Significance
It has remained an unresolved question whether microorganisms recovered from the most arid environments on Earth are thriving under such extreme conditions or are just dead or dying vestiges of viable cells fortuitously deposited by atmospheric processes. Based on multiple lines of evidence, we show that indigenous microbial communities are present and temporally active even in the hyperarid soils of the Atacama Desert (Chile). Following extremely rare precipitation events in the driest parts of this desert, where rainfall often occurs only once per decade, we were able to detect episodic incidences of biological activity. Our findings expand the range of hyperarid environments temporarily habitable for terrestrial life, which by extension also applies to other planetary bodies like Mars.
Keywords: habitat, aridity, microbial activity, biomarker, Mars
Abstract
Traces of life are nearly ubiquitous on Earth. However, a central unresolved question is whether these traces always indicate an active microbial community or whether, in extreme environments, such as hyperarid deserts, they instead reflect just dormant or dead cells. Although microbial biomass and diversity decrease with increasing aridity in the Atacama Desert, we provide multiple lines of evidence for the presence of an at times metabolically active, microbial community in one of the driest places on Earth. We base this observation on four major lines of evidence: (i) a physico-chemical characterization of the soil habitability after an exceptional rain event, (ii) identified biomolecules indicative of potentially active cells [e.g., presence of ATP, phospholipid fatty acids (PLFAs), metabolites, and enzymatic activity], (iii) measurements of in situ replication rates of genomes of uncultivated bacteria reconstructed from selected samples, and (iv) microbial community patterns specific to soil parameters and depths. We infer that the microbial populations have undergone selection and adaptation in response to their specific soil microenvironment and in particular to the degree of aridity. Collectively, our results highlight that even the hyperarid Atacama Desert can provide a habitable environment for microorganisms that allows them to become metabolically active following an episodic increase in moisture and that once it decreases, so does the activity of the microbiota. These results have implications for the prospect of life on other planets such as Mars, which has transitioned from an earlier wetter environment to today’s extreme hyperaridity.
The core region of the Atacama Desert is the most arid midlatitude desert on Earth and in the past has been devoid of precipitation for decades. A mean annual precipitation of <20 mm reduces weathering rates and leaching losses to levels below the accumulation rates of atmospheric salts and dust (1). Hence, atmospheric deposition over millions of years has resulted in high salt concentrations in the soils of the hyperarid area (2). The only documented microhabitats in the core region of the Atacama Desert are colonized by microbial communities thriving in surficial salt crusts, where microbial activity is enabled through deliquescence (3, 4). Even though there are traces of microbial life in the subsurface of the Atacama Desert (5), it remains unclear whether these environments support active microbial growth or whether the observed cells are sporadically introduced by atmospheric transport and continuously inactivated and degraded. To answer this question, in April of 2015 we sampled soils from the surface and near subsurface at six locations along a decreasing moisture gradient [coastal soil (CS), alluvial fan (AL), red sands (RS), Maria Elena (ME), Yungay (YU), and Lomas Bayas (LB)] (SI Appendix, Fig. S1) and characterized them and their microbial communities by using a broad suite of complementary methods. Since this sampling occurred shortly after an unexpected rain event, we repeated sampling in February 2016 and January 2017 to determine whether the detected microbial activity in 2015 was ongoing or episodic and related to the temporary increased availability of moisture.
Results
Environmental Setting.
The selected CS site has been occasionally subject to fog and rain, while sites further inland (ME, YU, and LB) are located in hyperarid areas (6, 7), where water content of surface soils is generally 1% by weight (SI Appendix, Fig. S2). Water activity is often below the threshold of 0.6 required to sustain metabolic activity (8). Relative humidity levels are generally below 30% and daily UV irradiation doses were ca. 30 J⋅m−2. Except for LB, where total organic carbon (TOC) reached 0.25% (wt/wt), TOC at all other sites was less than 0.1%. A prerequisite of our study was that the sampled sites are relatively pristine and little affected by human contamination, which was the case based on measured polycyclic aromatic hydrocarbon concentrations which are extremely low and generally in the microgram per kilogram range or lower. Soil minerals at all sites are dominated by alkali feldspar and plagioclase with minor amounts of quartz, chlorite, and amphibole, with some sites displaying a significant amount of anhydrite, bassanite, gypsum, and carbonates. Sites subject to higher levels of moisture (CS, AL, RS) contain large amounts of chlorides (e.g., halite), while soils obtained from the hyperarid areas (YU, ME, LB) mostly contain sulfates (e.g., gypsum or anhydrite), perchlorates (), and chlorates () (SI Appendix, Fig. S3). The first set of field samples was taken in April 2015 1 mo after a major El Niño triggered one of the rare rainfall events in the Atacama Desert (9). Eight millimeters of precipitation was recorded at Baquedana and 33 mm at Antofagasta, which was the highest amount of precipitation since the beginning of the official recording in 1978 (SI Appendix, Fig. S2C) and affected all study sites. The second and third sampling campaigns were conducted in February 2016 and January 2017, respectively, with only two minor rain events in between (each 6.7 mm, recorded at Antofagasta).
Microbial Diversity.
Metagenomic analyses of the DNA pool from topsoils revealed a high bacterial diversity at CS (nonpareil diversity index of 21.2 ± 0.5), similar to sandy soils, but considerably higher compared with the drier areas of YU and ME (nonpareil indexes of 19.5 ± 0.6 and 18.9 ± 0.1, respectively; SI Appendix, Fig. S4A). Phylogenetic profiles (SI Appendix, Fig. S4B) indicate that soils at YU were associated with microbiomes typical for sandy environments and desert soils, mainly consisting of Actinobacteria (5) with Corynebacteriales, Streptomycetales, and Micrococcales being the dominant suborders and a proportional decline of Rubrobacterales from the surface to the subsurface. In contrast, ME was dominated by Geodermatophilaceae, known to colonize hyperarid habitats (10) and tolerate high levels of oxidative stress, desiccation, salts, and metals (11). The same general trend of decreased biomass and diversity with increasing aridity was found for Archaea and Fungi, even though their proportion was lower than that of Bacteria. In all soil samples 200 fungal marker genes were detected, almost exclusively belonging to Ascomycota and Basidiomycota (SI Appendix, Fig. S5 A and B). At CS, Archaea (mainly halobacteria) reached a maximum and dominated the microbial community at a depth of 20–30 cm, while everywhere else they accounted for 4% of sequence reads (SI Appendix, Fig. S5C). Our DNA-based data were corroborated by phospholipid fatty acid profiles (Fig. 1 B and D), serving as biomarkers for living bacteria (12), and cultivation-based approaches both identifying Actinobacteria (e.g., Actinobacterium lienomycini, Kocuria sp., Pseudonocardina sp., Streptomyces sp.), Proteobacteria (e.g., Pseudomonas sp., Paracoccus marinus), and Firmicutes (e.g., Bacillus litoralis, Bacillus simplex, Halobacillus sp.).
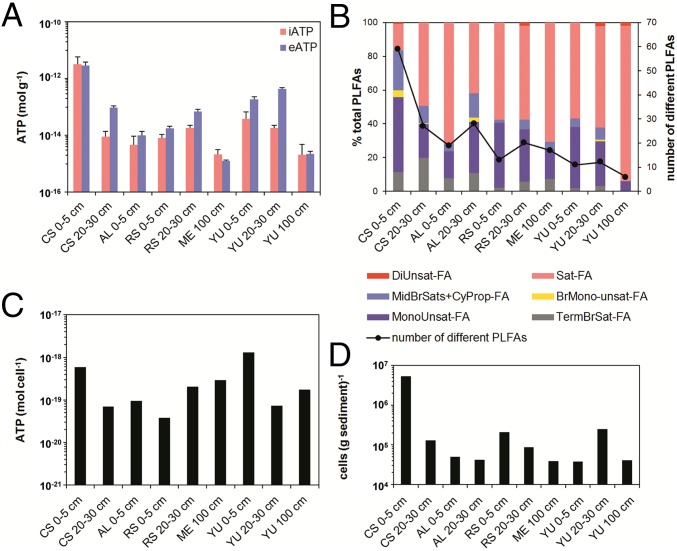
Fig. 1.
(A and B) Concentrations of intracellular ATP (iATP) and extracellular ATP (eATP) (both n = 3) (A) and PLFAs (B) in Atacama Desert soils. A decrease in the number of identified PLFAs indicates a decrease in diversity, which is related to increasing aridity. (C and D) Average cell-based ATP concentrations were obtained by relating iATP concentrations (C) to total biomass levels (D) measured at specific locations, which were obtained from PLFA analysis.
Abundance and Identification of Dead and Living Microorganisms.
A unique cell-separation technique (13) was used to differentiate between intracellular DNA (iDNA) indicating physically intact and potentially viable cells and extracellular DNA (eDNA) mainly representing preserved DNA from dead cells.
Quantitative PCR (qPCR) using universal bacterial primers was performed for both DNA pools, for all soil samples taken along the moisture gradients in 2015 and 2017, and at YU in 2016. After the rain event in 2015, the abundance of 16S rRNA genes in the iDNA pool (proxy of living bacterial biomass) was generally higher than in 2016/2017 (between ∼103 and ∼106 gene copies per gram soil; SI Appendix, Table S1), increasing with moisture (SI Appendix, Fig. S6). The copy numbers in the eDNA pool showed larger variations of between ∼101 and ∼107 gene copies per gram soil, and only at LB was no eDNA detectable with the primers applied in the qPCR. In contrast to the relatively high gene copy numbers in 2015, soil samples from the same sites, but taken 2 y after the rain event, revealed a drastic decrease in living cells (iDNA; equal to or less than 102 gene copies per gram soil). The same trend was also visible in the eDNA pool although gene copy numbers were somewhat higher than in the iDNA pool (SI Appendix, Table S1).
To provide a detailed characterization of living and dead microorganisms, high-throughput sequencing of 16S rRNA gene amplicons of both DNA pools was performed. In environments supporting an active microbial community, the eDNA pool is continuously replenished through biomass turnover of living cells (14). As indicated by the large number of shared operational taxonomic units (OTUs), this was likely the case at CS where relative humidity (RH) and nutrients were highest, constantly supplied by coastal fog. In contrast, OTUs identified at the hyperarid locations showed less overlap between eDNA and iDNA compared with CS, suggesting the dominance of autochthonous microbial taxa rather than of inactive transitory microorganisms periodically introduced by wind (Fig. 2A). Key organisms of these communities consisted of unclassified Acidimicrobiales, Actinobacteria, Alicyclobacillus, Burkholderia, Comamonadaceae, and Xanthomonadaceae. Although abundances differed among sites and soil depths, these characteristic taxa identified from iDNA were found in all samples from all sites and depths, suggesting a native and metabolically active desert core community. In general, as dryness increased, microbial diversity decreased, analogous to previous observations (15, 16).
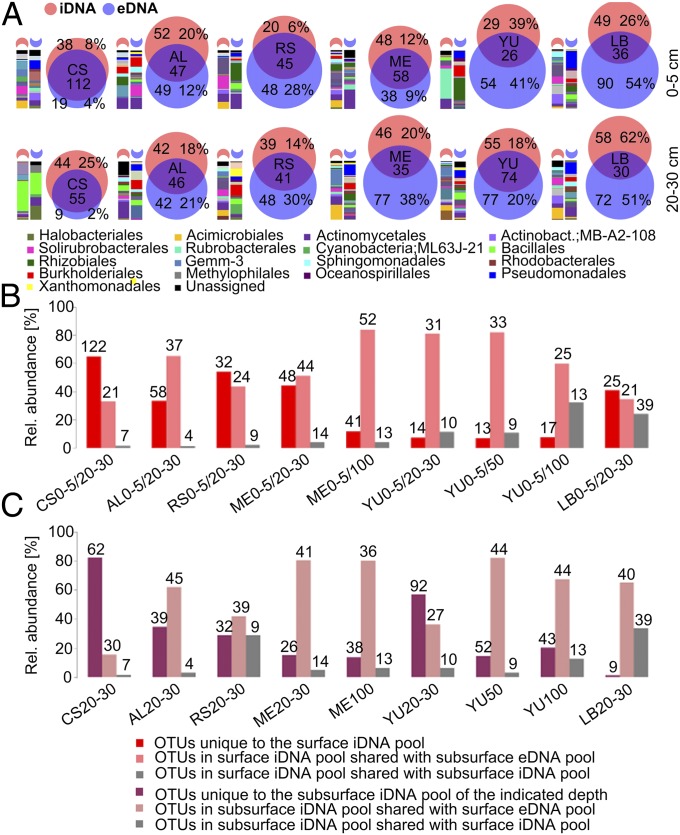
Fig. 2.
Microbial community structure and relationship between iDNA and eDNA pools at six soil sampling sites in the Atacama Desert: CS, AL, RS, ME, YU, and LB. (A) Venn diagrams of iDNA and eDNA OTU intersections for samples collected at 0–5 cm and 20–30 cm depth. Numbers indicate the numbers of different OTUs, and percentages refer to relative abundances of reads unique to iDNA or eDNA. Bars to the left of the Venn diagrams show relative abundances of bacterial orders in the subsets unique to the iDNA and eDNA pools of the indicated sampling depth. (B) Classification of iDNA pools from soil surface samples (0–5 cm) collected at the six sampling sites in comparison with the iDNA and eDNA pools in subsurface soil layers. (C) Classification of the subsurface iDNA pools (20–30 cm, 50 cm, 100 cm) in comparison with the iDNA and eDNA pools in the surface soils (0–5 cm). The bars show the percentages of OTU reads in the corresponding subsets, and numbers indicate the numbers of different OTUs.
Abundance of Endospores.
The dormant component of the bacterial communities was specifically assessed for the 2015 sampling period by quantifying endospores, which are characteristic of the phylum Firmicutes and stand out by their exceptional resistance to environmental stresses. Dipicolinic acid (DPA), a specific biomarker of intact endospores, was detected at all sites. Endospore concentrations in surface layers, however, decreased with increasing aridity by almost two orders of magnitude (7.7 × 105 to 1.5 × 104 spores per gram soil; SI Appendix, Table S2). The large size of this community suggests that an extensive and persistent Firmicutes seed bank remains available in the Atacama Desert, which is in agreement with the dominance of isolates from this phylum in our cultivation experiments. Importantly, the contribution of endospores to the iDNA pool was likely minor, because the conventional extraction methods that were used to extract iDNA from intact cells do not usually extract DNA from spores (17). Therefore, it is not surprising that the phylogenetic composition derived from iDNA sequencing does not reflect the abundance of endospores from the phylum Firmicutes (SI Appendix, Fig. S7).
Metabolic Activity.
As many of the cultivated bacteria were spore formers, we used independent complementary analytical approaches to obtain conclusive evidence for living microorganisms and their potential activity in hyperarid habitats.
First, evidence for microbial activity was obtained by a fluorescein diacetate hydrolysis assay to determine enzymatic activity (SI Appendix, Table S3). Enzymatic activity was highest at the surface of CS, ME, and LB sites, but still detectable at all other sites and shallow depths (0–5 cm and 20–30 cm, respectively), except for LB 20–30 cm, where it was below the detection limit of 10−3 nmol⋅g−1⋅h−1.
Second, our hypothesis of active, native microbial communities was supported by ATP analyses. These analyses allow the separation of intracellular ATP (iATP), indicative of viable cells, and extracellular ATP (eATP), indicative of ATP remaining in the soil after cell lysis. iATP levels were up to three orders of magnitude higher (3 × 10−12 mol⋅g−1) at CS compared with the sampling sites located in the driest desert core (e.g., 2 × 10−15 mol⋅g−1 at YU 100 cm depth; Fig. 1). Overall, ATP analyses supported the general trend of decreasing microbial activity with increasing aridity, both along the studied moisture transect (2015) and in YU surface soils from 2015 to 2017 (SI Appendix, Table S1). The ATP analyses provide evidence that even in the most arid sites of the Atacama core region native microorganisms can be at times metabolically active.
Third, the presence of metabolically active microorganisms was supported by the analyses of water-extractable metabolites via direct injection electrospray ionization Fourier-transform ion cyclotron resonance mass spectrometry [ESI(-) FT-ICR-MS], which allowed the accurate calculation of elemental formulas. On average, a rich signature of ∼1,600 elemental compositions (CHNOS) was detected in all samples, indicating a geochemical footprint (18) typical of natural organic matter superimposed by a biological footprint of fresh organic material (19) involving amino acids, small peptides, and fatty acids (SI Appendix, Fig. S8). Results suggest that water-soluble organic compounds consisted mainly of aliphatic carbohydrates and fatty acids (CHO), while nitrogen- and sulfur-containing compounds (CHNO and CHOS) were less abundant. The relative abundance of compounds, predominantly reflecting metabolic activity, decreased significantly along the aridity gradient from the coast to inland with constant low amounts of organic compounds at the hyperarid locations of ME, YU, and LB. This trend again suggests a marked decline in metabolic activity from moist to hyperarid soil habitats (SI Appendix, Fig. S9). Nevertheless, there was clear conservation of biosignatures even in the hyperarid locations, showing evidence of past and, most likely, recent metabolic activity, especially at a depth of 20–30 cm in YU (2015), where different types of metabolites were distinguishable (SI Appendix, Fig. S9F).
Fourth, the metagenomic analysis from the soil samples obtained in April 2015 indicated that microorganisms were active even in the driest soil samples. Sequence abundance of mycobacteriophages, gordoniaphages, and streptomycophages correlated positively with that of their respective hosts found in the different samples [Spearman coefficient correlations of at CS, at ME, and at YU (SI Appendix, Fig. S4 C–E)]. The detected virus–host relationships seem to be consistent with the observations that microbial blooms are followed by a bloom of phages specific for the microbes that dominated the microbial bloom (20, 21). However, while most identified viruses are phages persistent in the environment that have also been identified by other desert studies (22), the vast majority of the identified viruses (95+%) belong to the family Caudovirales, which includes both virulent and temperate members. Thus, we cannot exclude the presence of a large number of undetected prophages that might stay dormant for long periods of time.
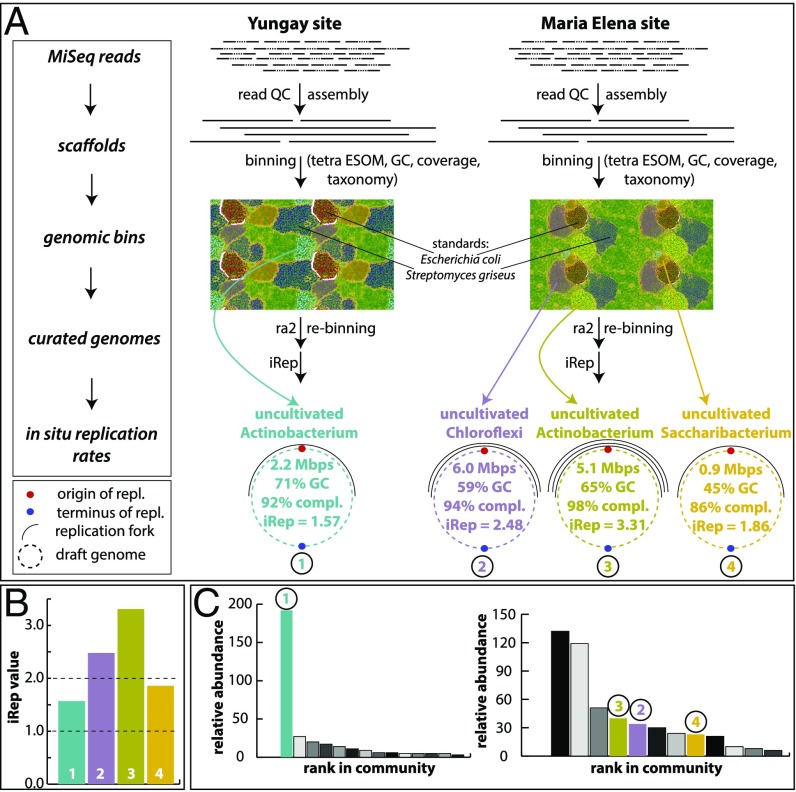
Finally, we investigated in situ genome replication rates of organisms via a genome-resolved metagenomics approach (23) of samples retrieved from YU and ME. For YU, we reconstructed a draft genome of the most dominant organism (uncultivated Actinobacteria) with an estimated completeness of 92% based on single-copy genes (Fig. 3). The iRep value (24) of the genome was 1.57, which is indicative of its slow replication. For the ME site, we retrieved three high-quality draft genomes belonging to members of the phyla Chloroflexi, Actinobacteria, and Saccharibacteria (completeness 86–98%; Fig. 3). The in situ replication rates of these genomes varied between 1.86 and 3.31. The lower replication rates compare with literature values of a wide array of organisms across multiple phyla, but the iRep value of 2.48 for the Chloroflexi indicates that each genome in the population has on average one bidirectional replication ongoing (24). The genome replication rate for the Actinobacteria was extremely high, which indicates that each genome of this population had several replication forks at the time of sampling, thus providing strong evidence for microbial activity.
Fig. 3.
Genome-resolved metagenomics analyses and results. (A) Work flow and main results from genome-resolved metagenomics. For details please see SI Appendix, Read-Based Metagenomics, Genome-Resolved Metagenomics and in Situ Replication Rates. Genome replication forks are symbolic. (B) Overview of iRep values retrieved from the four genomes from the YU and ME sites (color codes correspond to those in A). Dashed line at value 1 marks threshold at which no replication occurs. Dashed line at value 2 marks where each genome of a population has on average bidirectional replication taking place (24). (C) Rank-abundance curves based on rpS3 genes. Colored genomes correspond to those in A and B. For the YU site (Left rank-abundance curve) the most dominant organism was reconstructed, and all other genomes were fragmented. For the ME site (Right rank-abundance curve) three genomes were reconstructed. The three most abundant organisms were Actinobacteria, which were similar in GC and abundance.
A Transitory Habitat?
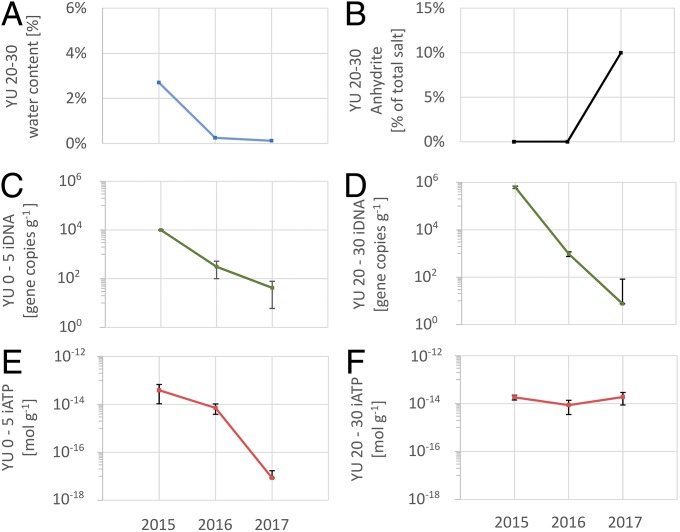
The continuous decline of nonstructural water at 20–30 cm depth at YU, from 2.7% by weight in 2015 to 0.2% and 0.1% in 2016 and 2017, respectively, suggests temporarily favorable conditions for the activity of specialized microorganisms after the rare precipitation event until water activity fell again beneath a critical threshold (Fig. 4A). Mineralogical data confirmed a desiccation process in the later years as some of the gypsum at YU 20–30 cm dehydrated to anhydrite (Fig. 4B). The steep decline of recovered iDNA by three to five orders of magnitudes (Fig. 4 C and D) indicates that the sampling campaign in 2015, shortly after a major and very rare rainfall event, tapped into a temporary, time-constrained habitat, rather than a permanent one. ATP analyses, indicative of active organisms, also support that assessment. The iATP values follow that trend, declining by more than three orders of magnitude in the YU surface soil (Fig. 4E), but remain constant in the deeper soil layer at YU (Fig. 4F), pointing to a longer retention of microbial activity. Possibly some water released by the desiccation of gypsum to anhydrite remains accessible to a specific part of the microbial community. This possibility is supported by isotopic evidence. The D values for the waters in the hydrous sulfate minerals suggest that small amounts of water accessible to microorganisms might be available even in these hyperarid soils (SI Appendix, Table S4), e.g., in the form of thin H2O films at mineral surfaces (25) or as a product of mineral–water exchange reactions (26).
Fig. 4.
Comparison of sampling events of April 2015, February 2016, and January 2017 at YU. (A) Available nonstructural water decreases significantly from 2015 to 2017. (B) Some of the gypsum at YU 20–30 desiccated and formed anhydrite. (C and D) Intracellular DNA amounts indicative of living organisms drop by several orders of magnitudes at 0–5 cm and 20–30 cm depths. (E and F) Intracellular ATP amounts indicative of active organisms drop by several orders of magnitudes at 0–5 cm, but stay constant at 20–30 cm depth.
Two other rainfall events occurred in August 2015 and June 2016 (each 6.7 mm at Antofagasta), but it is unclear how much rain fell at YU. No indication of that rainfall event was observed in our sharply declining iDNA and iATP values for the surface soils from April 2015 to January 2017. This suggests that both the August 2015 and June 2016 events were either insufficient to trigger temporarily habitable conditions and a microbial response or, since they occurred several months before our next sampling round, were too small and subsided by the time we resampled. Davis et al. (27) estimated that 2 mm or more precipitation are needed to provide free water for the support of biological activity in the soil. In previous studies, Navarro-Gonzalez et al. (7) were not able to recover any DNA from the YU site, and Warren-Rhodes et al. (28) reported on the virtual absence of hypolithic cyanobacteria. Certainly, the literature data can provide only a qualitative assessment about the presence or activity of microorganisms, because of both the methodologies used and the spatial heterogeneity of the sites. In contrast to our study, it appears that previous sampling campaigns did not tap into habitable conditions.
Discussion
The Atacama Desert soil microbiome has evolved as a result of the prevailing environmental conditions. While the soil surface was dominated by desiccation and UV-resistant species (Geodermatophilaceae and Rubrobacter), deeper layers with higher salt content were dominated by halophilic bacteria such as Betaproteobacteria (Comamonadaceae) or Firmicutes (Bacillaceae, Alicyclobacillaceae) (SI Appendix, Fig. S7) and halophilic Archaea (i.e., Halobacteria) at CS 20–30 cm (SI Appendix, Fig. S5C). Notably, microbiomes associated with the hyperarid soils were dominated by Bacteria rather than Archaea, consistent with an assessment of the global distribution of archaeal abundances in soils (29). The amount of unique OTUs in the iDNA pool of the surface and subsurface soils of the hyperarid localities ME and YU was much lower than at the moister sites. OTUs shared between iDNA recovered from the surface and eDNA from deeper layers (Fig. 2B) and between iDNA from the subsurface and eDNA from the surface (Fig. 2C) drastically increased compared with those in CS, indicating distinct microbial populations at different depths with increasing dryness. This indicates that selection pressures for microorganisms were much higher in the hyperarid surface soils than in the wetter coastal area, resulting in species well adapted to the extreme dryness and UV radiation. However, some salt-tolerant bacteria such as Acidimicrobiales, Comamonadaceae, and Bacillaceae potentially survive in deeper soil layers after being buried (SI Appendix, Fig. S7), e.g., by ongoing atmospheric deposition of salts and sediments or by halo- and thermoturbation of soils. Alternatively, microbial communities might have persisted in the subsurface since the onset of desertification, or an initial community successively changed over geological time to cope with altered environmental conditions (5). Our results suggest that incoming microbial “newcomers” have at least been exposed to passive environmental selection, but also maintain transient activity even in the deeper soil layers and sustain viability in the Atacama Desert for very long time periods. However, it remains questionable whether the organisms found in this environment are adapting to the harsh conditions present. Bacteria reaching the Atacama Desert by atmospheric processes have been exposed to desiccation and UV stress during aerial transport, possibly for extended periods (30). This suggests that environmental species filtering could be an important factor contributing to shaping the indigenous microbial communities. In line with this hypothesis, our shotgun metagenomics data revealed several genes associated with dehydration tolerance [e.g., groEL, dnaK, fadD, glgX-malZ, phaC (31)] and radiation/desiccation tolerance [e.g., recQ (32)]. Immunoassays corroborated these metagenomic results by detecting ATP synthase, GroEL, CspA, and DPS DNA-protecting proteins at YU (50 cm), CS, and RS, and metaproteomic analyses of samples taken at YU also confirmed the presence of ATP synthase and GroEL.
One additional challenge for microorganisms to persist at both surface and subsurface locations is the low organic matter content characteristic of hyperarid soils. A higher TOC and moisture content allowed a higher total microbial biomass and diversity (Fig. 1 and SI Appendix, Fig. S6). For example, chloromethane (CH3Cl) release during low-temperature thermolysis of surface soil samples, which is indicative of hetero-bonded methyl groups of organic matter, was highest for CS (∼300 ng⋅g−1) and much lower at the hyperarid sites (∼1–5 ng⋅g−1) (SI Appendix, Fig. S10). Stable hydrogen isotope analyses of the released chloromethane confirmed the biochemical origin of the methyl group. The emission profiles of CH3Cl are almost identical to observations made by the Curiosity rover on Mars (33), where even harsher environmental conditions prevail than in the hyperarid core of the Atacama Desert [lack of water, scarcity of organic matter, high UV irradiation, and high salt content in the soils including bassanite and perchlorates (34)]. No rain can fall from the Martian atmosphere today (35), but liquid water can be present near the Martian surface in the form of nightly snow storms/ice microbursts (36), fog (37), near-surface groundwater (38), and possibly also from mineral dehydration reactions (39). On Mars, the deeper soil layers with a higher water activity and reduced exposure to environmental stresses (e.g., UV irradiation, large daily temperature fluctuations) are expected to be more suitable for supporting life. At YU this was the case, with the gypsum-rich soil layer at a depth of 20–30 cm containing a higher biomass and microbial diversity (Figs. 1 B and D and 2) and also retaining a similar level of activity beyond 2015 for at least 2 y more. Thus, we observe in the hyperarid core of the Atacama Desert a transitory habitat with microorganisms that are active for short periods of time and which can serve as a reasonable working model for Mars.
Conclusions
Although both microbial biomass and diversity in the Atacama Desert decrease with increasing aridity, our study shows that even the lowest precipitation levels on Earth can sustain episodic incidences of microbial activity. There is no single agreed-upon method known to date reaching the bar of evidence for microbial activity for such low-biomass environments. However, using our complementary tool box of combining different methodologies, including unique genome-resolved metagenomics, we have addressed the question of microbial activity and can answer it positively for the sampling time after the major precipitation event in 2015. Thereafter, the biomarkers for microbial activity dropped dramatically, inferring that the transitory habitable conditions have ended until the next major rain event may occur, providing a sufficient amount of free water for the microbial biota. The insights gained from the hyperarid core of the Atacama Desert can serve as a working model for Mars, where environmental stresses are even harsher. If life ever evolved on Mars, the results presented here suggest that it could have endured the transition from the early aquatic stage, through increasing aridity cycles, and perhaps even found a subsurface niche beneath today’s severely hyperarid surface.
Materials and Methods
Detailed methods, including a description of the sampling sites with numerous figures and tables, are provided in SI Appendix. Two unique methods were used. The e/i-DNA methodology is described in detail in an appropriate subject journal (13), with some of the associated issues discussed elsewhere (40). The validity of the e/i-DNA method is further supported by a positive correlation between microbial biomass and the Shannon index (SI) calculated for iDNA (), but not with either calcium or sulfate concentrations. Conversely, the SI for eDNA correlated with calcium and sulfate soil concentrations ( and 0.61, respectively), but not with biomass (). The other state-of-the-art method was used to measure in situ replication rates of genomes by calculating the number of active replication forks reconstructed from the metagenome sequences (15) (see SI Appendix, Read-Based Metagenomics, Genome-Resolved Metagenomics and in Situ Replication Rates for more details). All data reported in this paper are compiled in SI Appendix and have been archived at GenBank/EMBL under BioProject ID PRJNA395196 and at EMBL-EBI under accession no. PRJEB20402 (sample IDs ERS1666624–ERS1666714).
Supplementary Material
Acknowledgments
D.S.-M. acknowledges support by the European Research Council Advanced Grant Habitability Of Martian Environments (339231), which provided base funding for the study, including sample collection. The fungal diversity assessment was supported by funding (to H.-P.G.) through the Leibniz Senatsausschuss Wettbewerb Project MycoLink and DFG Project Microprime (GR1540/23-1). F.K. received financial support from the German Science Foundation (DFG KE 884/8-2). The 16S rRNA gene amplicon (MiSeq) sequencing was financed through the Helmholtz Research Program “Geosystem–The Changing Earth” and the data were processed by Fabian Horn (GFZ German Research Center for Geosciences–Helmholtz Center Potsdam). The stable isotopic composition of water was carried out through the Europlanet 2020 Research Infrastructure supported by the European Union’s Horizon 2020 research and innovation program (654208). Endospore quantification benefited from funding by the Deep Carbon Observatory through a Pilot Project (L.W.). Part of the organic geochemical analyses were performed at Imperial College London, supported by United Kingdom Space Agency Grant ST/N000560/1. V.P. and D.C.’s work was supported by the Spanish Ministry of Economy and Competitiveness Grants ESP2015-69540-R and RYC-2014-19446, respectively. G.V. was supported by a Humboldt Research Fellowship for postdoctoral researchers. M.F. acknowledges support from National Institute of Food and Agriculture Hatch Project 1014527. The Leibniz Institute of Freshwater Ecology & Inland Fisheries (IGB) housed the workshop at which much of the presented work was coordinated. We thank M. Degebrodt for technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The metagenome sequences reported in this paper have been deposited in the EMBL-EBI database (accession no. PRJEB20402 with the sample IDs ERS1666624–ERS1666714) and in the GenBank/EMBL database (BioProject ID PRJNA395196).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714341115/-/DCSupplemental.
References
- 1.Ewing SA, et al. A threshold in soil formation at Earth’s arid-hyperarid transition. Geochim Cosmochim Acta. 2006;70:5293–5322. [Google Scholar]
- 2.Michalski G, Boehlke JK, Thiemens M. Long term atmospheric deposition as the source of nitrate and other salts in the Atacama Desert, Chile: New evidence from mass-independent oxygen isotopic compositions. Geochim Cosmochim Acta. 2004;68:4023–4038. [Google Scholar]
- 3.Davila AF, Schulze-Makuch D. The last possible outposts for life on Mars. Astrobiology. 2016;16:159–168. doi: 10.1089/ast.2015.1380. [DOI] [PubMed] [Google Scholar]
- 4.Wierzchos J, et al. Microbial colonization of Ca-sulfate crusts in the hyperarid core of the Atacama Desert: Implications for the search for life on Mars. Geobiology. 2011;9:44–60. doi: 10.1111/j.1472-4669.2010.00254.x. [DOI] [PubMed] [Google Scholar]
- 5.Crits-Christoph A, et al. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome. 2013;1:28. doi: 10.1186/2049-2618-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azua-Bustos A, Caro-Lara L, Vicuna R. Discovery and microbial content of the driest site of the hyperarid Atacama Desert, Chile. Environ Microbiol Rep. 2015;7:388–394. doi: 10.1111/1758-2229.12261. [DOI] [PubMed] [Google Scholar]
- 7.Navarro-Gonzalez R, et al. Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science. 2003;302:1018–1021. doi: 10.1126/science.1089143. [DOI] [PubMed] [Google Scholar]
- 8.Stevenson A, et al. Multiplication of microbes below 0.690 water activity: Implications for terrestrial and extraterrestrial life. Environ Microbiol. 2015;17:257–277. doi: 10.1111/1462-2920.12598. [DOI] [PubMed] [Google Scholar]
- 9.Bozkurt D, Rondanelli R, Garreaud R, Arriagada A. Impact of warmer eastern tropical Pacific SST on the March 2015 Atacama floods. Mon Weather Rev. 2016;144:4441–4460. [Google Scholar]
- 10.Normand P. Geodermatophilaceae fam. nov., a formal description. Int J Syst Evol Microbiol. 2006;56:2277–2278. doi: 10.1099/ijs.0.64298-0. [DOI] [PubMed] [Google Scholar]
- 11.Montero-Calasanz MdC, et al. Geodermatophilus tzadiensis sp. nov., a UV radiation-resistant bacterium isolated from sand of the Saharan desert. Syst Appl Microbiol. 2013;36:177–182. doi: 10.1016/j.syapm.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Ewing SA, et al. Rainfall limit of the N cycle on Earth. Glob Biogeochem Cycles. 2007;21:GB3009. [Google Scholar]
- 13.Alawi M, Schneider B, Kallmeyer J. A procedure for separate recovery of extra- and intracellular DNA from a single marine sediment sample. J Microbiol Methods. 2014;104:36–42. doi: 10.1016/j.mimet.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Levy-Booth DJ, et al. Cycling of extracellular DNA in the soil environment. Soil Biol Biochem. 2007;39:2977–2991. [Google Scholar]
- 15.Robinson CK, et al. Microbial diversity and the presence of algae in halite endolithic communities are correlated to atmospheric moisture in the hyper-arid zone of the Atacama Desert. Environ Microbiol. 2015;17:299–315. doi: 10.1111/1462-2920.12364. [DOI] [PubMed] [Google Scholar]
- 16.Finstad KM, et al. Microbial community structure and the persistence of cyanobacterial populations in salt crusts of the hyperarid Atacama desert from genome-resolved Metagenomics. Front Microbiol. 2017;8:1435. doi: 10.3389/fmicb.2017.01435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuske C, et al. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl Environ Microbiol. 1998;64:2463–2472. doi: 10.1128/aem.64.7.2463-2472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertkorn N, et al. Natural organic matter and the event horizon of mass spectrometry. Anal Chem. 2008;80:8908–8919. doi: 10.1021/ac800464g. [DOI] [PubMed] [Google Scholar]
- 19.Rossello-Mora R, et al. Metabolic evidence for biogeographic isolation of the extremophilic bacterium Salinibacter ruber. ISME J. 2008;2:242–253. doi: 10.1038/ismej.2007.93. [DOI] [PubMed] [Google Scholar]
- 20.Maslov S, Sneppen K. Population cycles and species diversity in dynamic Kill-the-Winner model of microbial ecosystems. Sci Rep. 2017;7:39642. doi: 10.1038/srep39642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–1328. [Google Scholar]
- 22.Fierer N, et al. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol. 2007;73:7059–7066. doi: 10.1128/AEM.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 24.Brown CT, Olm MR, Thomas BC, Banfield JF. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol. 2016;34:1256–1263. doi: 10.1038/nbt.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson SA, Bish DL. Formation of gypsum and bassanite by cation exchange reactions in the absence of free-liquid H2O: Implications for Mars. J Geophys Res. 2011;116:E09010. [Google Scholar]
- 26.Palacio S, Azorín J, Montserrat-Martí G, Ferrio JP. The crystallization water of gypsum rocks is a relevant water source for plants. Nat Commun. 2014;5:4660. doi: 10.1038/ncomms5660. [DOI] [PubMed] [Google Scholar]
- 27.Davis WL, de Pater I, McKay CP. Rain infiltration and crust formation in the extreme arid zone of the Atacama Desert, Chile. Planet Space Sci. 2010;58:616–622. [Google Scholar]
- 28.Warren-Rhodes KA, et al. Cyanobacterial ecology across environmental gradients and spatial scales in China’s hot and cold deserts. FEMS Microbiol Ecol. 2007;61:470–482. doi: 10.1111/j.1574-6941.2007.00351.x. [DOI] [PubMed] [Google Scholar]
- 29.Bates ST, et al. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith DJ, Griffin DW, Schuerger AC. Stratospheric microbiology at 20 km over the Pacific Ocean. Aerobiologia. 2010;26:35–46. [Google Scholar]
- 31.Rajeev L, et al. Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J. 2013;7:2178–2191. doi: 10.1038/ismej.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua X, Huang L, Tian B, Hua Y. Involvement of recQ in the ultraviolet damage repair pathway in Deinococcus radiodurans. Mutat Res. 2008;641:48–53. doi: 10.1016/j.mrfmmm.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Ming DW, et al. Volatile and organic compositions of sedimentary rocks in Yellowknife Bay, Gale crater, Mars. Science. 2014;343:1245267. doi: 10.1126/science.1245267. [DOI] [PubMed] [Google Scholar]
- 34.Neilson JW, et al. Arid soil microbiome: Significant impacts of increasing aridity. mSystems. 2017;2:e00195. doi: 10.1128/mSystems.00195-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Craddock RA, Lorenz RD. The changing nature of rainfall during the early history of Mars. Icarus. 2017;293:172–179. [Google Scholar]
- 36.Spiga A, et al. Snow precipitation on Mars driven by cloud-induced night-time convection. Nat Geosci. 2017;10:652–657. [Google Scholar]
- 37.Möhlmann DT, Niemand M, Formisano V, Savijrvi H, Wolkenberg P. Fog phenomena on Mars. Planet Space Sci. 2009;57:1987–1992. [Google Scholar]
- 38.Malin MC, Edgett KS. Evidence for recent groundwater seepage and surface runoff on Mars. Science. 2000;288:2330–2335. doi: 10.1126/science.288.5475.2330. [DOI] [PubMed] [Google Scholar]
- 39.Bish DL, Carey JW, Vaniman DT, Chipera SJ. Stability of hydrous minerals on the Martian surface. Icarus. 2003;164:96–103. [Google Scholar]
- 40.Vuillemin A, et al. Preservation and significance of extracellular DNA in ferruginous sediments from Lake Towuti, Indonesia. Front Microbiol. 2017;8:1440. doi: 10.3389/fmicb.2017.01440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.