We thank Kardos et al. (1) for their interest in our study (2), which is about the detection and quantification of inbreeding depression (ID) for complex traits from SNP data. Kardos et al. (1) make a number of points about the utility and interpretation of estimates of ID from runs of homozygosity (FROH) compared with frequency-based inbreeding measures, such as the FUNI measure (3) that estimates the correlation between uniting gametes.
First, Kardos et al. (1) say that FROH can be interpreted as a probability of identity-by-descent (IBD). However, in a random sample from a human population, the distribution of FROH will reflect both finite population size and occasionally pedigree inbreeding. In the absence of a well-defined base population with known allele frequencies [as in our study (2)], the estimate FROH from SNP data are not the same as the expected (IBD) inbreeding coefficient from a known pedigree. Therefore, the property that FROH lies between 0 and 1 will, per definition, lead to biased estimates of ID. Other reasons why FROH can yield biased estimates is the arbitrary definition of ROH, including a threshold for minimum length, when to ignore occasional heterozygotes in a run, and linkage disequilibrium pruning. In particular, defining ROH as genomic segments at least, for example, 1-Mb long ignores small IBD segments, which may still contribute to ID.
Second, Kardos et al. (1) claim that our reported upward biases of ID estimates from FROH are artifacts from our simulation assumptions. Our study addressed the question of estimating ID under polygenic genetic architectures, when ID results from directional dominance at causal variants randomly distributed across the entire genome. We addressed consequences of violating assumptions underlying FUNI in our study, including the loss of information due to imperfect linkage disequilibrium between causal variants and SNPs used to estimate ID. We showed that when causal alleles are rare compared with the SNPs used, FUNI yields downwardly biased ID estimates. Therefore, parameterizing the phenotype as a linear function of directional dominance at causal loci does not automatically guarantee unbiasedness.
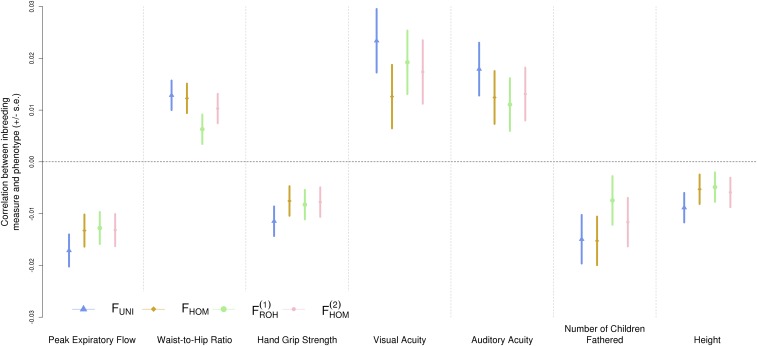
Third, Kardos et al. (1) say that regression slopes of traits on estimated inbreeding are not comparable because of differences in variances in their estimators. But the regression coefficients reflect the ratio of covariance between trait and inbreeding and variance of inbreeding, and correctly reflect the scale of interest. Kardos et al. propose to compare estimates from different measures of inbreeding using correlation coefficients instead of regression slopes. We agree that correlation is the right statistic to assess power of detection. Indeed, we found in theory, simulations and in real data that ID could be more often detected (more power) using the correlation approach with FUNI compared with FROH (table S2 of ref. 2) (Fig. 1). In random samples from human populations, variance of frequency-based estimators of inbreeding coefficients is expected and observed to be small (3).
Fig. 1.
Correlation between four inbreeding measures and seven phenotypes with significant inbreeding depression (2). FHOM (excess homozygosity), FUNI (correlation between uniting gametes), and (runs of homozygosity) are all defined as in Yengo et al. (2). Largest correlations across all traits are achieved with FUNI.
Our response reconfirms that FROH is not the panacea for estimating ID.
Footnotes
The authors declare no conflict of interest.
References
- 1.Kardos M, Nietlisbach P, Hedrick PW. How should we compare different genomic estimates of the strength of inbreeding depression? Proc Natl Acad Sci USA. 2018;115:E2492–E2493. doi: 10.1073/pnas.1714475115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yengo L, et al. Detection and quantification of inbreeding depression for complex traits from SNP data. Proc Natl Acad Sci USA. 2017;114:8602–8607. doi: 10.1073/pnas.1621096114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]