Significance
Invasive nontyphoidal Salmonella disease is a major and previously neglected tropical disease responsible for an estimated ∼390,000 deaths per year in Africa, largely caused by a variant of Salmonella Typhimurium called ST313. Despite the availability of >100,000 Salmonella genomes, it has proven challenging to associate individual SNPs with pathogenic traits of this dangerous bacterium. Here, we used a transcriptomic strategy to identify a single-nucleotide change in a promoter region responsible for crucial phenotypic differences of African S. Typhimurium. Our findings show that a noncoding nucleotide of the bacterial genome can have a profound effect upon the pathogenesis of infectious disease.
Keywords: Salmonella, noncoding genome, transcriptomics, evolution of virulence, host adaptation
Abstract
Salmonella enterica serovar Typhimurium ST313 is a relatively newly emerged sequence type that is causing a devastating epidemic of bloodstream infections across sub-Saharan Africa. Analysis of hundreds of Salmonella genomes has revealed that ST313 is closely related to the ST19 group of S. Typhimurium that cause gastroenteritis across the world. The core genomes of ST313 and ST19 vary by only ∼1,000 SNPs. We hypothesized that the phenotypic differences that distinguish African Salmonella from ST19 are caused by certain SNPs that directly modulate the transcription of virulence genes. Here we identified 3,597 transcriptional start sites of the ST313 strain D23580, and searched for a gene-expression signature linked to pathogenesis of Salmonella. We identified a SNP in the promoter of the pgtE gene that caused high expression of the PgtE virulence factor in African S. Typhimurium, increased the degradation of the factor B component of human complement, contributed to serum resistance, and modulated virulence in the chicken infection model. We propose that high levels of PgtE expression by African S. Typhimurium ST313 promote bacterial survival and dissemination during human infection. Our finding of a functional role for an extragenic SNP shows that approaches used to deduce the evolution of virulence in bacterial pathogens should include a focus on noncoding regions of the genome.
The bacterial pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) is one of the best-understood pathogens and a major cause of gastroenteritis globally. One sequence type of S. Typhimurium, ST313, is the primary cause of invasive nontyphoidal Salmonellosis (iNTS) across Africa, resulting in ∼388,000 deaths each year (1). Coinfection with HIV or malaria infection and young age (<5 y of age) are known risk factors for iNTS infection (1, 2).
Multidrug resistance has contributed to the expansion of S. Typhimurium ST313. Whole-genome sequence-based phylogenetics revealed clonal replacement of ST313 lineage 1 by lineage 2 in the mid-2000s, accompanied by the acquisition of chloramphenicol resistance (3). The ST313 clade has recently acquired resistance to ceftriaxone, a first-line antibiotic for multidrug-resistant bacterial infections (4). Genomic comparison between the classical gastroenteritis-associated S. Typhimurium ST19 and the African ST313 isolates shows that gene content and synteny are highly conserved, that ST313 has a distinct repertoire of plasmids and prophages, and carries 77 pseudogenes reflecting a degree of genome degradation (5, 6). ST313 and ST19 share >4,000 genes, and their core genomes differ by ∼1,000 SNPs (5). We have reported that 2.7% of the S. Typhimurium isolated from patients in England and Wales are ST313, but lack the characteristic prophages BTP1 and BTP5 that are signatures of African ST313 lineages (7).
Certain virulence-associated phenotypes have been examined in ST313 strains. Compared with the ST19 group of gastroenteritis-associated S. Typhimurium, ST313 is more resistant to complement-mediated killing by human sera (8, 9) and to macrophage-mediated killing (10). ST313 exhibits a stealth phenotype during macrophage infection, consistent with an immune evasion strategy that causes reduced levels of IL-1β cytokine production, apoptosis, and caspase-1–dependent macrophage death (10, 11). Strategies used by ST313 to evade the innate immune system include reduced expression of both FliC and the SPI1 effector protein SopE2 (11). Furthermore, ST313 does not express the SseI effector protein due to pseudogenisation of the sseI gene, allowing ST313 to cause increased systemic infection via dendritic cell-mediated dissemination to extraintestinal sites in the murine infection model (12).
We used a functional genomic approach to search for SNPs responsible for the increased virulence of S. Typhimurium ST313 lineage 2.
Results
The reference strain for S. Typhimurium ST313 lineage 2 is D23580, which was isolated from an HIV− Malawian child (5). The strain 4/74 was isolated from a calf in the United Kingdom and is a well-characterized representative of S. Typhimurium ST19. Our challenge was to identify which, if any, of the >1,000 SNPs that separate strains D23580 and 4/74 serve to differentiate the strains in terms of gene expression and phenotype. We investigated whether the emergence of the epidemic clade of S. Typhimurium ST313 was linked to the altered expression of a core genome-encoded virulence factor. Rather than focusing on a comparison of the core genome, we used comparative transcriptomics to identify transcripts that were both expressed at different levels and associated with a distinct SNP in the promoter region.
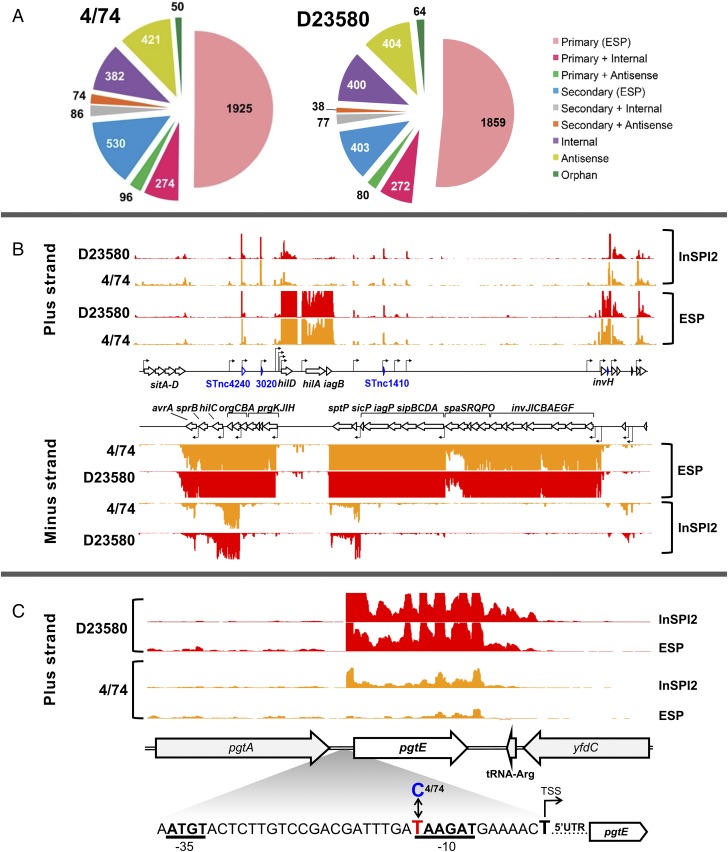
This study built upon the primary transcriptome of S. Typhimurium ST19 strain 4/74, which we determined using a combination of RNA-seq and differential RNA-seq (dRNA-seq) under infection-relevant growth conditions (13, 14). By working at the single-nucleotide level, we defined transcriptional start sites (TSS), and cataloged the transcripts expressed in the bacterial cell (15, 16). Here, we used the same approach to define the primary transcriptome and to identify the TSS of D23580, a representative strain of African S. Typhimurium ST313. RNA was isolated from in vitro growth conditions that reflect the extracellular and intracellular stages of infection, namely the early stationary phase (ESP) and the SPI2-inducing condition (InSPI2) (Materials and Methods). To find all relevant TSS, a pooled sample containing RNA from 16 environmental conditions was also analyzed (14) (Materials and Methods). TSS were identified by comparison of mapped sequence reads from each pair of dRNA-seq and RNA-seq samples, as described previously (13, 14, 16). We identified 3,597 TSS for S. Typhimurium strain D23580, revealing the active gene promoters across the genome of an ST313 isolate for the first time. Previously, we reported the locations of 3,838 TSS for the ST19 strain 4/74 (13). Categorization of the TSS into different classes showed that a similar proportion of transcription initiation sites of 4/74 and D23580 were designated as primary (61%) or antisense (11%) (Fig. 1A).
Fig. 1.
Primary transcriptome analysis of D23580 shows that virulence gene pgtE is highly expressed, and is associated with a SNP in the conserved −10 promoter motif. Classification of TSS of S. Typhimurium in 4/74 and D23580. (A) Categorization of TSS identified in S. Typhimurium 4/74 and D23580, respectively, into nine different promoter classes (16). (B) Visualization of mapped sequence reads of the SPI1 pathogenicity island in S. Typhimurium 4/74 and D23580, respectively [Integrated Genome Browser (IGB), scale 0–100 normalized reads for every sample]. Names of coding genes and sRNAs are labeled in black and blue, respectively. TSS are indicated by cornered arrows. (C) The sequence reads mapped to the pgtE locus were visualized in the IGB (67) (scale 0–100 normalized reads for every sample). Magnified region shows the pgtE promoter with −35/−10 promoter motifs in bold, and the TD23580 or C4/74 SNP highlighted.
We determined the level of conservation of transcriptional organization between D23580 and 4/74 by identifying the TSS shared between the two strains. The locations of the majority of the TSS defined for strain 4/74 were conserved in strain D23580. Specifically, of the 3,838 TSS of strain 4/74, 390 were absent from D23580 and included TSS located in 4/74-specific regions such as prophages sopEΦ and Gifsy-1 (Dataset S1, tab g). We identified 63 D23580-specific TSS, mainly located in the BTP1 and BTP5 prophages of D23580, which are absent from strain 4/74 (5, 6) (Dataset S1, tab d).
To benchmark the transcriptional architecture, we first focused on Salmonella pathogenicity islands SPI1 and SPI2, which are required for key aspects of Salmonella virulence (17). The locations of all TSS within the SPI1 and SPI2 islands were identical in strains D23580 and 4/74 (Fig. 1B and Dataset S1, tab c). In summary, two closely related S. Typhimurium strains that varied by 1,488 SNPs at the core genome level had a high level of conservation at the transcriptional level and shared 90% of promoter regions (Dataset S1, tab a).
To address our hypothesis that the level of expression of certain virulence genes varied between strains D23580 and 4/74 due to changes at the DNA sequence level, we cross-referenced the SNP differences between the two strains with the locations of the TSS (Datasets S1 and S2). We identified 19 TSS that were associated with nucleotide polymorphisms in the −40 to −1 region of the 2,211 primary TSS of D23580 (Dataset S3). We compared the expression level of each TSS between 4/74 and D23580, in two growth conditions, to identify the promoter SNPs responsible for transcriptional changes. No SNPs were identified in the vicinity of the −10 or −35 elements of the fliC or sopE2 gene promoters, indicating that the reported differential expression of FliC and SopE2 between ST313 and ST19 (11) is not caused by alterations to the promoter sequence (Datasets S1–S3). We found one SNP at the −12 position of the pgtE TSS, which was associated with an average 11-fold increase in TSS expression in D23580 compared with 4/74 (Dataset S3), and we investigated this experimentally.
Identification of a Nucleotide That Modulates Expression of the PgtE Virulence Factor.
PgtE is an outer membrane protease that belongs to the Omptin family of Gram-negative bacteria (18), cleaves and mediates resistance to α-helical antimicrobial peptides, and disrupts the human complement cascade by degrading complement components, such as factor B (19, 20). PgtE does not contribute to intramacrophage replication per se, but stimulates bacterial dissemination during murine infection (21) by facilitating extracellular survival upon release from host cells (22–25). Expression of the pgtE transcript is induced during intramacrophage replication (26, 27), controlled by the SPI2-associated regulators PhoPQ and SlyA (19, 28), and is activated by OmpR/EnvZ and SsrA/B (15).
The promoter and coding regions of the pgtE gene were compared between D23580 and 4/74 at the DNA sequence level, and differed by two SNPs. One SNP was identified in the coding region of pgtE at nucleotide location 2,530,498 in D23580 (2,504,548 in 4/74), generating a synonymous mutation [T54 (ACT) in 4/74 → T54 (ACC) in D23580]. The other SNP was located in the promoter region; the −12 nucleotide (relative to the +1 of the TSS) was C in 4/74 (C4/74) and T in D23580 (TD23580) (Fig. 1C). This T nucleotide in the −10 motif is a highly conserved element of highly expressed σ70-dependent promoters (14). We analyzed the functional role of TD23580 in the pgtE promoter region experimentally by replacing the TD23580 nucleotide with C4/74 by single-nucleotide exchange mutagenesis to generate strain D23580 PpgtE4/74. Whole-genome sequencing confirmed that the D23580 PpgtE4/74 strain only contained the intended single-nucleotide difference.
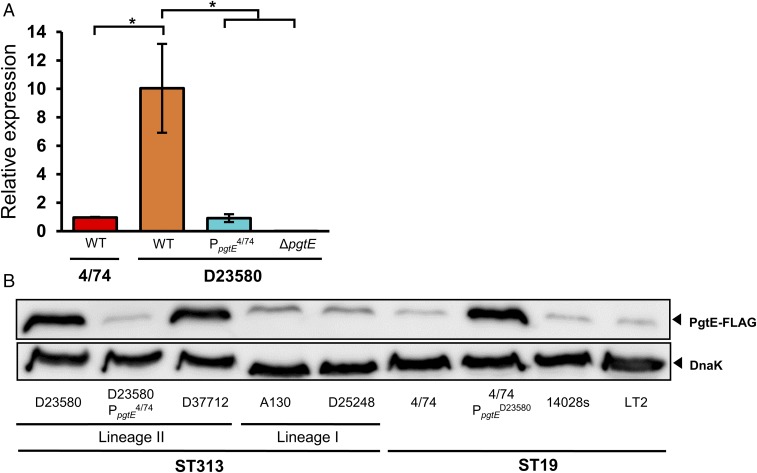
To determine the biological role of the TD23580 nucleotide, we assayed the level of pgtE transcription in 4/74, D23580, D23580 PpgtE4/74, and D23580 ΔpgtE strains using qRT-PCR (Fig. 2A). The high level of pgtE expression in D23580 was reduced 10-fold by the introduction of the single C4/74 nucleotide in the −10 region of the pgtE promoter, P < 0.01 (Fig. 2A). The level of the pgtE transcript expression in 4/74 and the D23580 PpgtE4/74 SNP mutant was similar.
Fig. 2.
The TD23580 SNP in the pgtE promoter of S. Typhimurium is associated with increased pgtE transcription and PgtE protein production. (A) The level of pgtE transcript was measured by qRT-PCR and the relative gene expression, normalized to endogenous control hns, was calculated using the ddCt algorithm (68) and is the average of three biological replicate experiments, with SEs. Significant differences were analyzed using an unpaired t test (*P < 0.01). (B) Immunodetection by Western blotting of FLAG-tagged PgtE in representative strains of ST313 and ST19. The status of the pgtE promoter (PpgtE) is only indicated for the strains with a mutated promoter sequence. Detection of DnaK served as loading control.
We hypothesized that the high level of pgtE transcription would increase PgtE protein production in D23580 compared with wild-type strain 4/74, and this was observed (Fig. 2B). A second lineage 2 isolate, D37712, expressed the same high level of PgtE. In contrast, low levels of PgtE were produced by the ST313 lineage 1 isolates A130 and D25248, and ST19 isolates 14028 and LT2 (Fig. 2B). The enhanced production of PgtE by D23580 was reduced to the level of the 4/74 strain by a single-nucleotide change in strain D23580 PpgtE4/74. To determine whether the TD23580 SNP would have a reciprocal effect in the S. Typhimurium ST19 genetic background, we introduced the T nucleotide to the −10 promoter motif of pgtE on the chromosome of strain 4/74 (Materials and Methods). The strain 4/74 PpgtED23580 produced increased levels of PgtE protein (Fig. 2B). Taken together, our data show that in D23580, the high-level expression of pgtE at the transcriptional and protein level is driven by the TD23580 nucleotide in the −10 region of the pgtE promoter.
The TD23580 SNP Increases Resistance to Human Serum Killing, Modulates Cleavage of Complement Factor B, and Interferes with Complement Deposition.
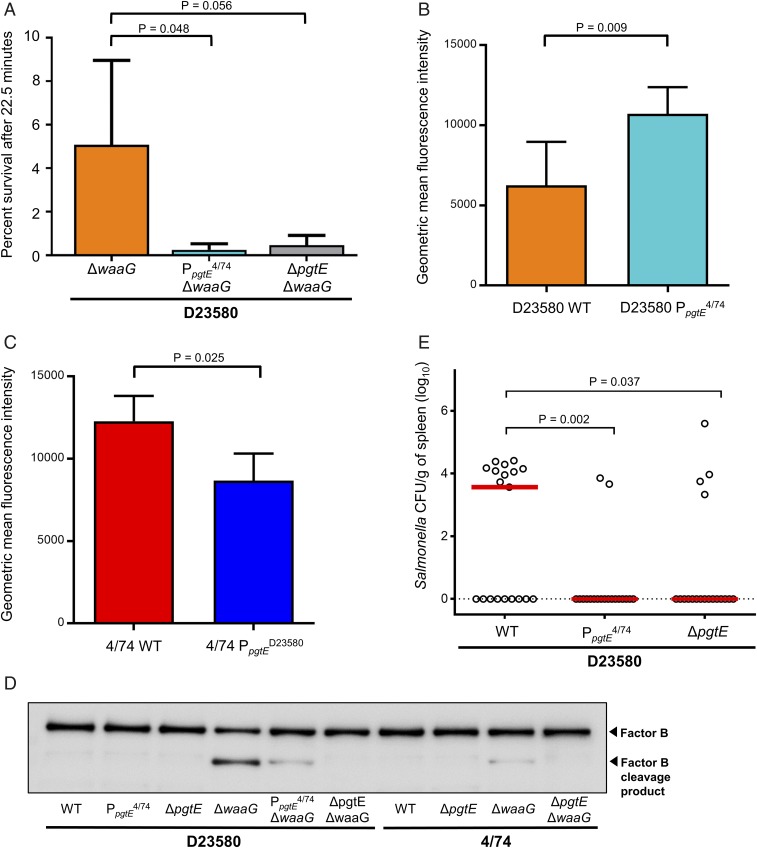
To determine the impact of the increased PgtE activity mediated by the promoter TD23580 SNP upon extracellular survival, we undertook serum bactericidal assays. Several bacterial factors contribute to the serum-resistance phenotype of Salmonella, including the long heterogenic O-antigen side chains of smooth lipopolysaccharide (LPS), which is the outermost component of the cell envelope of the Gram-negative cell (29–31). Therefore, we assayed resistance to human serum killing of in vitro-grown S. Typhimurium that lacked the LPS biosynthetic α1,3-glucosyltransferase enzyme WaaG. Following treatment with healthy human sera, the level of survival of D23580 ΔwaaG was significantly higher than D23580 ΔwaaG PpgtE4/74 (P ≤ 0.05) (Fig. 3A). No killing was observed following treatment with heat-inactivated sera that lacked active complement, proving that the observed bactericidal activity was complement-mediated. In summary, the promoter TD23580 SNP increases the resistance of D23580 to serum killing, and the low level of pgtE expression driven by the PpgtE4/74 promoter does not.
Fig. 3.
The pgtE promoter TD23580 SNP mediates increased resistance to human serum killing, decreases complement component 3 deposition, enhances cleavage of human complement factor B, and promotes virulence in the chicken infection model. (A) Sensitivity to pooled healthy human serum was assayed in a ΔwaaG background (truncated LPS), to observe only the effect of outer membrane proteases. D23580 ΔwaaG showed significantly greater serum resistance than D23580 PpgtE4/74 ΔwaaG (P = 0.048). D23580 (B) or 4/74 (C) strains were incubated with pooled human sera and C3 deposition was evaluated by flow cytometry. Data shown is the average of five (B) or three (C) biological replicates, with error bars representing SD. (B) D23580 WT (orange) displayed decreased geometric mean fluorescence intensity (C3 deposition) compared with D23580 PpgtE4/74 (light blue) after 5-min incubation with healthy human sera. (C) 4/74 WT (red) showed an increase of C3 deposition compared with 4/74 PpgtED23580 (dark blue) after 10-min incubation with healthy human sera. (D) PgtE-dependent cleavage of complement factor B detected by Western blotting. Polyclonal antibody against factor B was used. (E) Viable counts of S. Typhimurium D23580-derived strains as log CFU/g of spleen at 3-d postoral infection (108 CFU) of 7-d-old Lohmann Brown Layers. Data based on 19 individually sampled birds for each group; combined data for two separately repeated experiments. Each symbol represents the value for an individual chicken and the bars represent the median value for each group.
To confirm the phenotypic role of the TD23580 SNP in wild-type S. Typhimurium D23580, we determined levels of complement deposition by flow cytometry. Following incubation with human sera, lower levels of complement component 3b (C3b) were found bound to the surface of wild-type D23580 than D23580 PpgtE4/74 bacteria (Fig. 3B). Human serum killing of S. Typhimurium is mediated by activation of the classic complement pathway by the binding of specific IgG or IgM antibodies, followed by deposition of membrane attack complexes on the bacterial surface (32–34). We deduce that the TD23580 nucleotide carried by wild-type D23580 reduces the level of binding of C3b, and explains the increased resistance of D23580 ΔwaaG to serum-mediated killing described above.
As described above, moving the TD23580 SNP into strain 4/74 caused increased PgtE protein production (Fig. 2B). To investigate the function of the TD23580 SNP in the S. Typhimurium ST19 genetic background, we examined the role of the TD23580 SNP upon complement deposition. The 4/74 wild-type bacteria had higher surface-bound levels of C3b than 4/74 PpgtED23580 (Fig. 3C), showing that increased expression of PgtE interferes with the complement cascade in both S. Typhimurium ST19 and ST313.
PgtE disrupts the complement cascade by proteolytically cleaving the C3b protein (35). The mechanism of the serum-resistance phenotype was investigated by determining the ability of the S. Typhimurium strains to mediate PgtE-dependent cleavage of complement factor B (Fig. 3D). In agreement with the literature (36), no PgtE activity was detected in strains expressing smooth LPS [Fig. 3D, lanes 1–3 (D23580), lanes 7–8 (4/74)]. Because the results of the serum-resistance assay showed that short (rough) LPS was required to visualize PgtE activity, we repeated experiments in a ΔwaaG background (Fig. 3D, lanes 4–6 and 9–10), and discovered that the D23580 ΔwaaG mutant showed a high level of complement factor B cleavage. In contrast, 4/74 ΔwaaG and the D23580 ΔwaaG PpgtE4/74 strains showed a low level of complement factor B degradation.
We speculate that after the pathogen exits macrophages, the high level of expression of PgtE in S. Typhimurium ST313 strain D23580 interferes with opsonization and increases resistance to complement-mediated serum killing.
Assessment of PgtE-Mediated Virulence in the Chicken Infection Model.
Because S. Typhimurium ST313 has a hyperinvasive phenotype during chicken infection (37), this animal model was used to assess the virulence of the wild-type D23580 and the D23580 PpgtE4/74 strains. Following oral infection, the D23580 PpgtE4/74 SNP strain and the D23580 ΔpgtE strain showed significant attenuation in splenic colonization in comparison with D23580 wild-type (P = 0.0035 and P = 0.0379, respectively), based on two independent repeats of the experiment (Fig. 3E). The data show some bird-to-bird variation between all three tested isolates, which is likely a consequence of the oral route of infection and the use of a commercial outbred chicken line. We found that the D23580 PpgtE4/74 SNP strain, the D23580 ΔpgtE strain, and the wild-type D23580 strain colonized liver and cecum to similar levels (Fig. S1). Overall, the results showed that exchange of the TD23580 SNP to the C4/74 genotype resulted in lower median bacterial numbers and a reduced number of animals with splenic infection, equivalent to that seen in the absence of PgtE (10 of 19 for D23580 compared with 2 of 19 for D23580 PpgtE4/74 and 4 of 19 for D23580 ΔpgtE). We conclude that that the TD23580 SNP promotes systemic infection of the chicken, and that full virulence of D23580 requires the high level of expression of PgtE that is driven by the TD23580 nucleotide.
The pgtE Promoter SNP Is Only Carried by African ST313 Lineage 2, and Not Lineage 1.
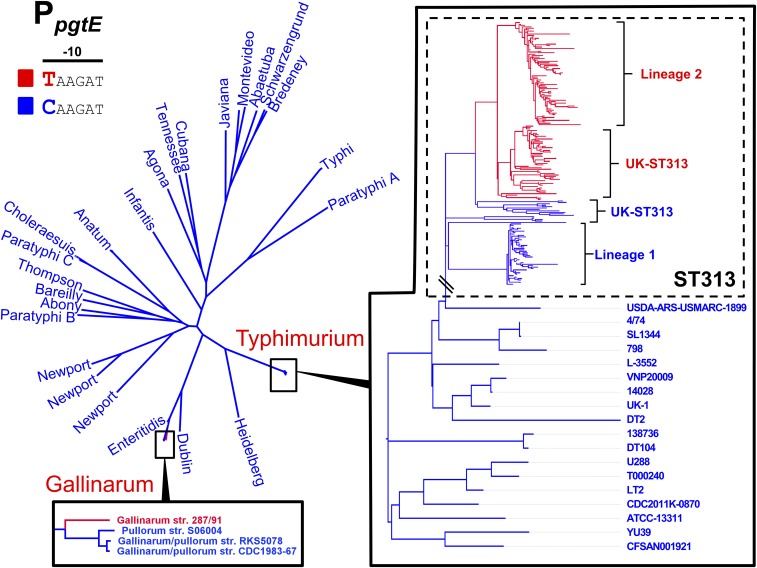
To determine if the pgtE promoter TD23580 SNP is a characteristic feature of iNTS, the pgtE promoter SNP was analyzed in the context of a phylogeny of 258 genomes of S. Typhimurium ST313 including isolates from Malawi, as well as recently described UK-ST313 genomes from the UK (7). The 228 genomes that carried the TD23580 SNP formed a monophyletic cluster that included lineage 2, as well as the UK-ST313 strains that share most recent common ancestry with lineage 2 (Fig. 4). The C4/74 SNP was found to be conserved in all 27 lineage 1 genomes analyzed and certain UK-ST313 genomes which shared more recent common ancestry with lineage 1. This suggests that the TD23580 SNP first arose in a common ancestor of lineage 2 and a subset of the UK-ST313 (Fig. S2).
Fig. 4.
Conservation of the pgtE promoter −10 TD23580 nucleotide across S. enterica subsp. enterica. Maximum-likelihood phylogenetic tree of 336 S. enterica subsp. enterica genomes including 258 ST313 genomes. Presence of C or T nucleotide at the −10 position of the pgtE promoter is indicated by blue and red, respectively. The T nucleotide is found in 100% of ST313 lineage two genomes surveyed, and in a subset of UK-ST313. Across S. enterica subsp. enterica subspecies 1, the C nucleotide is present in 23 serovars, and a T nucleotide was identified in a single genome (S. Gallinarum isolate 287/91).
The pgtE Promoter Is Highly Conserved in S. enterica.
To understand the wider distribution of the pgtE promoter SNP in the Salmonella genus, the conservation of the SNP was assessed in 83 published complete genomes representing the known genomic diversity of Salmonella. The pgtE promoter sequence was not conserved in the three genomes of Salmonella bongori included in the analysis (Dataset S4). Of 80 S. enterica genomes screened, 79 genomes carried the C4/74 genotype. The TD23580 SNP was only found in Salmonella Gallinarum strain 287/91, raising the possibility that the SNP has arisen independently in this serovar (Fig. 4). Apart from the −12 SNP present in S. Typhimurium ST313 and S. Gallinarum, the pgtE TSS −35 region was found to be 100% conserved in 75 of 80 S. enterica genomes, with only serovar Agona and subspecies Arizonae showing sequence divergence (Dataset S4).
In summary, the −10 C4/74 → TD23580 allele of the D23580 pgtE promoter causes an increase in transcription of the pgtE gene and the production of high levels of PgtE protein in D23580. The increased activity of PgtE in D23580 leads to increased resistance to human serum killing, reduced complement deposition, and enhanced degradation of complement factor B, which is required for activation of the alternative complement pathway. Importantly, the single SNP in the D23580 pgtE promoter drives the ability of D23580 to cause hyperinvasion in an avian infection model. Outside of ST313, only 1 of 80 complete S. enterica genomes, that of S. Gallinarum, carried the −12 TD23580 allele. S. Gallinarum is the causal agent of fowl typhoid, suggesting a putative link between the TD23580 SNP and the ability of S. enterica to cause systemic infection (38). We have shown that the pgtE promoter SNP is a signature of ST313 lineage 2, which clonally replaced ST313 lineage 1 in the early 2000s (3). All isolates of ST313 lineage 2 that we have analyzed carried the same TD23580 SNP in the −10 promoter motif of pgtE.
Discussion
Previous studies have identified SNP mutations associated with the host tropism of notorious pathogens such as Staphylococcus aureus (39) and Campylobacter jejuni (40). However, these examples involved SNP mutations located within coding genes, and functionally important SNP mutations have rarely been identified in intergenic regions of bacteria. As the expression of a gene is dependent on the −10 and −35 recognition motifs of σ70-dependent promoters (41), a single-nucleotide change can modulate promoter function. Examples include the C → T transition in the −10 promoter motif of the Mycobacterium tuberculosis eis gene, which increases eis expression to generate low-level resistance to Kanamycin (42). Similarly, a promoter SNP that affected expression of Escherichia coli succinate transporter dctA evolved to increase the utilization of citrate as a carbon source in one population of the Long-Term Evolution Experiment (43).
Here, we have identified a single SNP responsible for high levels of expression of the PgtE outer membrane protease that is linked to the virulence of African S. Typhimurium ST313. Our study has implications for bacterial genome-wide association studies, which should clearly include a focus on noncoding regions of the genome. The findings also emphasize the value of identifying all gene promoters in bacterial pathogens, to allow nucleotide differences to be correlated with the process of transcriptional initiation.
It is known that S. Typhimurium bacteria produce high levels of pgtE transcript inside host macrophages (26, 27), and that PgtE protease activity is high in S. Typhimurium bacteria released from infected macrophages (22). The steric hindrance caused by long O-antigen chains makes the modification of LPS a prerequisite for optimal PgtE activity. Such remodeling occurs during intramacrophage replication, resulting in shortening of the oligosaccharide chains (22). Accordingly, we used rough S. Typhimurium strains (ΔwaaG) that express short O-antigen chains to determine that the pgtE TD23580 SNP contributes to the resistance of ST313 D23580 to human serum killing.
PgtE is known to interfere with the complement cascade in several ways, including the cleavage of factors B and C3b (35). To demonstrate a role for the pgtE TD23580 SNP in wild-type (smooth) bacterial strains, we show that after incubation with healthy human serum, the pgtE TD23580 SNP caused decreased C3b deposition on the Salmonella cell surface in both the 4/74 and D23580 backgrounds. We speculate that the pgtE TD23580 SNP primes intracellular bacteria for an extracellular lifestyle, and aids the survival of complement-mediated attack by the innate immune system.
The opsonic activity of complement has been shown to be essential for phagocyte-mediated killing of Salmonella in the blood of African people (44), and therefore our data are consistent with the hypothesis that subversion of host innate immunity via interference with the complement cascade contributes to the pathogenesis of invasive nontyphoidal Salmonella in Africa. Because other factors have been reported to enhance the invasiveness of S. Typhimurium ST313 (11, 12, 45), our data suggest that the adaption of African ST313 to a systemic lifestyle is multifactorial. Clearly, further investigation into the interaction between the nucleotide changes responsible for the pseudogenisation of sseI and the pgtE promoter SNP is required to understand which alteration drove the adaptation to invasive disease, and which could represent “passenger” mutations.
We suggest that the high level of PgtE activity in D23580, together with the inactivated sseI effector gene (12) and the acquisition of chloramphenicol resistance (3), has been key to the success of S. Typhimurium ST313 lineage 2. This mechanism has a parallel in the evolution of Yersinia pestis, a pathogen that acquired the Pla Omptin protease, a PgtE homolog, at least 3,000 y ago (46). The Pla protein allowed Y. pestis to transition from a gastrointestinal to an extraintestinal lifestyle and cause plague (47). Here, we propose that the increased PgtE protease activity caused by the TD23580 SNP in the −10 motif of the pgtE promoter was an important evolutionary event that primed the emergence and clonal expansion of an epidemic S. Typhimurium lineage.
Materials and Methods
Bacterial Strains, Growth Conditions.
S. Typhimurium strain 4/74 (accession no. CP002487), a representative strain of nontyphoidal Salmonella sequence type 19, and D23580 (accession no. FN424405), a representative strain of nontyphoidal Salmonella sequence type 313 (ST313) were used in the study. Strain D23580 was isolated from an HIV− child from Malawi with blood stream infection, and use of this strain has been approved by the Malawian College of Medicine (COMREC ethics no. P.08/14/1614). Other wild-type strains belonging to ST19 and ST313 used in this study are listed in Table S1.
All environmental growth conditions were repeated exactly as previously described (13), with the exception that the “pool” sample was obtained by pooling RNA from 16 environmental conditions: early exponential phase (EEP), mid exponential phase (MEP), late exponential phase (LEP), ESP, late stationary phase (LSP), 25 °C, NaCl shock, bile shock, low Fe2+ shock, anaerobic shock, anaerobic growth, oxygen shock, non-SPI2, InSPI2, peroxide shock (InSPI2), and nitric oxide shock (InSPI2) (13).
When required, Lennox broth (LB) was supplemented with the following antibiotics: chloramphenicol (Cm) 25 µg/mL, kanamycin (Km) 50 µg/mL, tetracycline (Tc) 20 µg/mL, and gentamicin (Gm) 20 µg/mL.
Preparation of cDNA Libraries and Illumina Sequencing.
Before RNA-seq, total RNA was extracted using TRIzol, and treated with DNase I, as described previously (14). RNA integrity was inspected visually with the Bioanalyzer (Agilent Technologies). Contaminating DNA was removed using DNase I (Ambion) and RNA samples were not ribo-depleted before cDNA library construction. cDNA library construction, TEX-treatment for dRNA-seq (ESP, InSPI2, and pooled sample), and RNA-seq with the Illumina HiSeq platform was carried out by Vertis Biotechnologie, Germany. All protocols were identical to those used previously (13, 14). Sequence reads were mapped against the S. Typhimurium D23580 reference genome using Segemehl, with accuracy set to 100% (48, 49). RNA-seq and dRNA-seq data can be downloaded as raw reads (.fastq file format); the Gene Expression Omnibus database accession no. is GSE108104.
Identification of TSS by a Combination of RNA-Seq and dRNA-Seq.
Methods used to assign TSS in D23580 have been described previously (14). Briefly, a TSS was assigned when it was enriched in one of the dRNA-seq libraries (ESP, InSPI2, or Pool) compared with the corresponding RNA-seq library, and was linked to an expressed transcript. This analysis was followed by a second step of validation, in which the transcripts per million approach (50, 51) was used to calculate an expression value for the first 10 nucleotides associated with each TSS, designated the promoter usage value (PUV) (14, 27). A TSS was considered to be expressed when the PUV was ≥10. A TSS was defined as “conserved” between D23580 and 4/74 if the TSS nucleotide sequence was present in both strains, and the PUV value of the TSS was ≥10.
Identification of TSS-Located SNP Mutations Associated with Different Levels of Transcript Expression in D23580 and 4/74.
A list of SNP differences between the D23580 and 4/74 reference genomes (accession nos. FN424405 and CP002487) was generated using NUCmer (52) resulting in 1,488 SNPs and small indels (Dataset S2). We identified the SNPs located within the −40 to −1 region of primary TSSs in D23580 (primary = primary + primary/as + primary/internal) (Datasets S1 and S3). PUV values for each promoter in D23580 and 4/74 (14) were used to analyze the activity of the promoters associated with SNP differences in the −40 to −1 TSS region.
Bacterial Strain Construction Using λ Red Recombineering.
All of the bacterial strains and plasmids used and constructed in this study are described in Table S1 and the single-stranded DNA (ssDNA) oligonucleotides (primers) in Table S2.
The ΔpgtE, ΔwaaG and pgtE-FLAG mutations were constructed in S. Typhimurium ST19 and ST313 strains using the standard λ red recombination methodology (53). The heat-inducible λ red recombineering plasmid pSIM5-tet was used and the induction of the λ red operon was achieved by heat treatment (42 °C, 15 min) of bacterial cultures grown to midexponential phase (OD600 0.3–0.4 at 30 °C) in LB supplemented with Tc (53–55). After recombination (53–55), all of the genetic constructs (except the ΔwaaG::Kan mutation) were transferred into a clean wild-type background by phage transduction, using the P22 HT 105/1 int-201 (56), as previously described (6). When required, the antibiotic resistance cassettes were flipped-out using the FLP recombinase expression plasmid pCP20-TcR (57).
The pgtE gene was deleted in S. Typhimurium using PCR fragments generated with the primers DH95 and DH96 and the template plasmids pKD4 and pKD3. The resulting fragments, carrying, respectively, the Km resistance (Kan) or the Cm resistance (Cam) cassettes were, respectively, electroporated into D23580 and 4/74 carrying pSIM5-tet and recombinant ΔpgtE::Kan/Cam mutants were selected on Km or Cm LB agar plates. Finally, the mutations were transduced in the corresponding wild-type strain and the resistance cassette was removed, as described above. Similarly, waaG was inactivated using the primers del_waaG_F and del_waaG_R and pKD4 as template.
The FLAG-tagged strains were generated using a forward primer (DH93), which included the region homologous to pgtE end (′pgtE), the nucleotide sequence encoding the FLAG octa-peptide in frame with the pgtE coding region, the pgtE stop codon, and a region homologous to the resistance cassette (58). The ′pgtE-FLAG-Kan and ′pgtE-FLAG-Cam modules were amplified by PCR using, respectively, pKD4 and pKD3 and primers DH93/DH94. The resulting amplicons were, respectively, electroporated into S. Typhimurium ST313 (except A130) or ST19 (and A130) strains, carrying all pSIM5-tet, and recombinants were selected on Km or Cm LB agar plates. The insertions were then transduced into the corresponding wild-type strains and the resistance cassettes were removed, as described above.
Construction of Scarless Single-Nucleotide Substitution Mutants.
Two different methods were used to construct single-nucleotide mutants. The single-nucleotide T → C substitution in the pgtE promoter of S. Typhimurium D23580 was constructed using a ssDNA recombineering approach, as has been previously described (59). The protocol was identical to that used for construction of mutants using λ red recombineering (described above), except that 400 ng of the manufactured primer (DH90, HPLC purified) was used in the transformation reaction. After 2 h of recovery at 30 °C, dilutions of transformation were plated on LB agar (without selection). Clones were restreaked and screened by a stringent PCR with primers DH40 and DH41. Primer DH40 has full complementarity with the sought after mutant, representing one mismatch to the original strain (these two types could be distinguished using a stringent annealing temperature), while DH41 has full complementarity with both types of clone. The correct allele of the D23580 PpgtE4/74 strain was confirmed by whole-genome sequencing using Illumina technology (MicrobesNG, University of Birmingham). Variant-calling analysis confirmed that the D23580 PpgtE4/74 strain had the intended single-nucleotide difference compared with the wild-type strain.
The single-nucleotide C → T substitution in the pgtE promoter of S. Typhimurium 4/74 (chromosomal position 2504765) (4/74 PpgtED23580) was carried out by a scarless genome-editing technique based on the pEMG suicide plasmid, as previously described (6, 60). The pEMG derivative pNAW41 that carries the pgtE promoter region with the specific substitution was constructed as follows: the regions flanking the targeted nucleotide were PCR-amplified with the primers pairs NW_122/NW_123 and NW_124/NW_125, using 4/74 genomic DNA as template. The primers NW_123 and NW_124 encode for C → T substitution and are complementary to each other over a stretch of 20 nucleotides. The resulting PCR fragments (504 and 505 bp, respectively) were fused by overlap extension PCR and the resulting 989-bp fragment was digested and cloned into pEMG using the BamHI and EcoRI restriction sites. The pNAW41 suicide plasmid was mobilized from E. coli S17-1 λpir into S. Typhimurium 4/74 by conjugation and transconjugants that have integrated pNAW41 by homologous recombination were selected on minimal medium M9 agar supplemented with 0.2% of glucose and Km. The resulting merodiploids were resolved using the pSW-2 plasmid as previously described (6) and the C → T substitution was confirmed by PCR amplification and sequencing, using the primers NW_155 and NW_156.
qRT-PCR.
Total RNA was extracted from bacteria from midexponential (OD600 = 0.3) cultures of bacteria grown in PCN/InSPI2, DNase-treated with Turbo DNA-free kit (Ambion) and the RNA integrity was inspected visually using the Bioanalyzer. Complete DNA-removal was confirmed by a negative PCR with 40 cycles. Four-hundred nanograms of RNA were converted into cDNA using the GoScript Reverse Transcription System (Promega), with random primers according to the manufacturer’s instructions. The Sensifast SYBR Hi_ROX Kit (Bioline) was used for qRT-PCR with primer pairs DH54/DH55 and qPCR_hns_f/qPCR_hns_r. For each qRT-PCR, performed in duplicate, 26.66 ng cDNA was used in total reaction volumes of 20 µL. The amount of pgtE and hns mRNA was calculated using a standard curve based on 10-fold dilutions of genomic DNA (10–0.0001 ng/µL), included in each qRT-PCR run. The amount of pgtE mRNA was normalized to the amount of hns mRNA.
Western Blot Analysis.
Salmonella strains carrying the FLAG-tagged version of pgtE were grown in PCN/InSPI2 medium to OD600 = 0.3. Bacteria were harvested from 10 mL of culture by centrifugation (7,000 × g, 5 min, 4 °C). Cells were washed once with PBS and suspended in 67.5 µL of the same buffer. Seventy-five microliters of Laemmli buffer 2× [120 mM Tris⋅HCl pH 6.8, 4% (wt/vol) SDS, 20% (vol/vol) glycerol, Bromophenol blue 0.02% (wt/vol)] and 7.5 µL β-mercaptoethanol were subsequently added to the samples. The extracts were boiled for 10 min, chilled on ice, and cell debris was pelleted by centrifugation (20,000 × g, 5 min, 4 °C). Fifteen microliters of the samples (supernatant) were loaded on a SDS 10% polyacrylamide gel and proteins were separated for 80 min at 150 V in SDS/PAGE running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). Proteins were transferred onto a methanol-soaked PVDF membrane (Cat. No. 3 010 040; Roche) using a semiwet transfer system (#170–3940; Bio-Rad) for 2 h, 125 mA at 4 °C in transfer buffer (25 mM Tris, 192 mM glycine). The membrane was blocked for 15 h at 4 °C in Tris-buffered saline (TBS: 10 mM Tris⋅HCl pH 7.5, 0.9% NaCl) supplemented with 5% (wt/vol) of dry skimmed milk. Incubation with the antibodies (1 h at room temperature) and washing steps (two washes for 15 min each) were done in TBS containing 0.1% (vol/vol) Tween 20 and 0.5% (wt/vol) dry skimmed milk. The primary antibodies, monoclonal anti-FLAG M2 antibody (1:3,000 diluted, Sigma-Aldrich F3165) and anti-DnaK mAb 8E2/2 (1:10,000 diluted, ADI-SPA-880; Enzo Life Sciences), were used for the detection of PgtE-FLAG and DnaK (loading control), respectively. After incubation with the primary antibodies, the membrane was washed and incubated with the secondary antibody [goat anti-mouse IgG (H + L)-HRP, 1:2,500 diluted, # 172–1011; Bio-Rad]. After washes, the membrane was rinsed briefly in TBS before addition of Pierce ECL Western blotting substrate (32109; Thermo Scientific) and the chemiluminescence reaction was measured using the Image Quant LAS 4000 imager (GE Healthcare Life Sciences).
Complement Factor B Cleavage Assay.
The complement factor B cleavage assay was carried out as described previously (20), with the following modifications. Bacterial strains were grown overnight in LB at 37 °C. Overnight culture was pelleted by centrifugation and washed twice in PBS. Two OD600 (in 200 μL PBS containing 33 ng/μL of complement factor B) were incubated at 37 °C in a heat block with agitation (700 rpm) for 2 h and subsequently pelleted by centrifugation. Sixty-six nanograms of complement factor B were separated on an SDS/PAGE, before Western blotting, as described above.
Serum Bactericidal Assays.
Serum bactericidal assays were performed four times using a modification of the previously described method (61). Briefly, bacteria were grown to OD600 = 2 in 5 mL of LB at 37 °C and resuspended in PBS to a final concentration of 107 CFU/mL; 10 µL was then mixed with 90 µL of undiluted pooled healthy human sera at 37 °C with shaking (180 rpm), and viable counts determined. Killing was confirmed to be due to the activity of complement by using 56 °C heat-inactivated sera as a control.
Complement Deposition Assays.
Detection of C3 complement component deposition was done as previously described (32–34). In brief, overnight cultures of S. Typhimurium were grown to an OD600 of 2. Cells were washed twice with PBS and 5 μL of S. Typhimurium at 2 × 109 CFU/mL was mixed with 45 μL healthy human sera or PBS at room temperature for 5 min (D23580 strains) or 10 min (4/74 strains). Cells were then washed twice with 1 mL of PBS by 5 min centrifugation at 3,300 × g, and incubated with anti-human C3c FITC-conjugated antibody (Thermo Scientific) for 20 min at 4 °C. Samples were washed twice and fixed with 200 μL of 1% formaldehyde-PBS. Cells were acquired on a BD FACSAria and analyzed with FlowJo v10.4. Three to five replicate assays were conducted for each strain background and the average geometric mean fluorescence intensity for replicates is shown, along with the SD. An unpaired, single-tailed t test was used to assess the statistical significance of strain differences.
Chicken Infection Experiments.
All work was conducted in accordance with United Kingdom legislation governing experimental animals under project license PPL 40/3652 and was approved by the University of Liverpool ethical review process before the award of the license. All birds were checked a minimum of twice daily to ensure their health and welfare. Birds were housed in accommodation meeting United Kingdom legislation requirements. One-day-old Lohmann Brown Layers were obtained from a commercial hatchery, separated into groups on arrival, and given ad libitum access to water and a laboratory-grade vegetable protein-based pellet diet (SDS). Chicks were housed at a temperature of 30 °C. At 7 d of age, chickens were inoculated by oral gavage with 108 CFU of S. Typhimurium strains D23580, D23580 PpgtE4/74, or D23580 ∆pgtE. At 3 d postinfection, 10 birds from each group were killed for postmortem analysis. Samples from spleen, liver, and the caecal contents were removed aseptically from each bird and diluted 1:5 (wt/vol.) in sterile PBS (data from caecal content and liver are shown in Fig. S1). Tissues were then homogenized in a Colworth 80 microstomacher (A.J. Seward & Co. Ltd). Samples were serially diluted and dispensed onto Brilliant green agar (Oxoid) to quantify numbers of Salmonella, as described previously (62). A Mann–Whitney test was used to determine the statistical significance of strain differences.
Analysis of the Conservation of the pgtE Promoter SNP.
For analysis of the conservation of the pgtE promoter SNP, 258 genomes of S. Typhimurium ST313 isolates from Malawi and the United Kingdom (7) were assembled using the A5 pipeline (63) and Abacus (64), unless reference quality genomes were available (all strains and accession numbers are given in Dataset S4). Additionally, 83 reference genomes were downloaded from the National Center for Biotechnology Information that represent the known diversity of the Salmonella genus. Core genome SNPs were identified using the PanSeq package (65) and a maximum-likelihood phylogenetic tree was constructed from the concatenated SNP alignment using PhylML (66). BLASTn was used to identify the genotype of the entire pgtE TSS −35 region nucleotides in all genomes (Dataset S4).
Supplementary Material
Acknowledgments
We thank present and former members of the J.C.D.H. laboratory for helpful discussions, particularly Sathesh Sivasankaran and Aoife Colgan; and Paul Loughnane for his expert technical assistance. This work was supported by funding from a Wellcome Trust Senior Investigator award (to J.C.D.H.) (Grant 106914/Z/15/Z). D.L.H. was supported by the Wenner-Gren Foundation, Sweden. R.C. was supported by an EU Marie Curie International Incoming Fellowship (FP7-PEOPLE-2013-IIF, Project Reference 628450). N.W. was supported by an Early Postdoc Mobility Fellowship from the Swiss National Science Foundation (Project Reference P2LAP3_158684).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE108104).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714718115/-/DCSupplemental.
References
- 1.Ao TT, et al. Global burden of invasive nontyphoidal Salmonella disease, 2010(1) Emerg Infect Dis. 2015;21:941–949. doi: 10.3201/eid2106.140999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal Salmonella disease: An emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okoro CK, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44:1215–1221. doi: 10.1038/ng.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kariuki S, et al. Ceftriaxone-resistant Salmonella enterica serotype Typhimurium sequence type 313 from Kenyan patients is associated with the blaCTX-M-15 gene on a novel IncHI2 plasmid. Antimicrob Agents Chemother. 2015;59:3133–3139. doi: 10.1128/AAC.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kingsley RA, et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owen SV, et al. Characterization of the prophage repertoire of African Salmonella Typhimurium ST313 reveals high levels of spontaneous induction of novel phage BTP1. Front Microbiol. 2017;8:235. doi: 10.3389/fmicb.2017.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashton PM, et al. Public health surveillance in the UK revolutionises our understanding of the invasive Salmonella Typhimurium epidemic in Africa. Genome Med. 2017;9:92. doi: 10.1186/s13073-017-0480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goh YS, MacLennan CA. Invasive African nontyphoidal Salmonella requires high levels of complement for cell-free antibody-dependent killing. J Immunol Methods. 2013;387:121–129. doi: 10.1016/j.jim.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Siggins MK, et al. Differential timing of antibody-mediated phagocytosis and cell-free killing of invasive African Salmonella allows immune evasion. Eur J Immunol. 2014;44:1093–1098. doi: 10.1002/eji.201343529. [DOI] [PubMed] [Google Scholar]
- 10.Ramachandran G, Perkins DJ, Schmidlein PJ, Tulapurkar ME, Tennant SM. Invasive Salmonella Typhimurium ST313 with naturally attenuated flagellin elicits reduced inflammation and replicates within macrophages. PLoS Negl Trop Dis. 2015;9:e3394. doi: 10.1371/journal.pntd.0003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carden S, Okoro C, Dougan G, Monack D. Non-typhoidal Salmonella Typhimurium ST313 isolates that cause bacteremia in humans stimulate less inflammasome activation than ST19 isolates associated with gastroenteritis. Pathog Dis. 2015;73:ftu023. doi: 10.1093/femspd/ftu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carden SE, et al. Pseudogenization of the secreted effector gene sseI confers rapid systemic dissemination of S. Typhimurium ST313 within migratory dendritic cells. Cell Host Microbe. 2017;21:182–194. doi: 10.1016/j.chom.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kröger C, et al. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Kröger C, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci USA. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colgan AM, et al. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 2016;12:e1006258. doi: 10.1371/journal.pgen.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 17.Fàbrega A, Vila J. Salmonella enterica serovar Typhimurium skills to succeed in the host: Virulence and regulation. Clin Microbiol Rev. 2013;26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haiko J, Suomalainen M, Ojala T, Lähteenmäki K, Korhonen TK. Invited review: Breaking barriers—Attack on innate immune defences by omptin surface proteases of enterobacterial pathogens. Innate Immun. 2009;15:67–80. doi: 10.1177/1753425909102559. [DOI] [PubMed] [Google Scholar]
- 19.Guina T, Yi EC, Wang H, Hackett M, Miller SI. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J Bacteriol. 2000;182:4077–4086. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riva R, Korhonen TK, Meri S. The outer membrane protease PgtE of Salmonella enterica interferes with the alternative complement pathway by cleaving factors B and H. Front Microbiol. 2015;6:63. doi: 10.3389/fmicb.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramu P, et al. Activation of pro-matrix metalloproteinase-9 and degradation of gelatin by the surface protease PgtE of Salmonella enterica serovar Typhimurium. Int J Med Microbiol. 2008;298:263–278. doi: 10.1016/j.ijmm.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Lähteenmäki K, Kyllönen P, Partanen L, Korhonen TK. Antiprotease inactivation by Salmonella enterica released from infected macrophages. Cell Microbiol. 2005;7:529–538. doi: 10.1111/j.1462-5822.2004.00483.x. [DOI] [PubMed] [Google Scholar]
- 23.Pietilä TE, et al. Activation, cytokine production, and intracellular survival of bacteria in Salmonella-infected human monocyte-derived macrophages and dendritic cells. J Leukoc Biol. 2005;78:909–920. doi: 10.1189/jlb.1204721. [DOI] [PubMed] [Google Scholar]
- 24.Valls Serón M, Haiko J, DE Groot PG, Korhonen TK, Meijers JCM. Thrombin-activatable fibrinolysis inhibitor is degraded by Salmonella enterica and Yersinia pestis. J Thromb Haemost. 2010;8:2232–2240. doi: 10.1111/j.1538-7836.2010.04014.x. [DOI] [PubMed] [Google Scholar]
- 25.Yun TH, Cott JE, Tapping RI, Slauch JM, Morrissey JH. Proteolytic inactivation of tissue factor pathway inhibitor by bacterial omptins. Blood. 2009;113:1139–1148. doi: 10.1182/blood-2008-05-157180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 27.Srikumar S, et al. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 2015;11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarre WW, et al. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol. 2005;56:492–508. doi: 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 29.Grossman N, et al. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J Bacteriol. 1987;169:856–863, and erratum (1987) 169:2911. doi: 10.1128/jb.169.2.856-863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray GL, Attridge SR, Morona R. Regulation of Salmonella Typhimurium lipopolysaccharide O antigen chain length is required for virulence; Identification of FepE as a second Wzz. Mol Microbiol. 2003;47:1395–1406. doi: 10.1046/j.1365-2958.2003.03383.x. [DOI] [PubMed] [Google Scholar]
- 31.Rautemaa R, Meri S. Complement-resistance mechanisms of bacteria. Microbes Infect. 1999;1:785–794. doi: 10.1016/s1286-4579(99)80081-1. [DOI] [PubMed] [Google Scholar]
- 32.MacLennan CA, et al. The neglected role of antibody in protection against bacteremia caused by nontyphoidal strains of Salmonella in African children. J Clin Invest. 2008;118:1553–1562, and erratum (2011) 186:4527. doi: 10.1172/JCI33998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siggins MK, et al. Absent bactericidal activity of mouse serum against invasive African nontyphoidal Salmonella results from impaired complement function but not a lack of antibody. J Immunol. 2011;186:2365–2371. doi: 10.4049/jimmunol.1000284. [DOI] [PubMed] [Google Scholar]
- 34.Nyirenda TS, et al. Loss of humoral and cellular immunity to invasive nontyphoidal Salmonella during current or convalescent Plasmodium falciparum infection in Malawian children. Clin Vaccine Immunol. 2017;24:e00057-17. doi: 10.1128/CVI.00057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramu P, et al. The surface protease PgtE of Salmonella enterica affects complement activity by proteolytically cleaving C3b, C4b and C5. FEBS Lett. 2007;581:1716–1720. doi: 10.1016/j.febslet.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 36.Kukkonen M, Korhonen TK. The omptin family of enterobacterial surface proteases/adhesins: From housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int J Med Microbiol. 2004;294:7–14. doi: 10.1016/j.ijmm.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Parsons BN, et al. Invasive non-typhoidal Salmonella Typhimurium ST313 are not host-restricted and have an invasive phenotype in experimentally infected chickens. PLoS Negl Trop Dis. 2013;7:e2487. doi: 10.1371/journal.pntd.0002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson NR, et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 2008;18:1624–1637. doi: 10.1101/gr.077404.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viana D, et al. A single natural nucleotide mutation alters bacterial pathogen host tropism. Nat Genet. 2015;47:361–366. doi: 10.1038/ng.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheppard SK, et al. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci USA. 2013;110:11923–11927. doi: 10.1073/pnas.1305559110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lisser S, Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaunbrecher MA, Sikes RD, Jr, Metchock B, Shinnick TM, Posey JE. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2009;106:20004–20009. doi: 10.1073/pnas.0907925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gondwe EN, et al. Importance of antibody and complement for oxidative burst and killing of invasive nontyphoidal Salmonella by blood cells in Africans. Proc Natl Acad Sci USA. 2010;107:3070–3075. doi: 10.1073/pnas.0910497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrero-Fresno A, et al. The role of the st313-td gene in virulence of Salmonella Typhimurium ST313. PLoS One. 2014;9:e84566. doi: 10.1371/journal.pone.0084566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmussen S, et al. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell. 2015;163:571–582. doi: 10.1016/j.cell.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimbler DL, Schroeder JA, Eddy JL, Lathem WW. Early emergence of Yersinia pestis as a severe respiratory pathogen. Nat Commun. 2015;6:7487. doi: 10.1038/ncomms8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann S, et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLOS Comput Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson EJ, et al. Genome sequences of Salmonella enterica serovar Typhimurium, Choleraesuis, Dublin, and Gallinarum strains of well- defined virulence in food-producing animals. J Bacteriol. 2011;193:3162–3163. doi: 10.1128/JB.00394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 51.Wagner GP, Kin K, Lynch VJ. A model based criterion for gene expression calls using RNA-seq data. Theory Biosci. 2013;132:159–164. doi: 10.1007/s12064-013-0178-3. [DOI] [PubMed] [Google Scholar]
- 52.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Datta S, Costantino N, Court DL. A set of recombineering plasmids for Gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 55.Koskiniemi S, Pränting M, Gullberg E, Näsvall J, Andersson DI. Activation of cryptic aminoglycoside resistance in Salmonella enterica. Mol Microbiol. 2011;80:1464–1478. doi: 10.1111/j.1365-2958.2011.07657.x. [DOI] [PubMed] [Google Scholar]
- 56.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 57.Kintz E, et al. A BTP1 prophage gene present in invasive non-typhoidal Salmonella determines composition and length of the O-antigen of the lipopolysaccharide. Mol Microbiol. 2015;96:263–275. doi: 10.1111/mmi.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song H, et al. Modulation of the regulatory activity of bacterial two-component systems by SlyA. J Biol Chem. 2008;283:28158–28168. doi: 10.1074/jbc.M801058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sawitzke JA, et al. Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J Mol Biol. 2011;407:45–59. doi: 10.1016/j.jmb.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martínez-García E, de Lorenzo V. Engineering multiple genomic deletions in Gram-negative bacteria: Analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ Microbiol. 2011;13:2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x. [DOI] [PubMed] [Google Scholar]
- 61.Wells TJ, et al. Increased severity of respiratory infections associated with elevated anti-LPS IgG2 which inhibits serum bactericidal killing. J Exp Med. 2014;211:1893–1904. doi: 10.1084/jem.20132444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salisbury A-M, Bronowski C, Wigley P. Salmonella Virchow isolates from human and avian origins in England—Molecular characterization and infection of epithelial cells and poultry. J Appl Microbiol. 2011;111:1505–1514. doi: 10.1111/j.1365-2672.2011.05152.x. [DOI] [PubMed] [Google Scholar]
- 63.Coil D, Jospin G, Darling AE. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 64.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. ABACAS: Algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laing C, et al. Pan-genome sequence analysis using Panseq: An online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics. 2010;11:461. doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guindon S, Delsuc F, Dufayard J-F, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 67.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The integrated genome browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Rankin JD, Taylor RJ. The estimation of doses of Salmonella Typhimurium suitable for the experimental production of disease in calves. Vet Rec. 1966;78:706–707. doi: 10.1136/vr.78.21.706. [DOI] [PubMed] [Google Scholar]
- 70.Zinder ND, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64:679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella Typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Msefula CL, et al. Genotypic homogeneity of multidrug resistant S. Typhimurium infecting distinct adult and childhood susceptibility groups in Blantyre, Malawi. PLoS One. 2012;7:e42085. doi: 10.1371/journal.pone.0042085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.