Significance
Septacidin and its analogs are potential anticancer and pain-relief drugs. Hygromycin B is an anthelmintic agent practically used in swine and poultry farming. A common feature of these compounds is that they all have heptose moieties. Here we show that the heptoses of septacidin and hygromycin B are both derived from d-sedoheptulose-7-phosphate but are biosynthesized through different pathways. Septacidin producer, a gram-positive bacterium, shares the same ADP-heptose biosynthesis pathway with gram-negative bacterium lipopolysaccharide biosynthesis. These findings not only elucidate the biosynthesis mechanisms of septacidin and hygromycin B but enable opportunities for manipulation of their heptose moieties by combinatorial biosynthesis and for changing the structure of heptoses in gram-negative bacterium lipopolysaccharides.
Keywords: natural product, heptose, biosynthesis, lipopolysaccharide, Streptomyces
Abstract
Seven-carbon-chain–containing sugars exist in several groups of important bacterial natural products. Septacidin represents a group of l-heptopyranoses containing nucleoside antibiotics with antitumor, antifungal, and pain-relief activities. Hygromycin B, an aminoglycoside anthelmintic agent used in swine and poultry farming, represents a group of d-heptopyranoses–containing antibiotics. To date, very little is known about the biosynthesis of these compounds. Here we sequenced the genome of the septacidin producer and identified the septacidin gene cluster by heterologous expression. After determining the boundaries of the septacidin gene cluster, we studied septacidin biosynthesis by in vivo and in vitro experiments and discovered that SepB, SepL, and SepC can convert d-sedoheptulose-7-phosphate (S-7-P) to ADP-l-glycero-β-d-manno-heptose, exemplifying the involvement of ADP-sugar in microbial natural product biosynthesis. Interestingly, septacidin, a secondary metabolite from a gram-positive bacterium, shares the same ADP-heptose biosynthesis pathway with the gram-negative bacterium LPS. In addition, two acyltransferase-encoding genes sepD and sepH, were proposed to be involved in septacidin side-chain formation according to the intermediates accumulated in their mutants. In hygromycin B biosynthesis, an isomerase HygP can recognize S-7-P and convert it to ADP-d-glycero-β-d-altro-heptose together with GmhA and HldE, two enzymes from the Escherichia coli LPS heptose biosynthetic pathway, suggesting that the d-heptopyranose moiety of hygromycin B is also derived from S-7-P. Unlike the other S-7-P isomerases, HygP catalyzes consecutive isomerizations and controls the stereochemistry of both C2 and C3 positions.
Saccharides are common components found in bacterial natural products, which usually have a profound impact on the biological properties of the compounds (1, 2). A significant number of glycosylated bacterial natural products, such as erythromycin, doxorubicin, vancomycin, and avermectin, are practically used as drugs for the treatment of human and animal diseases, and it is well established that changing the carbohydrate moieties in these drugs can influence their pharmacological properties significantly (1, 2). A recent systematic analysis revealed that >20% of bacterial natural products are glycosylated with 344 distinct carbohydrates, including diverse pentoses, hexoses, and high-carbon-chain–containing sugars, which are usually derived from carbohydrate precursors generated by primary metabolism (2).
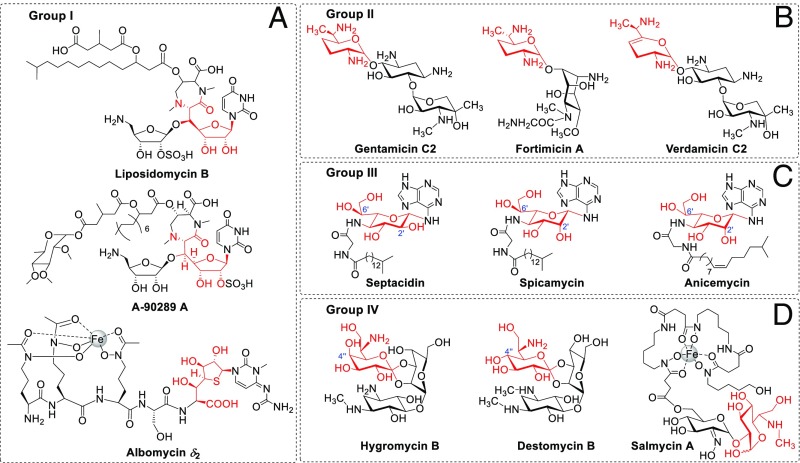
Among the sugar moieties of bacterial natural products, there are at least 11 seven-carbon-chain–containing carbohydrates (2), which can be classified into four groups structurally. Group I seven-carbon-chain–containing sugar has two members: a heptofuranose from the lipopeptidyl nucleoside family of translocase I inhibitors, represented by liposidomycin (3) and A-90289 (4), and a heptothiofuranose from the “Trojan horse” antibiotic albomycin (5) (Fig. 1A). A biosynthesis study of A-90289 revealed that the heptofuranose of its 5ʹ-C-glycyluridine moiety was formed by an l-threonine:uridine-5ʹ-aldehyde transaldolase, LipK, which extended the pentofuranose of uridine-5ʹ-aldehyde to heptofuranose via an l-threonine–dependent β-substitution reaction (6). Not surprisingly, a LipK homolog protein-encoding gene, abmH, is present in the albomycin biosynthetic gene cluster, indicating that the heptothiofuranose is formed through a similar carbon-chain extension process (5).
Fig. 1.
Representative bacterial natural products with seven-carbon-chain–containing carbohydrate structures. (A) Group I compounds with heptofuranoses or heptothiofuranose; (B) group II compounds with highly reduced heptopyranoses; (C) group III compounds with l-heptopyranoses; (D) group IV compounds with d-heptopyranoses. The seven-carbon-chain–containing carbohydrate moiety of each compound is shown in red.
Group II contains four highly reduced heptopyranoses from several clinically important aminoglycoside antibiotics, including gentamicin, verdamicin, and fortimicin (Fig. 1B) (7). Both in vivo and in vitro data support that a cobalamin-dependent radical SAM enzyme, GenK, catalyzes the C-6ʹ methylation of gentamicin X2, forming the 2ʹ-amino-2ʹ,7ʹ-dideoxy-α-d-gluco-heptose moiety of G418 (8–10), which is then converted to the highly reduced heptopyranoses in gentamicin or verdamicin by different tailoring processes. Similarly, a yet to be identified cobalamin-dependent methyltransferase was proposed to be responsible for the heptopyranose biosynthesis of fortimicin (11).
Group III contains at least two l-heptopyranoses: a 4ʹ-amino-4ʹ-deoxy-l-glycero-β-l-gluco-heptose (GGH) from septacidin (12) and its 2ʹ-epimer, 4ʹ-amino-4ʹ-deoxy-l-glycero-β-l-manno-heptose (LGMH), from spicamycin (13) (Fig. 1C). The structure of the l-heptopyranose from anicemycin is either LGMH or its 6ʹ-epimer, 4ʹ-amino-4ʹ-deoxy-d-glycero-β-l-manno-heptose (DGMH), which needs to be determined by further studies (14) (Fig. 1C). Septacidin, anicemycin, and spicamycin are all nucleoside antibiotics consisting of a unique 6-N-glycosylated heptosamine-adenine moiety and a side chain containing a glycyl group and varied fatty acyl groups (12–14). Septacidin, discovered as an antifungal and antitumor antibiotic (15, 16), was found to be an immunogenic cell death inducer recently (17); anicemycin is a newly identified inhibitor of anchorage-independent growth of tumor cells (14); spicamycin displays an excellent antitumor activity by inhibiting protein synthesis (18, 19), and one of its derivatives, KRN5500, is now an anticancer drug under clinical trial (20). An unexpected discovery during the cancer treatment process of KRN5500 is that it can relieve neuropathic pain of patients (21). Therefore, this compound is also under a phase II clinical trial as a drug for pain relief (21). Due to its importance, diverse synthesis routes were developed for spicamycin by different groups (22, 23). However, very little is known about the biosynthesis of the three nucleoside antibiotics and their special 4ʹ-amino-4ʹ-deoxy-l-heptoses, except that the septacidin heptose, GGH, was proposed to be derived from carbohydrates involved in pentose phosphate pathway based on an isotope-labeled feeding study (12).
Group IV contains three d-heptopyranoses from hygromycin B, destomycin, and salmycin (Fig. 1D) (2, 24). Hygromycin B and destomycin are aminoglycoside antibiotics inhibiting protein synthesis (25) and are used as anthelmintic agents in swine and poultry farming. Hygromycin B and its resistance gene are also a frequently used selection system in molecular biological research. Hygromycin B and destomycins A and C contain a common heptopyranose destomic acid; destomycin B contains the C-4ʹʹ epimer of destomic acid, named epi-destomic acid (26, 27). Although a hygromycin B gene cluster was deposited into GenBank as early as 2006, very little is known about the biosynthesis of hygromycin B and destomycin, let alone the origin of destomic acid and epi-destomic acid. Salmycin A is also a “Trojan horse” antibiotic containing a ferrioxamine (siderophore) core and an unusual aminodisaccharide moiety, which contains a 6ʹʹ-amino-6ʹʹ-deoxy-d-gluco-heptose and a 2ʹ-oximo-α-d-glucose (28). With the help of its siderophore structure, salmycin can be transported into cells by the iron uptake systems efficiently, which renders it very low inhibitory concentrations of both gram-positive and gram-negative bacteria (28). Total synthesis of danoxamine, the siderophore part of salmycin, was achieved (29); however, no chemical synthesis or biosynthesis study on the aminodisaccharide moiety has been reported.
To understand the biosynthesis of the special heptoses in groups III and IV and elucidate the biosynthetic mechanisms of the natural products containing those heptoses, we sequenced the genomes of the two Streptomyces strains producing septacidin and hygromycin B, identified the septacidin biosynthetic gene cluster, and discovered d-sedoheptulose-7-phosphate (S-7-P) as a common precursor for the heptose moieties of septacidin and hygromycin B in this work. We also discovered that the heptose is activated as an ADP-sugar in septacidin biosynthesis, exemplifying the involvement of ADP-sugar in microbial natural product biosynthesis.
Results
Identification of the Septacidin Biosynthetic Gene Cluster.
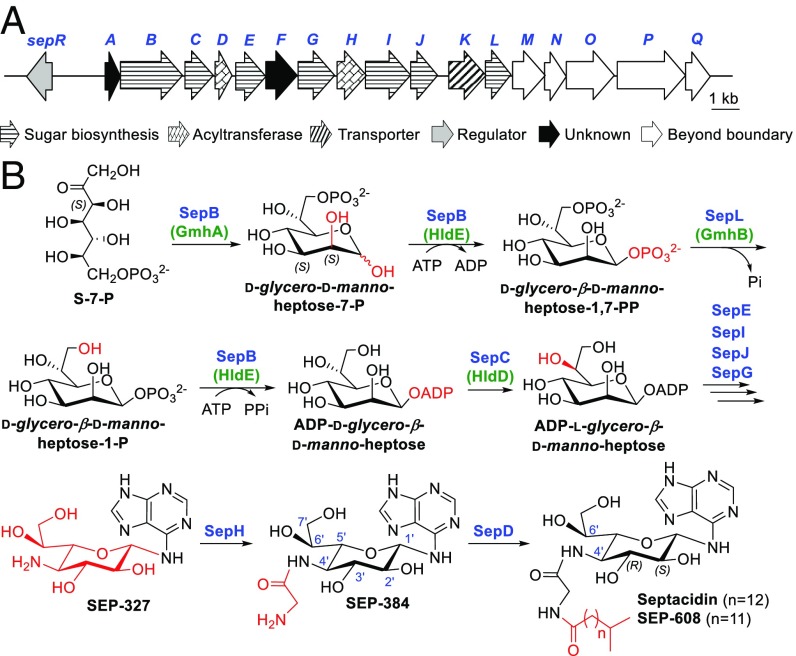
The genome of the septacidin producer Streptomyces fimbriatus CGMCC 4.1598 was draft sequenced and searched for the septacidin biosynthesis gene cluster using enzymes possibly involved in GGH biosynthesis as probes. After careful analysis, a DNA region, which contains genes encoding glycosyltransferase (SepE), aminotransferase (SepG), sugar isomerase (SepJ), and a tridomain protein (SepB) showing considerable similarities with enzymes involved in d-manno-heptose biosynthesis of Escherichia coli LPS (30), was chosen as the putative sep cluster (Fig. 2A and SI Appendix, Table S1). However, the production of septacidin in S. fimbriatus CGMCC 4.1598 is low and very unstable, prompting us to express this putative sep gene cluster in a heterologous host to confirm its identity and to facilitate the following studies.
Fig. 2.
The biosynthetic gene cluster and a proposed biosynthetic pathway of septacidin. (A) Genetic organization of the sep gene cluster from S. fimbriatus CGMCC 4.1598. (B) A proposed pathway for septacidin biosynthesis based on in vitro and in vivo experiments. The enzymes involved in gram-negative bacteria LPS heptose biosynthesis are bracketed and indicated in green.
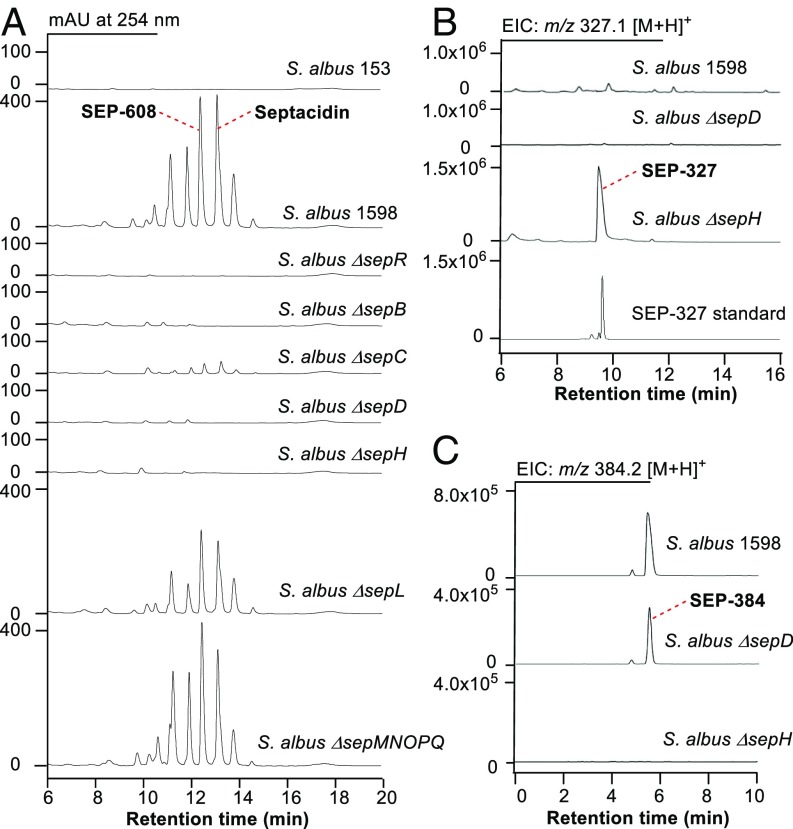
A 24-kb DNA fragment between two transposase genes (GenBank Accession No. MF372757), which contains all of the genes putatively involved in septacidin biosynthesis, was cloned into an E. coli-Streptomyces shuttle vector pSET153 to generate a plasmid pSET1598 via the newly developed Cas9-assisted targeting of chromosome segments method (31). Integration of pSET1598 and pSET153 into the chromosome of Streptomyces albus J1074 by conjugation respectively generated the sep gene cluster heterologous expression strain S. albus 1598 and a negative control strain S. albus 153. When cultured in the fermentation medium, S. albus 1598 produced a series of compounds having the same UV absorption as septacidin, while no such compound was produced by S. albus 153 (Fig. 3A). The presence of septacidin and its analogs in S. albus 1598 was further confirmed by MS analysis (SI Appendix, Fig. S1) and NMR analysis of one septacidin analog, SEP-608 (SI Appendix, Fig. S2 and Table S2). To confirm the structure of the heptose moiety, SEP-608 was hydrolyzed to generate the GGH-adenine moiety (named SEP-327), which was then carefully assigned by 1D and 2D NMR analyses (SI Appendix, Fig. S3 and Table S3). Overall, these results demonstrated that the 24-kb DNA fragment in pSET1598 contains a complete gene cluster encoding the biosynthesis of septacidin.
Fig. 3.
Metabolic profiles of the sep gene cluster heterologous expression strain S. albus 1598 and the sep gene in-frame deleted mutants upon HPLC and LC-MS analysis. (A) HPLC traces of the mycelia methanolic extracts of a negative control S. albus 153, S. albus 1598, and the sep gene mutants. (B) LC-MS–extracted ion count chromatograms of m/z 327.1 [M+H]+ in the supernatants of S. albus 1598, S. albus ∆sepD, and ∆sepH fermentations. (C) LC-MS extracted ion count chromatograms of m/z 384.2 [M+H]+ in the supernatants of S. albus 1598, S. albus ∆sepD, and ∆sepH fermentations. EIC, extracted ion count.
Determining the sep Gene Cluster Boundaries.
The 24-kb DNA fragment in pSET1598 contains 18 ORFs with an AraC family regulator encoding gene sepR at its left end. To determine the left boundary of the sep gene cluster, we constructed the sepR gene in-frame deletion mutant S. albus ∆sepR using λ RED-mediated PCR-targeting technology (SI Appendix, Fig. S4A), which avoided a possible polar effect caused by gene inactivation and was recruited in the following mutant strain constructions. Production of septacidins was totally abolished in S. albus ∆sepR (Fig. 3A), suggesting that sepR encodes a positive regulator controlling septacidin biosynthesis. To determine the right boundary of the sep gene cluster, several ORFs at its right ends were inactivated. Inactivation of the gene sepL (SI Appendix, Fig. S4B), which encodes a putative sugar phosphatase, reduced the production of septadicins considerably (Fig. 3A). In contrast, deletion of the region downstream of sepL, which contains five ORFs (sepM, sepN, sepO, sepP, and sepQ) (SI Appendix, Fig. S4C), showed no clear influence on septacidin production (Fig. 3A). Therefore, the sep gene cluster was narrowed down to a 16.5-kb region of DNA from sepR to sepL, which contains 13 genes encoding seven sugar biosynthesis enzymes (SepB, SepC, SepE, SepG, SepI, SepJ, and SepL), two acyltransferases (SepD and SepH), one transporter (SepK), one regulator (SepR), and two functional unknown proteins (SepA and SepF).
Conversion of S-7-P into ADP-d-Glycero-β-d-manno-heptose by SepB and SepL.
The 4ʹ-amino-4ʹ-deoxy-l-heptose of septacidin, GGH, was proposed to be derived from the pentose phosphate pathway in a previous isotope-labeled feeding study (12). Bioinformatic analysis suggested two genes, sepB and sepL, were most probably involved in the early stage of GGH biosynthesis. As previously mentioned, SepB is a tridomain protein with its N-terminal part (1–220 aa) displaying 23.9% identity with GmhA, an S-7-P isomerase (32), and its rest part (221–707 aa) displaying 30.3% identity with HldE, a di-domain protein with kinase and nucleotidyltransferase activities (33). SepL shows 25% identity with a phosphatase, GmhB (34). GmhA, GmhB, and HldE are enzymes involved in the biosynthesis of heptoses of the core region of E. coli LPS, and they catalyze a four-reaction relay converting S-7-P into ADP-d-glycero-β-d-manno-heptose (Fig. 2B) (30).
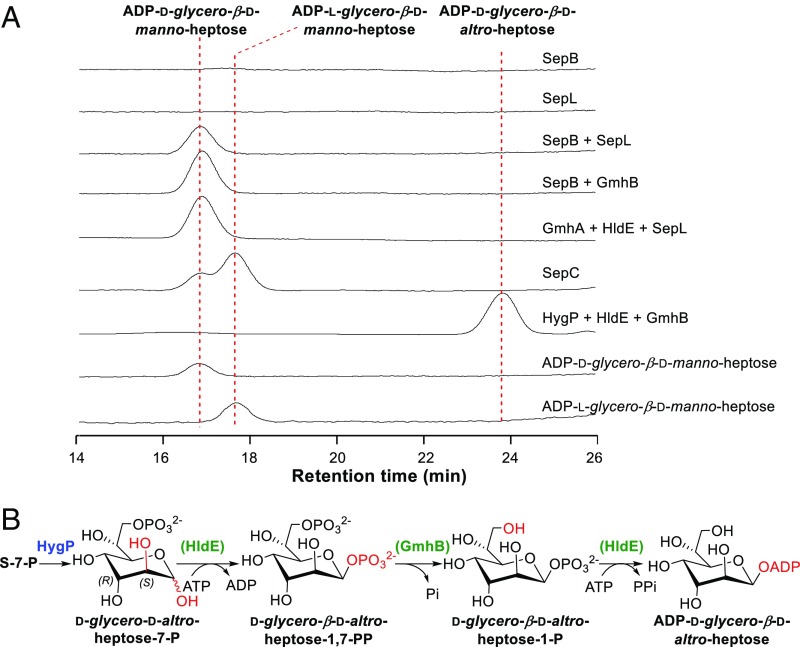
To study the function of SepB, we constructed the sepB gene in-frame deletion mutant S. albus ∆sepB (SI Appendix, Fig. S4D), in which the production of septacidin was totally abolished, and no accumulation of septadicin-related intermediate was observed (Fig. 3A), implying that sepB is involved in the early stage of septacidin biosynthesis. We then overexpressed SepB and SepL in E. coli, purified them as N-His6–tagged proteins (SI Appendix, Fig. S5A), and incubated them with S-7-P, ATP (as a phosphate donor for kinase activity), and other NTPs (as a possible sugar acceptor for nucleotidyltransferase activity). To our delight, a product was detected by HPLC when only S-7-P and ATP were used as substrates (Fig. 4A). Addition of the other NTPs had no influence on the reaction (SI Appendix, Fig. S6), suggesting that ATP acted as both a phosphate donor and a sugar acceptor here. The product was purified and determined to be ADP-d-glycero-β-d-manno-heptose by high resolution (HR)-MS analysis (m/z 618.0841 for [M-H]−, C17H27N5O16P2, cacld 618.0855) (SI Appendix, Fig. S5B) and HPLC coinjections with authentic standards prepared by the GmhA + GmhB + HldE assay and by organic synthesis (SI Appendix, Figs. S7–S9 and Tables S4 and S5). In addition, we tested the catalytic activities of two hybrid enzyme sets, SepB + GmhB and GmhA + HldE + SepL, and both of them could convert S-7-P into ADP-d-glycero-β-d-manno-heptose efficiently (Fig. 4A), confirming that SepB is a trifunctional protein with isomerase, kinase, and nucleotidyltransferase activities and SepL is a heptose phosphatase (Fig. 2B).
Fig. 4.
Representative assays of the septacidin and hygromycin B heptose biosynthesis enzymes. (A) HPLC analysis of enzymatic assays of SepB, SepC, SepL, and HygP. (B) A proposed ADP-d-glycero-β-d-altro-heptose biosynthetic pathway catalyzed by HygP, HldE, and GmhB. The enzymes from gram-negative bacteria LPS heptose biosynthesis are bracketed and indicated in green.
SepC Is an ADP-d-Glycero-β-d-manno-heptose 6ʹ-Epimerase.
The sepC gene encodes a protein displaying 26.7% identity with HldD, an NAD+-dependent 6ʹ-epimerase converting ADP-d-glycero-β-d-manno-heptose to ADP-l-glycero-β-d-manno-heptose in E. coli LPS biosynthesis (35). S. albus ∆sepC, the sepC gene in-frame deletion mutant, was constructed via PCR-targeting technology (SI Appendix, Fig. S4E). HPLC analysis revealed that production of septacidins was reduced dramatically in S. albus ∆sepC (Fig. 3A), confirming the involvement of sepC in septacidin biosynthesis. The ∆sepC mutant still produced a few amount of septacidins, indicating that an alternative epimerase in the cell could complement SepC reaction. Another possibility, which cannot be excluded, is that the enzymes in the later steps can tolerate the according substrates with 6ʹ-R configurations and form septacidin derivatives with a 4ʹ-amino-4ʹ-deoxy-d-glycero-β-l-gluco-heptose moiety. To verify the function of SepC, we overexpressed sepC in E. coli, purified the N-His6–tagged SepC (SI Appendix, Fig. S10A), and tested its catalytic activity toward ADP-d-glycero-β-d-manno-heptose. After incubation with SepC for 2 h, ADP-d-glycero-β-d-manno-heptose was converted to another compound (Fig. 4A), which was determined to be ADP-l-glycero-β-d-manno-heptose by HR-MS analysis (m/z 618.0855 for [M-H]−, C17H27N5O16P2, cacld 618.0855) (SI Appendix, Fig. S10B) and HPLC coinjection with a standard prepared by the HldD assay. An HPLC analysis of the extracts of denatured SepC showed that this enzyme uses NAD+ as a cofactor binding tightly via noncovalent bonds (SI Appendix, Fig. S10C). Overall, these results suggested that SepC is an ADP-d-glycero-β-d-manno-heptose 6ʹ-epimerase functionalizing immediately after SepB and SepL (Fig. 2B).
SepD and SepH Are Acyltransferases Forming the Side Chain of Septacidin.
In the sep gene cluster, there are two putative acyltransferase encoding genes, sepD and sepH, which might be responsible for the formation of the glycyl-fatty acyl side chain. We constructed the S. albus ∆sepD and ∆sepH mutants (SI Appendix, Fig. S4 F and G) and checked their metabolic profiles carefully. When their mycelia were extracted with methanol and analyzed by HPLC, production of septacidins was not detected (Fig. 3A). Considering that the intermediates accumulated in S. albus ∆sepD and ∆sepH should lose the fatty acyl chain and become much more hydrophilic than septacidins, we analyzed the supernatants of their fermentation broths by LC-MS. An intermediate was accumulated in S. albus ∆sepH (Fig. 3B), and it was determined to be SEP-327 by MS analysis and HPLC coinjection with the GGH-adenine standard, which suggested that SepH catalyzes the addition of the N-4ʹ-glycyl group. Interestingly, compound SEP-384, with a molecular formula C14H21N7O6 (HR-MS, m/z 384.1639 for [M + H]+, calcd 384.1626), was detected in both S. albus 1598 and the ∆sepD mutant (Fig. 3C) but was absent in the ∆sepH mutant. This compound was assigned as N-4ʹ-glycyl-GGH-adenine by careful MS, NMR, and hydrolysis analyses (SI Appendix, Figs. S11 and S12 and Table S6), implying that SepD is a fatty acyl transferase catalyzing the last step of septacidin biosynthesis (Fig. 2B).
S-7-P Is a Precursor for Destomic Acid of Hygromycin B.
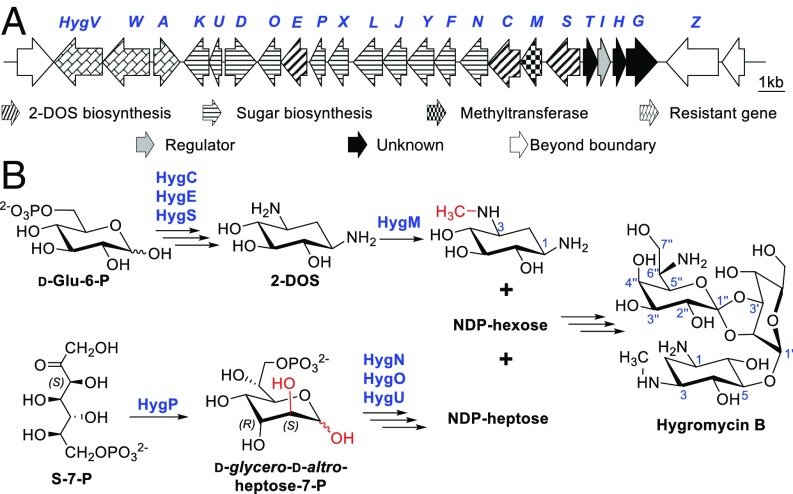
A hygromycin B gene cluster (Accession No. AJ628642) from Streptomyces hygroscopicus DSM 40578 was deposited into GenBank by Piepersberg et al. in 2006. We sequenced the genome of S. hygroscopicus DSM 40578 and found that the hyg gene cluster contains the only set of genes (hygC, hygE, and hygS) responsible for the formation of 2-deoxystreptamine (2-DOS), the featured structure of 2-DOS containing aminoglycosides (7), which further confirmed the identity of the hyg gene cluster (Fig. 5A and SI Appendix, Table S7). Notably, two genes in this cluster, hygP and hygU, encode proteins sharing moderate similarities with the isomerase domain of SepB (1–220 aa, 25.9% identity) and the phosphatase SepL (20.8% identity), leading us to propose that destomic acid, the heptose of hygromycin B, may also be derived from S-7-P. In addition to hygP and hygU, two more genes, hygN and hygO, are very likely involved in the early biosynthesis steps of destomic acid. HygN displays 25.3% identity with HddA, a kinase catalyzing the phosphorylation of d-glycero-d-manno-heptose-7-P to generate d-glycero-α-d-manno-heptose-1,7-diphosphate in Aneurinibacillus thermoaerophilus DSM 10155 (36). HygO is a putative nucleotidyltransferase displaying moderate similarities with a number of hypothetical galactose-1-phosphate uridylyltransferases.
Fig. 5.
The biosynthetic gene cluster and a proposed biosynthetic pathway of hygromycin B. (A) Genetic organization of the hyg gene cluster from S. hygroscopicus DSM 40578. (B) A proposed pathway for hygromycin B biosynthesis.
We envisaged that S-7-P may also be used as a precursor for the biosynthesis of hygromycin B and is converted to an NDP-heptose by HygP (isomerase), HygN (kinase), HygU (phosphatase), and HygO (nucleotidyltransferase) by a similar means as that of SepB and SepL (Fig. 5B). Since S. hygroscopicus DSM 40578 is genetically intractable, we sought to determine the precursor for destomic acid by in vitro enzymatic studies. We tried to express the four proteins HygP, HygN, HygU, and HygO in E. coli BL21. Unfortunately, only N-His6–tagged HygP was obtained as a soluble protein after exhaustive testing of various protein expression conditions (SI Appendix, Fig. S13A). A hybrid assay was then tested by incubating HygP together with the E. coli ADP-heptose synthesis enzymes, GmhB (phosphatase) and HldE (bifunctional kinase/nucleotidyltransferase), using S-7-P and ATP as substrates. To our delight, the three enzymes converted S-7-P to another compound, which has the same molecular formula, C17H27N5O16P2 (HR-MS, m/z 618.0854 for [M-H]−, cacld 618.0855) (SI Appendix, Fig. S13B), but a clearly different HPLC retention time than ADP-d-glycero-β-d-manno-heptose (Fig. 4A). The enzymatic synthesis was therefore conducted on a preparative scale to obtain this compound, and it was determined to be ADP-d-glycero-β-d-altro-heptose by careful analysis of its NMR data (SI Appendix, Fig. S14 and Table S8). These results suggested that HygP converts S-7-P to d-glycero-d-altro-heptose-7-P through consecutive isomerizations and controls the stereochemistry of both C2 and C3 positions (Fig. 4B and SI Appendix, Fig. S13D), which is very rare for sugar isomerases. In addition, GmhB and HldE can tolerate the 3-epimers of their natural substrates and generate ADP-d-glycero-β-d-altro-heptose efficiently (Fig. 4B).
Discussion
As previously mentioned, the seven-carbon-chain–containing carbohydrates in bacterial natural products can be divided into four groups structurally. The biosynthetic mechanisms of heptoses belonging to the first two groups have been well elucidated in compounds like A-90289 (group I) and gentamicin (group II). However, very little is known about the biosynthesis of heptoses belonging to the last two groups. Based on the results shown in this work, we propose that S-7-P is a common precursor for the heptoses of septacidin (group III) and hygromycin B (group IV) and is converted to NDP-heptoses through similar biosynthetic pathways in those compounds. Although S-7-P is a known precursor for some natural products (37)[e.g., it can be converted to 2-epi-5-epi-valiolone and then incorporated into acarbose (38) or validamycin (39)], these are rare examples of incorporating S-7-P into natural products as sugar components.
In the case of septacidin biosynthesis, we propose that S-7-P is converted to d-glycero-β-d-manno-heptose-1-phosphate, activated as an ADP-sugar and epimerized to ADP-l-glycero-β-d-manno-heptose by SepB, SepL, and SepC through a five-step pathway (Fig. 2B), which has been well delineated in the gram-negative bacterium LPS biosynthesis previously (30). LPS is the major component of the outer leaflet of the outer membrane of gram-negative bacteria. It is surprising to discover this pathway in the septacidin producer S. fimbriatus CGMCC 4.1598, a gram-positive filamentous bacterium, since a big difference between gram-positive and gram-negative bacteria is that the former have a thicker peptidoglycan layer but lack the LPSs containing outer membrane. In gram-positive bacteria, the only biosynthetically characterized NDP-heptose originated from S-7-P is GDP-d-glycero-α-d-manno-heptose, which is incorporated into a disaccharide involved in S-layer protein decoration in A. thermoaerophilus DSM 10155 (36). In gram-negative bacteria, the ADP-activated l-glycero-β-d-manno-heptose is usually transferred to 3-deoxy-d-manno-oct-2-ulosonic acid to form the inner core region of LPSs (40), while in S. fimbriatus CGMCC 4.1598, the heptose becomes the carbohydrate moiety of a secondary metabolite, septacidin. Discovery of the ADP-l-glycero-β-d-manno-heptose biosynthetic pathway in the septacidin producer presents a rare example of sharing the same biosynthesis logic in gram-negative bacterium primary metabolism and gram-positive bacterium secondary metabolism. Worthy of particular note is the involvement of ADP-sugar in microbial natural product biosynthesis, which indicates that, besides GDP-, CDP-, UDP-, and dTDP-sugar, ADP-sugar may also serve as a donor for natural product glycosylation.
Besides SepB, SepC, and SepL, four more enzymes (SepE, SepG, SepI, and SepJ) are likely to be involved in the formation of the GGH-adenine moiety. Based on the results of bioinformatic analysis, we propose that the glycosyltransferase SepE transfers heptose onto the 6-N position of adenine to form the unusual N-glycosidic bond; the oxidoreductase SepI and aminotransferase SepG are responsible for the formation of 4ʹ-amino-4ʹ-deoxy-heptose by converting 4ʹ-OH of heptose into 4ʹ-keto (SepI) and then into 4ʹ-NH2 (SepG); and the putative isomerase SepJ catalyzes the 3ʹ,5ʹ-epimerization reaction converting d-heptose into its l-configuration. GGH-adenine is then decorated by SepH and SepD to add the glycyl and fatty acyl groups sequentially to generate diverse septacidins (Fig. 2B).
Spicamycin and anicemycin have very similar structures as septacidin. Spicamycin is the 2ʹ-epimer of septacidin and its heptose moiety is LGMH, the 2ʹ-epimer of GGH. Encouraged by the S-7-P isomerase HygP, which can generate d-glycero-d-altro-heptose-7-P, the 3-epimer of d-glycero-d-manno-heptose-7-P involved in septacidin biosynthesis, we propose that the biosynthesis of spicamycin may be initiated by an isomerase, which can generate d-glycero-d-gluco-heptose-7-P, the 2-epimer of d-glycero-d-manno-heptose-7-P, as an intermediate. Another possibility is that a 2ʹ-epimerase, which can convert septacidin analog to its 2ʹ-epimer, is involved in spicamycin biosynthesis. Unlike septacidin, anicemycin has an LGMH or DGMH moiety and an unsaturated fatty acyl chain. The draft genome sequence of the anicemycin producer Streptomyces sp. TP-A0648 was reported recently (41). However, no anicemycin biosynthesis gene was found when we inspected this genome using the septacidin genes as probes. A higher-quality genome sequence of Streptomyces sp. TP-A0648 may be necessary for elucidation of the biosynthesis mechanism of anicemycin.
In the case of hygromycin B biosynthesis, we propose that HygC, HygE, and HygS convert d-glucose-6-phosphate to 2-DOS through the same biosynthesis pathway as that of the well-characterized 2-DOS containing aminoglycoside antibiotics (7). The only methyltransferase in the hyg cluster, HygM, may catalyze the N-methylation of 2-DOS to generate 3-N-methyl-2-DOS (42), which is then decorated to form the disaccharide containing a d-gulose and a destomic acid to form hygromycin B (Fig. 5B). Based on the fact that HygP, GmhB, and HldE can convert S-7-P to ADP-d-glycero-β-d-altro-heptose, we propose that the precursor of destomic acid is S-7-P, which is converted to d-glycero-d-altro-heptose-7-P by HygP. Unlike GmhA and the isomerase domain of SepB, which catalyze one isomerization step and control the stereochemistry of only the C2 position, HygP is a special heptose isomerase that catalyzes consecutive isomerizations and determines the stereochemistry of both C2 and C3 positions. d-Glycero-d-altro-heptose-7-P may be modified by the kinase HygN, the phosphatase HygU, and the nucleotidyltransferase HygO sequentially to form an NDP-heptose, which will be further converted to destomic acid by the following tailoring enzymes (Fig. 5B). The high structure similarities between destomycins and hygromycin B imply that destomycins are biosynthesized in a similar manner as hygromycin B and the heptose of destomycin B, epi-destomic acid, is also derived from S-7-P.
In conclusion, we showed that the special seven-carbon–containing sugars of septacidin and hygromycin B are both derived from S-7-P, a heptose in the pentose phosphate pathway. The heptose biosynthesis in septacidin surprisingly shares the same logic with LPS heptose biosynthesis at its early stage and recruits ADP-sugars as intermediates. HygP was characterized as an interesting isomerase that initiates destomic acid biosynthesis in hygromycin B by converting S-7-P to d-glycero-d-altro-heptose-7-P through consecutive isomerizations. The findings in this study set the stage for the generation of novel septacidin derivatives by combinatorial biosynthesis. In addition, generation of ADP-d-glycero-β-d-altro-heptose by HygP, GmhB, and HldE enables interesting opportunities for structural modification of the heptoses in the LPS inner core by replacing the gmhA gene with hygP in gram-negative bacterium. Both avenues are currently being actively pursued in our laboratory.
Materials and Methods
Detailed descriptions of materials and methods, including bacterial strains, plasmids, and primers used in this study (SI Appendix, Tables S9 and S10); construction of mutant strains; isolation and characterization of compounds; and enzymatic assays and other procedures used are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Jinwei Ren and Dr. Yaxin Zhu, Institute of Microbiology, Chinese Academy of Sciences (CAS), for technical support. The study was supported by Ministry of Science and Technology of China Grants 2015CB150600 and 2013CB734000 and National Natural Science Foundation of China Grants 31370095 and 31522001. Y.C. is an awardee of the “Hundred Talents Program” of CAS. Z.G. is an awardee of the Youth Innovation Promotion Association of CAS (2017124).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. MF372757).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711665115/-/DCSupplemental.
References
- 1.Thibodeaux CJ, Melançon CE, 3rd, Liu HW. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed Engl. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elshahawi SI, Shaaban KA, Kharel MK, Thorson JS. A comprehensive review of glycosylated bacterial natural products. Chem Soc Rev. 2015;44:7591–7697. doi: 10.1039/c4cs00426d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ubukata M, Isono K, Kimura K, Nelson CC, McCloskey JA. The structure of liposidomycin B, an inhibitor of bacterial peptidoglycan synthesis. J Am Chem Soc. 1988;110:4416–4417. [Google Scholar]
- 4.Fujita Y, et al. A-90289 A and B, new inhibitors of bacterial translocase I, produced by Streptomyces sp. SANK 60405. J Antibiot (Tokyo) 2011;64:495–501. doi: 10.1038/ja.2011.38. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Y, et al. Biosynthesis of albomycin δ(2) provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates. ACS Chem Biol. 2012;7:1565–1575. doi: 10.1021/cb300173x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnard-Britson S, et al. Amalgamation of nucleosides and amino acids in antibiotic biosynthesis: Discovery of an L-threonine:uridine-5′-aldehyde transaldolase. J Am Chem Soc. 2012;134:18514–18517. doi: 10.1021/ja308185q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busscher GF, Rutjes FPJT, van Delft FL. 2-Deoxystreptamine: Central scaffold of aminoglycoside antibiotics. Chem Rev. 2005;105:775–791. doi: 10.1021/cr0404085. [DOI] [PubMed] [Google Scholar]
- 8.Hong W, Yan L. Identification of gntK, a gene required for the methylation of purpurosamine C-6′ in gentamicin biosynthesis. J Gen Appl Microbiol. 2012;58:349–356. doi: 10.2323/jgam.58.349. [DOI] [PubMed] [Google Scholar]
- 9.Karki S, Kim JY, Park SH, Kwon HJ. Gene inactivation study on gntK, a putative C-methyltransferase gene in gentamicin biosynthesis from Micromonospora echinospora. J Korean Soc Appl Biol Chem. 2012;55:439–442. [Google Scholar]
- 10.Kim HJ, et al. GenK-catalyzed C-6′ methylation in the biosynthesis of gentamicin: Isolation and characterization of a cobalamin-dependent radical SAM enzyme. J Am Chem Soc. 2013;135:8093–8096. doi: 10.1021/ja312641f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzuyama T, Seki T, Dairi T, Hidaka T, Seto H. Nucleotide sequence of fortimicin KL1 methyltransferase gene isolated from Micromonospora olivasterospora, and comparison of its deduced amino acid sequence with those of methyltransferases involved in the biosynthesis of bialaphos and fosfomycin. J Antibiot (Tokyo) 1995;48:1191–1193. doi: 10.7164/antibiotics.48.1191. [DOI] [PubMed] [Google Scholar]
- 12.Price NPJ, et al. Biosynthesis of 4-aminoheptose 2-epimers, core structural components of the septacidins and spicamycins. J Antibiot (Tokyo) 2014;67:405–414. doi: 10.1038/ja.2014.15. [DOI] [PubMed] [Google Scholar]
- 13.Sakai T, et al. Absolute configuration of spicamycin, an antitumor antibiotic produced by Streptomyces alanosinicus. J Antibiot (Tokyo) 1995;48:899–900. doi: 10.7164/antibiotics.48.899. [DOI] [PubMed] [Google Scholar]
- 14.Igarashi Y, et al. Anicemycin, a new inhibitor of anchorage-independent growth of tumor cells from Streptomyces sp. TP-A0648. J Antibiot (Tokyo) 2005;58:322–326. doi: 10.1038/ja.2005.40. [DOI] [PubMed] [Google Scholar]
- 15.Dutcher JD, Vonsaltza MH, Pansy FE. Septacidin, a new antitumor and antifungal antibiotic produced by Streptomyces fimbriatus. Antimicrob Agents Chemother (Bethesda) 1963;161:83–88. [PubMed] [Google Scholar]
- 16.Acton EM, Ryan KJ, Luetzow AE. Antitumor septacidin analogues. J Med Chem. 1977;20:1362–1371. doi: 10.1021/jm00221a002. [DOI] [PubMed] [Google Scholar]
- 17.Sukkurwala AQ, et al. Screening of novel immunogenic cell death inducers within the NCI mechanistic diversity set. OncoImmunology. 2014;3:e28473. doi: 10.4161/onci.28473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayakawa Y, et al. Spicamycin, a new differentiation inducer of mouse myeloid leukemia cells (Ml) and human promyelocytic leukemia cells (HL-60) Agric Biol Chem. 1985;49:2685–2691. [Google Scholar]
- 19.Kamishohara M, et al. Antitumor activity of a spicamycin derivative, KRN5500, and its active metabolite in tumor cells. Oncol Res. 1994;6:383–390. [PubMed] [Google Scholar]
- 20.Gadgeel SM, et al. A phase I clinical trial of spicamycin derivative KRN5500 (NSC 650426) using a phase I accelerated titration “2B” design. Invest New Drugs. 2003;21:63–74. doi: 10.1023/a:1022972427532. [DOI] [PubMed] [Google Scholar]
- 21.Weinstein SM, et al. A spicamycin derivative (KRN5500) provides neuropathic pain relief in patients with advanced cancer: A placebo-controlled, proof-of-concept trial. J Pain Symptom Manage. 2012;43:679–693. doi: 10.1016/j.jpainsymman.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Kamishohara M, et al. Structure-antitumor activity relationship of semi-synthetic spicamycin analogues. J Antibiot (Tokyo) 1993;46:1439–1446. doi: 10.7164/antibiotics.46.1439. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki T, et al. Total synthesis of spicamycin. J Org Chem. 2002;67:2874–2880. doi: 10.1021/jo010925c. [DOI] [PubMed] [Google Scholar]
- 24.Flatt PM, Mahmud T. Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds. Nat Prod Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- 25.Brodersen DE, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 26.Kondo S, Iinuma K, Naganawa H, Shimura M, Sekizawa Y. Structural studies on destomycins A and B. J Antibiot (Tokyo) 1975;28:79–82. doi: 10.7164/antibiotics.28.79. [DOI] [PubMed] [Google Scholar]
- 27.Shimura M, Sekizawa Y, Iinuma K, Naganawa H, Kondo S. Destomycin C, a new member of destomycin family antibiotics. J Antibiot (Tokyo) 1975;28:83–84. doi: 10.7164/antibiotics.28.83. [DOI] [PubMed] [Google Scholar]
- 28.Wencewicz TA, Möllmann U, Long TE, Miller MJ. Is drug release necessary for antimicrobial activity of siderophore-drug conjugates? Syntheses and biological studies of the naturally occurring salmycin “Trojan Horse” antibiotics and synthetic desferridanoxamine-antibiotic conjugates. Biometals. 2009;22:633–648. doi: 10.1007/s10534-009-9218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roosenberg JM, 2nd, Miller MJ. Total synthesis of the siderophore danoxamine. J Org Chem. 2000;65:4833–4838. doi: 10.1021/jo000050m. [DOI] [PubMed] [Google Scholar]
- 30.Kneidinger B, et al. Biosynthesis pathway of ADP-L-glycero-β-D-manno-heptose in Escherichia coli. J Bacteriol. 2002;184:363–369. doi: 10.1128/JB.184.2.363-369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W, et al. Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat Commun. 2015;6:8101. doi: 10.1038/ncomms9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooke JS, Valvano MA. Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. J Biol Chem. 1996;271:3608–3614. doi: 10.1074/jbc.271.7.3608. [DOI] [PubMed] [Google Scholar]
- 33.McArthur F, Andersson CE, Loutet S, Mowbray SL, Valvano MA. Functional analysis of the glycero-manno-heptose 7-phosphate kinase domain from the bifunctional HldE protein, which is involved in ADP-L-glycero-D-manno-heptose biosynthesis. J Bacteriol. 2005;187:5292–5300. doi: 10.1128/JB.187.15.5292-5300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, et al. Divergence of biochemical function in the HAD superfamily: D-glycero-D-manno-heptose-1,7-bisphosphate phosphatase (GmhB) Biochemistry. 2010;49:1072–1081. doi: 10.1021/bi902018y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding L, Seto BL, Ahmed SA, Coleman WG., Jr Purification and properties of the Escherichia coli K-12 NAD-dependent nucleotide diphosphosugar epimerase, ADP-L-glycero-D-mannoheptose 6-epimerase. J Biol Chem. 1994;269:24384–24390. [PubMed] [Google Scholar]
- 36.Kneidinger B, Graninger M, Puchberger M, Kosma P, Messner P. Biosynthesis of nucleotide-activated D-glycero-D-manno-heptose. J Biol Chem. 2001;276:20935–20944. doi: 10.1074/jbc.M100378200. [DOI] [PubMed] [Google Scholar]
- 37.Osborn AR, Kean KM, Karplus PA, Mahmud T. The sedoheptulose 7-phosphate cyclases and their emerging roles in biology and ecology. Nat Prod Rep. 2017;34:945–956. doi: 10.1039/c7np00017k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahmud T, et al. Biosynthetic studies on the α-glucosidase inhibitor acarbose in Actinoplanes sp.: 2-epi-5-epi-valiolone is the direct precursor of the valienamine moiety. J Am Chem Soc. 1999;121:6973–6983. [Google Scholar]
- 39.Dong H, Mahmud T, Tornus I, Lee S, Floss HG. Biosynthesis of the validamycins: Identification of intermediates in the biosynthesis of validamycin A by Streptomyces hygroscopicus var. limoneus. J Am Chem Soc. 2001;123:2733–2742. doi: 10.1021/ja003643n. [DOI] [PubMed] [Google Scholar]
- 40.Czyzyk DJ, Liu C, Taylor EA. Lipopolysaccharide biosynthesis without the lipids: Recognition promiscuity of Escherichia coli heptosyltransferase I. Biochemistry. 2011;50:10570–10572. doi: 10.1021/bi201581b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komaki H, Hosoyama A, Kimura A, Ichikawa N, Igarashi Y. Draft genome sequence of an anicemycin producer, Streptomyces sp. TP-A0648. Genome Announc. 2017;5:e01468-16. doi: 10.1128/genomeA.01468-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker JB. Enzymatic synthesis of aminoglycoside antibiotics: Novel adenosylmethionine:2-deoxystreptamine N-methyltransferase activities in hygromycin B- and spectinomycin-producing Streptomyces spp. and uses of the methylated products. Appl Environ Microbiol. 2002;68:2404–2410. doi: 10.1128/AEM.68.5.2404-2410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.