Significance
Penicillin-binding proteins (PBPs) synthesize the bacterial cell wall. These enzymes are important therapeutic targets, but little is known about how their activity is controlled to promote proper cell morphogenesis. Uncovering these regulatory mechanisms promises to reveal new approaches for disrupting cell wall biogenesis for antibiotic development. Here, we identify MacP as a factor required for PBP activity in the pathogen Streptococcus pneumoniae. We further show that MacP is a substrate of the kinase and global cell cycle regulator StkP and that phosphorylation is required for PBP activation by MacP. Our results support a model in which StkP and MacP form part of the regulatory network that coordinates cell wall synthesis with other morphogenetic activities during the cell cycle.
Keywords: cell wall, peptidoglycan, PBP, cell division, bacterial physiology
Abstract
Most bacterial cells are surrounded by an essential cell wall composed of the net-like heteropolymer peptidoglycan (PG). Growth and division of bacteria are intimately linked to the expansion of the PG meshwork and the construction of a cell wall septum that separates the nascent daughter cells. Class A penicillin-binding proteins (aPBPs) are a major family of PG synthases that build the wall matrix. Given their central role in cell wall assembly and importance as drug targets, surprisingly little is known about how the activity of aPBPs is controlled to properly coordinate cell growth and division. Here, we report the identification of MacP (SPD_0876) as a membrane-anchored cofactor of PBP2a, an aPBP synthase of the Gram-positive pathogen Streptococcus pneumoniae. We show that MacP localizes to the division site of S. pneumoniae, forms a complex with PBP2a, and is required for the in vivo activity of the synthase. Importantly, MacP was also found to be a substrate for the kinase StkP, a global cell cycle regulator. Although StkP has been implicated in controlling the balance between the elongation and septation modes of cell wall synthesis, none of its substrates are known to modulate PG synthetic activity. Here we show that a phosphoablative substitution in MacP that blocks StkP-mediated phosphorylation prevents PBP2a activity without affecting the MacP–PBP2a interaction. Our results thus reveal a direct connection between PG synthase function and the control of cell morphogenesis by the StkP regulatory network.
Most bacterial cells are surrounded by an exoskeleton-like matrix of peptidoglycan (PG) made of long glycan chains with short peptides that are used to form an interconnected network of cross-links. This material forms a continuous 3D lattice that encases the cytoplasmic membrane, specifies cell shape, and protects the bacterium from osmotic rupture. Cell growth and morphogenesis are therefore critically dependent on the expansion and division of the PG matrix.
PG biogenesis requires two reactions, glycan chain polymerization catalyzed by PG glycosyltransferases (GTs) and peptide cross-linking catalyzed by transpeptidases. The class A penicillin-binding proteins (aPBPs) possess both activities in a single enzyme. For many years, the GT domain of aPBPs was the only type of PG polymerase known. However, the highly conserved SEDS (shape, elongation, division, and sporulation) family protein RodA was recently shown to have PG polymerase activity (1). SEDS proteins are often found associated with class B PBPs (bPBPs) that possess transpeptidase activity (2). It has therefore been proposed that SEDS–bPBP complexes serve as a second type of bifunctional PG synthase in bacteria (1, 2).
Rod-shaped model organisms like Escherichia coli and Bacillus subtilis carry out two modes of cell wall synthesis, elongation and division, each promoted by different synthetic machineries (3). Elongation requires the Rod system, which consists of RodA and PBP2 as the SEDS–bPBP PG synthase pair along with several additional membrane proteins of poorly characterized function (MreC, MreD, and RodZ) (3, 4). These factors are organized into complexes by filaments of the actin-like protein MreB that are dispersed throughout the cell cylinder (3). These synthetic machines have been found to move circumferentially around the long axis of the cell and movement is thought to reflect the polymerization of new cell wall material (3). The aPBPs are not required for Rod system function and appear to support cell elongation and cell growth while working apart from these cytoskeletally organized complexes (2).
PG synthesis at the division site is promoted by a distinct complex called the divisome. This system is organized by polymers of the tubulin-like FtsZ protein, which assemble to form a ring-shaped structure called the Z-ring at midcell. Many factors are recruited to the Z-ring to catalyze cytokinesis, including known or predicted PG synthases like aPBPs and the SEDS–bPBP pair FtsW-PBP3 (3). The interplay between these different synthases at the division site and their respective roles in building the cell septum and eventual daughter cell poles is currently unclear.
Ovococci like the Gram-positive pathogen Streptococcus pneumoniae also grow using elongation and division modes of PG synthesis. However, unlike rod-shaped cells, all new synthesis occurs at midcell (4). S. pneumoniae lacks MreB, but retains homologs of the other Rod system components (4, 5). In ovococci, cell elongation proceeds via the zonal incorporation of new side-wall PG material at midcell (5). In addition, cell wall synthesis at midcell is also directed inward to form the cell septum (cross-wall) that separates the nascent daughters (6). The septal PG is eventually remodeled to form the new cell poles. Different subsets of synthases have been associated with elongation and septal modes of PG synthesis in S. pneumoniae, but the number of distinct machineries that are needed to carry out these processes is not completely clear (5–7).
Newborn rod-shaped cells and ovococci initially undergo elongation as the sole mode of PG biogenesis and then transition to a division mode of growth (8). This switch in growth phase is unlikely to be absolute, with some level of cell wall elongation proceeding during septal PG synthesis (8). Nevertheless, the transition between these growth modes and the balance between elongation and septation are likely to be tightly controlled to maintain proper cell size and shape. The mechanisms which govern these transitions are poorly understood in all cell types, but the eukaryotic-like serine/threonine kinase StkP has emerged as a key mediator of growth mode control in S. pneumoniae (9, 10). Several morphogenetic proteins, including DivIVA, FtsA, FtsZ, and MapZ (LocZ), have been found to be StkP substrates (11–16). Moreover, disrupting the normal phosphorylation of some StkP substrates causes imbalances in PG biogenesis and morphological defects (10, 14, 17). For example, in S. pneumoniae strain R800, a DivIVA variant that cannot be phosphorylated displays an elongated cell phenotype indicative of problems initiating septum synthesis (11). Furthermore, a phosphomimetic version of the substrate EloR (Jag/KhpB) was recently found to overstimulate elongation (17–19). Thus, StkP appears to modulate the balance between elongation and division modes of PG biogenesis in S. pneumoniae and potentially the switch to the septation growth mode through changes in the phosphorylation status of its substrates. However, to date none of the known targets of StkP phosphorylation have been found to be directly involved in modulating the activity of PG synthases like the aPBPs.
Here, we report the identification of MacP (SPD_0876) as a membrane-anchored cofactor of S. pneumoniae PBP2a, a class A PBP enzyme. We show that MacP localizes to the division site of S. pneumoniae, forms a complex with PBP2a, and is required for the in vivo activity of the synthase. Importantly, we report that MacP is a substrate for the kinase StkP, which phosphorylates MacP at residue T32. Blocking phosphorylation of MacP with a phosphoablative T32A substitution prevented PBP2a activity without disrupting the MacP–PBP2a interaction. Our results thus reveal a long sought after connection between PG synthase activity and the StkP regulatory network.
Results
A Genetic Screen Identifies MacP as a Cofactor Required for PBP2a Function.
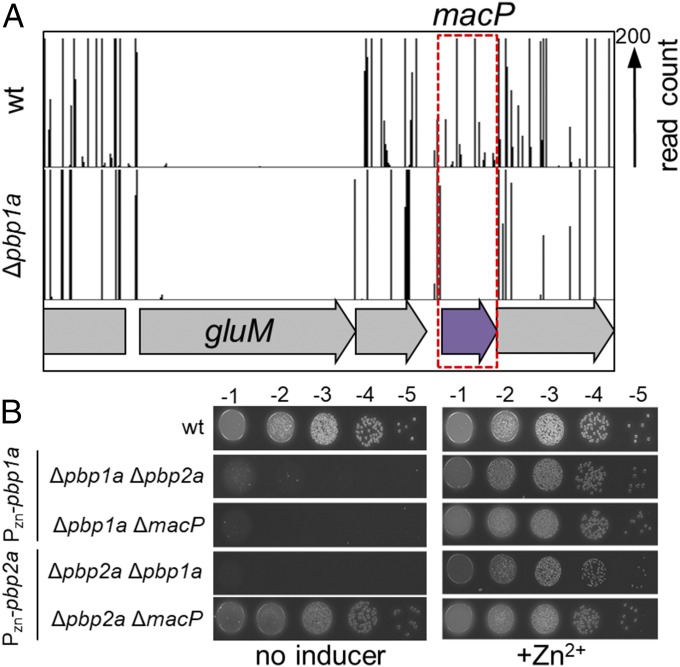
S. pneumoniae encodes three aPBPs: PBP1a, PBP1b, and PBP2a. No single enzyme is essential for growth (20, 21). However, a strain lacking both PBP1a and PBP2a could not be isolated, suggesting these enzymes form a synthetically lethal pair (20). Given this genetic relationship, we reasoned that genes encoding factors required for PBP2a activity could be identified by screening for mutants synthetically lethal with a pbp1a deletion. We therefore generated large transposon libraries in an unencapsulated variant of S. pneumoniae D39 (D39 Δcps) and an isogenic derivative lacking PBP1a (Δpbp1a). The global distribution of transposon insertions in the libraries were then determined using transposon sequencing (Tn-Seq) and compared with identify genes with insertion profiles that differed between them (Fig. S1) (22). Using this approach, we previously found that cozE had few transposon insertions in wild-type cells and appeared to be essential while it was readily inactivated by insertions in cells lacking PBP1a (23). This Tn-Seq profile suggested that CozE functions to restrain PBP1a activity to prevent its lethal malfunction, and we confirmed this to be the case. Here, we focused on the gene spd_0876, which had a Tn-Seq profile consistent with it being nonessential in the wild-type background, but essential in Δpbp1a cells (Fig. 1A and Fig. S1). The synthetic lethal phenotype suggested that spd_0876 encodes a factor required for PBP2a activity in vivo. Based on the results described below, we have renamed the gene macP for membrane-anchored cofactor of PBP2a.
Fig. 1.
macP is essential in cells lacking PBP1a. (A) Transposon insertion profiles for a select region of the chromosome for wild-type and the ∆pbp1a mutant. The height of each line in the profile represents the number of sequencing reads corresponding to a transposon insertion at the indicated genome position. Transposon insertions in the macP gene were significantly (P < 0.0002) underrepresented in the ∆pbp1a mutant relative to wild-type. Virtually no insertions were mapped to the essential gene gluM in either strain. Tn-seq output and statistical data for all libraries used in this study are shown in Fig. S1. (B) Spot dilutions of wild-type D39 ∆cps (wt) and the indicated derivatives. Cells were grown to exponential phase in the presence of 200 µM ZnCl2. The resulting cultures were normalized to an OD600 of 0.2, 10-fold serially diluted, and spotted (5 µL) onto TSA 5%SB plates in the presence or absence of 200 µM ZnCl2. Plates were incubated at 37 °C in 5% CO2 and imaged.
To verify the genetic relationship between macP and pbp1a, we constructed strains deleted for either pbp1a or pbp2a and harboring a zinc-inducible copy of the corresponding gene at an ectopic locus. As expected, a strain lacking PBP1a required PBP2a production for growth and viability, and a strain lacking PBP2a was similarly dependent on PBP1a expression (Fig. 1B and Fig. S2). Cells lacking MacP also required pbp1a induction for growth, but were not affected by the status of pbp2a expression (Fig. 1B and Fig. S2C). These results are consistent with our Tn-Seq analysis and suggest that MacP is required for PBP2a activity.
Inactivation of aPBPs Results in a Lethal Cell Size Defect.
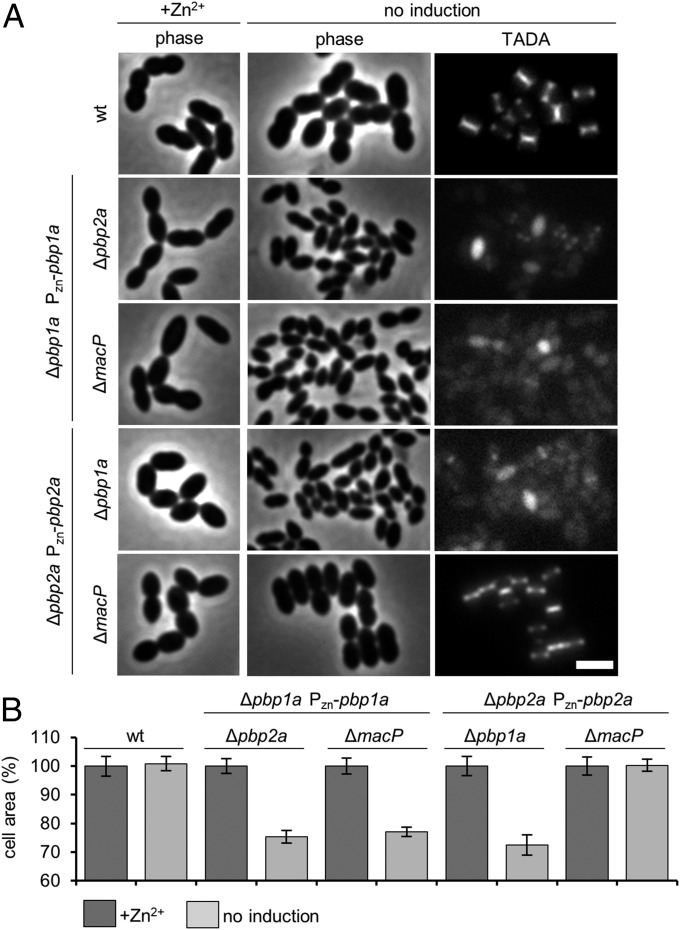
The terminal phenotype resulting from the simultaneous inactivation of PBP1a and PBP2a has not been previously described. We therefore monitored the effect of PBP1a depletion on morphology and cell wall synthesis in cells lacking PBP2a. Upon PBP1a depletion, the Δpbp2a cells displayed a progressive reduction in cell size before ultimately lysing (Fig. 2 and Movie S1). Cell area was reduced by roughly 30% before lysis, which corresponds to a 65% reduction in average cell volume. Cell wall synthesis was also dramatically reduced in these cells as assessed by labeling with the fluorescent D-amino acid 3-amino-d-alanine (TADA) (Fig. 2A). Similar phenotypes were observed in Δpbp1a cells upon depletion of PBP2a (Fig. 2 and Movie S1). Importantly, depletion of PBP1a in cells inactivated for MacP phenocopied the morphological and cell wall synthesis defects observed for PBP1a depletion in Δpbp2a cells (Fig. 2 and Movie S1). On the other hand, cells lacking MacP were unaffected by the depletion of PBP2a (Fig. 2 and Movie S1). Similar results were obtained when MacP was depleted in cells lacking either PBP1a or PBP2a (Movie S2). Collectively, these experiments indicate that aPBP enzymes in S. pneumoniae are responsible for maintaining cell size, presumably due to their contributions to both cell elongation and division activities, and that MacP is specifically required for PBP2a function in vivo.
Fig. 2.
Loss of essential aPBP activity results in a lethal cell size defect. (A) Representative phase-contrast and fluorescence images of the indicated strains. Midexponential phase cultures were diluted to an OD600 of 0.025 in the presence or absence of 200 µM ZnCl2 inducer. Cells were grown at 37 °C in 5% CO2 for 5 h 45 min. Cells were labeled with TADA for 15 min before imaging on 2% agarose pads. n ≥ 3. (Scale bar, 3 µm.) Additional controls and time-lapse movies can be found in Fig. S2A and Movie S1, respectively. (B) Cells depleted for aPBP activity show similar reductions in cell area. Strains were grown and imaged as described in A. Cell areas were calculated from cell meshes generated in MicrobeTracker (40): 750 cells were measured in total for each condition, 250 per experiment n = 3. Error bars show SD around the mean of each experiment.
MacP Is an Integral Membrane Protein Recruited to Midcell Independently from PBP2a.
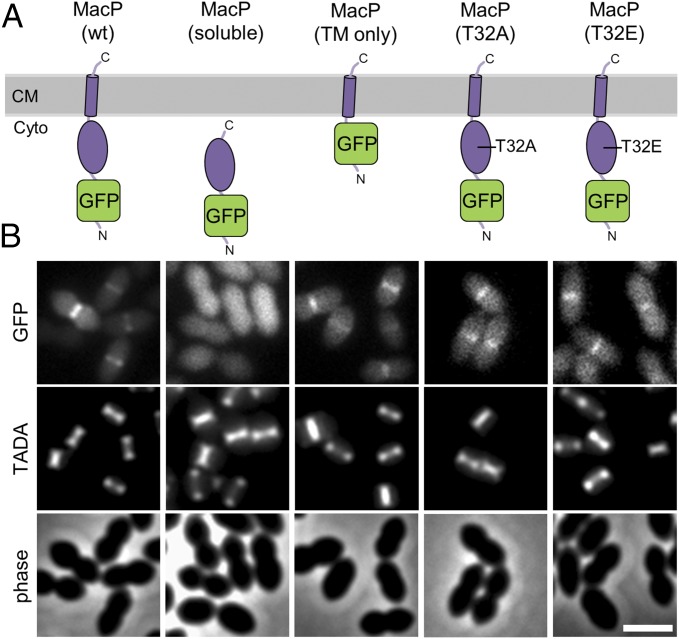
MacP is a small 104-amino acid protein with a single predicted transmembrane (TM) domain at its C terminus (Fig. 3A and Fig. S3A). Its N-terminal domain is predicted to reside on the cytoplasmic side of the membrane, and this topology was confirmed using a protease protection assay (Fig. S3 B and C). To investigate MacP function and localization, we generated an N-terminal GFP fusion (GFP–MacP) that supported wild-type cell morphology and growth rates in cells lacking PBP1a (Fig. 3A and Figs. S4A and S5D). Consistent with a role for MacP in cell wall biogenesis, GFP–MacP was enriched at midcell where the bulk of PG synthesis in S. pneumoniae takes place (Fig. 3 and Fig. S4 B and C). To investigate the functions of the N- and C-terminal domains, truncated GFP–MacP derivatives were generated and tested for their ability to localize and complement the growth of ΔmacP cells depleted of PBP1a. A fusion lacking the C-terminal transmembrane domain did not complement the ΔmacP defect nor did it localize to midcell (Fig. 3 and Fig. S5). However, the soluble N-terminal domain was not well expressed (Fig. S5A), so its functional status is unclear. Although nonfunctional, the GFP-fusion to the TM domain alone was produced at levels similar to GFP–MacP and was sufficient for midcell localization (Fig. 3 and Fig. S5). We therefore conclude that MacP has a topology with its N-terminal domain in the cytoplasm and that its TM domain is sufficient for midcell recruitment but not full MacP function.
Fig. 3.
MacP localizes to sites of new PG synthesis at midcell. (A) Schematic representation of GFP–MacP and truncations used for domain analysis. The domain architecture and membrane topology of MacP are based on protease protection data and in silico predictions shown in Fig. S3. Only the full-length GFP–MacP fusion was functional as assayed by the ability to complement ∆pbp1a ∆macP synthetic lethality (Figs. S4 and S5). The MacP soluble domain was taken as residues 1–85 and the TM segment residues 86–104. Immunoblot analysis of fusion proteins is shown in Fig. S5A. (B) Representative fluorescent and phase-contrast images of the fusions shown in A. GFP–MacP was enriched at sites of new PG synthesis at midcell. The fusions shown in A were expressed from a fucose-inducible promoter in a ΔmacP mutant inserted at the native locus. Strains were grown in Todd Hewitt broth containing 0.5% yeast extract (THY) in the presence of 0.4% fucose at 37 °C in 5% CO2 to an OD600 ∼ 0.2. Cells were labeled with TADA for 15 min before imaging on 2% agarose pads. n = 3. (Scale bar, 2 µm.)
We next investigated the requirement of MacP for the midcell localization of PBP2a and vice versa. Neither GFP–PBP1a nor GFP–PBP2a recruitment to midcell was affected by the loss of MacP (Fig. S6). Similarly, GFP–MacP localization was unaffected by the inactivation of either PBP1a or PBP2a (Fig. S4B). We therefore conclude that MacP and the aPBPs are recruited to midcell by independent mechanisms. Thus, rather than affecting its localization, MacP is likely to be required for promoting PBP2a function once it is recruited to the division site.
MacP Interacts with PBP2a.
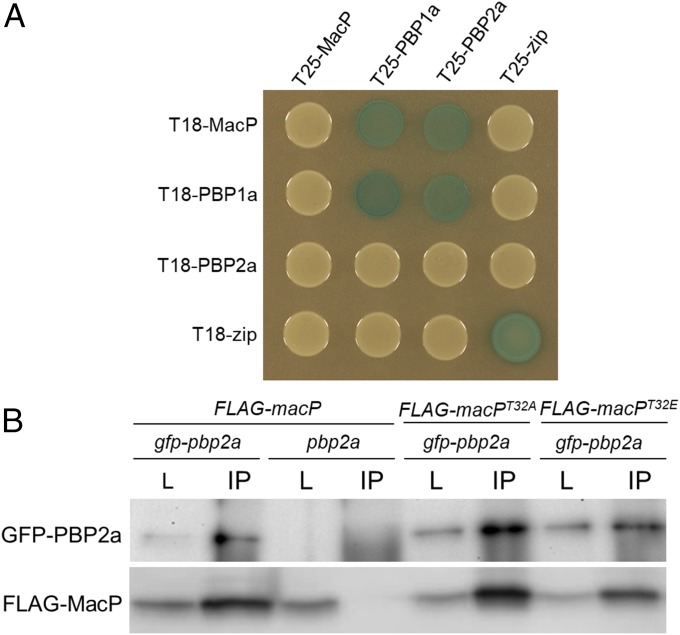
To investigate whether MacP binds PBP2a to promote its function, we used the E. coli bacterial two-hybrid (BACTH) system to screen for interactions between MacP and PBP2a, as well as other components of the S. pneumoniae PG synthetic machinery. Fusions to the T18 and T25 fragments of adenylate cyclase were constructed and expressed in E. coli cells lacking the endogenous adenylate cyclase gene. Positive interactions that generated elevated adenylate cyclase activity were detected as increased expression of a lacZ reporter gene (24). Our analysis revealed a BACTH interaction between MacP and both PBP1a and PBP2a (Fig. 4A). The observed interaction signals are likely to be specific because MacP did not have positive interactions with other membrane spanning S. pneumoniae cell wall synthetic factors: MreC, MreD, and CozE or membrane-associated E. coli control proteins (Fig. S7).
Fig. 4.
MacP interacts with both PBP2a and PBP1a. (A) BACTH interactions between MacP and aPBP enzymes. E. coli strain BTH101 (Δcya) expressing protein fusions to domains (T25 and T18) of adenylate cyclase. Strains were grown to stationary phase and 5 µL spotted on LB agar plates containing X-gal, incubated at 30 °C, and imaged. The “zip” fusions are to a leucine zipper domain and serve as both positive and negative controls. Additional controls are provided in Fig. S7. n = 3. (B) Coimmunoprecipitation of GFP–PBP2a and a functional FLAG–MacP fusion. Each fusion was expressed from the Pzn promoter using 400 µM ZnCl2. Digitonin-solubilized membrane preparations from the indicated strains were incubated with anti-GFP resin, washed, and eluted in sample buffer. Immunoblots show matched samples of solubilized membrane fractions (L) and immunoprecipitate (IP). IP samples are 20× concentrated relative to load. Fractions were probed with anti-GFP and anti-FLAG antibodies. Representative blots are shown, n = 3. Evidence of FLAG–MacP functionality, antibody specificity, additional controls, and full immunoblot analysis are provided in Fig. S8.
To further investigate the interaction between MacP and PBP2a, we performed coimmunoprecipitations using detergent-solubilized membrane preparations from S. pneumoniae cells. Specifically, we immunoprecipitated GFP–PBP2a using anti-GFP antibody resin from cells coexpressing a functional FLAG–MacP fusion. In agreement with the BACTH results, FLAG–MacP efficiently coprecipitated with GFP–PBP2a, but not in control preparations derived from cells harboring an untagged PBP2a (Fig. 4B and Fig. S8). A low but detectable amount of FLAG–MacP was also found to coprecipitate with GFP–PBP1a (Fig. S8). We conclude that MacP resides in a complex with PBP2a and to a much lesser extent with PBP1a. Because the interaction was detected in E. coli with these factors being the only S. pneumoniae proteins present in the cells, we infer that this interaction is likely to be direct. Given the weak interaction observed with PBP1a in the coimmunoprecipitation assays and the lack of any in vivo evidence for a functional relationship between PBP1a and MacP (Figs. 1B and 2), the significance of the MacP–PBP1a interaction is unclear.
MacP Is a Substrate of the StkP Kinase and Phosphorylation Is Required for Its Activity.
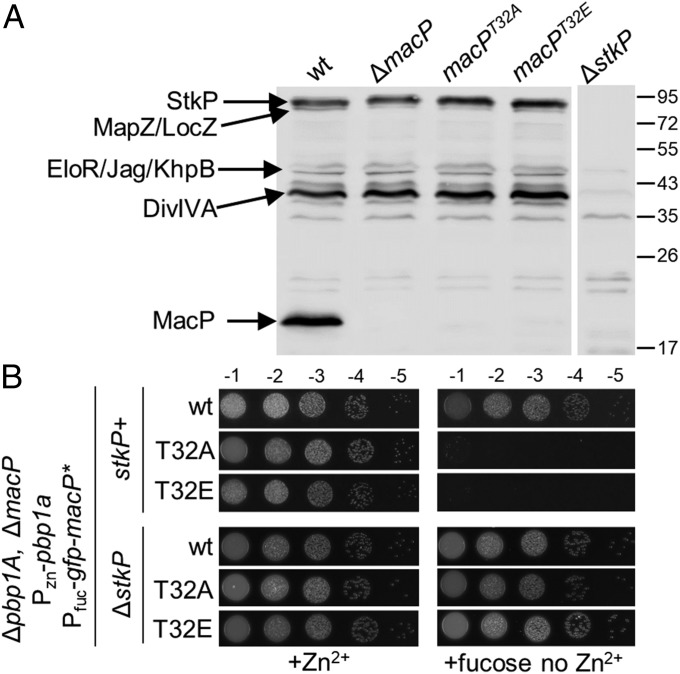
Analysis of the S. pneumoniae phosphoproteome revealed that MacP is phosphorylated at residue T32 (15). Notably, a strong StkP-dependent signal with a molecular mass of ∼15 kDa has been observed in antiphospho-threonine immunoblots (Fig. 5A) (14). The size of this protein is similar to MacP. Accordingly, we repeated the immunoblot analysis using lysates derived from cells lacking MacP or producing MacP with a phosphoablative T32A substitution. Strikingly, the StkP-dependent signal at ∼15 kDa was absent in these mutants without affecting other StkP-dependent phosphorylated signals (Fig. 5A). We conclude that MacP is a substrate of StkP and that the kinase phosphorylates it on residue T32.
Fig. 5.
The in vivo function of MacP requires phosphorylation by StkP. (A) Antiphospho-threonine immunoblot analysis of whole-cell lysates from the indicated strains. Phosphorylated StkP and its substrates are indicated based on work from previous studies (18, 19, 29). The positions of protein markers are indicated in kiloDaltons. (B) GFP–MacP(T32A)/(T32E) do not support growth of ∆pbp1A cells depleted of MacP. The indicated strains were grown to exponential phase in the presence of 200 µM ZnCl2, normalized to an OD600 of 0.2, and 5 µL of serially dilutions were spotted onto TSAII 5%SB plates in the presence of 0.2% fucose or 200 µM ZnCl2. Plates were incubated at 37 °C in 5% CO2 and imaged. Microscopy images of lethal cell size phenotype of MacP T32A and T32E on depletion of PBP1a are shown in Fig. S5C. Growth curves of each strain are provided in Fig. S5D.
To investigate whether phosphorylation for MacP was important for its function, we tested the functionality of GFP–MacP(T32A). Although this variant was stably produced, it was unable to support the growth of ΔmacP cells depleted of PBP1a or to prevent their lethal cell size phenotype (Fig. 5B and Fig. S5). However, and importantly, the fusion retained its ability to localize to midcell and to interact with PBP2a (Figs. 3A and 4B and Fig. S7C). We therefore infer that MacP phosphorylation is required for its ability to promote PBP2a function rather than for the formation of a MacP–PBP2a complex. We next tested the functionality of a phospho-mimetic GFP–MacP(T32E) fusion protein (Fig. 3). This fusion was stably produced and retained its ability to interact with PBP2a but, like MacP(T32A), was unable to support growth or prevent the lethal cell size phenotype in cells depleted of PBP1a (Figs. 3A and 4B and Fig. S7C). Although it is uncertain whether the glutamic acid substitution fully mimicked phosphorylation at threonine 32, the phenotypes raise the possibility that PBP2a may be subject to additional regulatory control.
The requirement for StkP-dependent phosphorylation of MacP suggested that the inactivation of StkP should result in a synthetic lethal phenotype with Δpbp1a that mimics those observed for ΔmacP, macP(T32A), and macP(T32E) mutants. However, we did not observe a significant growth defect in cells lacking MacP and StkP upon depletion of PBP1a (Fig. 5B). Furthermore, we found that deletion of stkP could suppress the synthetic lethality of ΔmacP cells depleted of PBP1a (Fig. 5B). These results suggest that MacP is critical for PBP2a activity when StkP is functional and other substrates of the kinase are also phosphorylated (Discussion).
Discussion
The PBPs are one of our oldest and most important therapeutic targets. It is therefore surprising how little is known about how bacterial cells control the activity of these key cell wall synthases. Several years ago, outer membrane lipoprotein activators were found to be required for the activity of aPBPs in the model Gram-negative bacterium E. coli (25, 26). LpoA is needed to activate PBP1a, whereas the unrelated lipoprotein LpoB is needed for PBP1b function. The role of Lpo factors as aPBP activators appears to be conserved in several related Gram-negative species (3, 25). To date, however, similar activators of aPBPs in Gram-positive organisms have not been described. Here, we identify MacP as one such activator required for the function of the aPBP PBP2a in S. pneumoniae. Similar to the Lpo proteins, MacP is narrowly distributed in bacteria. Among the Firmicutes family it is well conserved in within the Streptococci (Fig. S9). Like lpo mutants, deletion of macP results in phenotypes that mimic the inactivation of its cognate PBP, which for macP is synthetic lethality with PBP1a and a progressive reduction in cell size before ultimately lysing when PBP1a is depleted. In addition, MacP also interacts directly with its partner PBP like Lpo factors (25). However, unlike the Lpo factors, which are embedded in the outermembrane and bind specific regulatory subdomains on their target PBP, MacP is oriented such that it likely associates with PBP2a by binding its TM domain or N-terminal cytoplasmic tail (Fig. 6). In vitro studies have shown that these domains increase PBP2a activity (27). However, how MacP binding facilitates PBP2a function in vivo is not clear. Because PBP2a localizes normally in the absence of MacP, the complex is not required for proper PBP2a recruitment to midcell for it to participate in PG synthesis. One possibility is that MacP binding to the N-terminal tail could activate PBP2a by releasing it from inhibition by another factor or by inducing a conformational change in the PBP that stimulates its PG synthase activity. Alternatively, the enzymatic activity of PBP2a may be unaffected by MacP but instead the formation of the MacP–PBP2a complex may facilitate the association of PBP2a with other components of the PG synthetic machinery needed for its proper function. Additional studies will be required to distinguish among these possibilities.
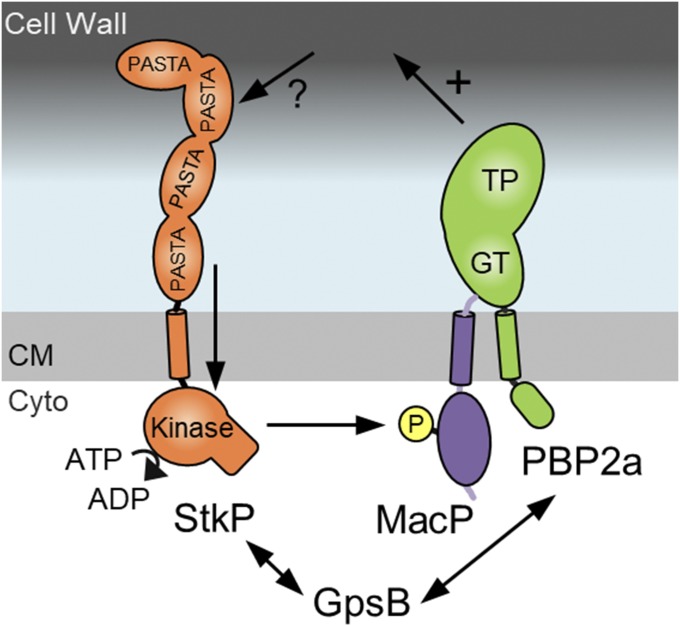
Fig. 6.
StkP-dependent phosphorylation of MacP is required for PBP2a activity in vivo. Schematic model of StkP-dependent regulation of PBP2A. The protein shapes and domains are adapted from published structural information (PDB ID codes 3PY9 and 1O6Y) and data from Fig. S3. StkP senses an unknown signal through its extracellular PASTA repeats triggering its kinase domain to phosphorylate MacP at T32. Phosphorylated MacP in complex with PBP2a then stimulates PBP2a-dependent cell wall synthesis (+). GpsB interacts with StkP and is required to maintain full kinase activity and is also required for PBP2a activity. GpsB data adapted from ref. 30. CM, cell membrane; Cyto, cytoplasm.
Another intriguing difference between MacP and the Lpo factors is that MacP activity is dependent on phosphorylation and therefore its ability to promote PBP activity is subject to kinase regulation. MacP was found to be phosphorylated at residue T32 in an StkP-dependent manner such that it is likely to be a direct substrate of this serine-threonine kinase. Disrupting phosphorylation by introduction of a T32A substitution results in a nonfunctional MacP that is stable and capable of associating with PBP2a in vivo, but apparently cannot activate PBP2a.
Surprisingly, a ΔstkP mutation, which should also block MacP phosphorylation and therefore prevent PBP2a activation, was not found to be synthetically lethal with PBP1a inactivation. There are several possible explanations for this result. Reports in the literature differ in their assessment of ΔstkP cell-division defects (9, 10, 28, 29). Therefore, it is possible that ΔstkP mutations are easily suppressed (9) and that strains inactivated for StkP contain a suppressor mutation that bypasses the MacP requirement for PBP2a function. This explanation is in close agreement with recent work in the literature, where suppressor mutations in ΔstkP strains are thought to bypass the function of other essential cell-division processes (30). Alternatively, MacP could be required to overcome an inhibitory activity of StkP or one of its substrates when it is phosphorylated. Thus, inactivation of StkP would relieve PBP2a inhibition and bypass its requirement for MacP. In the context of this model, MacP and its modulation by StkP would connect PBP2a activity with the function of other factors controlled by StkP phosphorylation. A better understanding of the mechanism by which MacP promotes PBP2a activity should help clarify the genetic relationships between stkP and macP.
How phosphorylation affects MacP activity is not known, but it likely provides a means of integrating PBP2a activation within the larger StkP regulatory network. StkP is the only known serine/threonine kinase in S. pneumoniae and global phosphoproteomic analysis has identified >70 proteins that are phosphorylated on serine or threonine residues in vivo (15). While only a very small number of these proteins have thus far been validated as functionally relevant targets, the emerging view is that StkP-dependent phosphorylation promotes a shift from the elongation to the division mode of cell wall synthesis. In the R800 S. pneumoniae strain background, a DivIVA variant that cannot be phosphorylated by StkP displays an elongated cell phenotype indicative of problems initiating septum synthesis (13, 14), while MapZ phosphorylation may be important for septum closure (11, 12). Here, we show that StkP-dependent phosphorylation of MacP is critical for controlling the cell wall synthetic activity of PBP2a. In S. pneumoniae, PBP1a and PBP2a are functionally redundant and can each compensate for the absence of the other. However, recent work on PBP1a suggests that its primary role may be in zonal elongation as it resides in a complex with MreC, MreD, and CozE (23, 31, 32). Accordingly, we speculate that PBP2a activity may be more closely tied to division. If correct, StkP-mediated phosphorylation of MacP would function in concert with DivIVA in promoting a shift toward the septal mode of PG synthesis. It remains an open question whether StkP phosphorylation similarly impacts the predicted cell wall synthesis activity of the SEDS–bPBP pair FtsW–PBP2x that plays an essential role in building the cross wall.
This work defines a direct connection between StkP kinase activity and PBP2a function mediated by MacP (Fig. 6). Interestingly, recent studies suggest that a second factor, GpsB, may be intimately involved in this regulatory pathway. Evidence suggests that GpsB interacts with StkP (14) and is required to maintain full StkP activity (14, 30). Furthermore, genetic analysis suggests that GpsB is required for PBP2a activity (30). The GpsB ortholog from Listeria monocytogenes directly interacts with the N-terminal cytoplasmic domain of the aPBP PBP A1 (33). Thus, one possibility is that in S. pneumoniae, GpsB bound to PBP2a functions to recruit StkP to MacP and stimulate substrate phosphorylation (Fig. 6). Future experiments will be aimed at establishing whether GpsB and MacP function in the same or parallel regulatory pathways.
In Gram-positive bacteria, StkP, and other serine/threonine kinases play important roles in bacterial cell biology (34, 35). These bitopic membrane proteins have a cytoplasmic kinase domain and an extracellular domain composed of PASTA (penicillin-binding protein and serine/threonine kinase associated) repeats (36). Where examined, these kinases act as global mediators/regulators of morphogenesis (35), differentiation (34), and PG remodeling (37). Work on several family members indicate that the PASTA repeats recognize muropeptides or uncross-linked PG (37–39). In S. pneumoniae, the PASTA domains on StkP are required for function (10) and the current thinking is that they recognize some aspect of PG synthesis or remodeling that triggers kinase activity. A central challenge for the future is to define what specific feature of PG or the PG assembly process is sensed by these repeats to stimulate StkP and ultimately trigger the activation of septal PG synthesis. Equally important will be to identify the full suite of functionally relevant StkP phosphorylation targets and establish how their phosphorylation might shift the balance between elongation and division modes of PG synthesis. Answers to these questions will provide a deeper understanding of cell morphogenesis and help identify new ways of disrupting cell wall biogenesis for future antibiotic development.
Experimental Procedures
All S. pneumoniae strains were derived from D39 Δcps. Cells were grown in Todd Hewitt broth containing 0.5% yeast extract at 37 °C with 5% CO2. Tn-seq and coimmunoprecipitation assays were carried out as described previously (23). Fluorescence and phase-contrast microscopy was performed on a Nikon Eclipse Ti-E inverted microscope. Detailed protocols are provided in SI Experimental Procedures. Strains, plasmids, and oligonucleotides used in this study are listed in Table S1.
Supplementary Material
Acknowledgments
We thank all members of the T.G.B. and D.Z.R. supergroup for helpful advice and discussions; Malcolm Winkler, Jan-Willem Veening, and Don Morrison for strains, reagents, and technical assistance; the Microscopy Resources on the North Quad (MicRoN) core at Harvard Medical School for help with imaging and analysis; and Céline Freton from the C.G. laboratory for expert technical assistance. This work was supported by National Institutes of Health Grants R01AI083365 (to T.G.B.), CETR U19 AI109764 (to T.G.B. and D.Z.R.), and GM073831 (to D.Z.R.). C.G. was supported by grants from the CNRS, the University of Lyon, the Agence National de la Recherche (ANR-15-CE32-0001-01), and the Bettencourt-Schueller Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715218115/-/DCSupplemental.
References
- 1.Meeske AJ, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho H, et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol. 2016;1:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Massidda O, Nováková L, Vollmer W. From models to pathogens: How much have we learned about Streptococcus pneumoniae cell division? Environ Microbiol. 2013;15:3133–3157. doi: 10.1111/1462-2920.12189. [DOI] [PubMed] [Google Scholar]
- 5.Tsui HT, et al. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol Microbiol. 2014;94:21–40. doi: 10.1111/mmi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sham L-T, Tsui H-CT, Land AD, Barendt SM, Winkler ME. Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Curr Opin Microbiol. 2012;15:194–203. doi: 10.1016/j.mib.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Land AD, et al. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol Microbiol. 2013;90:939–955. doi: 10.1111/mmi.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol Microbiol. 2011;82:1096–1109. doi: 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- 9.Beilharz K, et al. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Natl Acad Sci USA. 2012;109:E905–E913. doi: 10.1073/pnas.1119172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleurie A, et al. Mutational dissection of the S/T-kinase StkP reveals crucial roles in cell division of Streptococcus pneumoniae. Mol Microbiol. 2012;83:746–758. doi: 10.1111/j.1365-2958.2011.07962.x. [DOI] [PubMed] [Google Scholar]
- 11.Fleurie A, et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature. 2014;516:259–262. doi: 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holečková N, et al. LocZ is a new cell division protein involved in proper septum placement in Streptococcus pneumoniae. MBio. 2014;6:e01700–e01714. doi: 10.1128/mBio.01700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fadda D, et al. Streptococcus pneumoniae DivIVA: Localization and interactions in a MinCD-free context. J Bacteriol. 2007;189:1288–1298. doi: 10.1128/JB.01168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleurie A, et al. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 2014;10:e1004275. doi: 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, et al. Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J Proteome Res. 2010;9:275–282. doi: 10.1021/pr900612v. [DOI] [PubMed] [Google Scholar]
- 16.Giefing C, Jelencsics KE, Gelbmann D, Senn BM, Nagy E. The pneumococcal eukaryotic-type serine/threonine protein kinase StkP co-localizes with the cell division apparatus and interacts with FtsZ in vitro. Microbiology. 2010;156:1697–1707. doi: 10.1099/mic.0.036335-0. [DOI] [PubMed] [Google Scholar]
- 17.Stamsås GA, et al. Identification of EloR (Spr1851) as a regulator of cell elongation in Streptococcus pneumoniae. Mol Microbiol. 2017;105:954–967. doi: 10.1111/mmi.13748. [DOI] [PubMed] [Google Scholar]
- 18.Ulrych A, et al. Characterization of pneumococcal Ser/Thr protein phosphatase phpP mutant and identification of a novel PhpP substrate, putative RNA binding protein Jag. BMC Microbiol. 2016;16:247. doi: 10.1186/s12866-016-0865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng JJ, Perez AJ, Tsui HT, Massidda O, Winkler ME. Absence of the KhpA and KhpB (JAG/EloR) RNA-binding proteins suppresses the requirement for PBP2b by overproduction of FtsA in Streptococcus pneumoniae D39. Mol Microbiol. 2017;106:793–814. doi: 10.1111/mmi.13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoskins J, et al. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J Bacteriol. 1999;181:6552–6555. doi: 10.1128/jb.181.20.6552-6555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paik J, Kern I, Lurz R, Hakenbeck R. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J Bacteriol. 1999;181:3852–3856. doi: 10.1128/jb.181.12.3852-3856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Opijnen T, Bodi KL, Camilli A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenton AK, Mortaji LE, Lau DTC, Rudner DZ, Bernhardt TG. CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat Microbiol. 2016;2:16237. doi: 10.1038/nmicrobiol.2016.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karimova G, Pidoux J, Ullmann A, Ladant D, Adant DAL. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Typas A, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paradis-Bleau C, et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helassa N, Vollmer W, Breukink E, Vernet T, Zapun A. The membrane anchor of penicillin-binding protein PBP2a from Streptococcus pneumoniae influences peptidoglycan chain length. FEBS J. 2012;279:2071–2081. doi: 10.1111/j.1742-4658.2012.08592.x. [DOI] [PubMed] [Google Scholar]
- 28.Osaki M, et al. The StkP/PhpP signaling couple in Streptococcus pneumoniae: Cellular organization and physiological characterization. J Bacteriol. 2009;191:4943–4950. doi: 10.1128/JB.00196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nováková L, et al. Identification of multiple substrates of the StkP Ser/Thr protein kinase in Streptococcus pneumoniae. J Bacteriol. 2010;192:3629–3638. doi: 10.1128/JB.01564-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rued BE, et al. Suppression and synthetic-lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin-binding protein interactions in Streptococcus pneumoniae D39. Mol Microbiol. 2017;103:931–957. doi: 10.1111/mmi.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsui HCT, et al. Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Mol Microbiol. 2016;100:1039–1065. doi: 10.1111/mmi.13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Land AD, Winkler ME. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J Bacteriol. 2011;193:4166–4179. doi: 10.1128/JB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rismondo J, et al. Structure of the bacterial cell division determinant GpsB and its interaction with penicillin-binding proteins. Mol Microbiol. 2016;99:978–998. doi: 10.1111/mmi.13279. [DOI] [PubMed] [Google Scholar]
- 34.Pereira SFF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manuse S, Fleurie A, Zucchini L, Lesterlin C, Grangeasse C. Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis. FEMS Microbiol Rev. 2016;40:41–56. doi: 10.1093/femsre/fuv041. [DOI] [PubMed] [Google Scholar]
- 36.Yeats C, Finn RD, Bateman A. The PASTA domain: A β-lactam-binding domain. Trends Biochem Sci. 2002;27:438–440. doi: 10.1016/s0968-0004(02)02164-3. [DOI] [PubMed] [Google Scholar]
- 37.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mir M, et al. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011;7:e1002182. doi: 10.1371/journal.ppat.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maestro B, et al. Recognition of peptidoglycan and β-lactam antibiotics by the extracellular domain of the Ser/Thr protein kinase StkP from Streptococcus pneumoniae. FEBS Lett. 2011;585:357–363. doi: 10.1016/j.febslet.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.