Indeed, results from a total of 11 samples/group showed that BrdUrd-positive cells in the basal layer of microscopically comparable epidermis were ≈2.6-fold higher in hPS1 rescue mice as compared with littermate controls (68 ± 6.3 vs. 26 ± 4.2 nuclei/mm, respectively).
Figure 6.
Activation of cyclin D1 and altered cell-cycle control in PS1-deficient epidermis. (A) Overexpression of cyclin D1 in the skin of hPS1 rescue neonates and tumor samples. WT and Rescue, newborn epidermis from WT and rescue pups, respectively; Cyclin D1, 293 cells transfected with cyclin D1; Tumor, tumor sample taken from a representative adult hPS1 rescue mouse. β-Actin staining serves as loading control. (B). BrdUrd incorporation in WT (Upper) and hPS1 rescue (R, Lower) newborn skin. BrdUrd-positive cells are mostly localized to the hair follicles and basal cell layer (highlighted by arrows) of the epidermis, with rescue epidermis showing a higher labeling index. Scale bar = 100 μm. Cumulative (C) and representative (D) FACS analysis shows that there is accelerated G1 to S and G2/M phase transition in the absence of PS1. Keratinocytes isolated from WT (open bars) and hPS1 rescue (R, filled bars) were subjected to FACS analysis, and the percentages of cells in G1, S, and G2/M phases (mean ± SD of eight independent measurements) were plotted.
The effect of cyclin D1 activation was further determined by FACS using primary keratinocytes. Consistent with the BrdUrd incorporation result, there was a significant reduction in the percentage of cells in the G1 phase and a corresponding increase in the S and G2/M phases of the cell cycle in hPS1 rescue culture as compared with the controls (Fig. 6 C and D).
Discussion
PS1 has been shown to be essential in Notch signaling and mammalian development. In addition, PS1 is expressed throughout adulthood, and mutations in PS1 are linked to age-related familial Alzheimer's disease. It is, therefore, important to determine whether PS1 has additional physiological function in adult tissues. However, the perinatal lethal phenotype of the PS1 null mouse excludes this aspect of the analysis. In the present study, we took advantage of our established “rescue” system and present findings that loss of PS1 in skin leads to the development of epidermal hyperplasia and skin tumors in adult mice. Thus, PS1 plays an important role in regulating epidermal cell proliferation.
Deregulation of β-catenin/LEF signaling has been associated with various malignancies, including skin cancers, in human and transgenic mice (26, 27). As we have shown that PS1 interacts with β-catenin and negatively regulates its stability in vitro (30), we examined various stages of β-catenin signaling pathway using primary keratinocyte cultures as well as tissues from skin tumor samples. Our results showed that, in the absence of PS1: (i) soluble β-catenin protein, representing the signaling pool of β-catenin, was significantly higher; (ii) nuclear β-catenin protein was present in tumor cells; (iii) β-catenin/Lef-dependent signaling was elevated; and (iv) the downstream target cyclin D1 was activated. Further, up-regulated β-catenin/LEF signaling could be reversed by introducing WT PS1 but not by a PS1 deletion mutant defective in β-catenin binding. These results suggest that activated β-catenin pathway contributes to the skin tumor phenotype caused by PS1 deficiency. That PS1 plays a functional role in regulating β-catenin signaling is consistent with our in vitro finding using PS1 null fibroblast cultures (30). It is also in line with recent publications showing that Drosophila presenilin dPS was identified as an Armadillo/β-catenin modifier in a genetic screen and that loss of dPS in Drosophila resulted in the accumulation of Armadillo/β-catenin in the cytoplasm (37, 38).
It is important to note that cyclin D1 activation was detected at the time of birth before any overt clinical pathology. The notion that the cyclin D1 increase is caused by activated β-catenin signaling rather than nonspecific effect as a result of hyperproliferation is supported by our data showing similar increases in cyclin D1, but not cyclin A and cdc2, two related genes not subject to β-catenin regulation, in PS1−/− fibroblasts (30). Furthermore, PS1 down-regulates the transcriptional activity mediated by the cyclin D1 promoter, but not that of cyclin E and cyclin A, in a dose-dependent manner (30). The reduction of percentage of cells in the G1 phase and corresponding increase in the S and G2/M phases of the cell cycle in PS1-deficient keratinocytes is consistent with the documented role of cyclin D1 being a rate-limiting factor in G1 to S phase transition. In vivo, cyclin D1 has been shown to be a target of activated β-catenin in breast cancer, and its overexpression correlates with tumor progression (39). Thus, a precedent exists whereby enhanced transcription of cyclin D1 because of abnormally high levels of β-catenin seems to be an important contributor to tumor formation.
The involvement of cyclin D1 in skin tumor has been documented by numerous published reports showing that overexpression of cyclin D1 is an early event in mouse skin tumorigenesis and that it contributes directly to the tumorigenic progression (40–42). There are likely other events in addition to cyclin D1 activation that contribute to the tumor pathology, as transgenic mice overexpressing cyclin D1 in epidermis leads to hyperplasia without tumor development (43).
We did not detect a significant activation of the other β-catenin/LEF target, c-myc, suggesting that it may not be subject to the regulation by β-catenin in our system (data not shown). Besides skin tumors, we have not observed other tumor types, noticeably colorectal polyposis in hPS1 rescue mice, suggesting that PS1–β-catenin interaction may be cell-type specific. Alternatively, the PS1 homolog, PS2, may compensate for the loss of PS1 in certain tissues.
It is interesting to note that transgenic mice expressing an amino-terminal truncated and hence stabilized form of β-catenin driven by the K-14 keratin promoter (K14-ΔN87βcat) exhibit de novo hair follicle morphogenesis and develop skin and hair tumors (26). In our mice, the hair follicle morphology was normal (data not shown), and the tumor types were different (trichofolliculoma and pilomatricoma as opposed to squamous cell carcinomas) (26). These could be contributed by differences in the timing, the expression pattern, and the level of β-catenin activation present in the two systems. Whereas the K14 keratin promoter delivers a high level of stable β-catenin specifically to the basal cell layer of the epidermis and follicle outer root sheath, the absence of PS1 likely induces a broader but lesser degree of cellular elevation of β-catenin.
In addition to interacting with β-catenin, PS1 has been shown to bind to E-cadherin and promote cytoskeletal cadherin/catenin complexes (5, 44). It is, therefore, conceivable that loss of PS1 may lead to destabilization of the cadherin/catenin complex, which may contribute to the skin tumor phenotype. However, total β-catenin is not changed in PS1 rescue keratinocytes, suggesting that the membrane-bound, cadherin-associated β-catenin pool is intact (data not shown). In addition, there is strong evidence to show that β-catenin involved in cell adhesion and Wnt signaling are independently regulated (18). However, it remains possible that destabilization of the cadherin/catenin complex contributes to the increased cytosolic β-catenin in the absence of PS1.
It has been established that PS1 is required for Notch processing and activation (8, 12, 45). There is evidence to suggest that the Notch and Wnt pathways can be mutually inhibitory (46). Therefore, it can be argued that deregulation of β-catenin/LEF signaling and tumorigenic phenotype in the absence of PS1 is the result of defective Notch signaling. We believe that it represents an unlikely scenario as we have shown that a PS1 allele defective in β-catenin binding, while retaining full Notch processing activity, cannot suppress the elevated β-catenin signaling caused by PS1 deficiency (Fig. 4). Thus, PS1 function in modulating β-catenin pathway is mediated through direct interaction between the two molecules and can be separated from its activity in Notch processing. The fact that the same mutant, in contrast to WT PS1, is also defective in reversing the accelerated cell cycle in PS1 null fibroblasts strongly suggests that activated β-catenin signaling directly contributes to the hyperproliferative phenotype (30). In addition, the oncogenic activity of Notch seems to be mainly caused by activated Notch alleles rather than Notch deficiency (47, 48). However, studies described above do not exclude the possibility that a concomitant deficiency in Notch signaling may also contribute to the skin tumor progression. Definitive proof that the tumor phenotype is unrelated to Notch can be obtained only by analyzing PS1 mutant animals that are normal in Notch signaling but defective for β-catenin regulation in all PS1-expressing tissues.
In summary, the current study revealed that PS1 plays an important role in regulating keratinocyte cell cycle and skin tumorigenesis. It documents a physiological function of PS1 in adult tissues. Molecular analysis suggests that deregulation of β-catenin/LEF signaling contributes to the skin tumor progression. Whether PS1 in β-catenin regulation is involved in Alzheimer's disease pathogenesis is an issue not addressed in the present study, although it is interesting to note that PS1 containing the FAD mutations was inactive in modulating β-catenin signaling (30). This may not be sufficient to cause skin tumor in PS1 FAD patients, as there is a WT allele present and we have shown that 50% of normal PS1 is able to maintain a normal proliferative cycle in mice. Because the current therapeutic strategy of reducing amyloid load targets the PS1 proteolytic activity, a mechanism that is independent from its regulation of β-catenin, our results suggest that partial and specific inhibition of PS1 is not likely to disrupt the normal cell cycle. However, total inhibition of PS1 may lead to hyperproliferative side effect.
Acknowledgments
We thank D. Trinh and H. Tang for expert technical assistance, Dr. W. He for advisement in culturing keratinocytes, P. Wang and Dr. W.-K. Shum for insightful suggestions, and Dr. J. Reed for reviewing the histopathology. We are grateful to Drs. G. Thinakaran and M. van de Wetering for the gifts of antibodies and reporter constructs, respectively. This work was supported by Merck Research Labs and by National Institutes of Health Grants NS 40039 (to H.Z.) and CA73687 (to X.W.). H.Z. is a New Scholar of the Ellison Medical Foundation.
Abbreviations
- PS1
presenilin 1
- APP
β-amyloid precursor protein
- FAD
familial Alzheimer's disease
- LEF
lymphoid enhancer factor-1
- WT
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 10522.
References
- 1.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D J. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 3.De Strooper B, Beullens M, Contreras B, Levesque L, Craessaerts K, Cordell B, Moechars D, Bollen M, Fraser P, George-Hyslop P S, Van Leuven F. J Biol Chem. 1997;272:3590–3598. doi: 10.1074/jbc.272.6.3590. [DOI] [PubMed] [Google Scholar]
- 4.Annaert W G, Levesque L, Craessaerts K, Dierinck I, Snellings G, Westaway D, George-Hyslop P S, Cordell B, Fraser P, De Strooper B. J Cell Biol. 1999;147:277–294. doi: 10.1083/jcb.147.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georgakopoulos A, Marambaud P, Efthimiopoulos S, Shioi J, Cui W, Li H C, Schutte M, Gordon R, Holstein G R, Martinelli G, et al. Mol Cell. 1999;4:893–902. doi: 10.1016/s1097-2765(00)80219-1. [DOI] [PubMed] [Google Scholar]
- 6.Thinakaran G, Borchelt D R, Lee M K, Slunt H H, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, et al. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 7.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 8.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, et al. Nature (London) 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 9.Brown M S, Ye J, Rawson R B, Goldstein J L. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 10.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D J, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 11.Wong P C, Zheng H, Chen H, Becher M W, Sirinathsinghji D J, Trumbauer M E, Chen H Y, Price D L, Van der Ploeg L H, Sisodia S S. Nature (London) 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 12.Struhl G, Greenwald I. Nature (London) 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 13.Ye Y, Lukinova N, Fortini M E. Nature (London) 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 14.Qian S, Jiang P, Guan X M, Singh G, Trumbauer M E, Yu H, Chen H Y, Van de Ploeg L H, Zheng H. Neuron. 1998;20:611–617. doi: 10.1016/s0896-6273(00)80999-x. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Liyanage U, Medina M, Ho C, Simmons A D, Lovett M, Kosik K S. NeuroReport. 1997;8:2085–2090. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]
- 16.Murayama M, Tanaka S, Palacino J, Murayama O, Honda T, Sun X, Yasutake K, Nihonmatsu N, Wolozin B, Takashima A. FEBS Lett. 1998;433:73–77. doi: 10.1016/s0014-5793(98)00886-2. [DOI] [PubMed] [Google Scholar]
- 17.Yu G, Chen F, Levesque G, Nishimura M, Zhang D M, Levesque L, Rogaeva E, Xu D, Liang Y, Duthie M, et al. J Biol Chem. 1998;273:16470–16475. doi: 10.1074/jbc.273.26.16470. [DOI] [PubMed] [Google Scholar]
- 18.Willert K, Nusse R. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 19.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 21.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 22.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tetsu O, McCormick F. Nature (London) 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 24.Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, et al. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 25.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 26.Gat U, DasGupta R, Degenstein L, Fuchs E. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 27.Chan E F, Gat U, McNiff J M, Fuchs E. Nat Genet. 1999;21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Hartmann H, Do V M, Abramowski D, Sturchler-Pierrat C, Staufenbiel M, Sommer B, van de Wetering M, Clevers H, Saftig P, et al. Nature (London) 1998;395:698–702. doi: 10.1038/27208. [DOI] [PubMed] [Google Scholar]
- 29.Kang D E, Soriano S, Frosch M P, Collins T, Naruse S, Sisodia S S, Leibowitz G, Levine F, Koo E H. J Neurosci. 1999;19:4229–4237. doi: 10.1523/JNEUROSCI.19-11-04229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soriano S, Kang D E, Fu M, Pestell R, Chevallier N, Zheng H, Koo E H. J Cell Biol. 2001;152:785–794. doi: 10.1083/jcb.152.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beham A, Regauer S, Soyer H P, Beham-Schmid C. Adv Anat Pathol. 1998;5:269–280. [PubMed] [Google Scholar]
- 32.Gordon J W, Chesa P G, Nishimura H, Rettig W J, Maccari J E, Endo T, Seravalli E, Seki T, Silver J. Cell. 1987;50:445–452. doi: 10.1016/0092-8674(87)90498-3. [DOI] [PubMed] [Google Scholar]
- 33.Saura C A, Tomita T, Soriano S, Takahashi M, Leem J Y, Honda T, Koo E H, Iwatsubo T, Thinakaran G. J Biol Chem. 2000;275:17136–17142. doi: 10.1074/jbc.M909624199. [DOI] [PubMed] [Google Scholar]
- 34.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 35.Baldin V, Lukas J, Marcote M J, Pagano M, Draetta G. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 36.Pestell R G, Albanese C, Reutens A T, Segall J E, Lee R J, Arnold A. Endocr Rev. 1999;20:501–534. doi: 10.1210/edrv.20.4.0373. [DOI] [PubMed] [Google Scholar]
- 37.Cox R T, McEwen D G, Myster D L, Duronio R J, Loureiro J, Peifer M. Genetics. 2000;155:1725–1740. doi: 10.1093/genetics/155.4.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noll E, Medina M, Hartley D, Zhou J, Perrimon N, Kosik K S. Dev Biol. 2000;227:450–464. doi: 10.1006/dbio.2000.9925. [DOI] [PubMed] [Google Scholar]
- 39.Lin S Y, Xia W, Wang J C, Kwong K Y, Spohn B, Wen Y, Pestell R G, Hung M C. Proc Natl Acad Sci USA. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. . (First Published April 4, 2000; 10.1073/pnas.060025397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bianchi A B, Fischer S M, Robles A I, Rinchik E M, Conti C J. Oncogene. 1993;8:1127–1133. [PubMed] [Google Scholar]
- 41.Robles A I, Conti C J. Carcinogenesis. 1995;16:781–786. doi: 10.1093/carcin/16.4.781. [DOI] [PubMed] [Google Scholar]
- 42.Robles A I, Rodriguez-Puebla M L, Glick A B, Trempus C, Hansen L, Sicinski P, Tennant R W, Weinberg R A, Yuspa S H, Conti C J. Genes Dev. 1998;12:2469–2474. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Puebla M L, LaCava M, Conti C J. Cell Growth Differ. 1999;10:467–472. [PubMed] [Google Scholar]
- 44.Baki L, Marambaud P, Efthimiopoulos S, Georgakopoulos A, Wen P, Cui W, Shioi J, Koo E, Ozawa M, Friedrich V L, Robakis N K. Proc Natl Acad Sci USA. 2001;98:2381–2386. doi: 10.1073/pnas.041603398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Struhl G, Greenwald I. Proc Natl Acad Sci USA. 2001;98:229–234. doi: 10.1073/pnas.011530298. . (First Published December 26, 2000; 10.1073/pnas.011530298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Axelrod J D, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Science. 1996;271:1826–1832. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- 47.Robbins J, Blondel B J, Gallahan D, Callahan R. J Virol. 1992;66:2594–2599. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pear W S, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]