Significance
The evolution of nonrecombining chromosomes such as sex chromosomes involves degeneration leading to loss of genetic information. We do not know, however, what happens during the incipient stages of such chromosomes, before appreciable degeneration. We studied this process in white-throated sparrows, a species that occurs in two alternative behavioral phenotypes determined by a nonrecombining autosomal rearrangement. We report that this rearrangement shows few signs of large-scale genetic degeneration. Instead, substantial changes have evolved at the level of gene expression, some of them consistent with adaptive evolution. Our work with this chromosome reveals that rapid changes in gene expression and dosage compensation, not necessarily large-scale genetic degeneration, characterize the early evolution of heteromorphic chromosomes and the associated divergent phenotypes.
Keywords: suppression of recombination, genetic degeneration, regulatory evolution, dosage compensation, sex chromosomes
Abstract
In the white-throated sparrow (Zonotrichia albicollis), the second chromosome bears a striking resemblance to sex chromosomes. First, within each breeding pair of birds, one bird is homozygous for the standard arrangement of the chromosome (ZAL2/ZAL2) and its mate is heterozygous for a different version (ZAL2/ZAL2m). Second, recombination is profoundly suppressed between the two versions, leading to genetic differentiation between them. Third, the ZAL2m version is linked with phenotypic traits, such as bright plumage, high aggression, and low parental behavior, which are usually associated with males. These similarities to sex chromosomes suggest that the evolutionary mechanisms that shape sex chromosomes, in particular genetic degeneration of the heterogametic version due to the suppression of recombination, are likely important in this system as well. Here, we investigated patterns of protein sequence evolution and gene expression evolution between the ZAL2 and ZAL2m chromosomes by whole-genome sequencing and transcriptome analyses. Patterns of protein evolution exhibited only weak signals of genetic degeneration, and few genes harbored signatures of positive selection. We found substantial evidence of transcriptome evolution, such as significant expression divergence between ZAL2 and ZAL2m alleles and signatures of dosage compensation for highly expressed genes. These results suggest that, early in the evolution of heteromorphic chromosomes, gene expression divergence and dosage compensation can prevail before large-scale genetic degeneration. Our results show further that suppression of recombination between heteromorphic chromosomes can lead to the evolution of alternative (sex-like) behavioral phenotypes before substantial genetic degeneration.
The white-throated sparrow (Zonotrichia albicollis) occurs in two alternative morphs, white-striped and tan-striped (hereafter referred to as white and tan) (Fig. 1A), that differ in both plumage and social behavior (1–5). The morphs are completely linked to a large rearrangement on the second chromosome, called ZAL2m (6). White birds are heterozygous for ZAL2m, whereas tan birds are homozygous for a different version of the chromosome, ZAL2, which shares little genetic exchange with ZAL2m. Intriguingly, nearly all matings in the white-throated sparrow occur between birds of different morphs (7, 8). Because of this disassortative mating, ZAL2m is rarely homozygous and is, like the W and Y chromosomes in diverse species (9), excluded from homologous recombination. A group of closely linked loci that control a polymorphic phenotype, such as those found on sex chromosomes, is sometimes referred to as a “supergene” (10–16). The ZAL2/ZAL2m system represents an intriguing example of a non–sex-linked chromosome functioning as a supergene responsible for phenotypic divergence.
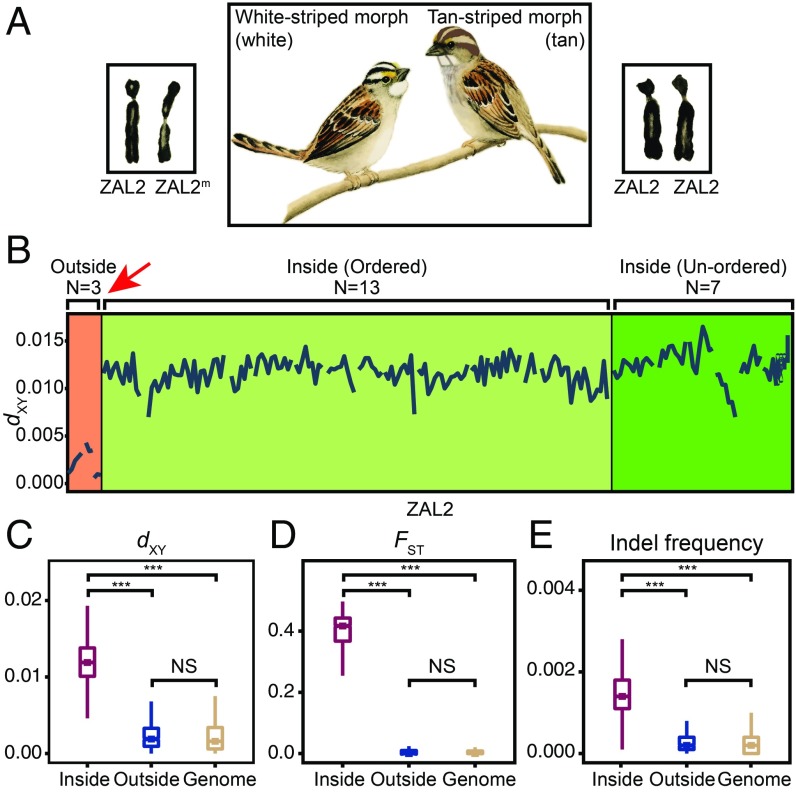
Fig. 1.
Genetic divergence between the ZAL2 and ZAL2m chromosomes in the white-throated sparrow. (A) Two plumage morphs in the white-throated sparrow, white and tan, are completely linked to the presence or absence of a rearrangement in the second pair of chromosomes (karyotypes adapted from ref. 6, with permission from the Society for the Study of Evolution). (B) Nucleotide divergence measures per 500-kb windows between ZAL2 and ZAL2m are shown for scaffolds longer than 1 Mbp. A red arrow indicates the inferred major breakpoint (18). Scaffolds residing outside the rearrangement (n = 3, orange background) show little divergence between the two chromosomes. Divergence is elevated in regions within the rearrangement. Scaffolds with known, precise locations (n = 13) are shown on a light green background. Additional scaffolds (n = 7) that reside within the rearrangement are shown on a dark green background. (C–E) Pairwise nucleotide divergences (dXY), degrees of population differentiation (FST), and indel frequencies, all measured in 10-kb nonoverlapping windows, were significantly higher in scaffolds within the rearrangement than in those outside it (Mann–Whitney U test, ***P < 0.001; NS, not significant). Including all scaffolds that are putatively on the second chromosome in the calculation yielded highly similar results (SI Appendix, Fig. S1).
Evolutionary analyses have established that the ZAL2m rearrangement arose at most 2–3 million years ago (7, 17, 18). Indeed, putatively neutral sites in the two versions have differentiated by only 1% over that time period (17, 18). Thus, this system affords a rare look at the early evolution of heteromorphic chromosomes. Previously, Thomas et al. (18) utilized both population genetic and cytogenetic mapping approaches to investigate this system and revealed an ∼100-Mb rearrangement consisting of at least two inversions. Subsequent studies have found strong linkage disequilibrium as well as reduced genetic diversity within the rearranged ZAL2m region compared with ZAL2 counterparts for dozens of loci (19, 20), which is consistent with suppressed recombination within the rearranged region. Interestingly, despite the evidence of reduced recombination, Davis et al.’s (20) study of 62 genes did not find any pseudogenization or truncation of proteins in the rearranged region of ZAL2m. Recently, Tuttle et al. (7) characterized the divergence between ZAL2 and ZAL2m chromosomes by low-coverage pooled sequencing of heterozygous individuals and reported slightly reduced levels of genetic diversity and an excess of nonsynonymous polymorphisms on the ZAL2m. However, reliably distinguishing ZAL2 and ZAL2m alleles in heterozygous individuals was challenging, which limited the interpretation of the results (7).
In this study, we overcame the challenges associated with distinguishing ZAL2m and ZAL2 alleles in heterozygous individuals (7, 18) and offer a comprehensive genome-wide comparison of the two chromosomes. We took advantage of a fortuitous finding of a rare individual homozygous for the ZAL2m chromosome (21). This “superwhite” female, a product of an extremely rare mating between white morphs, exhibited brighter plumage and higher aggression compared with both white and tan birds (21), consistent with the role of the ZAL2m chromosome in alternative phenotypes in this species. We sequenced the genome of this rare bird to compare the ZAL2 sequence with the ZAL2m sequence. Moreover, we extended our genome comparison with study expression divergence by integrating deep-sequencing data from the superwhite female with brain transcriptome data from behaviorally characterized tan and white birds (22). We then proceeded to look for evidence of genetic degeneration, which would be predicted for nonrecombining chromosomes (23–26), as well as evidence of divergence at the level of gene expression. Interestingly, comparison of protein-coding sequences between ZAL2 and ZAL2m suggests that a large-scale degeneration is yet to occur in ZAL2m. On the other hand, gene expression divergence between ZAL2 and ZAL2m is extensive.
Results
Genomic Differentiation of ZAL2 and ZAL2m.
We generated ∼35× coverage sequence data from a rare superwhite female homozygous for ZAL2m as described by Horton et al. (21) [Sequence Read Archive (SRA) accession no. SRR4191732]. We mapped these data to the existing Z. albicollis reference assembly from a tan individual (ZAL2/ZAL2) (7) and identified scaffolds that are likely to reside on the ZAL2/2m chromosomes on the basis of previous fluorescent in situ hybridization (FISH) studies (7, 18, 20) and interchromosomal homology in birds (27–30) (SI Appendix). Divergence measures between ZAL2 and ZAL2m alleles were computed using the zebra finch (Taeniopygia guttata) (29) as an outgroup. We were able to distinguish ZAL2 scaffolds that reside within versus outside of the rearrangement based upon the number of nucleotide substitutions per site (dXY) between ZAL2 and ZAL2m, population differentiation (FST) between the tan and white morphs, and the presence of putatively fixed differences between ZAL2 and ZAL2m in the 21 white-throated sparrows used in this study (Materials and Methods and SI Appendix).
We identified 35 (105.96 Mb) ZAL2 scaffolds within the rearrangement and five ZAL2 scaffolds (5.38 Mb) outside the rearrangement (SI Appendix, Table S1). The average dXY between ZAL2 and ZAL2m inside the rearrangement was 1.202 ± 0.006% (Fig. 1 B and C). The variability of regions outside the rearrangement was statistically indistinguishable from that of the genomic background (Fig. 1C), reflecting the level of intraspecies polymorphism. We did not observe clear clustering of nucleotide divergence, or so-called “evolutionary strata” (31), within the rearranged region. Population differentiation between the morphs was reflected in an average FST of 0.381 ± 0.006 within the rearranged region (Fig. 1D). Regions outside the rearrangement, as well as in the rest of the genome, showed little differentiation, as expected (Fig. 1D). The rearranged regions in ZAL2 and ZAL2m also differed with respect to insertion-deletion (indel) frequencies; regions inside the rearrangement exhibited significantly more indels compared with regions outside or in the rest of the genome (Fig. 1E). These results confirm the high degree of genetic differentiation between the rearranged ZAL2m region and ZAL2.
Weak Signatures of Degeneration and Positive Selection in Protein-Coding Sequences.
We examined protein-coding sequences for signatures of genetic degeneration, such as nonsense mutations and frame-shift mutations. Of 1,007 protein-coding loci linked to the rearranged ZAL2m, only 28 contained premature stop codons or had lost start or stop codons (SI Appendix, Table S2). We found no frame-shift mutations or deletions of genes from the rearranged portion of ZAL2m (SI Appendix).
Several genome-scale features of the ZAL2m-linked loci were consistent with a subtle level of genetic degeneration. Rates of nonsynonymous substitution (dN), radical amino acid substitution (dR), and promoter (defined as 1 kb upstream from transcription start site) substitution (dP), the ratio of radical-to-conservative amino acid substitution rates (dR/dC), as well as the ratio of nonsynonymous-to-synonymous substitution rates (dN/dS), were all elevated on ZAL2m compared with ZAL2 (Fig. 2A). These results could be explained by the accumulation of slightly deleterious mutations on the ZAL2m chromosome. On the other hand, rates of synonymous substitution (dS) and whole-genome divergence (dXY) did not show chromosomal bias (Fig. 2A). The numbers of putatively fixed substitutions across nonsynonymous sites, synonymous sites, and 5′ and 3′ untranslated transcribed regions (UTRs) varied in concordance with the degree of purifying selection (Fig. 2B). Synonymous sites showed the highest level of divergence. The level of divergence was progressively lower in 3′ UTRs, 5′ UTRs, and nonsynonymous sites. The number of fixed substitutions thus correlated inversely with the expected degree of purifying selection (e.g., ref. 32). Rates of conservative amino acid substitution (dC) were also similar between the two chromosomes, consistent with strong purifying selection (Fig. 2A).
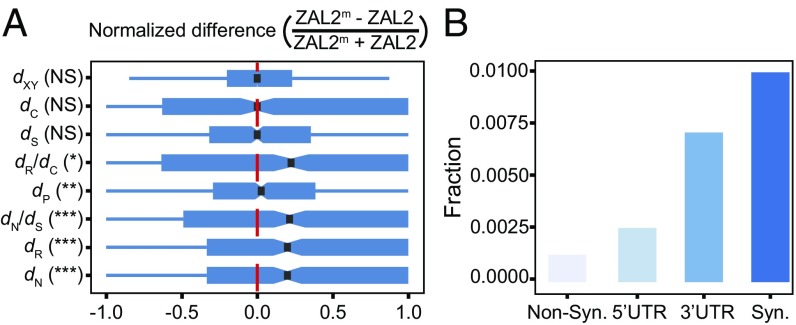
Fig. 2.
Substitution bias and proportions of putatively fixed differences suggest weak degeneration of the ZAL2m chromosome. (A) Substitution bias differs between putatively functional versus neutral sites. The normalized difference in substitution rates between ZAL2 and ZAL2m is shown. This value is expected to be zero when there are equal numbers of substitutions on ZAL2 and ZAL2m (thus, no bias), positive when there are more substitutions on ZAL2m, and negative when there are more ZAL2 substitutions. Deviation from zero was tested using the paired Mann–Whitney U test. The measures presented include nucleotide divergence (dXY), conservative amino acid substitution rate (dC), synonymous substitution rate (dS), the ratio of radical-to-conservative amino acid substitution rates (dR/dC), promoter (defined as 1 kb upstream from transcription start site) substitution rate (dP), the ratio of nonsynonymous-to-synonymous substitution rates (dN/dS), radical amino acid substitution rate (dR), and nonsynonymous substitution rate (dN), presented in the order of decreasing P values. Paired Mann–Whitney U test, *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. (B) Distribution of putatively fixed differences according to the type of functional site. Synonymous sites (Syn.) show the highest fraction (∼1%) of putatively fixed differences, which is similar to the genome-wide nucleotide divergence. Nonsynonymous sites (Non-Syn.) show the smallest fraction. This pattern correlates inversely with presumed selective constraints (32).
Using a branch-site model in PAML (33) with sequences from 13 other Passeriformes and a simulation-based approach (34) (Materials and Methods), we examined signatures of positive selection in the coding sequences. We found signs of positive selection for three genes on ZAL2 and two genes on ZAL2m by using a cutoff of false discovery rate (FDR)-adjusted Q < 0.2 (SI Appendix, Table S3). Because incomplete lineage sorting may result in discordance between gene trees and the species tree and therefore interfere with identification of positively selected genes, we constructed maximum-likelihood gene trees for the five genes and reran PAML. The signatures of positive selection remained for all five genes after Bonferroni correction (Bonferroni-corrected P < 0.05). We also tested whether genes in specific functional categories were evolving under different selective constraints (35, 36) (Materials and Methods). Interestingly, some categories of genes showed evidence of significantly different selection pressure between the two chromosomes (SI Appendix, Table S4), for example, rRNA processing [Gene Ontology (GO): 0006364] on ZAL2 and positive regulation of neuron projection development (GO: 0010976) and the Wnt-signaling pathway (GO: 0016055) on ZAL2m.
Differential Allelic Expression Between ZAL2 and ZAL2m.
Genetic differentiation of regulatory regions could cause divergence of gene expression even in the absence of coding sequence divergence (37). We examined gene expression divergence between ZAL2 and ZAL2m alleles using RNA-seq data of 9 tan and 10 white individuals from two brain regions implicated in social behavior in birds: the hypothalamus and nucleus taeniae (called medial amygdala in some publications) (22). These data came from breeding birds that had been confirmed to exhibit the typical morph differences in behavior. We first tested for allele-specific expression of ZAL2 and ZAL2m alleles in white birds using DESeq2 (38) (Materials and Methods and SI Appendix). We found 335 and 375 genes (41.41 and 46.53% of all ZAL2/ZAL2m-linked genes examined) with significant allele-specific expression in the hypothalamus and nucleus taeniae, respectively (FDR-adjusted Q < 0.05; Table 1). Among all of the genes with allele-specific expression, 236 were common between the two brain regions, a significant enrichment of shared allele-specific expression (hypergeometric test, P < 0.001). This supports the idea that allele-specific expression of ZAL2 and ZAL2m alleles is driven by divergence of regulatory sequences. As expected, there was a bias toward reduced ZAL2m expression (one-tailed, paired Mann–Whitney U test, hypothalamus: P = 0.066 and nucleus taeniae: P = 0.049).
Table 1.
Patterns of morph-biased expression for genes showing allele-specific expression
| Brain region | tan > white | white > tan | Other (tan ≈ white) | |
| Hypothalamus | ZAL2 > ZAL2m | 63 (87) | 1 (2) | 111 (118) |
| ZAL2m > ZAL2 | 0 (0) | 109 (146) | 51 (40) | |
| Nucleus taeniae | ZAL2 > ZAL2m | 54 (86) | 1 (1) | 153 (136) |
| ZAL2m > ZAL2 | 0 (0) | 99 (123) | 68 (65) |
Among genes that show allele-specific expression between ZAL2 and ZAL2m in white birds, some also show morph-biased expression. Above, morph-biased genes are separated into tan > white and white > tan. Genes that do not exhibit morph-biased expression are shown in the “Other” category. Differences were assessed by DESeq2, and the numbers of genes with FDR-corrected Q < 0.05 (or Q > 0.05 for the “Other” category) are shown in the table, with P < 0.05 (or P > 0.05 for the “Other” category) in parentheses.
Potential Dosage Compensation of Highly Expressed Genes.
We then asked whether allele-specific expression in white birds has led to differential expression between the tan and white morphs (henceforth referred to as “morph-biased gene expression”). For example, if morph-biased expression of a given gene is caused entirely by allele-specific expression, reduced expression of the ZAL2m allele compared with the ZAL2 allele (ZAL2 > ZAL2m) could lead to lower total expression in the white birds (ZAL2/ZAL2m) relative to tan birds (ZAL2/ZAL2) (Fig. 3 A and B). Selection could further favor increased expression of ZAL2 compared with the degenerated ZAL2m allele, leading to increased expression in tan birds (23, 39). Conversely, genes for which ZAL2m expression is elevated compared with ZAL2 (ZAL2m > ZAL2) could exhibit higher expression in white birds compared with tan birds (SI Appendix, Fig. S2).
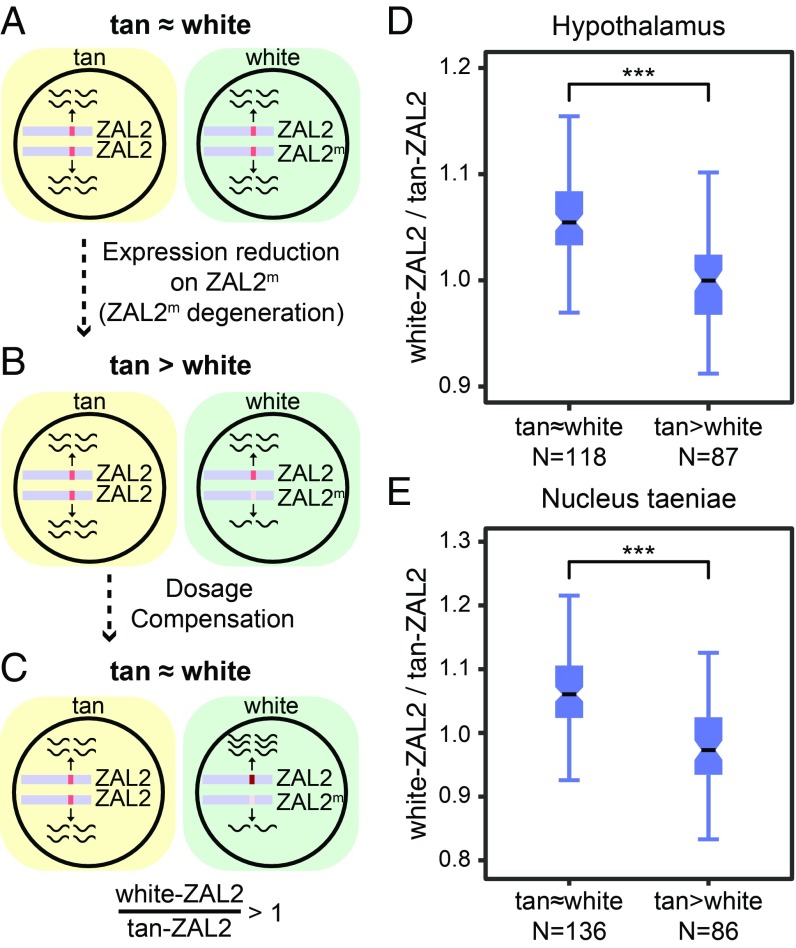
Fig. 3.
Potential dosage compensation in the ZAL2/2m system. (A–C) A potential scenario for dosage compensation. (A) Initially, expression dosage (shown as black waves) is similar between ZAL2 and ZAL2m and between tan and white. (B) If the expression of ZAL2m alleles is reduced and there is no dosage compensation, heterozygous (white) individuals should show reduced expression. (C) Dosage between the morphs may be rebalanced via overexpression of the ZAL2 allele in white birds. Consequently, expression of the ZAL2 allele should be greater in white than in tan birds. (D and E) Levels of compensation (white-ZAL2/tan-ZAL2) are significantly elevated for tan ≈ white (dosage-compensated) genes compared with nondosage-compensated genes (tan > white). The definitions of tan ≈ white and tan > white genes are based on P values from DESeq2 (Table 1). Using FDR-corrected Q values yielded similar results (SI Appendix, Fig. S5). Mann–Whitney U test, ***P < 0.001.
Interestingly, a large number of genes that exhibit significant allele-specific expression were similarly expressed between the morphs (Table 1). We tested the hypothesis that a dosage-rebalancing mechanism may be in play for these genes. In other words, disruption of ZAL2m allelic expression could be compensated by changes in ZAL2 expression in white birds to bring total levels of expression close to those seen in tan birds (Fig. 3C). In this scenario, ZAL2 allelic expression in white birds should be higher than that in tan birds (white-ZAL2/tan-ZAL2 > 1; Fig. 3C), a result that we observed in our data (Mann–Whitney U test, P < 0.001 for both brain regions; Fig. 3 D and E). Likewise, an up-regulation of ZAL2m allelic expression could be compensated by decreased ZAL2 expression in white birds (SI Appendix, Fig. S2).
Recent studies have demonstrated that dosage compensation may be limited to a subset of genes (40–43), in which highly expressed genes are particularly enriched (44–47). To determine whether dosage rebalancing is observed preferentially for highly expressed genes, we compared expression levels of candidate dosage-compensated genes (tan ≈ white and ZAL2 > ZAL2m or tan ≈ white and ZAL2m > ZAL2) with those of the background (i.e., genes that do not exhibit allele-specific or morph-biased expression patterns). For ZAL2 > ZAL2m genes, candidate dosage-compensated genes displayed significantly higher expression levels relative to the background (SI Appendix, Fig. S3). For ZAL2m > ZAL2 genes, this effect was weak and limited to the nucleus taeniae (SI Appendix, Fig. S4).
Allele-Specific and Morph-Biased Genes Play Central Roles in Gene Coexpression Networks.
We previously identified weighted coexpression networks from our RNA-seq data (22). Genes in two large modules, namely the “black” module in the hypothalamus (containing 511 genes, 226 genes on ZAL2/ZAL2m) and the “green-yellow” module in nucleus taeniae (containing 157 genes, 115 genes on ZAL2/ZAL2m), were significantly correlated with territorial singing, a behavior that differs between the morphs (22). In other words, expression of these genes was morph-biased. We hypothesized that genes that are allele-specific and morph-biased, compared with genes that are only morph-biased, play more central roles in the gene coexpression networks. Consistent with our hypothesis, genes with allele-specific expression exhibited significantly higher intramodule connectivities compared with other genes in the modules (Fig. 4).
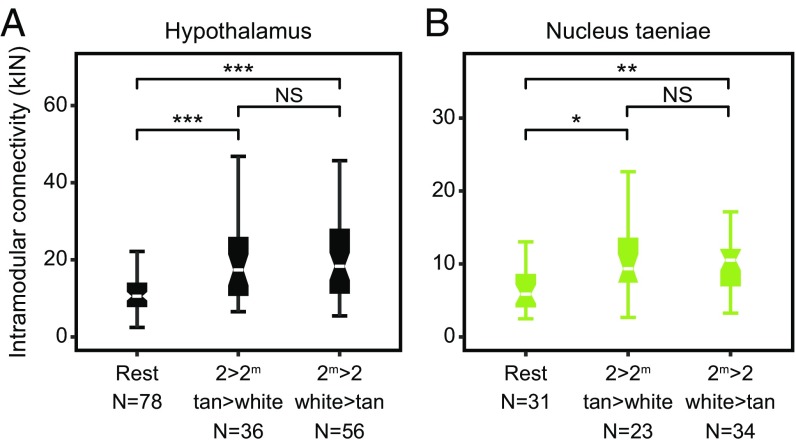
Fig. 4.
Allele-specific and morph-biased genes occupy more central positions in gene coexpression networks. Intramodular connectivity (kIN) for genes exhibiting allele-specific and morph-biased expression compared with genes in the rest of the module (Rest) (22) for (A) the hypothalamus and (B) the nucleus taeniae. Mann–Whitney U test, *P < 0.05; **P < 0.01; ***P < 0.001; NS: P > 0.05.
Discussion
In sex chromosomal supergenes, the heterogametic chromosome (e.g., the Y or W chromosome) is prevented from recombining because it is transmitted nearly always via heterozygotes. Lack of recombination reduces the effective population size of the heterogametic chromosome, leading to its degeneration (24, 26). Here, we have investigated the early evolution of a chromosomal system with striking similarity to sex chromosomes. In white-throated sparrows, the ZAL2m is almost always heterozygous due to strong disassortative mating (6, 7, 21, 48, 49). Consequently, it has been hypothesized that the ZAL2m chromosome may undergo genetic degeneration (7, 18). Previous analyses have been inconclusive in this regard. Karyotyping studies did not identify any signs of heterochromatinization in the ZAL2m chromosome (6, 18). Population genetic analyses have revealed slightly reduced levels of genetic diversity within ZAL2m-linked loci (7, 17, 19). A recent genome-wide study further reported a weak but significant excess of nonsynonymous polymorphisms on ZAL2m (7). However, other signs of degeneration, such as pseudogenization and accumulation of repetitive sequences, have not been observed (20). Our genome-wide comparison of protein-coding sequences between ZAL2 and ZAL2m shows that the degree of degeneration of ZAL2m is weak at most, consistent with a young supergene system. Despite the pronounced phenotypic differentiation associated with ZAL2m, we did not observe a massive accumulation of pseudogenes or evidence of a substantial increase in repetitive sequences on this chromosome (SI Appendix).
The low level of degeneration is probably due to the infrequent yet present recombination between ZAL2m chromosomes in rare homozygotes. Unlike other non–sex-linked supergenes in fire ants (Solenopsis invicta) and ruffs (Philomachus pugnax) (10–12, 50), the ZAL2m is not homozygous-lethal. Healthy adult ZAL2m/ZAL2m homozygotes of both sexes have been observed in nature (6, 21, 49), indicating that ZAL2m is functional. The rarity of ZAL2m/ZAL2m homozygotes can be explained by the disassortative mating system. According to Tuttle et al. (7), 0.8% of total matings occur between white birds; thus, the expected frequency of ZAL2m/ZAL2m homozygotes is ∼0.2% of total offspring. Based upon the five reports of ZAL2m/ZAL2m homozygotes (6, 7, 21, 48, 49), the frequencies of such individuals range between 0 and 1%, and the frequency based on all data is 0.2% (6 of 3,057 birds genotyped to date), exactly the same as the expected frequency. The more pressing question, then, is why and how the disassortative mating has evolved in this species. It is possible that, because the two morphs complement each other with respect to territorial aggression and parenting, same-morph pairs are not as successful (1).
While we did not find evidence for substantial degeneration in protein-coding regions, there were robust signals of regulatory evolution in substantial numbers of genes. More than 40% of ZAL2/ZAL2m-linked genes exhibited allele-specific expression in white birds. One explanation for this finding could be that ZAL2m allelic expression is disrupted due to degeneration of regulatory sequences. Indeed, we observed an overall reduction of ZAL2m allelic expression compared with ZAL2, although this trend was weak and not significant in the hypothalamus (Table 1). Importantly, a large proportion of the genes showing allele-specific expression are shared between the two brain regions that we sampled, suggesting that the allele-specific expression could be driven by genetic divergence of regulatory regions (37, 51, 52). Alternatively, the differences in allelic expression could be driven by relatively small changes in the protein sequences of important transcription factors (53).
Despite substantial allele-specific expression of some genes, many genes exhibited similar expression levels between the morphs. This result is reminiscent of dosage compensation in sex chromosomes (9, 54–56). Dosage compensation may evolve as a response to reduced allelic dose (23, 39), particularly for highly expressed genes (44–47). Dosage compensation can also be achieved without selective up- or down-regulation of specific genes through passive feedback or other buffering in regulatory networks (57–59). This compensation may potentially occur via epigenetic mechanisms such as DNA methylation (60). Interestingly, we observed that within the ZAL2m down-regulated genes (ZAL2 > ZAL2m), those that are dosage-compensated tend to be more highly expressed than the background genes. In addition, the proportion of genes that appear to be dosage-compensated is significantly higher for ZAL2m down-regulated genes compared with ZAL2m up-regulated genes (Table 1; proportional test, P < 0.001 for both brain regions). These observations are consistent with the idea that degeneration of the ZAL2m chromosome drives dosage compensation (23, 39). The fact that we observe gene expression changes resembling dosage compensation is particularly notable given that avian sex chromosomes generally exhibit weak levels of dosage compensation (61, 62).
Supergenes that are formed via chromosomal rearrangements are emerging as key determinants for alternative reproductive phenotypes in diverse taxa (e.g., fire ant, ruff, and some plants) (10–12, 15, 50). The pivotal step in the evolution of a supergene is the suppression of recombination (13, 14, 63). The best-understood consequence of suppression of recombination is genetic degeneration of the nonrecombining region (24, 64). Previous studies of white-throated sparrows, such as the recent work by Tuttle et al. (7), have shown evidence consistent with degeneration. Those signals were weak, however, which was viewed at odds with the pervasive phenotypic differentiation between the morphs. Our work shows that the phenotypic differentiation may be driven by extensive expression divergence, which is already present in this system, without massive genetic degeneration of the nonrecombining chromosome. In the newly evolving sex chromosomes of Drosophila albomicans, a chromosome-wide down-regulation of gene expression was observed in a nonrecombining chromosome without obvious degeneration of protein-coding sequences (65). In our system, down-regulation of ZAL2m-linked alleles is subtle. In contrast, there is substantial allele-specific expression and potential signs of dosage compensation.
Expression biases that favor one allele or morph (Table 1) could be caused by natural selection, genetic drift, or both. Mutations that confer regulatory changes beneficial for the tan morph could accumulate on the ZAL2 chromosome, similar to the accumulation of female-beneficial mutations on the X chromosome (e.g., ref. 66). Similarly, mutations that cause up-regulation beneficial to white birds could accumulate on the ZAL2m because of its white morph-limited transmission. Consistent with this idea, we observe an excess of white morph-biased genes when ZAL2m is up-regulated (compared with tan-morph bias when ZAL2m is down-regulated; Table 1). Alternatively, such mutations on ZAL2m alleles could spread by genetic drift, as long as the deleterious effect of expression change is on par with the inverse of the effective population size of ZAL2m (67). Additional data on the variability of regulatory regions in the two chromosomes will help us distinguish the evolutionary mechanisms at play for this newly differentiating chromosome pair.
Materials and Methods
Detailed descriptions of materials and methods are provided in SI Appendix.
Sequencing.
We sequenced a “superwhite” bird homozygous for the rearrangement (ZAL2m/ZAL2m) (21). High-molecular-weight genomic DNA was extracted from liver and sequenced using HiSeq2500 at the Roy Carver Genome Center of the University of Illinois. Approximately 240 million reads of 150 bp were generated, which are available from the SRA database (accession no. SRR4191732).
Identification of Scaffolds on the Second Chromosome.
We mapped scaffolds from the genome of a tan bird in National Center for Biotechnology Information (NCBI) onto that of the zebra finch using LASTZ 1.03.73 (68) (more details in SI Appendix). We identified ZAL2 scaffolds using homology to the corresponding zebra finch chromosome [commonly referred to as TGU3 due to its homology to chicken chromosome 3 (29)], previous FISH studies (7, 18, 20), as well as homology with two other passerine birds with chromosome-level assemblies, the collared flycatcher (Ficedula albicollis) and the great tit (Parus major) (28, 30).
Genetic Divergence Between ZAL2 and ZAL2m Alleles.
To measure the divergence between the ZAL2 and ZAL2m chromosomes, we called variants (SNPs and indels) by following best practices for the Genome Analysis Toolkit (GATK) for variant calling in genome-sequencing data (69–71). To calculate FST, we called variants with available transcriptome data from the hypothalamus and nucleus taeniae of 9 tan and 11 white individuals (22) following the GATK best practices for variant calling in RNA-seq data (69–71). A variant was considered as putatively fixed in the sampled individuals if (i) the variant was biallelic; (ii) tan individuals were homozygous for the reference allele (AA); (iii) white individuals (ZAL2/ZAL2m) were heterozygous (Aa); and (iv) the superwhite (ZAL2m/ZAL2m) individual was homozygous for the alternative allele (aa). Scaffolds residing inside versus outside the rearrangement were identified on the basis of distinctive patterns of genetic divergence (more details in SI Appendix).
Analyses of Protein-Coding Sequences.
For each ZAL2-linked gene, we extracted the longest transcript and constructed the ZAL2m version with the putatively fixed differences. Additionally, we utilized all available genome annotations in NCBI for 13 other avian species in the order of Passeriformes (the same order as the white-throated sparrow) as outgroup species (SI Appendix, Fig. S6). We obtained codon alignments for a total of 800 genes. We calculated dN and dS for the ZAL2 and ZAL2m branches with a free-ratio model using codeml from the PAML 4.8 package (33). The Hon-New package (72), which adopts the amino acid classification system that considers charge and polarity, was used to estimate dR and dC. To test for positive selection, a branch-site model was run with codeml in PAML, which was then compared with the null model following the simulation approach used by Nielsen et al. (34). Positively selected functional categories were identified by first assigning genes according to the PANTHER Classification System version 10 (73). For each category with more than 10 genes, the difference between the cumulative distributions of P for genes in the category, versus that of genes not in that category, was examined using a one-tailed Mann–Whitney U test (35).
Gene Expression.
We used RNA-Seq data from 9 tan and 10 white males (22) to detect morph-biased and allele-specific gene expression. To account for potential mapping bias toward the reference (ZAL2/ZAL2) genome caused by mismatches between ZAL2 and ZAL2m, we N-masked (74) putatively fixed differences in the reference. We additionally checked potential leftover bias by aligning whole-genome sequences of three white birds described by Tuttle et al. (7) to this N-masked genome. ZAL2 and ZAL2m alleles exhibited roughly equal coverage per site, as expected if there is little mapping bias (SI Appendix, Fig. S7). Read counts per gene at the morph or allele level were calculated by htseq-count 0.9.1 (75). Morph-biased and allele-specific expression analyses were performed using DESeq2 1.12.3 (38) (more details in SI Appendix).
Supplementary Material
Acknowledgments
We thank the Roy Carver Genome Center of the University of Illinois for help with sequencing; Dr. Kathleen Grogan, members of the S.V.Y. laboratory, Brian Charlesworth, and two anonymous reviewers for comments on the manuscript; and Xiulan Pan and Chen Feng for help with illustration. This work was supported by National Institutes of Health Grants R01 MH082833 (to D.L.M.) and 1R21 NIMH102677 (to D.L.M. and S.V.Y.) and by Georgia Tech (S.V.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Sequence Read Archive database (accession no. SRR4191732).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717721115/-/DCSupplemental.
References
- 1.Tuttle EM. Alternative reproductive strategies in the white-throated sparrow: Behavioral and genetic evidence. Behav Ecol. 2003;14:425–432. [Google Scholar]
- 2.Maney DL, Horton BM, Zinzow-Kramer WM. Estrogen receptor alpha as a mediator of life-history trade-offs. Integr Comp Biol. 2015;55:323–331. doi: 10.1093/icb/icv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maney DL. Endocrine and genomic architecture of life history trade-offs in an avian model of social behavior. Gen Comp Endocrinol. 2008;157:275–282. doi: 10.1016/j.ygcen.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 4.Horton BM, Moore IT, Maney DL. New insights into the hormonal and behavioural correlates of polymorphism in white-throated sparrows, Zonotrichia albicollis. Anim Behav. 2014;93:207–219. doi: 10.1016/j.anbehav.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton BM, et al. Estrogen receptor α polymorphism in a species with alternative behavioral phenotypes. Proc Natl Acad Sci USA. 2014;111:1443–1448. doi: 10.1073/pnas.1317165111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorneycroft HB. A cytogenetic study of white-throated sparrow, Zonotrichia albicollis (Gmelin) Evolution. 1975;29:611–621. doi: 10.1111/j.1558-5646.1975.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 7.Tuttle EM, et al. Divergence and functional degradation of a sex chromosome-like supergene. Curr Biol. 2016;26:344–350. doi: 10.1016/j.cub.2015.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowther JK. Polymorphism in the white-throated sparrow, Zonotrichia albicollis (Gmelin) Can J Zool. 1961;39:281–292. [Google Scholar]
- 9.Graves JA. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet. 2016;17:33–46. doi: 10.1038/nrg.2015.2. [DOI] [PubMed] [Google Scholar]
- 10.Küpper C, et al. A supergene determines highly divergent male reproductive morphs in the ruff. Nat Genet. 2016;48:79–83. doi: 10.1038/ng.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamichhaney S, et al. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax) Nat Genet. 2016;48:84–88. doi: 10.1038/ng.3430. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, et al. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013;493:664–668. doi: 10.1038/nature11832. [DOI] [PubMed] [Google Scholar]
- 13.Charlesworth D. Evolution of recombination rates between sex chromosomes. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160456. doi: 10.1098/rstb.2016.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlesworth D. The status of supergenes in the 21st century: Recombination suppression in Batesian mimicry and sex chromosomes and other complex adaptations. Evol Appl. 2015;9:74–90. doi: 10.1111/eva.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson MJ, Jiggins CD. Supergenes and their role in evolution. Heredity (Edinb) 2014;113:1–8. doi: 10.1038/hdy.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwander T, Libbrecht R, Keller L. Supergenes and complex phenotypes. Curr Biol. 2014;24:R288–R294. doi: 10.1016/j.cub.2014.01.056. [DOI] [PubMed] [Google Scholar]
- 17.Huynh LY, Maney DL, Thomas JW. Chromosome-wide linkage disequilibrium caused by an inversion polymorphism in the white-throated sparrow (Zonotrichia albicollis) Heredity (Edinb) 2011;106:537–546. doi: 10.1038/hdy.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas JW, et al. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics. 2008;179:1455–1468. doi: 10.1534/genetics.108.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh LY, Maney DL, Thomas JW. Contrasting population genetic patterns within the white-throated sparrow genome (Zonotrichia albicollis) BMC Genet. 2010;11:96. doi: 10.1186/1471-2156-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis JK, et al. NISC Comparative Sequencing Program Haplotype-based genomic sequencing of a chromosomal polymorphism in the white-throated sparrow (Zonotrichia albicollis) J Hered. 2011;102:380–390. doi: 10.1093/jhered/esr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horton BM, et al. Behavioral characterization of a white-throated sparrow homozygous for the ZAL2(m) chromosomal rearrangement. Behav Genet. 2013;43:60–70. doi: 10.1007/s10519-012-9574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zinzow-Kramer WM, et al. Genes located in a chromosomal inversion are correlated with territorial song in white-throated sparrows. Genes Brain Behav. 2015;14:641–654. doi: 10.1111/gbb.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlesworth B. Model for evolution of Y chromosomes and dosage compensation. Proc Natl Acad Sci USA. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi S, Charlesworth B. Contrasting patterns of molecular evolution of the genes on the new and old sex chromosomes of Drosophila miranda. Mol Biol Evol. 2000;17:703–717. doi: 10.1093/oxfordjournals.molbev.a026349. [DOI] [PubMed] [Google Scholar]
- 26.Bachtrog D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shetty S, Griffin DK, Graves JA. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 1999;7:289–295. doi: 10.1023/a:1009278914829. [DOI] [PubMed] [Google Scholar]
- 28.Laine VN, et al. Great Tit HapMap Consortium Evolutionary signals of selection on cognition from the great tit genome and methylome. Nat Commun. 2016;7:10474. doi: 10.1038/ncomms10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 31.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 32.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark AG, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 36.Park J, et al. Comparative analyses of DNA methylation and sequence evolution using Nasonia genomes. Mol Biol Evol. 2011;28:3345–3354. doi: 10.1093/molbev/msr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohno S. Sex Chromosomes and Sex-Linked Genes. Springer; Berlin: 1967. [Google Scholar]
- 40.Lin F, Xing K, Zhang J, He X. Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc Natl Acad Sci USA. 2012;109:11752–11757. doi: 10.1073/pnas.1201816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uebbing S, Künstner A, Mäkinen H, Ellegren H. Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol Evol. 2013;5:1555–1566. doi: 10.1093/gbe/evt114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White MA, Kitano J, Peichel CL. Purifying selection maintains dosage-sensitive genes during degeneration of the threespine stickleback Y chromosome. Mol Biol Evol. 2015;32:1981–1995. doi: 10.1093/molbev/msv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimmer F, Harrison PW, Dessimoz C, Mank JE. Compensation of dosage-sensitive genes on the chicken Z chromosome. Genome Biol Evol. 2016;8:1233–1242. doi: 10.1093/gbe/evw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gout JF, Kahn D, Duret L. Paramecium Post-Genomics Consortium The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 2010;6:e1000944, and erratum (2010) 6, 10.1371/annotation/c55d5089-ba2f-449d-8696-2bc8395978db. doi: 10.1371/journal.pgen.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makanae K, Kintaka R, Makino T, Kitano H, Moriya H. Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Genome Res. 2013;23:300–311. doi: 10.1101/gr.146662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pessia E, Engelstädter J, Marais GA. The evolution of X chromosome inactivation in mammals: The demise of Ohno’s hypothesis? Cell Mol Life Sci. 2014;71:1383–1394. doi: 10.1007/s00018-013-1499-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GAB. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci USA. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michopoulos V, Maney DL, Morehouse CB, Thomas JW. A genotyping assay to determine plumage morph in the white-throated sparrow (Zonotrichia albicollis) Auk. 2007;124:1330–1335. [Google Scholar]
- 49.Falls JB, Kopachena JG. 2010. White-throated sparrow (Zonotrichia albicollis). The Birds of North America, ed Rodewald PG (Cornell Lab of Ornithology, Ithaca, NY), Retrieved from the Birds of North America.
- 50.Pracana R, Priyam A, Levantis I, Nichols RA, Wurm Y. The fire ant social chromosome supergene variant Sb shows low diversity but high divergence from SB. Mol Ecol. 2017;26:2864–2879. doi: 10.1111/mec.14054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 52.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 54.Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muyle A, et al. Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biol. 2012;10:e1001308. doi: 10.1371/journal.pbio.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papadopulos AS, Chester M, Ridout K, Filatov DA. Rapid Y degeneration and dosage compensation in plant sex chromosomes. Proc Natl Acad Sci USA. 2015;112:13021–13026. doi: 10.1073/pnas.1508454112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnold AP, Itoh Y, Melamed E. A bird’s-eye view of sex chromosome dosage compensation. Annu Rev Genomics Hum Genet. 2008;9:109–127. doi: 10.1146/annurev.genom.9.081307.164220. [DOI] [PubMed] [Google Scholar]
- 58.Mank JE. Sex chromosome dosage compensation: Definitely not for everyone. Trends Genet. 2013;29:677–683. doi: 10.1016/j.tig.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 59.Acar M, Pando BF, Arnold FH, Elowitz MB, van Oudenaarden A. A general mechanism for network-dosage compensation in gene circuits. Science. 2010;329:1656–1660. doi: 10.1126/science.1190544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keller TE, Yi SV. DNA methylation and evolution of duplicate genes. Proc Natl Acad Sci USA. 2014;111:5932–5937. doi: 10.1073/pnas.1321420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itoh Y, et al. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ellegren H, et al. Faced with inequality: Chicken do not have a general dosage compensation of sex-linked genes. BMC Biol. 2007;5:40. doi: 10.1186/1741-7007-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity (Edinb) 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 65.Zhou Q, Bachtrog D. Chromosome-wide gene silencing initiates Y degeneration in Drosophila. Curr Biol. 2012;22:522–525. doi: 10.1016/j.cub.2012.01.057. [DOI] [PubMed] [Google Scholar]
- 66.Vicoso B, Charlesworth B. Evolution on the X chromosome: Unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 67.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; Cambridge, UK: 1983. [Google Scholar]
- 68.Harris RS. 2007. Improved pairwise alignment of genomic DNA. PhD thesis (Pennsylvania State Univ, State College, PA)
- 69.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J Mol Evol. 2000;50:56–68. doi: 10.1007/s002399910007. [DOI] [PubMed] [Google Scholar]
- 73.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krueger F, Andrews SR. SNPsplit: Allele-specific splitting of alignments between genomes with known SNP genotypes. F1000 Res. 2016;5:1479. doi: 10.12688/f1000research.9037.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anders S, Pyl PT, Huber W. HTSeq–A python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.