Significance
To understand how RNA polymerase (RNAP) can synthesize or degrade RNA, and how these processes are regulated, it is necessary to elucidate the catalytic mechanisms of these reactions. Catalysis is sensitive to external factors that modulate the rate of polymerization or hydrolytic attack on the RNA. Here we report that a ubiquitous cellular metabolite, inorganic phosphate, stimulates RNAP exonuclease transcript cleavage activity nearly 2,000-fold. This finding represents a stunning example of how simple cellular molecules can reprogram an enzyme’s active center by providing functional groups and suggests an additional important role for inorganic phosphate in cell biology.
Keywords: phosphate, vanadate, arsenate, transcript cleavage, RNA polymerase
Abstract
Inorganic Pi is involved in all major biochemical pathways. Here we describe a previously unreported activity of Pi. We show that Pi and its structural mimics, vanadate and arsenate, enhance nascent transcript cleavage by RNA polymerase (RNAP). They engage an Mg2+ ion in catalysis and activate an attacking water molecule. Pi, vanadate, and arsenate stimulate the intrinsic exonuclease activity of the enzyme nearly 2,000-fold at saturating concentrations of the reactant anions and Mg2+. This enhancement is comparable to that of specialized transcript cleavage protein factors Gre and TFIIS (3,000- to 4,000-fold). Unlike these protein factors, Pi and its analogs do not stimulate endonuclease transcript cleavage. Conversely, the protein factors only marginally enhance exonucleolytic cleavage. Pi thus complements cellular protein factors in assisting hydrolytic RNA cleavage by extending the repertoire of RNAP transcript degradation modes.
Pi, the simplest soluble form of phosphorus, plays central roles in cellular energetics and metabolism, as well as in biological structure and regulation (for review see ref. 1). It is believed that Pi participated in the principal processes that led to the appearance of life on Earth (1–4). Disruption of phosphate homeostasis is associated with human disorders (5). The high intracellular concentration of Pi, 50–500 mM (5, 6), underscores its biological significance. In the cell, phosphate forms anhydrides (e.g., pyrophosphate, acylphosphates, and nucleoside-5′-di- and triphosphates) and esters with the hydroxyl groups of a variety of cellular metabolites (e.g., sugars, lipids, and proteins) as well as DNA and RNA diesters.
RNA, synthesized by RNA polymerase (RNAP) from a DNA template, encodes the genetic information that directs cell functioning. Although RNA synthesis is the principal activity of RNAP, the enzyme can also degrade RNA in the reverse reaction of pyrophosphorolysis or by hydrolytic cleavage. In the present study we report that Pi strongly catalyzes RNA hydrolysis by RNAP, suggesting an additional important role for this anion in cell biology.
RNA synthesis is carried out through ordered polymerization of NTPs by RNAP using one DNA strand as template. During NTP polymerization by transcription elongation complex (TEC; shown in Fig. 1A) the RNA product remains transiently bound to template DNA strand, forming an RNA:DNA hybrid 9–10 bp long (7). Advancement of TEC along the template is frequently delayed by pausing. Some pauses are due to backtracking, that is, reversible back-translocation of transcribing RNAP (7, 8). When RNAP is backtracked the RNA 3′ segment is disengaged from the RNA:DNA hybrid and extruded into the secondary channel (Fig. 1A), thus occluding the active center and preventing transcript elongation. Backtracking is promoted by occasional incorporation of noncognate RNA residues, DNA damage, and roadblocks conferred by DNA binding proteins. Bacterial factors GreA and GreB (9–11) and eukaryotic factor TFIIS (12) rescue backtracked complexes by stimulating an RNAP endonuclease activity that removes the disengaged RNA segment by hydrolytic cleavage. RNAP is also able to eliminate the 3′ terminal RNA residue in pretranslocated TEC by exonucleolytic cleavage (13). Cleavage is stimulated by noncomplementary nucleotides (13). Unlike endonucleolytic cleavage, the physiological role of exonucleolytic RNA degradation is not readily apparent and remains to be explored.
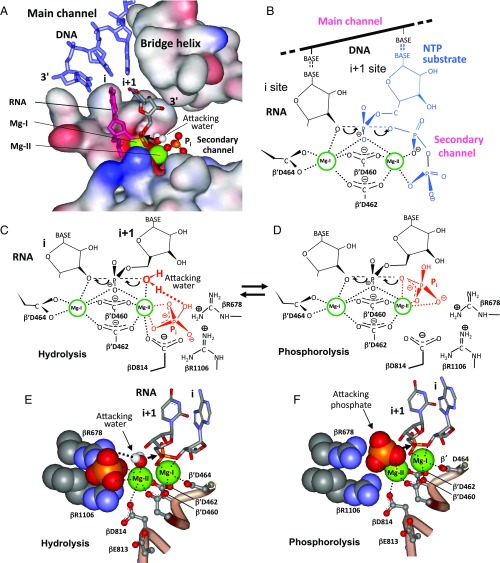
Fig. 1.
Location and models of action for the RNAP active center. (A) The active center performing Pi-stimulated RNA hydrolysis. Catalytic Mg2+ ions are indicated. (B) Nucleotidyl transfer reaction. The dotted lines represent coordination bonds. Arrows indicate the migration of electron density. (C and D) Models for Pi action in hydrolytic RNA cleavage and RNA phosphorolysis, respectively. (E and F) Structural models for C and D, respectively.
Both RNA synthesis and degradation by RNAP occur in the same active center (14), situated at the cross-section of the RNAP main channel that secludes the RNA–DNA hybrid and the secondary channel (Fig. 1A). The latter supplies the NTP substrates for transcript synthesis and also accommodates the disengaged RNA segment in RNAP backtracked complexes. The channels are divided by the bridge helix, a long β′ subunit α-helix.
All RNAP reactions proceed through an Sn2 mechanism (13–15), which involves two Mg2+ ions (Fig. 1) chelated by an invariant aspartate triad of β′ subunit residues (D460, D462, and D464). One Mg2+ ion (Mg-I) saturates the active center, whereas the other (Mg-II) binds transiently and must be recruited through additional coordination for each round of catalysis (13). In the nucleotidyl transfer reaction Mg-II is stabilized by chelation with phosphate groups of the NTP substrate. In transcript cleavage it is coordinated with carboxylate residues of Gre or TFIIS transcription cofactors, which approach the active center through the secondary channel, or by the triphosphate residue of noncognate NTP substrate bound at the E-site of the active center (13). In RNA synthesis Mg-I activates the 3′-hydroxyl of the terminal RNA residue and the α-phosphate of an NTP substrate for nucleophilic attack, whereas Mg-II promotes release of pyrophosphate (Fig. 1B). The two Mg2+ ions switch roles for RNA hydrolytic cleavage. Thus, Mg-II activates the attacking water and Mg-I aids in release of the leaving group (Fig. 1C). In addition to the above-mentioned aspartate triad, two neighboring acidic residues, βE813 and βD814, as well as a basic cluster (βK1073, βR678, βR1106, and β′R731), are engaged in both substrate binding and catalysis (11, 13, 16).
RNAP catalytic activity is regulated by small compounds, as exemplified by the inhibitor tagetitoxin (17) and noncognate NTPs (13), which bind to the active center and change Mg-II coordination. The analogous action mechanism of Pi and its structural mimics on transcription reported in this study represents a stunning example of how simple cellular molecules can reprogram the RNAP active center by providing additional functional groups.
Results
Pi Stimulates Exonuclease Transcript Cleavage in TEC.
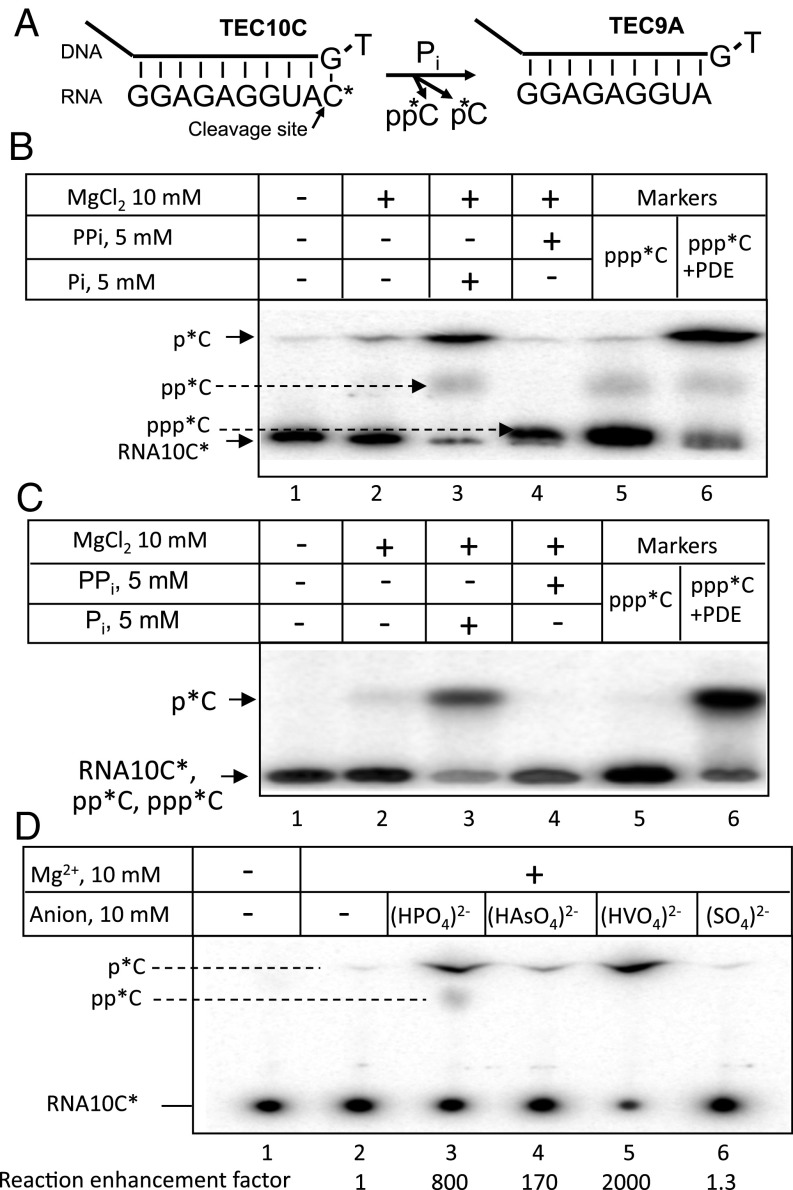
In experiments exposing TEC to different factors we noticed nascent transcript cleavage that was dependent upon the presence of phosphate in the reaction mixture. Such cleavage had not been previously reported, and we therefore decided to investigate this reaction in detail. TEC10C (18) was constructed bearing an RNA carrying a 3′-labeled C (Fig. 2A) and incubated at ambient temperature. As seen from ion-exchange (Fig. 2B) and silicagel TLC analyses (Fig. 2C), the major phosphate-induced RNA degradation product comigrated with [α-32P]CMP. We thus identify the product as [α-32P]CMP originating from 3′exonuclease transcript cleavage. Exonucleolytic cleavage was also observed in TEC12C assembled on a natural T7 A1 promoter template using an incomplete set of NTP substrates (Fig. S1). A secondary radioactive RNA degradation product comigrated with CDP, suggesting that this product originates from RNA phosphorolysis by nucleophilic attack of phosphate on the phosphodiester bond. Pi failed to induce exonuclease cleavage of a terminal mismatched RNA residue (Fig. S2).
Fig. 2.
Stimulatory action of Pi and its structural mimics on hydrolytic RNA cleavage. (A) Scheme of exonuclease RNA hydrolysis in TEC10C. (B) Ion-exchange TLC analysis of the RNA degradation products in TEC10C under various incubation conditions. (C) Analysis of the samples from B by silica gel TLC. (D) RNA degradation in TEC10C in the presence of orthophosphate, arsenate, vanadate, or sulfate.
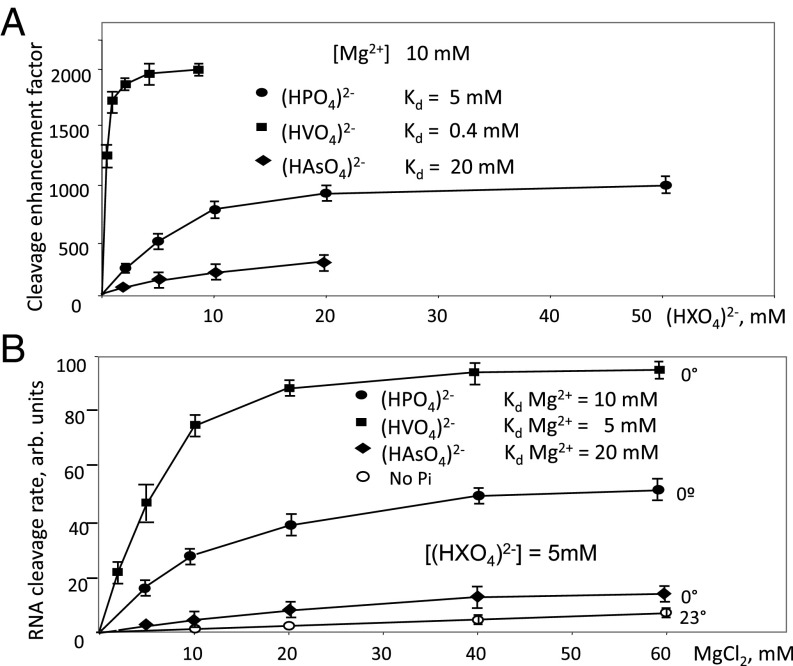
We next tested compounds structurally related to phosphate: arsenate and vanadate (Fig. 2D). As seen from Fig. 2D, vanadate, and to a lesser extent arsenate, stimulated hydrolytic cleavage, but the close structural analog, sulfate, did not. The latter can be explained by a reduced number of ionized groups and their more acidic character in sulfate anion. The fold enhancement under these conditions (10 mM reactants [HXO4]2− and 10 mM Mg2+) was about 800, 2,000, and 170 for phosphate, vanadate, and arsenate, respectively. To further characterize their RNAP exonuclease-enhancing activities TEC10C was incubated with various concentrations of the above anions. The dependence of the reaction rate on anion concentration (Fig. 3A) revealed saturation curves with apparent Km values equal to 5 mM, 0.4 mM, and 20 mM for phosphate, vanadate, and arsenate, respectively. Transcript cleavage experiments were performed at 0 °C due to high rate of the reaction at 37 °C (60- to 80-fold higher than at 0°).
Fig. 3.
Effects of phosphate, vanadate, arsenate, and Mg2+ concentration on RNA cleavage in TEC10. (A) Dependence of the reaction rate on phosphate, vanadate, and arsenate concentration. (B) Dependence of the reaction rate on Mg2+ concentration. Calculated Kd values are presented. Error bars represent SD determined from three independent experiments.
Pi and Its Mimics Increase Retention of the Catalytic Mg-II Required for Transcript Cleavage.
The rate of the exonuclease reaction is enhanced by factors that increase retention of the catalytic Mg-II ion that is required for hydrolytic cleavage, and which is weakly bound in the active center (13). We thus determined the effect of Mg2+ ion concentration on the RNA cleavage rate in the presence and absence of Pi and its structural mimics. As shown in Fig. 3B, the anions tested dramatically reduced the Mg2+ dependence of the reaction, suggesting that they enhance Mg2+ binding. Indeed, the calculated Kd values for Mg2+ were ∼10 mM, 5 mM, and 20 mM in the presence phosphate, vanadate, and arsenate, respectively, and >100 mM for the nonstimulated reaction.
From these results we calculate the maximal reaction enhancement factor for all anions tested (at saturating concentration of both the reactant anions and Mg2+) to be about 2,000. Remarkably, the magnitude of this enhancement approaches that for prokaryotic Gre (11) and eukaryotic TFIIS (19) factors in the transcript endonucleolytic cleavage reaction (3,000- to 4,000-fold).
Pi Does Not Stimulate Endonucleolytic Transcript Cleavage or Endonucleolytic Phosphorolysis.
Only pretranslocated TEC can support exonuclease activity. Endonuclease activity is observed in backtracked TEC. We asked whether Pi stimulates endonucleolytic cleavage by comparing Pi to GreA, a factor known to promote this reaction. For these experiments we have generated the backtracking-prone TEC11A. Upon incubation in transcription buffer the RNAP intrinsic endonuclease reaction released 3′-terminal dinucleotide pCpA from RNA11A. However, Pi had little, if any, effect on this reaction (Table 1 and Fig. S3, lane 5). Conversely, in contrast to Pi, GreA only marginally stimulated exonuclease cleavage of TEC10 (Table 1) while strongly enhancing endonucleolytic cleavage of TEC11 (Table 1 and Fig. S3, lane 6).
Table 1.
Rate constants for transcript cleavage reactions with WT and mutant RNAP at 0° C in TEC10 and TEC11 at 10 mM MgCl2, in the absence or presence of 2.5 mM Pi or 0.1 µM GreA
| Nuclease reaction in WT or mutant TEC | k, h−1/enhancement factor |
| Exo, TEC10, WT | 0.0047 |
| Exo, TEC10, WT, Pi | 1.2 |
| Exo,TEC10, WT, Pi enhancement | 255 |
| Exo, TEC10, βR1106A | 0.332 |
| Exo, TEC10, βR1106A, Pi | 1.0 |
| Exo, TEC10, βR1106A, Pi enhancement | 3.3 |
| Exo, TEC10, βE813, D814/AA | 0.0028 |
| Exo, TEC10, βE813, D814/AA, Pi | 0.014 |
| Exo, TEC10, βE813, D814/AA, Pi enhancement | 5.0 |
| Exo, TEC10, GreA | 0.007 |
| Exo, TEC10, GreA enhancement | 1.4 |
| Endo, TEC11 | 0.14 |
| Endo, TEC11, GreA | 4.1 |
| Endo, TEC11, GreA enhancement | 29 |
| Endo, TEC11, Pi | 0.18 |
| Endo, TEC11, Pi enhancement | 1.3 |
The data represent the average of three independent measurements.
Effect of pH on Pi-Stimulated Exonuclease.
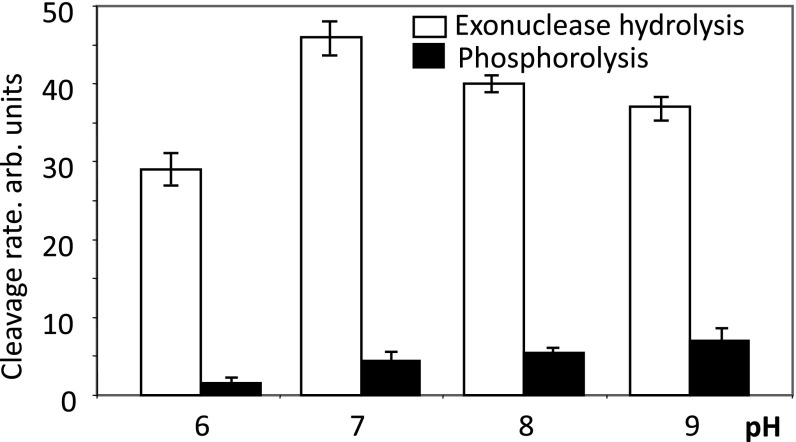
Deprotonation of the attacking water at high pH stimulates nuclease activity; this stimulation decelerates as the pH reaches and exceeds the pKa value of bound water. However, the effect of pH on hydrolytic RNA cleavage is complex, since Mg-II binding is affected by pH (13). Indeed, retention of Mg-II can be enhanced through additional coordination by the neighboring carboxylate residue of the active center, βD814, and possibly βE813 (Fig. 1C). At physiological pH, however, βD814 is salt-bridged with the closely positioned βR1106 and is not available for coordination (Fig. 1C). Deprotonation of βR1106 can explain the abrupt upturn in the exonuclease pH-dependence curve at the pH range 9–10 by releasing the carboxylate side chain(s) for additional Mg-II coordination (13). The above two effects of pH on hydrolytic cleavage can be dissected by elimination of the salt bridge through the R1106A substitution (discussed below), which makes Mg-II binding pH-independent. This allows calculation of the pKa of the attacking water (9.0) by measuring the reaction rate at various pH with mutant TEC (13).
Fig. 4 shows the effect of pH on Pi-stimulated RNA cleavage. Raising the pH from 6 to 7 only marginally stimulated cleavage, and there was no further increase above pH 7. An apparent decrease in the reaction rate within the pH interval 7–9 was due to competing phosphorolytic cleavage. The cleavage rate was about half of the plateau value at about pH 6, suggesting that this is the pKa for the attacking water. This is in striking contrast to nonstimulated exonuclease cleavage, where the apparent pKa of the attacking water is about 9.0 (13). Therefore, in addition to enhancing Mg-II binding, Pi additionally facilitates hydrolytic attack by increasing the fraction of reactive ionized water at physiological pH. The status of Pi ionization can be expected to contribute to the rate of RNA hydrolysis, since protonation of the Pi dianion (pKa 7.2) would compromise Mg2+ coordination. This could account for the observed change in the hydrolysis rate within the pH range 6–7. However, a further increase in the hydrolytic efficiency should be observed upon a pH change from 7 to 9 due to ionization of the attacking water, if the latter is not already activated (as observed for the same reaction in the absence of Pi). This increase is not seen in the presence of Pi, suggesting that the attacking water is already ionized. Therefore, the observed pH profile for the reaction rate in the presence of Pi is best explained by Pi-mediated activation of the attacking water.
Fig. 4.
Effect of pH on RNA degradation in TEC10C. Relative rates of Pi-induced transcript hydrolytic cleavage and phosphorolysis at various pH. Error bars represent SD determined from three independent experiments. arb, arbitrary.
Effect of Active Center Mutations on Pi-Induced RNA Cleavage.
As noted above, enhancement of Mg-II binding in the exonuclease reaction at elevated pH through additional coordination by the βD814, and possibly βE813, can also be achieved by eliminating the salt bridge between βD814 and βR1106 by a βR1106A mutation. Consistent with the additional coordination of Mg-II by the carboxylate residue(s), the double alanine substitution βE813A/βD814A abolished the increased retention of the catalytic metal ion at elevated pH (13). To examine the effect of these active center mutations on Pi-stimulated exonuclease we assembled TEC10C with these RNAP mutants. We asked whether the βR1106A mutation and Pi independently stimulate exonuclease reaction through increasing Mg-II retention. As seen from Table 1 the high basal exonuclease activity of βR1106A mutant was stimulated only threefold by Pi. We propose, therefore, that the mutation reduces Pi binding or forces it into a suboptimal orientation. This conclusion is consistent with the results of molecular modeling described below and presented in Fig. 1E, which suggest direct involvement of the βR1106 residue in Pi binding. In accord with previous studies (13) the βE813A/βD814A mutant was slightly less active (approximately twofold) than WT enzyme in a nonstimulated cleavage reaction. Strikingly, these mutations dramatically (about 50-fold) ablated Pi enhancement of exonucleolytic activity (Table 1). This implies that these residues mediate the bulk of the Pi stimulation. The substitution can reduce Mg-II retention through eliminating a carboxylate ligand from the metal coordination sphere (Fig. 1E), or affect Pi binding at the active center, as described below (Modeling of the Phosphate-Induced Reactions).
Two Modes of Pi Action.
Our experiments indicate that Pi stimulates 3′ terminal exonucleolytic hydrolysis and phosphorolysis by distinct mechanisms, which display different responses to pH (Fig. 4). These reactions are competitive and proceed from alternative Pi binding modes in TEC (Fig. 1 C–F). Since both reactions are likely to proceed through the same Sn2 mechanism, the attacking water in hydrolytic cleavage must be in the same position relative to the scissile phosphodiester group as the oxygen atom of the Mg-II–coordinated phosphate in phosphorolysis (Fig. 1 C and D, respectively). Therefore, the hydrolytic coordination mode for the phosphate is achieved by “phosphorolytic” state isomerization, which frees an Mg-II valence for binding the attacking water (Fig. 1 C–F). Both these activities of Pi are mechanistically and functionally related to those observed for pyrophosphate: pyrophosphorolysis and pyrophosphate-induced exonuclease RNA hydrolysis characterized in our previous study (13).
Modeling of the Phosphate-Induced Reactions.
We have constructed a working model to explain Pi-stimulated transcript degradation (Fig. 1 E and F). The atomic coordinates for the modeling were those of yeast backtracked TEC (Protein Data Bank ID code 3GTG) (20). We chose this structure over an apparently more relevant X-ray structure (1I6V) of pretranslocated TEC (the exonuclease substrate) because the latter depicts a catalytically inactive state in which the contacts of the 3′ terminal RNA residue in the active center are not established (21). To generate the structure of a functional pretranslocated TEC we deleted the 3′ disengaged RNA residue from the 3GTG atomic model. We used the following structural constraints for modeling: (i) Bound Pi must be within coordination bond distance to Mg-II of the active center, (ii) one coordination valence of the chelated Mg-II must remain available for the attacking water, and (iii) the hydrolytic attack must proceed codirectionally with the displacement of the leaving group (Fig. 1C). The arrangement of the catalytic Mg-II ion relative to the reactant groups, predicated by the Sn2 mechanism, was as in our previous modeling (13). These constraints in the context of the X-ray structure placed Pi unequivocally in the active center.
In the resulting model (Fig. 1 C and E) Pi fits into a small cleft between the βR1106 and βR678 side chains and Mg-II. This forms a sandwich-like structure, in which the Arg residues flank the Pi ligand. Pi stacks to the βR1106 side chain in this cavity so that three hydroxyl groups of the Pi are within salt-bridging distance to three guanidinium-group nitrogens, among which the positive charge of the group is distributed. Two of these Pi hydroxyl groups simultaneously salt-bridge with βR678. This orientation poses the fourth Pi oxygen within hydrogen-bonding distance to the attacking water, thus explaining the observed enhanced dissociation constant for the reactive water (discussed above). The interaction network between Pi and the Arg residues poses two Pi hydroxyl groups to coordinate Mg-II, which explains observed enhanced metal ion retention in the presence of Pi. Moreover, the remaining coordination valence of Mg-II can be used to chelate D814 (Fig. 1E), which is consistent with the involvement of D814 in Mg-II coordination under conditions where the salt bridge between this residue and R1106 is eliminated (discussed above). Such Pi docking also accounts for the effect of the active center mutations on RNA cleavage. Thus, the βR1106A substitution reduces the Pi binding by eliminating the bulk of Pi salt-bridging interactions and by widening the cavity in which the ligand binds. The βD813, D814/AA mutation can affect Pi binding by changing the orientation of the βR1106 side chain involved in interactions with Pi. In fact, the βD814 residue is in immediate contact with βR1106 (Fig. 1E), which is reinforced by salt-bridging between these two residues. Therefore, substitution of Asp by a smaller Ala side chain simultaneously creates a gap in the active center and eliminates the salt bridge.
For modeling RNA phosphorolytic cleavage (Fig. 1 D and F) we postulate that the Pi orientation in this reaction is the same as that for the attacking phosphate part of a pyrophosphate molecule in the related pyrophosphorolysis reaction. We suggest that these alternate phosphate coordination modes at Mg-II exist in equilibrium. Transition from a hydrolytic to a phosphorolytic complex is achieved by rotation of Pi around its two Mg-II coordination bonds. This is accompanied by displacement of the coordinated attacking water by a Pi oxygen, which was originally hydrogen-bound to the water (Fig. 1E). As suggested by molecular modeling, this transition would distance Pi from βR1106 and βR678 (Fig. 1F), thus destabilizing the complex. The equilibrium between the two Pi coordination modes is therefore expected to favor the more stable hydrolytic orientation. This is consistent with significantly higher efficiency of Pi-induced RNA hydrolytic cleavage (ca.10-fold) compared with Pi phosphorolysis (Figs. 2 and 4).
Discussion
The most striking finding reported here is the dramatic stimulation by orthophosphate and its structural mimics, arsenate and vanadate, of hydrolytic exonuclease RNA cleavage by RNAP in a TEC. We demonstrate that these compounds act by recruiting the Mg-II ion of the active center, which, unlike Mg-I, is delivered by substrates or cofactors for each act of catalysis. Stimulation of exonuclease by E site-bound noncognate NTP substrates likewise results from recruitment of Mg2+ (13). Thus, our work suggests that RNAP catalysis is regulated in general by modulation of the coordination of active center Mg2+ ion. The magnitude of stimulation by Pi and its structural mimics is remarkable—comparable to that observed for specialized transcript cleavage protein factors Gre and TFIIS. Despite the mechanistic analogy, Pi and Gre enhance nuclease cleavage in opposite modes. The former stimulates exonuclease and the latter endonuclease cleavage reactions, as explained in our previous studies (16). Recall that the substrate for exonuclease is pretranslocated TEC, whereas endonuclease acts on backtracked TEC. As follows from molecular modeling, the disengaged 3′ RNA segment in a backtracked complex blocks the Pi binding site. In contrast, Gre-mediated catalysis in backtracked TEC relies on interactions of the first 3′ disengaged RNA residue in the active center. These interactions set the active center for catalysis by attracting Mg-II in intrinsic and Gre-assisted reactions (16). We also observe some small enhancing action of GreA factor on exonuclease RNA cleavage. This is consistent with the effect of Gre and TFIIS factor on exonuclease hydrolysis observed for yeast and Escherichia coli enzymes (22, 23).
The biological role of the two Pi-mediated processes characterized in this study is not known. Nevertheless, the high intracellular Pi concentration (50–500 mM) and reasonable Km (5 mM) for Pi in RNA phosphorolysis and hydrolytic cleavage suggest that this metabolite can efficiently execute both reactions in vivo. We demonstrated that Pi does not support exonuclease cleavage of 3′ misincorporated RNA residue, which excludes the role of Pi in proofreading. Also, Pi does not stimulate endophosphorolytic RNA cleavage in backtracked TEC. However, a related pyrophosphate (PPi)-induced cleavage was observed in yeast Pol-II backtracked TEC (24). This implies that Pi can act as a PPi analog in eukaryotes, thus assisting proofreading. Since the intracellular Pi concentration is about 1,000-fold greater than that of PPi, Pi-promoted nucleolytic cleavage might have a physiological role in eukaryotes by complementing TFIIS factor in hydrolytic RNA cleavage. In addition, Pi might act in conjunction with transcription termination factors that lock TEC in the pretranslocated state (18). By stimulating exonuclease RNA cleavage in such complexes Pi would shorten the RNA:DNA hybrid, thereby facilitating TEC decay and transcript release. Finally, active center-bound orthophosphate can suppress RNAP backtracking and transcriptional arrest by stabilizing the pretranslocated TEC state through phosphodiester coordination (Fig. 1C).
Taking into account the average intracellular concentrations of Pi and NTP, as well as their Km in the reactions of RNA degradation and extension by RNAP, it is expected that Pi can affect the rate of transcription by competing with NTP substrate binding at the active center. However, NTP and other chelating molecules in the cell can scavenge Mg2+, whose concentration in unbound form is very low (25), thus reducing the concentration of Pi–Mg2+ complex, which is likely the active center-binding form of Pi. This should favor RNA extension, since NTPs bind Mg2+ stronger than Pi. Also, nucleotidyl transfer reaction proceeds with significantly higher maximal rate than RNA hydrolysis. Nevertheless, a regulatory role of Pi on transcription via competition with NTP substrates cannot be excluded. This mechanism could operate in starved cells, when Pi content in the cell is high and the NTP concentration is low (26).
Vanadate and arsenate have attracted considerable attention. Thus, vanadate was used as a mimic for the transition phosphate state in various enzymatic reactions (27), while arsenate has been validated as a phosphate analog in some enzymatic systems (28, 29). In this study we report a previously unknown activity of these compounds, that is, their strong stimulation of RNA degradation in transcription complexes. The ability of arsenate and vanadate to greatly accelerate hydrolytic RNA cleavage by RNAP is a striking example of their potential biological activity.
Materials and Methods
All chemicals were from Sigma-Aldrich. His6-tagged RNAP was purified from the RL721 E. coli strain. Mutant RNAPs were obtained as previously described (13). Ribonucleoside-5′-triphosphates were from Pharmacia. [α-32P]CMP was obtained from [α-32P]CTP, 3,000 Ci/mmol (MP Biomedicals) by treatment with snake venom phosphodiesterase (New England BioLabs). Oligonucleotides were from Oligos. Radioactive products of RNA degradation reaction by RNAP were resolved by electrophoresis, or by TLC on PEI cellulose or silicagel plates (Sigma-Aldrich) and quantified by phosphoimagery using a Molecular Dynamics device (GE Healthcare). Molecular modeling was performed using WebLab ViewerLight 4.0 (Molecular Simulations Inc.). TEC9 was assembled essentially as previously described (18). Detailed protocol is provided in SI Materials and Methods.
Effect of Phosphate, Vanadate, Arsentate, and Sulfate on Exonuclease Transcript Cleavage.
Reaction mixtures (8 µL) containing labeled TEC10C in 20 mM Tris⋅HCl, pH 8.0, and 0.1 M NaCl were supplemented with 1 µL of 0.1 M sodium phosphate, vanadate, arsentate, or sulfate with pH adjusted to 8.0. The reaction was started by addition of 1 µL of 0.1 M MgCl2 and carried out at 0 °C. After 20-min incubation the reaction was quenched and analyzed by PEI cellulose TLC as described above.
Determination of the Kd for Phosphate, Vanadate, and Arsentate in Exonuclease RNA Cleavage.
TEC10C in transcription buffer (TB) lacking Mg2+ was supplemented with orthophosphate or its mimics to final concentrations indicated in Fig. 3A. The reaction was started by addition of MgCl2 to a final concentration of 10 mM and, after incubation at 0 °C, quenched and analyzed as described above. Incubation time was 10 min, 20 min, and 40 min for vanadate, orthophosphate, and arsenate, respectively The reactions were quenched and analyzed as described above.
Determination of the Kd for Mg2+ in the Reaction of Exonuclease RNA Cleavage in the Presence of Phosphate, Vanadate, and Arsentate.
The reaction was carried out at 0 °C as described above in the presence of 5 mM Pi or its mimics. The concentration of Mg2+ in the reaction mixtures is indicated in Fig. 3B. The incubation time was 5, 10, and 40 min for vanadate, phosphate, and arsenate respectively. The control reaction was performed under the same conditions but in the absence of the reactant anions.
Comparative Effect of Pi and GreA on Exonuclease and Endonuclease Transcript Cleavage.
Reactions were performed at 0 °C with labeled TEC10C or TEC11A in TB in the presence of 2.5 mM Pi or 0.1 µM GreA. The incubation time was 10 min and the reactions were quenched as previously described.
Effect of Active Center Mutations in RNAP on Pi-Induced Transcript Cleavage.
TEC9A with mutant enzymes was assembled as for WT RNAP. Elongation of TEC9A with [α-32P]CTP was performed at 20 °C for 1 h, followed by washing with TB lacking MgCl2. TECs were supplemented with 10 mM MgCl2 and 2.5 mM Pi and kept at 0 °C. Incubation times were 15 h for βE814A/βD814A, 1 h for βR1106A, and 15 min for WT RNAP TECs. The reaction products were resolved by TLC on PEI cellulose in 0.1 M potassium phosphate (pH 4.1) and detected by phosphoimaging.
Effect of pH on Pi-Induced Transcript Degradation.
Reactions were performed with TEC10C obtained as described above. The desired pH was created by using the following buffers: MES pH 6.0, Hepes pH 7.0, Tris pH 8.0, and taurine pH 9.0 at 20 mM concentration. The mixtures were supplemented with 10 mM MgCl2 and 2.5 mM Pi and kept for 20 min at 0 °C. The reaction products were analyzed by TLC on PEI cellulose in 0.1 M potassium phosphate (pH 4.1).
Supplementary Material
Acknowledgments
This work was supported by NIH Grant 2 R01 GM037219-26 (to M.E.G.) and by a Public Health Research Institute research support grant (to A.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720370115/-/DCSupplemental.
References
- 1.Pasek MA, Kee TP. On the origin of phosphorylated biomolecules. In: Egel R, Lankenau D-D, Mulkidjanian AY, editors. Origins of Life: The Primal Self-Organization. Springer; Berlin: 2011. pp. 57–84. [Google Scholar]
- 2.Gull M. Prebiotic phosphorylation reactions on early Earth. Challenges. 2014;5:193–212. [Google Scholar]
- 3.Ruiz-Mirazo K, Briones C, de la Escosura A. Prebiotic systems chemistry: New perspectives for the origins of life. Chem Rev. 2014;114:285–366. doi: 10.1021/cr2004844. [DOI] [PubMed] [Google Scholar]
- 4.Westtheimer FH. Why nature chose phosphates. Science. 1987;235:1173–1178. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 5.Bugg NC, Jones JA. Hypophosphataemia. Pathophysiology, effects and management on the intensive care unit. Anaesthesia. 1998;53:895–902. doi: 10.1046/j.1365-2044.1998.00463.x. [DOI] [PubMed] [Google Scholar]
- 6.Libanati CM, Tandler CJ. The distribution of the water-soluble inorganic orthophosphate ions within the cell: Accumulation in the nucleus. Electron probe microanalysis. J Cell Biol. 1969;42:754–765. doi: 10.1083/jcb.42.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 8.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 9.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 10.Stebbins CE, et al. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature. 1995;373:636–640. doi: 10.1038/373636a0. [DOI] [PubMed] [Google Scholar]
- 11.Sosunova E, et al. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reines D, Ghanouni P, Li QQ, Mote J., Jr The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage. J Biol Chem. 1992;267:15516–15522. [PMC free article] [PubMed] [Google Scholar]
- 13.Sosunov V, et al. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sosunov V, et al. The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucleic Acids Res. 2005;33:4202–4211. doi: 10.1093/nar/gki688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 16.Sosunova E, Sosunov V, Epshtein V, Nikiforov V, Mustaev A. Control of transcriptional fidelity by active center tuning as derived from RNA polymerase endonuclease reaction. J Biol Chem. 2013;288:6688–6703. doi: 10.1074/jbc.M112.424002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vassylyev DG, et al. Structural basis for transcription inhibition by tagetitoxin. Nat Struct Mol Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vitiello CL, Kireeva ML, Lubkowska L, Kashlev M, Gottesman M. Coliphage HK022 Nun protein inhibits RNA polymerase translocation. Proc Natl Acad Sci USA. 2014;111:E2368–E2375. doi: 10.1073/pnas.1319740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weilbaecher RG, Awrey DE, Edwards AM, Kane CM. Intrinsic transcript cleavage in yeast RNA polymerase II elongation complexes. J Biol Chem. 2003;278:24189–24199. doi: 10.1074/jbc.M211197200. [DOI] [PubMed] [Google Scholar]
- 20.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Hawley DK. Identification of a 3′–>5′ exonuclease activity associated with human RNA polymerase II. Proc Natl Acad Sci USA. 1993;90:843–847. doi: 10.1073/pnas.90.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Severinov K, Goldfarb A. Topology of the product binding site in RNA polymerase revealed by transcript slippage at the phage λ PL promoter. J Biol Chem. 1994;269:31701–31705. [PubMed] [Google Scholar]
- 24.Rudd MD, Izban MG, Luse DS. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci USA. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milo R, Phillips R. 2015 Cell Biology by the Numbers (Garland, New York). Available at book.bionumbers.org/what-are-the-concentrations-of-different-ions-in-cells/. Accessed February 20, 2018.
- 26.Mason PW, Carbone DP, Cushman RA, Waggoner AS. The importance of inorganic phosphate in regulation of energy metabolism of Streptococcus lactis. J Biol Chem. 1981;256:1861–1866. [PubMed] [Google Scholar]
- 27.Crans DC, Smee JJ, Gaidamauskas E, Yang L. The chemistry and biochemistry of vanadium and the biological activities exerted by vanadium compounds. Chem Rev. 2004;104:849–902. doi: 10.1021/cr020607t. [DOI] [PubMed] [Google Scholar]
- 28.Németi B, Gregus Z. Mechanism of thiol-supported arsenate reduction mediated by phosphorolytic-arsenolytic enzymes: I. The role of arsenolysis. Toxicol Sci. 2009;110:270–281. doi: 10.1093/toxsci/kfp112. [DOI] [PubMed] [Google Scholar]
- 29.Krebs HA, Eggleston LV, Knivett VA. Arsenolysis and phosphorolysis of citrulline in mammalian liver. Biochem J. 1955;59:185–193. doi: 10.1042/bj0590185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.