Significance
Studies of mammalian development are mostly performed in the mouse model, where reverse genetics is most advanced. Recent developments in gene editing, including CRISPR-Cas9, facilitated functional studies of specific genes in other mammalian species and revealed important differences (e.g., between human and murine development). In this study, we generated loss-of-function mutations of the OCT4 gene in bovine fibroblasts and produced embryos by nuclear transfer cloning. We demonstrated that, similar to human development but in contrast to mouse development, OCT4 is required for maintaining NANOG-positive epiblast cells in the inner cell mass of blastocysts. Our study outlines a general strategy for dissecting the roles of specific genes in preimplantation development in domestic species.
Keywords: OCT4, NANOG, GATA6, embryo, bovine
Abstract
Mammalian preimplantation development involves two lineage specifications: first, the CDX2-expressing trophectoderm (TE) and a pluripotent inner cell mass (ICM) are separated during blastocyst formation. Second, the pluripotent epiblast (EPI; expressing NANOG) and the differentiated primitive endoderm (PrE; expressing GATA6) diverge within the ICM. Studies in mice revealed that OCT4/POU5F1 is at the center of a pluripotency regulatory network. To study the role of OCT4 in bovine preimplantation development, we generated OCT4 knockout (KO) fibroblasts by CRISPR-Cas9 and produced embryos by somatic cell nuclear transfer (SCNT). SCNT embryos from nontransfected fibroblasts and embryos produced by in vitro fertilization served as controls. In OCT4 KO morulae (day 5), ∼70% of the nuclei were OCT4 positive, indicating that maternal OCT4 mRNA partially maintains OCT4 protein expression during early development. In contrast, OCT4 KO blastocysts (day 7) lacked OCT4 protein entirely. CDX2 was detected only in TE cells; OCT4 is thus not required to suppress CDX2 in the ICM. Control blastocysts showed a typical salt-and-pepper distribution of NANOG- and GATA6-positive cells in the ICM. In contrast, NANOG was absent or very faint in the ICM of OCT4 KO blastocysts, and no cells expressing exclusively NANOG were observed. This mimics findings in OCT4-deficient human blastocysts but is in sharp contrast to Oct4-null mouse blastocysts, where NANOG persists and PrE development fails. Our study supports bovine embryogenesis as a model for early human development and exemplifies a general strategy for studying the roles of specific genes in embryos of domestic species.
During mammalian preimplantation development, two lineage specifications occur. First, the trophectoderm (TE) differentiates, leading to blastocyst formation; and subsequently, two lineages diverge within the inner cell mass (ICM): the pluripotent epiblast (EPI) and the differentiated primitive endoderm (PrE) or hypoblast (HB), which is the PrE equivalent in bovine embryos (1). Genes and mechanisms controlling these lineage-specification events have been studied extensively in mouse embryos. It is now established that the transcription factor OCT4/POU5F1 is at the center of a pluripotency regulatory network (2), although it is neither necessary for the first lineage segregation into the TE and ICM nor for the initiation of toti- or pluripotency (3–5). Oct4-null mouse embryos show normal development until late blastocyst stage (day 3.5), reflected by unchanged cell numbers in the TE and ICM and by repression of TE-specific genes in the ICM (3, 5, 6). Precursor cells of the PrE and EPI show a mutually exclusive salt-and-pepper distribution of the lineage-specific markers GATA6 and NANOG, according to the situation in wild-type embryos. With further development, GATA6-positive cells disappear from the ICM of Oct4-null embryos, and the proportion of cells expressing neither GATA6 nor NANOG increases until day 4.25, when almost no GATA6-positive cells are present. Activation of PrE-specific gene expression fails, and there is no PrE development (4, 7). GATA6 expression in Oct4-null embryos is lost, because OCT4 is required to initiate FGF4 secretion from EPI cells and to activate the expression of PrE genes cell-autonomously (4). In wild-type embryos, addition of exogenous FGF4 during culture induces expression of PrE genes, resulting in expression of GATA6 in all cells of the ICM (8). In contrast, blastocysts lacking FGF4 have an ICM entirely made up of NANOG-positive cells (9).
Similar to early mouse embryo development, maternal OCT4 transcripts are present in the bovine oocyte and decrease in abundance until the 8- to 16-cell stage, when major embryonic genome activation occurs (10, 11). OCT4 has been detected in all nuclei of bovine morula-stage embryos (11–13). While OCT4 is extinguished in the TE of day 3.5 mouse blastocysts, which may allow rapid differentiation of the TE and implantation of the embryo, bovine embryos coexpress CDX2 and OCT4 in the TE until day 11 (14). At day 7, the ICM of bovine blastocysts shows the same salt-and-pepper distribution of GATA6- and NANOG-positive cells as the ICM of mouse blastocysts (15, 16). However, the role of FGF4 in bovine embryo development differs from the observations in mouse embryos, as inhibition of FGF/MAPK signaling only partially blocks GATA6 expression. Therefore, in bovine embryos, FGF4 signaling is not essential for GATA6 expression (16).
Substantial differences in preimplantation development between murine and bovine embryos regarding the OCT4–CDX2 interaction and the role of FGF/MAPK signaling during the second lineage differentiation highlight the need for developmental studies of different mammalian species. This is also emphasized by recent findings in OCT4-mutant human embryos (17). In the present study, we addressed the role of OCT4 in bovine embryogenesis by mutating OCT4 using CRISPR-Cas9 in fibroblasts and producing bovine embryos with OCT4 loss-of-function mutations by somatic cell nuclear transfer (SCNT) (Fig. 1A). This approach revealed important similarities between bovine and human embryonic development and provides a general strategy for studying the roles of specific genes in preimplantation embryo development in domestic species.
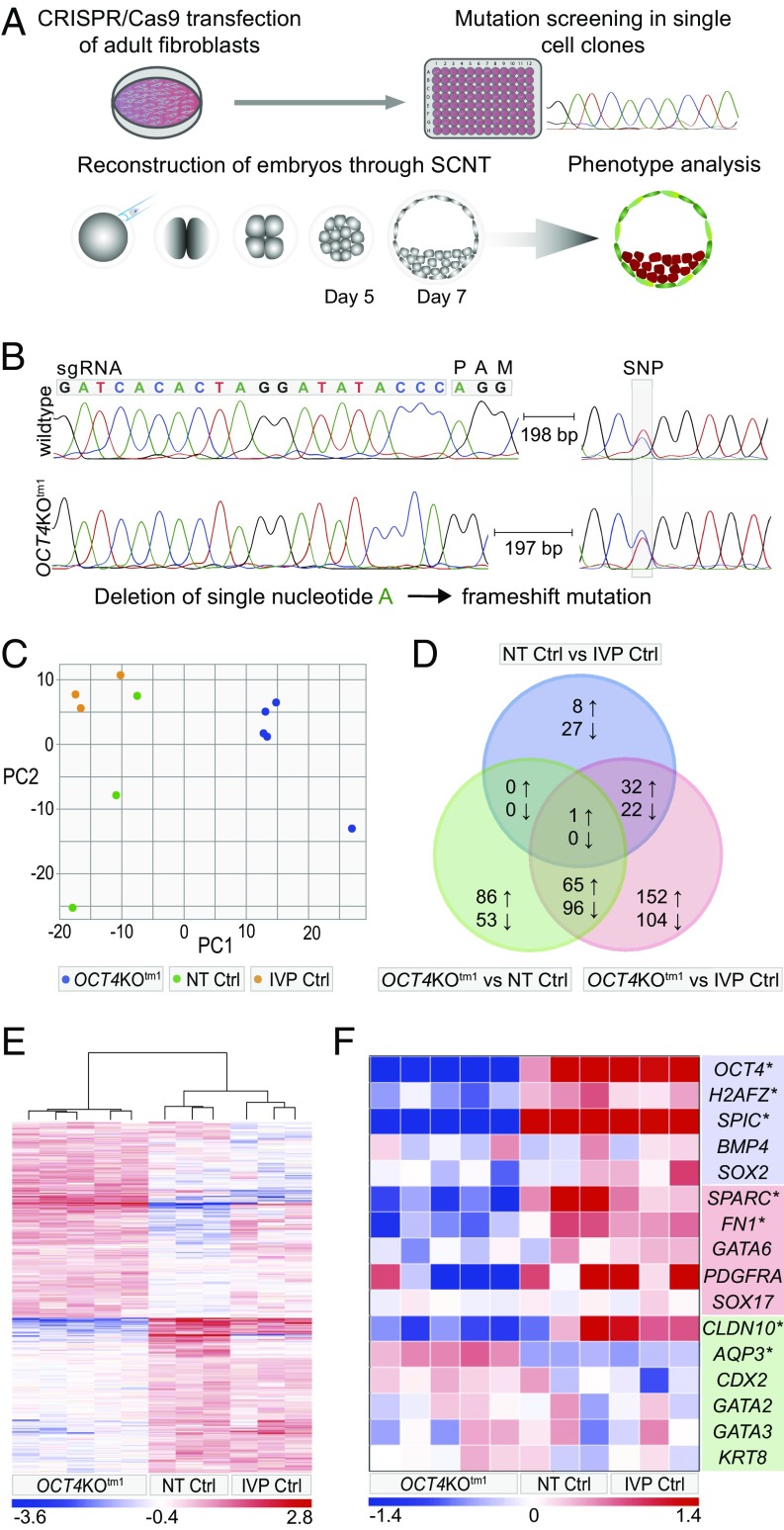
Fig. 1.
(A) Experimental procedure to produce OCT4 KO embryos through SCNT. (B) Single guide RNA (sgRNA) design to mutate exon 2 of OCT4; biallelic deletion of single nucleotide in OCT4KOtm1; and maintained SNP. (C) PCA of transcriptome profiles from individual day 7 OCT4KOtm1 (n = 5), NT Ctrl (n = 3), and IVP Ctrl blastocysts (n = 3). PC1, principal component 1; PC2, principal component 2. (D) Venn diagram of differentially abundant transcripts (DATs) identified by DESeq2 analysis of the NT Ctrl vs. IVP Ctrl, OCT4KOtm1 vs. NT Ctrl, and OCT4KOtm1 vs. IVP Ctrl blastocyst transcriptome data. (E) Heat map of DATs from DESeq2 [n = 625; adjusted P value (Padj) < 0.05]. (F) Heat map of genes specific for EPI (blue), HB (red), and TE (green); asterisks indicate DATs at Padj < 0.05.
Results
Mutation of OCT4 in Bovine Fibroblasts and Generation of OCT4 KO SCNT Embryos.
Fibroblasts from a PGK-EGFP transgenic bull (18, 19) were nucleofected with plasmids encoding Cas9 and a single guide RNA targeting exon 2 of the OCT4 gene. Of 156 single-cell clones analyzed, 4 (2.6%) had mutations in OCT4. One of the mutant cell clones had a homozygous deletion of 1 bp in exon 2, leading to a frameshift and the introduction of a premature termination codon in exon 4, located 91 bp upstream of the following exon–exon junction. The mutant transcripts are thus expected to undergo nonsense-mediated decay (20) (Fig. 1B and Fig. S1). The identical biallelic deletion was confirmed by a single-nucleotide polymorphism (SNP) located 200 bp downstream of the protospacer adjacent motif (PAM). In addition, this cell clone had a monoallelic modification in a known OCT4 pseudogene located within intron 1 of the ETF1 gene (21). Interestingly, 8 out of 22 characterized single-cell clones (36%) were mutated at this ETF1 region. To exclude effects of this off-target mutation on embryo development, we examined a single-cell clone (ETF1muttm1) that carried exactly the same mutation as OCT4KOtm1 at ETF1 (Fig. S2) but no mutation in OCT4. To exclude cell clone-specific effects of OCT4KOtm1, we examined another OCT4 KO cell clone (OCT4KOtm2) generated of female adult fibroblasts using the same OCT4-specific CRISPR-Cas9 system. This cell clone had deletions of two and three nucleotides on the respective alleles at the target site and deletions on both alleles in the OCT4 pseudogene within ETF1 (Figs. S2 and S3). OCT4KOtm1, OCT4KOtm2, and ETF1muttm1 cell clones, as well as the unmodified parental cells of OCT4KOtm1 [nuclear transfer (NT) Ctrl] were used as donors for SCNT. In total, 741 OCT4KOtm1, 272 OCT4KOtm2, 315 ETF1muttm1, and 439 NT Ctrl embryos were produced in 21, 5, 6, and 18 SCNT experiments, respectively.
OCT4 Mutagenesis Has Significant Effects on the Transcriptome of Blastocysts.
To examine the effect of loss of OCT4 on the transcriptome, we performed RNA sequencing on individual day 7 OCT4KOtm1 (n = 5), NT Ctrl (n = 3), and in vitro produced (IVP) Ctrl (n = 3) blastocysts. Principal component analysis (PCA) of the transcriptome profiles showed that the NT Ctrl and IVP Ctrl blastocysts clustered closely together, whereas OCT4KOtm1 blastocysts formed a distant cluster (Fig. 1C). Accordingly, differential gene expression analysis using DESeq2 revealed fewer differentially abundant transcripts (DATs) when comparing “NT Ctrl vs. IVP Ctrl” (n = 90) than in the comparisons of “OCT4KOtm1 vs. IVP Ctrl” (n = 472) and “OCT4KOtm1 vs. NT Ctrl” (n = 301). OCT4 transcripts in OCT4KOtm1 embryos were reduced to about 10% of the levels detected in NT Ctrl or IVP Ctrl embryos, while the abundance of ETF1 transcripts was not affected by the off-target mutation compared with all other SCNT-derived embryos but increased ∼1.5-fold in OCT4KOtm1 vs. IVP Ctrl embryos (Dataset S1). No common elements among the transcripts with reduced abundance and only one shared transcript with increased abundance (BOK, BCL2 family apoptosis regulator) show that effects on the transcriptome are mainly attributable to loss of OCT4 and not to the SCNT procedure (Fig. 1D). This is supported by the nonsupervised clustering of samples in the heat map produced from the DATs (Fig. 1E). To examine if genes specific to EPI, HB, or TE were affected by loss of OCT4, we compared the DATs in OCT4 KO embryos to sets of genes reported to be lineage specific in mouse, human, or bovine embryos (4, 6, 17, 22–31). Only genes with different transcript abundance in both OCT4KOtm1 vs. IVP Ctrl and OCT4KOtm1 vs. NT Ctrl, but not in NT Ctrl vs. IVP Ctrl, were considered to be affected by the loss of OCT4. In OCT4 KO blastocysts, the transcript levels of H2AFZ, SPIC (EPI), SPARC, FN1 (HB), and CLDN10 (TE) were significantly reduced, while the abundance of AQP3 transcripts (TE) was significantly increased (Fig. 1F and Dataset S1). Overall, the read count of NANOG transcripts was relatively low; NANOG-specific reads were detected in one of three NT Ctrl blastocysts and in two of three IVP Ctrl blastocysts, but in none of the OCT4 KO blastocysts. There were no significant differences between the three embryo groups in the transcript levels of GATA6 or of SOX17, another HB-specific gene (22).
Effects of OCT4 and/or ETF1 Mutagenesis on Embryo Development.
There was no significant difference between OCT4KOtm1 and NT Ctrl embryos regarding cleavage rate. However, development to the blastocyst stage was significantly decreased in OCT4KOtm1, OCT4KOtm2, and ETF1muttm1 embryos compared with NT Ctrl embryos (Table 1). In addition, we determined the total cell numbers of blastocysts and the numbers of TE cells after staining for the TE-specific marker CDX2. While SCNT embryos had, on average, 23% fewer cells than IVP Ctrl embryos, the proportion of TE cells was ∼60% in all experimental groups. In OCT4 KO blastocysts, the total cell numbers and the proportions of TE cells per blastocyst did not differ from NT Ctrl embryos, indicating that embryonic OCT4 has no effect on the quantitative allocation of cells to either ICM or TE during the first lineage differentiation (Table 2).
Table 1.
Developmental rates of SCNT embryos
| Experimental group | OCT4KOtm1 | OCT4KOtm2 | ETF1muttm1 | NT Ctrl |
| No. of SCNT experiments | 21 | 5 | 6 | 18 |
| No. of fused constructs | 741 | 272 | 315 | 439 |
| No. cleaved (cleavage rate,* %) | 516 (69.5 ± 2.9a) | 152 (53.4 ± 4.2ab) | 165 (51.8 ± 5.4b) | 266 (64.4 ± 3.1ab) |
| No. of day 7 blastocysts (blastocyst rate,* %) | 125 (16.8 ± 2.2a) | 39 (13.8 ± 2.8a) | 65 (18.7 ± 3.1a) | 135 (32.1 ± 2.6b) |
Data presented as mean ± SE. Different superscript letters within a row indicate significant differences (P < 0.05, one-way ANOVA with Tukey multiple comparison test).
Table 2.
Total cell numbers and percentages of CDX2-positive cells
| Experimental group | OCT4KOtm1 | OCT4KOtm2 | NT Ctrl | IVP Ctrl |
| No. of CLSM-analyzed day 7 blastocysts | 24 | 8 | 20 | 40 |
| No. of total cells* | 89.6 ± 5.6a | 105 ± 5.8ab | 96.3 ± 8.5a | 125.8 ± 5.8b |
| CDX2-positive cells,* % | 56.8 ± 2.4 | 61.1 ± 2.8 | 62.5 ± 2.1 | 59.3 ± 1.5 |
Data presented as mean ± SE. Different superscript letters within a row indicate significant differences (P < 0.05, one-way ANOVA with Tukey multiple comparison test).
Maternal OCT4 Transcripts Are Sufficient to Partially Maintain OCT4 in Day 5 OCT4 KO Morulae.
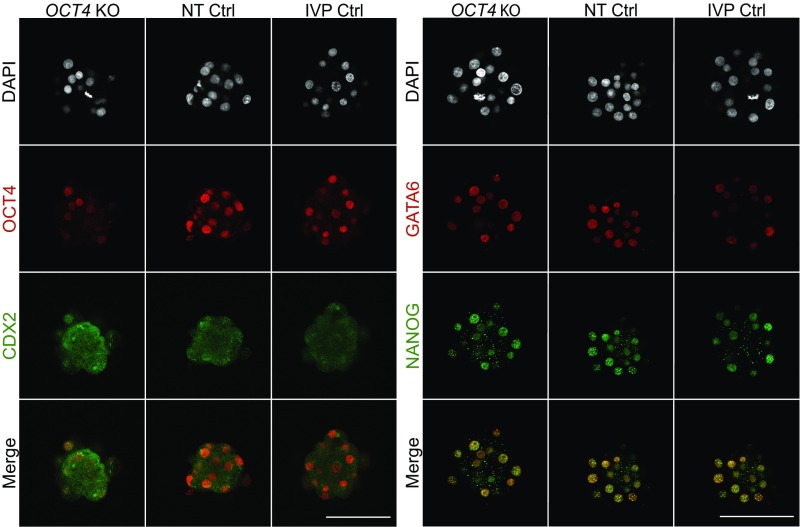
We have previously shown that maternal OCT4 RNA is present in bovine oocytes and early embryos and that embryonic expression of OCT4 starts at the eight-cell stage (10, 13). To address the question if maternal RNA alone can maintain OCT4 protein abundance in OCT4 KO embryos before the blastocyst stage, we analyzed day 5 morulae by immunofluorescence staining using OCT4-specific antibodies. While the nuclei of all NT Ctrl (n = 3) and IVP Ctrl (n = 10) morulae stained positive for OCT4, only 68 ± 5% of the nuclei of OCT4KOtm1 (n = 6) morulae were OCT4 positive, indicating that maternal OCT4 mRNA is sufficient to maintain OCT4 protein in a proportion of blastomeres up to the morula stage. Staining for CDX2 revealed no clear differences among IVP Ctrl, NT Ctrl, and OCT4KOtm1 morulae (Fig. 2). The proportion of GATA6-positive cells (60 ± 5%) in OCT4KOtm1 (n = 9) morulae was significantly (P < 0.05) decreased compared with NT Ctrl (n = 14; 93 ± 2%) and IVP Ctrl morulae (n = 7; 90 ± 2%). While the proportion of NANOG-positive cells was significantly (P < 0.05) higher in NT Ctrl morulae (93 ± 2%), there was no difference between OCT4KOtm1 (81 ± 3%) and IVP Ctrl morulae (85 ± 4%).
Fig. 2.
Representative confocal planes of day 5 morulae stained for OCT4/CDX2 (Left) and NANOG/GATA6 (Right) from OCT4KOtm1, NT Ctrl, and IVP Ctrl embryos. Sample sizes of OCT4/CDX2 and NANOG/GATA6 were n = 6, 3, and 10 and n = 9, 14, and 7 for OCT4KOtm1, NT Ctrl, and IVP Ctrl, respectively. (Scale bars, 100 µm.)
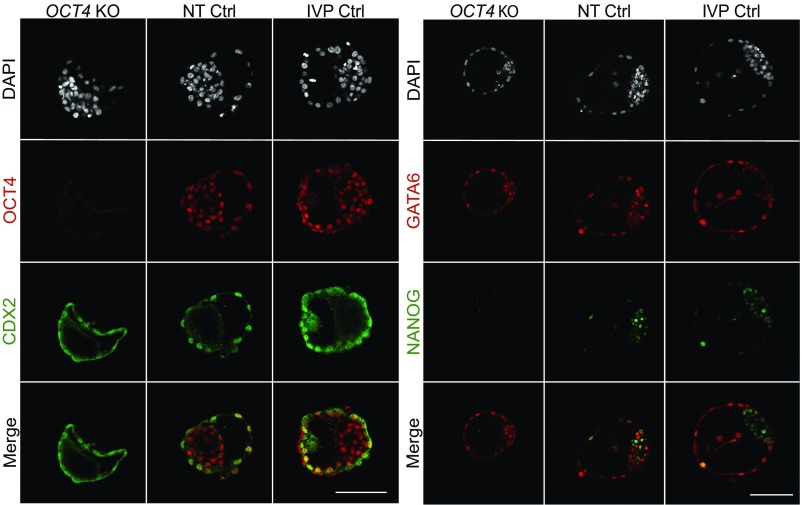
Absent or Markedly Reduced NANOG in Day 7 OCT4 KO Blastocysts Lacking OCT4.
While NT Ctrl (n = 20) and IVP Ctrl (n = 40) blastocysts presented ubiquitous expression of OCT4, staining for OCT4 was negative in blastocysts derived from OCT4KOtm1 (n = 24) and OCT4KOtm2 (n = 8) cells (Fig. 3 and Fig. S3). In all groups of blastocysts, CDX2 expression was restricted to the TE cells, indicating that OCT4 is initially not required to suppress CDX2 expression in the ICM of early blastocysts.
Fig. 3.
Representative confocal plane of day 7 blastocysts stained against OCT4/CDX2 (Left) and NANOG/GATA6 (Right) from OCT4KOtm1, NT Ctrl, and IVP Ctrl embryos. Sample sizes of OCT4/CDX2 and NANOG/GATA6 were n = 24, 20, and 40 and n = 21, 23, and 9 for OCT4KOtm1, NT Ctrl, and IVP Ctrl, respectively. (Scale bars, 100 µm.)
To investigate the role of OCT4 during the second lineage differentiation, we stained day 7 blastocysts for the EPI- and HB-specific markers NANOG and GATA6, respectively. Day 7 NT Ctrl (n = 23) and IVP Ctrl blastocysts (n = 9) already showed the typical salt-and-pepper distribution of NANOG- and GATA6-positive cells in the ICM, but the presence of these proteins was not mutually exclusive in all cells, in line with previous reports (15, 16, 32). Expression of NANOG and GATA6 was not restricted to the ICM, as cells from the TE also still stained positive for both markers, although NANOG staining was already faint. In OCT4KOtm1 blastocysts (n = 21), NANOG staining was absent or very faint, and no cells expressing exclusively NANOG were observed, indicating that maintenance of EPI cells at the beginning of the second lineage differentiation fails in the absence of OCT4 (Fig. 3). This was confirmed in blastocysts from cell clone OCT4KOtm2 (n = 5; Fig. S3), while embryos derived from OCT4-intact ETF1muttm1 cells showed a normal distribution of NANOG and GATA6-positive cells in the ICM (n = 4; Fig. S4). The staining pattern of GATA6 in OCT4 KO blastocysts was similar to NT Ctrl and IVP Ctrl blastocysts, with a few cells exhibiting higher staining intensity in the ICM. There were no GATA6-negative cells in the TE or ICM of OCT4 KO blastocysts, while OCT4-intact blastocysts had cells expressing NANOG exclusively (Fig. 3).
Discussion
The role of Oct4 has been studied extensively in mouse preimplantation embryos, but several reports demonstrated that insights from the mouse model often could not be transferred to other mammalian species. With the development of highly efficient gene editing tools, it is now possible to study key mechanisms of early development, such as maintenance of pluripotency and early differentiation, in species other than mouse, including in humans (17).
Studies on FGF/MAPK signaling during the second lineage differentiation (16) and expression patterns of OCT4 and CDX2 (12, 14, 33) revealed that bovine embryogenesis resembles early human development better than the mouse model. Moreover, assisted reproduction techniques, such as in vitro fertilization (IVF) and SCNT, are well established in bovines and facilitate reverse genetics studies.
Although SCNT embryos have limitations as a model for normal development, we decided to mutate OCT4 in somatic cells and produce embryos by cloning. In contrast to CRISPR-Cas9–mediated editing of zygotes, this approach guarantees consistent modification of all cells of the embryo and allows efficient screening for off-target effects. We used one cell line with a PGK-EGFP reporter construct, to enable later chimeric complementation studies of OCT4-deficient blastomeres (34). To exclude possible effects of the reporter construct, we also used a wild-type fibroblast cell line.
Importantly, in our study, NT Ctrl embryos did not differ from IVP Ctrl embryos in any of the examined parameters, except for the transcriptome profile determined by RNA sequencing that identified 90 genes with significantly different transcript abundance. Nevertheless, PCA of the transcriptome dataset revealed that NT Ctrl and IVP Ctrl blastocysts clustered closely together, whereas OCT4 KO blastocysts formed a distant cluster. Moreover, the numbers of DATs in OCT4 KO blastocysts compared with NT Ctrl (301 DATs) or IVP Ctrl blastocysts (472 DATs) were substantially higher than in NT Ctrl vs. IVP Ctrl blastocysts (90 DATs). This indicates that the loss of OCT4 has a much greater effect than the NT procedure per se. Among the transcripts that are considered to be specific for the EPI, HB, or TE lineages, only AQP3 mRNA was more abundant in OCT4 KO than in NT Ctrl and IVP Ctrl blastocysts. In mouse blastocysts, Aqp3 transcripts (encoding the water channel protein aquaporin 3) were specifically detected in the TE (27). The transcript abundance of several other lineage-specific genes was significantly reduced in OCT4 KO compared with NT Ctrl and IVP Ctrl blastocysts. Among them were the EPI-expressed genes H2AFZ, coding for H2A histone family member Z, and SPIC that encodes the Spi-C transcription factor. In addition, the transcript levels of the HB-expressed genes SPARC (coding for a cysteine-rich acidic matrix-associated protein) and FN1 (encoding fibronectin 1) and of the TE-expressed gene CLDN10, which codes for the tight junction protein claudin 10, were significantly decreased in OCT4 KO blastocysts. Collectively, these findings suggest that similar to OCT4-deficient murine (4) and human (17) embryos, there is no conversion of bovine OCT4 KO blastocysts toward one particular lineage, but rather an overall decrease in relevant gene expression in all three lineages, which eventually leads to developmental failure.
The efficiency of OCT4 mutagenesis in fibroblasts was relatively low (2.6% and 1.7%) in the two cell lines tested. In comparison, the mutation rate of the OCT4 pseudogene sequence in ETF1 was much higher (36%). This may be related to the fact that OCT4 in differentiated cells is silenced and condensed (35), while the ETF1 locus is active and may be more accessible for gene editing (36). Although we were not able to generate OCT4-deficient embryos without a mutation in the OCT4 pseudogene sequence in the ETF1 locus, the key findings of this study could be clearly attributed to the loss of OCT4 since they were not present in embryos derived from OCT4-intact ETF1muttm1 cells.
While the proportion of development to blastocysts was higher in the NT Ctrl group than in the groups from gene-edited single-cell clones, this is unlikely a consequence of the OCT4 loss-of-function mutation, as the developmental potential of OCT4-intact ETF1muttm1 embryos was in the same range. The reduced development to blastocyst in the modified groups is more likely due to the procedures involved in generating gene-edited single-cell clones (i.e., transfection with plasmids and clonal expansion). This is in line with observations that the loss of maternal and zygotic OCT4 did not affect the proportion of Oct4-null mouse blastocysts, which remained at the expected mendelian frequency of ∼25% (3–5, 7).
The analysis of day 5 morulae revealed that maternal OCT4 mRNA is sufficient to maintain OCT4 protein in the majority of the nuclei of OCT4 KO embryos, while in NT Ctrl and IVP Ctrl embryos, all nuclei were OCT4 positive. In day 7 blastocysts produced from OCT4 KO cell clones, OCT4 was entirely absent. Nevertheless, CDX2 was only detected in the TE of these blastocysts, demonstrating that OCT4 is not required to repress CDX2 in the ICM at the time of blastocyst formation, as was also observed in mouse early blastocysts (23).
NANOG transcripts are not present in matured oocytes, and embryonic NANOG expression does not start before the eight-cell stage (10, 32). Because OCT4 KO embryos showed NANOG expression at the day 5 morula stage, we conclude that NANOG activation is not dependent on embryonic activation of OCT4. However, absence or very low levels of the EPI marker NANOG in day 7 OCT4 KO blastocysts indicate that maintenance of EPI cells at the beginning of the second lineage differentiation fails in the absence of OCT4. As we also did not detect any GATA6-negative cells in OCT4 KO blastocysts, it seems that progressive extinction of GATA6 from a subset of ICM cells is dependent on a functional activation of NANOG.
The failure of NANOG expression and maintenance of EPI cells in bovine OCT4 KO blastocysts is in sharp contrast to mouse preimplantation development, where NANOG persists in Oct4-null blastocysts while development of the PrE fails (4, 7, 17). A very recent study inactivating OCT4 in human embryos by microinjecting CRISPR-Cas9 into zygotes revealed that loss of OCT4 resulted in reduced expression of EPI-associated genes in the blastocyst, including NANOG. Immunofluorescence analysis additionally showed that NANOG was absent in OCT4-null blastocysts (17), which is consistent with our findings in OCT4-deficient bovine blastocysts.
Although the mouse is the classical model organism for mammalian developmental biology, recent studies of rabbit, porcine, and bovine embryos revealed that important features of development in these species reflect human embryo development better than mouse embryos do. Examples are the coexpression of OCT4 and CDX2 in the TE (14, 37–39) and the regulation of the second lineage differentiation (16, 40). The present study shows that bovine embryos share with human embryos the essential role of OCT4 for normal NANOG expression, which is not the case in mouse embryo development. The specific features of mouse development may have evolved to enable fast implantation and a short gestation period (14).
Our experimental approach for the functional analysis of OCT4 provides a general strategy for studying the roles of specific genes in mammalian preimplantation embryos and shows that bovine embryos are an interesting model for early human development.
Materials and Methods
CRISPR-Cas9–Mediated KO of OCT4 in Adult Fibroblasts.
OCT4 gene function was disrupted by inducing frameshift-causing mutations in exon 2 of the gene using target sites predicted by CHOPCHOP software (41). Plasmids expressing the fusion of the CRISPR RNA (crRNA) and trans-activating crRNA (synthesized by Invitrogen/Thermo Fisher) and Cas9 (42) were transiently transfected into adult fibroblasts with the Nucleofector Device (Lonza) according to the manufacturer’s instructions, and single-cell clones were produced as described previously (43). Gene editing-induced modifications in the OCT4 alleles and naturally occurring SNPs were examined by Sanger sequencing using the primers (synthesized by Biomers) shown in Table S1.
Production and Analysis of SCNT and IVP Embryos.
SCNT and IVP procedures were performed as described previously (44). At 5 or 7 d after activation of fused complexes or IVF, respectively, the zona pellucida was removed, and embryos were fixed in 2% paraformaldehyde (34) or stored at −80 °C until RNA extraction.
Immunofluorescence Staining and Confocal Laser Scanning Microscope.
Before staining, embryos were incubated for 1 h at room temperature in a blocking solution containing 0.5% Triton X-100 and 5% donkey serum (Jackson ImmunoResearch). Simultaneous staining for either OCT4 and CDX2 or NANOG and GATA6 was achieved by incubation overnight at 4 °C in primary antibody solution and transfer to secondary antibody solution at 37 °C for 1 h after washing three times. For OCT4/CDX2 staining, dilutions of goat anti-human OCT4 polyclonal antibodies (SC8628; Santa Cruz) and rabbit anti-human CDX2 polyclonal antibodies (ab88129; Abcam) were 1:500 and 1:250, respectively. The secondary antibodies donkey anti-rabbit Alexa Fluor 555 (ab150074; Abcam) and donkey anti-goat Alexa Fluor 633 (A212082; Thermo Fisher) were both diluted 1:800. Staining of NANOG/GATA6 was performed with rabbit anti-human NANOG (500-P236, 1:500; Peprotech) and goat anti-human GATA6 (AF1700, 1:250; R&D Systems) and the secondary antibodies donkey anti-rabbit Alexa Fluor 555 (1:500) and donkey anti-goat Alexa Fluor 633 (1:400). Labeled embryos were mounted in Vectashield mounting medium containing DAPI (Vector Laboratories) in a manner that conserves the 3D structure of the specimen (13). Stacks of optical sections were recorded using an LSM710 Axio Observer confocal laser scanning microscope (Zeiss) with an interval of 2.5 µm using a 25× water immersion objective (LD LCI Plan-Apochromat 25×/0.8 Imm Korr DIC M27) and a pinhole of 32 µm. DAPI, Alexa Fluor 555, and Alexa Fluor 633 were excited with laser lines of 405 nm, 561 nm, and 633 nm, respectively, and detection ranges were set to 410 to 562 nm, 582 to 631 nm, and 638 to 747 nm, respectively.
Generation of RNA-Sequencing Libraries, Sequencing, and Data Analysis.
RNA was isolated following manufacturer’s instructions using the MicroPrep Kit (Zymo Research) and analyzed with an Agilent RNA 6000 Pico Chip on a bioanalyzer (Agilent). After digestion with DNase I, RNase-free (Thermo Scientific), 500 pg of purified RNA was used to generate cDNA using the Ovation RNA-Seq System V2 Kit (Nugen) according to the manufacturer’s protocol. RNA sequencing libraries were generated with tagmentation technology of the Nextera XT kit (Illumina) following the manufacturer’s manual. Libraries were quantified on the bioanalyzer and finally sequenced on a HiSeq1500 machine (Illumina). Reads were mapped to the bovine reference genome (bosTau7) with a STAR RNA sequence read mapper, and differential gene expression analysis was performed by using DeSeq2.
Supplementary Material
Acknowledgments
We thank Eva-Maria Jemiller and Tuna Güngör for their excellent technical assistance and Ulrike Gaul and Christophe Jung for access to confocal microscopy. This study was supported in part by the Bayerische Forschungsstiftung (Grant AZ-1031-12).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718833115/-/DCSupplemental.
References
- 1.Artus J, Chazaud C. A close look at the mammalian blastocyst: Epiblast and primitive endoderm formation. Cell Mol Life Sci. 2014;71:3327–3338. doi: 10.1007/s00018-014-1630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu G, Schöler HR. Role of Oct4 in the early embryo development. Cell Regen (Lond) 2014;3:7. doi: 10.1186/2045-9769-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 4.Frum T, et al. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev Cell. 2013;25:610–622. doi: 10.1016/j.devcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu G, et al. Establishment of totipotency does not depend on Oct4A. Nat Cell Biol. 2013;15:1089–1097. doi: 10.1038/ncb2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ralston A, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- 7.Le Bin GC, et al. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development. 2014;141:1001–1010. doi: 10.1242/dev.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- 9.Kang M, Piliszek A, Artus J, Hadjantonakis A-K. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development. 2013;140:267–279. doi: 10.1242/dev.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graf A, et al. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc Natl Acad Sci USA. 2014;111:4139–4144. doi: 10.1073/pnas.1321569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurosaka S, Eckardt S, McLaughlin KJ. Pluripotent lineage definition in bovine embryos by Oct4 transcript localization. Biol Reprod. 2004;71:1578–1582. doi: 10.1095/biolreprod.104.029322. [DOI] [PubMed] [Google Scholar]
- 12.Kirchhof N, et al. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol Reprod. 2000;63:1698–1705. doi: 10.1095/biolreprod63.6.1698. [DOI] [PubMed] [Google Scholar]
- 13.Wuensch A, et al. Quantitative monitoring of pluripotency gene activation after somatic cloning in cattle. Biol Reprod. 2007;76:983–991. doi: 10.1095/biolreprod.106.058776. [DOI] [PubMed] [Google Scholar]
- 14.Berg DK, et al. Trophectoderm lineage determination in cattle. Dev Cell. 2011;20:244–255. doi: 10.1016/j.devcel.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Denicol AC, et al. The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB J. 2014;28:3975–3986. doi: 10.1096/fj.14-253112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuijk EW, et al. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development. 2012;139:871–882. doi: 10.1242/dev.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogarty NME, et al. Genome editing reveals a role for OCT4 in human embryogenesis. Nature. 2017;550:67–73. doi: 10.1038/nature24033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichenbach M, et al. Germ-line transmission of lentiviral PGK-EGFP integrants in transgenic cattle: New perspectives for experimental embryology. Transgenic Res. 2010;19:549–556. doi: 10.1007/s11248-009-9333-5. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann A, et al. Generation of transgenic cattle by lentiviral gene transfer into oocytes. Biol Reprod. 2004;71:405–409. doi: 10.1095/biolreprod.104.028472. [DOI] [PubMed] [Google Scholar]
- 20.Popp MW, Maquat LE. Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell. 2016;165:1319–1322. doi: 10.1016/j.cell.2016.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiffmacher AT, Keefer CL. CDX2 regulates multiple trophoblast genes in bovine trophectoderm CT-1 cells. Mol Reprod Dev. 2013;80:826–839. doi: 10.1002/mrd.22212. [DOI] [PubMed] [Google Scholar]
- 22.Negrón-Pérez VM, Zhang Y, Hansen PJ. Single-cell gene expression of the bovine blastocyst. Reproduction. 2017;154:627–644. doi: 10.1530/REP-17-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 24.Blakeley P, et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3151–3165. doi: 10.1242/dev.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagatomo H, et al. Transcriptional wiring for establishing cell lineage specification at the blastocyst stage in cattle. Biol Reprod. 2013;88:158. doi: 10.1095/biolreprod.113.108993. [DOI] [PubMed] [Google Scholar]
- 26.Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 27.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Kurimoto K, et al. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rugg-Gunn PJ, et al. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brinkhof B, et al. A mRNA landscape of bovine embryos after standard and MAPK-inhibited culture conditions: A comparative analysis. BMC Genomics. 2015;16:277. doi: 10.1186/s12864-015-1448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madeja ZE, et al. Changes in sub-cellular localisation of trophoblast and inner cell mass specific transcription factors during bovine preimplantation development. BMC Dev Biol. 2013;13:32. doi: 10.1186/1471-213X-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan DR, et al. Expression of pluripotency master regulators during two key developmental transitions: EGA and early lineage specification in the bovine embryo. PLoS One. 2012;7:e34110. doi: 10.1371/journal.pone.0034110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niakan KK, Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev Biol. 2013;375:54–64. doi: 10.1016/j.ydbio.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Simmet K, Reichenbach M, Reichenbach H-D, Wolf E. Phytohemagglutinin facilitates the aggregation of blastomere pairs from Day 5 donor embryos with Day 4 host embryos for chimeric bovine embryo multiplication. Theriogenology. 2015;84:1603–1610. doi: 10.1016/j.theriogenology.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Kimura H, Tada M, Nakatsuji N, Tada T. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol Cell Biol. 2004;24:5710–5720. doi: 10.1128/MCB.24.13.5710-5720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isaac RS, et al. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. eLife. 2016;5:e13450. doi: 10.7554/eLife.13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobolak J, et al. Promoter analysis of the rabbit POU5F1 gene and its expression in preimplantation stage embryos. BMC Mol Biol. 2009;10:88. doi: 10.1186/1471-2199-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cauffman G, Van de Velde H, Liebaers I, Van Steirteghem A. Oct-4 mRNA and protein expression during human preimplantation development. Mol Hum Reprod. 2005;11:173–181. doi: 10.1093/molehr/gah155. [DOI] [PubMed] [Google Scholar]
- 39.Kuijk EW, et al. Differences in early lineage segregation between mammals. Dev Dyn. 2008;237:918–927. doi: 10.1002/dvdy.21480. [DOI] [PubMed] [Google Scholar]
- 40.Piliszek A, Madeja ZE, Plusa B. Suppression of ERK signalling abolishes primitive endoderm formation but does not promote pluripotency in rabbit embryo. Development. 2017;144:3719–3730. doi: 10.1242/dev.156406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richter A, et al. Potential of primary kidney cells for somatic cell nuclear transfer mediated transgenesis in pig. BMC Biotechnol. 2012;12:84. doi: 10.1186/1472-6750-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bauersachs S, et al. The endometrium responds differently to cloned versus fertilized embryos. Proc Natl Acad Sci USA. 2009;106:5681–5686. doi: 10.1073/pnas.0811841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.