Significance
The HIV-1 latent reservoir cannot be eradicated by antiretroviral therapy (ART). The reservoir is a major barrier to cure. To characterize the mechanisms that contribute to persistence of the latent reservoir, we examined clonally expanded cell populations carrying replication-competent HIV-1 and followed them longitudinally. Expanded clones harboring replication-competent HIV-1 were identified in all study participants, but these clones emerge and wane on a time scale of years. A similar pattern was identified in viruses sampled from residual viremia. The findings suggest that the latent reservoir is likely to be maintained through expansion driven by antigens and cytokines, and that the expansion is balanced with a constant cell loss.
Keywords: HIV persistence, latent reservoir, clonal expansion
Abstract
The latent reservoir for HIV-1 in resting CD4+ T cells is a major barrier to cure. Several lines of evidence suggest that the latent reservoir is maintained through cellular proliferation. Analysis of this proliferative process is complicated by the fact that most infected cells carry defective proviruses. Additional complications are that stimuli that drive T cell proliferation can also induce virus production from latently infected cells and productively infected cells have a short in vivo half-life. In this ex vivo study, we show that latently infected cells containing replication-competent HIV-1 can proliferate in response to T cell receptor agonists or cytokines that are known to induce homeostatic proliferation and that this can occur without virus production. Some cells that have proliferated in response to these stimuli can survive for 7 d while retaining the ability to produce virus. This finding supports the hypothesis that both antigen-driven and cytokine-induced proliferation may contribute to the stability of the latent reservoir. Sequencing of replication-competent proviruses isolated from patients at different time points confirmed the presence of expanded clones and demonstrated that while some clones harboring replication-competent virus persist longitudinally on a scale of years, others wax and wane. A similar pattern is observed in longitudinal sampling of residual viremia in patients. The observed patterns are not consistent with a continuous, cell-autonomous, proliferative process related to the HIV-1 integration site. The fact that the latent reservoir can be maintained, in part, by cellular proliferation without viral reactivation poses challenges to cure.
The latent reservoir for HIV-1 in resting memory CD4+ T cells persists even in patients on optimal antiretroviral therapy (ART) and is the major barrier to cure (1–3). The reservoir is established early in infection (4, 5) and is extremely stable, with an estimated half-life of 44 mo (6–8), making cure with ART alone unlikely. One strategy to eliminate the latent reservoir is termed shock-and-kill (9, 10). It depends on inducing proviral expression in latently infected cells to allow their elimination by immune mechanisms or viral cytopathic effects.
Previous studies suggested that the proliferation of infected cells might contribute to HIV-1 persistence (11–19). At least three mechanisms may explain the proliferation of infected cells: antigen-driven T cell proliferation (16), homeostatic proliferation (13), and proliferation driven by effects related to the site of HIV-1 integration (14, 15). Proliferation of infected cells driven by antigen or cytokines is unexpected because these stimuli also induce viral gene expression in latently infected cells (20, 21), which exposes the cells to viral cytopathic effects and immune clearance. In vivo, most productively infected cells have a short half-life (22, 23). Interestingly, it has been shown in a model system that latently infected CD4+ T cells can proliferate in response to cytokines such as IL-2 and IL-7 without viral reactivation (24).
Early evidence for the in vivo proliferation of HIV-1–infected cells came from detailed phylogenetic studies showing the presence in plasma of viruses with identical sequences, termed predominant plasma clones (PPCs), in some infected individuals on prolonged ART (11, 12). In treated patients who have suppression of viremia to below the limit of detection of clinical assays, this residual viremia can be detected with very sensitive assays (25–28). The residual viremia consists of archival viral sequences that are sensitive to the current drug regimen and do not show evolution over time (12, 29–32). Numerous studies have shown that levels of residual viremia cannot be reduced by treatment intensification (33–35). All of these results suggest that residual viremia results from the daily activation of a small number of latently infected cells rather than ongoing cycles of replication. The clonal nature of residual viremia suggested that it is produced by a limited subset of infected cells carrying identical HIV-1 sequences, likely arising as a result of cellular proliferation (11, 12).
Further support for the role of T cell proliferation in HIV-1 reservoir dynamics came from a landmark study that defined the distribution of latent HIV-1 among the central memory, transitional memory, and effector memory CD4+ T cell subsets and the role of cytokine-driven homeostatic proliferation in reservoir stability (13).
More recent studies used HIV-1 integration site analysis to provide direct and definitive evidence for clonal expansion in infected cells in individuals on long-term ART (14, 15, 36). A large fraction of the infected cells appear to have undergone clonal expansion based on the presence of identical HIV-1 integration sites in different infected peripheral blood lymphocytes (14, 15, 36). One study described identical viral sequences integrated at exactly the same position in the human genome in multiple cells, consistent with infected cell proliferation (15). Two studies suggested that HIV-1 integration into genes associated with cell growth and survival might drive proliferation of infected cells (14, 15).
While clonal expansion of infected CD4+ T cells is prevalent in infected individuals, most HIV-1 proviruses are defective (37, 38). Since integration site analysis captures only a small part of the HIV-1 genome, it is likely that many of the expanded cellular clones detected by this method harbor defective proviruses (36). However, a highly expanded clonal population of CD4+ T cells harboring a replication-competent provirus was recently characterized in an infected individual with metastatic squamous cell carcinoma, suggesting that cells carrying replication-competent proviruses can expand and persist (16). In addition, three recent studies demonstrated that clonal CD4+ T cell populations carrying replication-competent viruses are common in infected individuals, accounting for over 50% of the latently infected cells examined (17–19). Therefore, it is crucial to evaluate how cells containing replication-competent provirus proliferate. This information may help us to understand the forces that shape and preserve the latent reservoir.
To address these issues, we asked whether infected CD4+ T cells harboring replication-competent provirus could proliferate in response to T cell receptor (TCR) stimulation or cytokines that are known to drive homeostatic proliferation and, if so, whether this proliferation can occur without viral reactivation. In addition, we examined the in vivo dynamics of clonal populations of infected cells carrying replication-competent HIV-1 to determine whether and how expanded clones harboring replication-competent provirus persist over time.
Results
CD4+ T Cells Containing Replication-Competent Virus Can Proliferate in Response to TCR Activation and Cytokine Treatment.
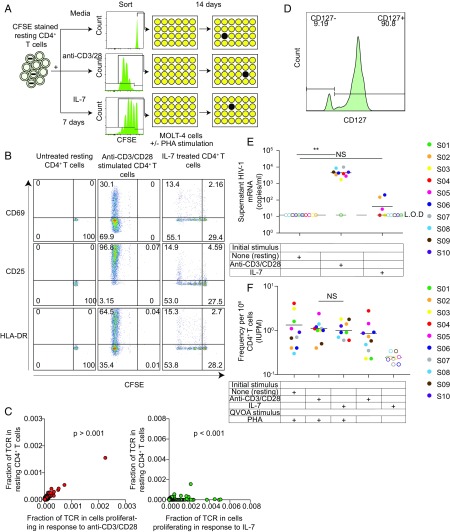
The immune system maintains normal levels of T cells, in part, through a process of homeostatic proliferation. For CD4+ T cells, the gamma chain cytokine IL-7 is an important stimulus for homeostatic proliferation in vivo (39, 40). A previous study evaluated the role of IL-7–driven proliferation in HIV-1 reservoir stability and demonstrated a relationship between plasma IL-7 concentration and reservoir size (13). Bosque et al. (24) used a primary cell model of HIV-1 latency to demonstrate that latently infected central memory T cells could proliferate in the presence of IL-7 without viral reactivation. To explore the stimuli and conditions associated with proliferation of infected cells in vivo, we purified resting CD4+ T cells from 10 HIV-1–infected donors on ART and stained the cells with carboxyfluorescein succinimidyl ester (CFSE) dye. The cells were then treated with anti-CD3/CD28 antibodies or with an optimal concentration of IL-7 (24) for 7 d in the presence of antiretroviral drugs (Fig. 1A). IL-2 was included in all of the cultures to maintain cell viability and match the conditions described by Bosque et al. (24). Most cells proliferated in response to anti-CD3/CD28 antibodies, while proliferation induced by IL-7 was more limited (Fig. 1B). We also examined the activation status of cells cultured for 7 d with anti-CD3/CD28 or IL-7 by flow cytometry. Cells were stained for CD69, CD25, and HLA-antigen D related (DR). With anti-CD3/CD28 stimulation, most cells were positive for CD25, while only 20% of the cells treated with IL-7 were positive for CD25 (Fig. 1B).
Fig. 1.
Proliferation of infected cells in response to TCR stimulation and cytokines. (A) Experimental setup with assay time course. Resting CD4+ T cells were isolated from participants on ART; stained with CFSE; and then cultured for 7 d with media alone, cultured with anti-CD3/CD28, or treated with IL-7. On day 7, cells that had proliferated in response to anti-CD3/CD28 or IL-7 were isolated based on CFSE dilution. Half of the sorted cells were then plated in a limiting dilution QVOA with PHA and irradiated allogeneic PBMCs. The other half of the cells were plated at the same dilutions without PHA or irradiated allogeneic PBMCs. After 24 h, PHA was removed and MOLT-4/CCR5+ cells were added to all culture wells to expand virus released from infected cells. On day 20, a p24 ELISA was performed to quantify viral outgrowth. Cells treated with media alone were cultured without sorting. (B) Activation marker expression and proliferation induced by anti-CD3/CD28 and IL-7 stimulations. Resting CD4+ T cells were stained with CFSE before stimulation. CFSE dilution and activation marker expression were quantitated by flow cytometry 7 d after stimulation. (C) TCR sequence analysis. TCRs of cells that proliferated with anti-CD3/CD28 stimulation or IL-7 stimulation were sequenced, and the percentage of each TCR was then calculated in each sample. (D) Resting CD4+ T cell expression of IL-7 receptor (CD127). Freshly isolated resting CD4+ T cells were stained with CD127 to quantify IL-7 receptor expression level before any treatment. (E) Induction of virus production by anti-CD3/CD28 and IL-7. Resting CD4+ T cells from individuals on ART (n = 10) were left untreated or simulated with anti-CD3/CD28 or IL-7 for 7 d. HIV-1 RNA levels in culture supernatants were measured by qRT-PCR. **P < 0.01. L.O.D., limit of detection; NS, P > 0.05. (F) Frequency of latently infected cells that proliferated in response to anti-CD3/CD28 or IL-7 as measured by QVOA. Sorted cells were analyzed by QVOA with or without an activating stimulus (PHA and allogeneic PBMCs). The frequency of cells producing replication-competent virus was determined by limiting dilution statistics 14 d later.
The cells receiving each stimulus were then sorted based on whether they had proliferated in response to anti-CD3/CD28 or IL-7. This allowed us to examine the properties and fate of infected cells that had recently gone through the cell cycle. We sequenced the TCRs in the sorted CFSE low cell populations that proliferated in response to each treatment and compared the sequences with those in the resting T cell population. With anti-CD3/CD28 stimulation, the proportions of cells with particular TCRs remained the same as in unstimulated resting CD4+ T cells (P > 0.001), while some clones achieved a higher frequency in the population that proliferated in response to IL-7 compared with that in unstimulated resting CD4+ T cells (P < 0.001) (Fig. 1C). To explore the nonuniform expansion induced by IL-7, we then analyzed IL-7 receptor (CD127) expression on resting CD4+ T cells from infected individuals and found that 90% of the cells expressed CD127 (Fig. 1D). This observation indicates that not all resting CD4+ T cells are able to proliferate in response to cytokines, which may provide a partial explanation for the preferential expansion of certain clones in response to IL-7 (Fig. 1C). In any event, the sorting of cells that had proliferated in response to anti-CD3/CD28 or IL-7 allowed us to directly examine the clonal expansion of HIV-1–infected cells.
We next asked whether either stimulation induced rapid virus release from freshly isolated resting CD4+ T cells. We collected culture supernatants from resting CD4+ T cells from 10 infected individuals after 7 d of stimulation with media alone, anti-CD3/CD28, or IL-7. Anti-CD3/CD28 induced virus production, as measured by HIV-1 mRNA in the supernatants. IL-7 treatment induced detectable HIV-1 mRNA in the supernatant of cells from five of 10 infected individuals, but the HIV-1 mRNA levels for those patients were at least two logs lower than those observed with anti-CD3/CD28 (Fig. 1E). The finding that latently infected CD4+ T cells treated with IL-7 can proliferate with little or no virus production suggests that homeostatic proliferation induced by cytokines could potentially expand the latent reservoir without exposing the infected cells to immune clearance. Anti-CD3/CD28 induced much higher levels of T cell proliferation and latency reversal.
Most infected cells harbor defective proviruses that may not interfere with clonal expansion (37, 38). To determine whether cells harboring replication-competent HIV-1 genomes can proliferate in response to these stimuli, we plated the sorted cells that had proliferated in response to anti-CD3/CD28 or IL-7 in a limiting dilution quantitative viral outgrowth assay (QVOA) (Fig. 1A). In this assay, the cells are stimulated with phytohemagglutinin (PHA) and γirradiated allogeneic peripheral blood mononuclear cells (PBMCs) to reverse latency as previously described (41, 42). Infectious virus particles released following reversal of latency are detected by adding MOLT4/CCR5+ cells to the culture. These cells lack class II MHC expression and do not cause allogeneic stimulation of patient cells (42). However, they are highly susceptible to infection and allow exponential growth of infectious virus released from patient cells. With this assay, we detected viral outgrowth by p24 ELISA of supernatants from cultures of unstimulated resting CD4+ T cells or cell populations that had proliferated in response to stimulation with either anti-CD3/CD28 or IL-7 (Fig. 1F). The frequencies of cells that were able to produce replication-competent virus were similar in all three cases (Fig. 1F). Thus, cells harboring replication-competent virus can proliferate in response to either TCR stimulation or cytokines, survive for at least 7 d, and retain the ability to produce infectious virus upon subsequent stimulation.
In the same experiment, cell populations that had proliferated in response to either anti-CD3/CD28 stimulation or cytokine treatment were also plated in the QVOA without additional PHA stimulation. Viral outgrowth was observed in cultures set up with MOLT-4 cells and cells that had proliferated in response to anti-CD3/CD28 without another round of PHA activation, suggesting that CD4+ T cells carrying replication-competent provirus can proliferate with anti-CD3/CD28 stimulation and continue to produce virus for at least 7 d. However, no viral outgrowth was observed in cultures set up with cells that had proliferated in response to IL-7 unless PHA and irradiated allogeneic PBMCs were added (Fig. 1F). This result suggests that the concentrations of cytokines used in this culture assay were sufficient to induce proliferation of some resting CD4+ T cells without latency reversal. The progeny cells could produce virus with additional stimulation. We have previously shown that CD4+ T cells harboring replication-competent provirus can proliferate in response to global T cell activation without producing virus while retaining the ability to do so upon subsequent stimulation (18, 37). Thus, both antigen and cytokines provide a potential explanation for the presence of expanded cellular clones in the latent reservoir.
Some Expanded Clones Harboring Replication-Competent Virus Persist Over Time, While Others Wax and Wane.
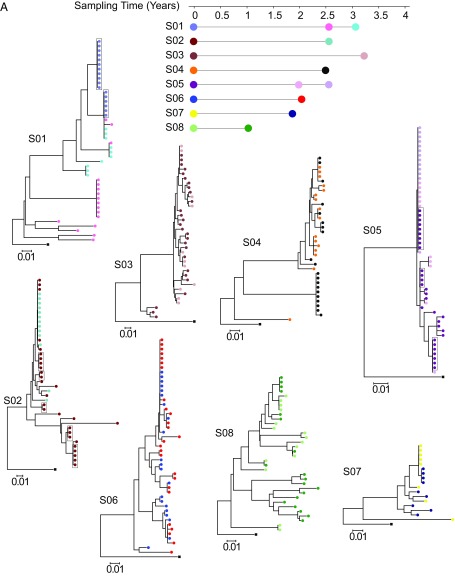
Three recent studies showed that expanded cellular clones containing replication-competent HIV-1 are common in infected individuals (17–19). However, little is known about the dynamics of these clonal populations. To examine whether these clones persist, we recovered the infectious virus from eight treated individuals at two or three time points spanning 2–3 y (Fig. 2A). Limiting dilutions of resting CD4+ T cells were subjected to stimulation in the QVOA with the T cell mitogen PHA and γ-irradiated allogeneic PBMCs. MOLT-4/CCR5+ cells were added on day 2 of the culture to expand virus released from cells in which latency was reversed, and the culture supernatants were tested for virus production by p24 ELISA on day 14 (42). We amplified the highly variable V3–V4 region of the env gene by reverse transcriptase (RT)-PCR from viral RNA in the supernatants of all p24+ wells from the QVOA. As expected from the limiting dilution format of the QVOA, sequences from individual p24+ wells should represent independent isolates of replication-competent virus. Sequences from each individual were compared by phylogenetic analysis. All eight individuals had one or more sets of independent isolates with identical env sequences at two or three time points (Fig. 2A). To determine whether these isolates with identical env sequences were identical throughout the genome, a previously described clonal prediction score was used (43). The env amplicon had a clonal prediction score of 96, indicating that 96% of the sequences identical in this region are identical throughout the entire HIV-1 genome. In addition, we previously established using full-genome sequencing that a subset of these sequences obtained at the first time point were identical throughout the entire HIV-1 genome (18) (Fig. 2A). Phylogenetic analysis established that identical proviruses in these patients reflect expanded cellular clones rather than infection of a large number of cells by a dominant viral species (18). Therefore, each set of identical env sequences is very likely to represent a clonal population of infected cells derived from a single initially infected cell by extensive in vivo proliferation.
Fig. 2.
Expanded clones carrying replication-competent HIV-1 emerge and wane over time. (A) Phylogenetic trees of env sequences of independent isolates of replication-competent virus from eight subjects on ART (S01–S08) are shown. Sequencing was performed on genomic viral RNA in supernatants of p24+ wells. Different colors correspond to viruses recovered from different time points as indicated under the time line. Groups of identical sequences are indicated by symbols present on the same vertical “rake.” Sequences for the first time point were included in a previous study (18). Sequences that were previously shown to be identical by full-genome sequencing are grouped in boxes (18). The time scale indicates time in years from study entry. All patients were on suppressive ART for >6 mo before study entry. Black squares indicate the reference sequence HXB2. (B) Dynamics of expanded clones containing replication-competent HIV-1. Each pie figure shows how all of the replication-competent viruses (n) collected at a specific time point (shown on the x axis) are divided into clonal populations, with distinct colors representing different clones. Clones marked by M were identified at multiple time points. Starred lines indicate samples that are significantly different according to a test for difference in clone proportions when the null model is a random partition of the aggregated samples (Materials and Methods) (*P < 0.05; **P < 0.01; ***P < 0.001. NS, P > 0.05).
Longitudinal sampling over a time span of 1–3 y allowed us to address the question of whether clones harboring replication-competent virus persist over time. In seven of eight individuals, we observed sequences that were present and prevalent at one time point at other time points (Fig. 2 A and B), suggesting some cellular clones persisted on a time scale of years and comprised a substantial fraction of the total population of resting CD4+ T cells with replication-competent proviruses over these time periods. However, in seven of eight individuals, we also found clonal populations carrying different replication-competent viruses at time point 2 and time point 3 that were not present at time point 1 (Fig. 2 A and B). In addition to the appearance of new clones, we observed the disappearance of other clones. In subject S01, one large expanded clone was only observed at time point 1 and then disappeared, while another expanded clone appeared and was only seen at time point 2 (Fig. 2 A and B). The same dramatic appearance and disappearance pattern was observed in subject S04 (Fig. 2 A and B). Of all of the sequences collected, 65.6% were seen only at one time point. Of 17 clones observed at two or more time points, 10 clones showed an increasing frequency over time, while seven clones decreased in frequency over time. Overall, these results are consistent with a homeostatic process in which individual clones increase and decrease in frequency, while the size of the total pool of latently infected cells decreases only very slowly (1, 7, 8).
Due to the low frequency of cells harboring inducible replication-competent proviruses, the number of infected cells sampled at any given time point is limited. We therefore analyzed whether a difference in observed clone frequencies between time points within the same patient could be attributed to sampling error alone (Fig. 2B). In most comparisons between pairs of time points, the observed difference in clone frequencies was highly significant, supporting the notion that the difference in clone frequencies cannot be accounted for by sampling alone (Fig. 2B). These findings demonstrate that clonally expanded cells harboring replication-competent virus wax and wane on a time scale of years. Our results do not support a process of continuous expansion.
Clonal Populations of Free Plasma Virus Change Over Time.
To provide further insight into the dynamics of expanded cellular clones carrying replication-competent HIV-1, we analyzed residual viremia in patients on suppressive ART over time. Most of the defects found in HIV-1 proviruses are major defects that would preclude the production of virus particles (37, 38, 44). Therefore, the dynamics of the cells that produce the residual viremia are likely to reflect the dynamics of cells that carry replication-competent proviruses. However, analysis of residual viremia provides additional information because virus production requires latency reversal.
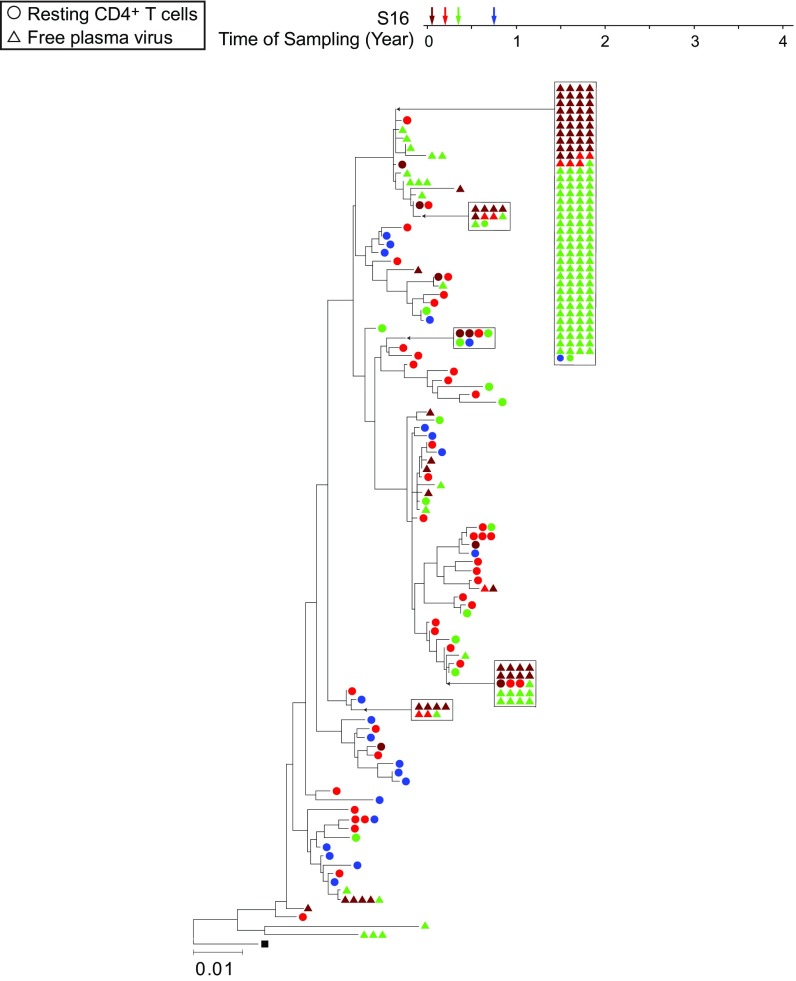
Fig. 3 shows phylogenetic analysis of env gene sequences from plasma virus and from proviruses in resting CD4+ T cells from a representative patient on ART. This phylogenetic tree illustrates previously described features of residual viremia, including an intermingling of plasma and cellular sequences (12), the lack of temporal structure (degree of divergence is not correlated with time of sampling) (45), the presence of PPCs (12), and a lack of correlation between the frequency of clonal sequences in plasma and resting CD4+ T cells (12). All of these features are consistent with the hypothesis that a stable reservoir of HIV-1 in resting CD4+ T cells contributes to the residual viremia as cells in the reservoir become activated. One large clonal population was detected at time point 1 and persisted through time point 3, which occurred 5 mo later, but very few matching proviral sequences were found. The lack of correlation between the frequency of clonal sequences in plasma and resting CD4+ T cells reflects the fact that most of the proviruses in resting CD4+ T cells are defective and incapable of producing plasma virus (37, 38). Therefore, extensive sampling is required to find the matching proviral sequences. The presence of a large clonal population in plasma reflects not only the proliferation of a clone of infected cells but also the activation of at least some of those cells, presumably by some antigen, to a degree that reverses latency.
Fig. 3.
Neighbor-joining phylogenetic tree constructed with env sequences from the plasma and resting CD4+ T cells from subject S16. Samples were taken at the indicated times after study entry while the plasma HIV-1 RNA level was <50 copies per milliliter. Samples were processed for analysis of HIV-1 RNA in plasma (triangles) and proviral DNA in resting CD4+ T cells (circles).
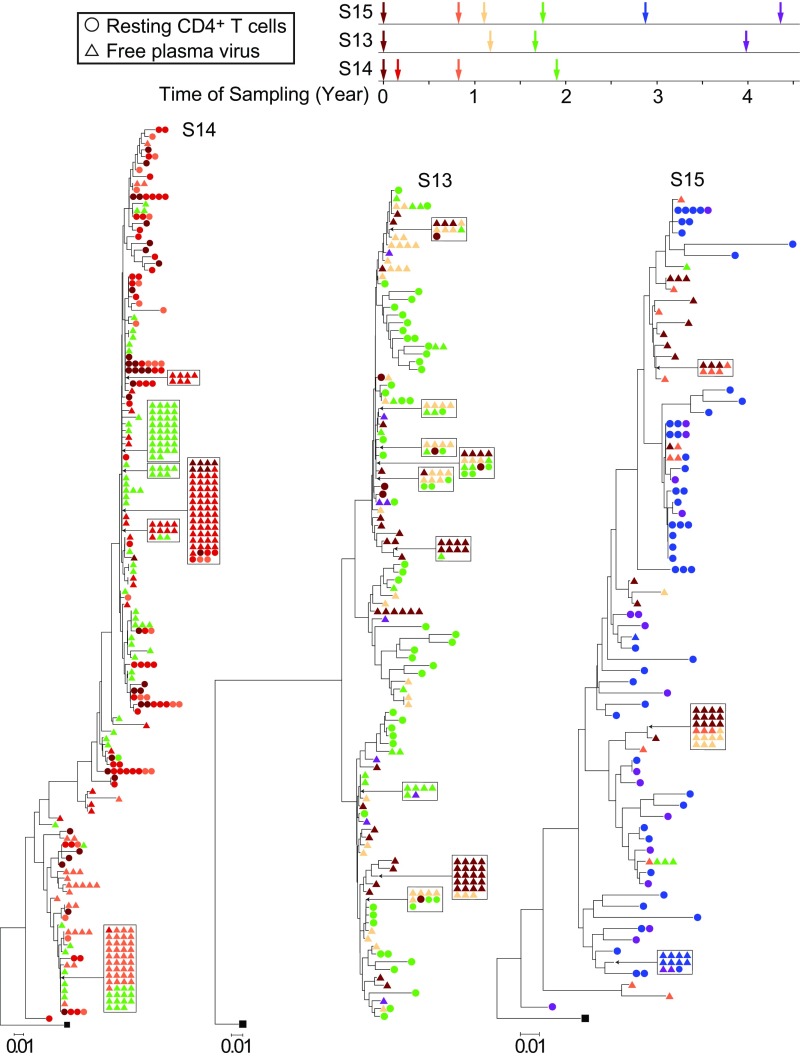
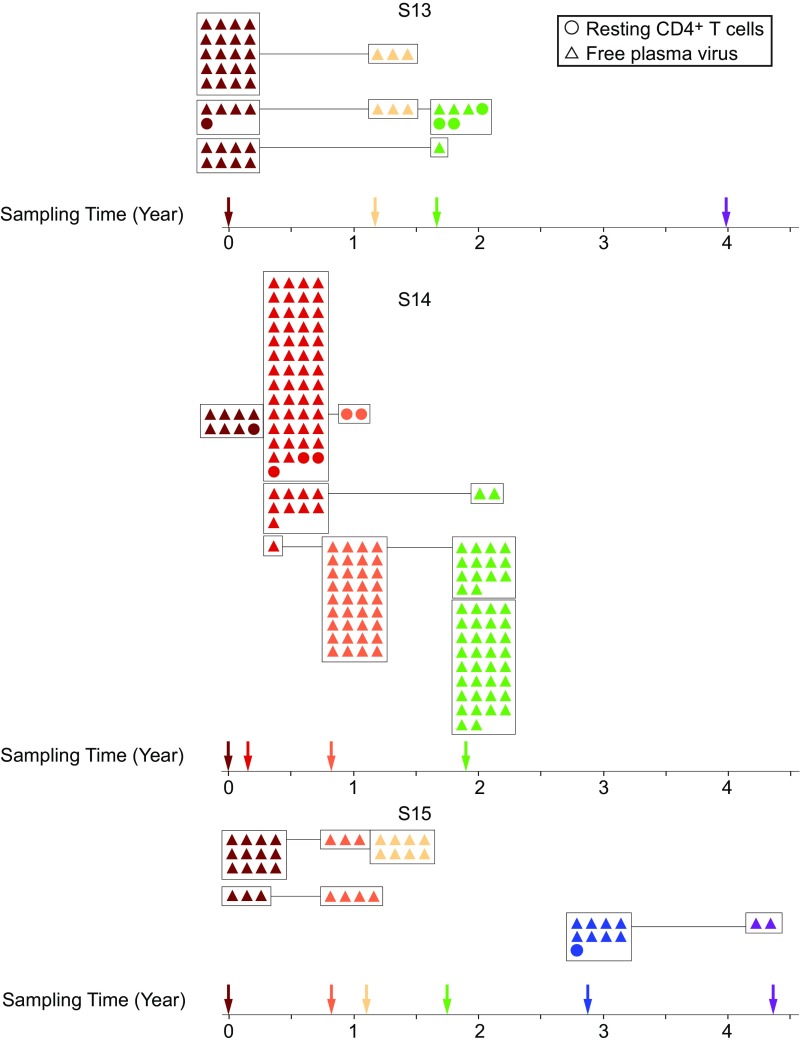
In four of eight patients sampled, we found dominant plasma virus populations, as evidenced by the identical viral sequences obtained in multiple, independent, single-genome amplifications. Interestingly, we observed the same pattern of appearance and disappearance of these plasma virus clones as we observed with proviruses in resting CD4+ T cells (Fig. 4). Subject 14 was studied at four time points over a 2-y period. We identified one large clonal population that was first detected at time point 2, became dominant at time point 3, and persisted through time point 4, which occurred 1.2 y later (Figs. 4 and 5). Another distinct clonal population was dominant at time point 2 but was not seen in the plasma at later time points. Another distinct clonal population of plasma virus was identified only at the last time point in this patient. This study participant also had populations of clonal proviruses from resting CD4+ T cells. However, we only found matching proviral sequences for one of the clonal plasma viruses (Fig. 4), consistent with the fact that the vast majority of proviral sequences are defective. These results show that clonal populations of infected cells producing residual viremia wax and wane on a time scale of years. This likely reflects changes in the frequency of individual clones, as indicated by viral outgrowth studies (Fig. 2). This, in turn, may reflect changes in the exposure to cognate antigens that could drive both proliferation and virus production.
Fig. 4.
Plasma virus clones wax and wane over a time scale of years. Neighbor-joining phylogenetic trees constructed with env sequences from the plasma and resting CD4+ T cells from subjects S14, S13, and S15. Samples were taken at the indicated times after study entry while the plasma HIV-1 RNA level was <50 copies per milliliter. Samples were processed for analysis of HIV-1 RNA in plasma (triangles) and proviral DNA in resting CD4+ T cells (circles).
Fig. 5.
Composite graph illustrating the appearance and longevity of clonal populations of free plasma virus and provirus derived from resting CD4+ T cells. Colors correspond to time of sampling and are derived directly from the corresponding phylogenies.
In subject 13, we sampled plasma viruses at four points over a 4-y period (Figs. 4 and 5). We observed multiple distinct clonal populations of plasma viruses, the abundance of which changed over time. In subject 15, we sampled plasma viruses at six time points over a 4-y period (Fig. 4). Three distinct clonal populations of plasma viruses were identified. Two populations were observed at earlier time points but not after the third time point. Another population was only observed at time point 5 and time point 6 (Figs. 4 and 5). The pattern of emergence and disappearance of clonal populations of plasma virus is summarized in Fig. 5. This pattern is consistent with our finding that clonal populations of replication-competent provirus wax and wane over time and supports a model of release of virus from clonally expanded cells in the reservoir.
Discussion
The stable latent reservoir for HIV-1 in resting CD4+ T cells is a major barrier to cure (1). Proliferation of infected cells could explain the stability of the latent reservoir. However, some mechanisms that drive proliferation of infected cells also induce proviral expression so that infected cells can be eliminated by immune mechanisms or die from viral cytopathic effects (20, 21). Despite the fact that productively infected cells have a very short half-life (22, 23), accumulating evidence suggests that infected cells harboring replication-competent virus can undergo clonal expansion in vivo (16–19). Sequencing of plasma virus in subjects on ART provided additional evidence for clonal expansion of cells carrying replication-competent virus, as residual viremia is often dominated by identical viral sequences (11, 12). There is great current interest in understanding the stimuli driving clonal expansion.
Studies in a model system suggested that infected cells can proliferate in response to IL-7 and IL-2 without viral reactivation (24). Therefore, we examined whether infected cells carrying replication-competent virus from patients on ART could expand through cytokine-driven homeostatic proliferation and whether clonally expanded populations persist over time in vivo. Using patient samples, we demonstrate here that infected cells harboring replication-competent HIV-1 can proliferate through TCR or cytokine-driven mechanisms. We show that cells carrying replication-competent HIV-1 can proliferate in response to cytokine treatment. They can do so without producing infectious virus, while retaining the ability to produce virus following a subsequent stimulation. It is possible that IL-7 could induce latently infected cells to express low amounts of viral antigen even if the cells do not go on to produce infectious virus. It is worth noting that the level of proliferation induced by cytokine treatment is related to IL-7 receptor expression. The IL-7 receptor α-chain (CD127) is expressed at higher levels in central memory and transitional memory CD4+ T cells than in effector memory CD4+ T cells (13, 39, 46). We have previously shown that latently infected cells carrying replication-competent proviruses can also proliferate in response to mitogen stimulation without producing virus (18). These findings are consistent with the observation of clonal expansion in vivo and help explain the large number of latently infected cells carrying identical intact viral sequences (16–19).
Although it is clear that the latent reservoir is dominated by clonal populations, the dynamics of these populations have been unclear and could differ from those of clonal populations of infected cells carrying defective proviruses. We show here that some clonal populations of resting CD4+ T cells carrying replication-competent HIV-1 wax and wane in vivo on a time scale of years, while others persist over the time period examined. We also demonstrate that clonal populations of plasma viruses emerge and disappear on a time scale of years in participants on ART. The observations on plasma virus are consistent with the presence of expanded clones of latently infected CD4+ T cells and show that at least a fraction of the cells are constantly activated to produce virus. The waxing and waning of predominant plasma viral populations are likely a reflection of the underlying proliferative mechanisms that maintain the latent reservoir, as well as changes in the stimuli that drive infected cells to produce virus. The original demonstration of the clonal nature of residual viremia (11, 12) suggested that it is produced by a cell population that is capable of clonal expansion, and the demonstration of expanded clones of CD4+ T cells carrying replication-competent proviruses (16–19) is consistent with the idea that residual viremia reflects virus release from latently infected CD4+ T cells that have become activated. These findings do not exclude the possibility that virus also persists in other reservoirs, some of which (e.g., macrophages, microglial cells) are not known to undergo extensive clonal expansion (47, 48).
Overall, these results support the hypothesis that the latent reservoir in CD4+ T cells is maintained by clonal proliferation. This is important, as eradication of HIV-1 infection necessitates elimination of cells carrying replication-competent latent proviruses, and the potential of these cells to undergo enormous clonal expansions makes eradication more challenging. Therefore, it is important to determine the mechanisms responsible for clonal expansion of cells carrying replication-competent proviruses. The observation that expanded clones wax and wane over time suggests that proliferation of infected cells is balanced by a significant amount of cell loss. The dynamic changes support antigen and cytokines as potential drivers of clonal expansion, but they do not support a cell-autonomous proliferation driven by effects related to the site of integration, as such a mechanism would be more likely to result in progressive expansion.
Materials and Methods
Study Subjects.
Participants were HIV-1–infected individuals who had suppression of viremia to less than 20 copies of HIV-1 RNA per milliliter on ART for more than 6 mo. The Johns Hopkins Institutional Review Board approved this study, and written informed consent was obtained from all subjects.
Resting CD4+ T Cell Isolation and Plasma Sample Processing.
A total of 180 mL of blood was collected at each study visit using an acid-citrate-dextrose anticoagulant and processed as previously described (41). Briefly, plasma and cells were separated using a Ficoll density gradient. The plasma layer was quickly removed, centrifuged to remove any contaminating cells, and immediately frozen and stored at −80 °C until further use. The buffy coat layer was subsequently removed, and resting CD4+ T cells were purified from total PBMCs via magnetic bead depletion as previously described (42).
Cell Culture Conditions.
Approximately 10 × 106 cells were cultured untreated or treated with anti-CD3/CD28 Dynabeads (25 μL per million cells; Thermo Fisher Scientific) + IL-2 (30 U/mL) or with IL-7 (10 ng/mL; BioLegend) + IL-2 (30 U/mL) in RPMI plus 10% FBS for 7 d in the presence of tenofovir disoproxil fumarate (10 μM) and emtricitabine (10 μM).
TCR Sequencing.
Cells proliferating in response to anti-CD3/CD28 or IL-7 were sorted using a Sony SH800 cell sorter based on CFSE dilution (discussed below). DNA was extracted from aliquots of resting CD4+ T cells and sorted cell populations (Qiagen). Adaptive Biotechnologies performed TCR sequencing on extracted DNA.
CFSE Dilution and Activation Marker Staining.
Total resting CD4+ T cells were stained with 5 μM CFSE (Life Technologies) before stimulation with anti-CD3/CD28 or IL-7 as described above. The dilution of CFSE was analyzed 1 wk later by flow cytometry on a FACSCanto II cytometer (BD Biosciences). Unstimulated resting CD4+ T cells cultured for the period of time served as a negative control. Expression of activation markers was analyzed 1 wk after initial stimulation. An aliquot of cells from each culture was stained with anti-CD25 (PerCP-cyanine5.5), anti-CD69 (allophycocyanin), and anti–HLA-DR (Pacific Blue) antibodies (BioLegend) at 4 °C for 15 min and analyzed by flow cytometry on the FACSCanto II cytometer.
QVOA.
The QVOA was performed on cultured, unsorted, resting CD4+ T cells and on sorted cells that had proliferated in response to anti-CD3/CD28 or IL-7. The cells were plated at a limiting dilution for viral outgrowth (200,000 cells in 2 mL of media per well) (37). Cells were activated with 0.5 μg/mL PHA and γ-irradiated allogeneic PBMCs from healthy donors (1). On day 2, media with PHA were removed and replaced with fresh media, and 106 MOLT-4/CCR5+ cells were added to each well to allow expansion of virus released from cells in which latency had been reversed (42). MOLT-4/CCR5+ cells were obtained from the NIH AIDS Reagent Program. CCR5 receptor expression on MOLT-4/CCR5+ cells was routinely tested by flow cytometry. On day 5, 0.75 mL of media was removed and cells in each well were resuspended. Media were changed on days 5 and 9. A p24 ELISA (PerkinElmer) was performed on the supernatant on day 14 of the culture. Limiting dilution maximum likelihood statistics were used to calculate the frequency of latently infected cells (silicianolab.johnshopkins.edu/) (49).
RNA Isolation, cDNA Synthesis, and Amplification of the env Gene from Proviruses in Resting CD4+ T Cells.
Viral RNA isolation was performed on 200 μL of the supernatant from each p24+ well using a ZR-96 Viral RNA Kit (Zymo Research Corporation). Isolated RNA was treated with DNase (Thermo Fisher Scientific) and utilized for cDNA synthesis using a qScript cDNA Supermix Kit (Quanta Biosciences). We then ran a nested PCR assay on cDNA from each p24+ well targeting the V3–V4 region of env. The nested PCR assay was performed using 500 ng of cDNA and primers ES7 (5′-CTGTTAAATGGCAGTCTAGC-3′) and ES8 (5′-CACTTCTCCAATTGTCCCTCA-3′) for the outer reaction. The outer PCR products were then diluted 1:50, and 5 μL of this diluted outer PCR product was used for the inner PCR with primers Nesty8 (5′-CATACATTGCTTTTCCTACT-3′) and DLoop (5′-GTCTAGCAGAAGAAGAGG-3′). Primers were obtained from Integrated DNA Technologies. Detailed amplification conditions were as follows: 94 °C for 30 s; 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 2 min for 39 cycles; and then 68 °C for 5 min. PCR products were visualized on 1% agarose gels, and bands were extracted using a QIAquick Gel Extraction Kit (Qiagen). Extracted DNA was analyzed directly by Sanger sequencing at Genewiz, Inc. (MG680752–MG680913).
Amplification and Sequencing of the env Gene from Plasma Virus.
Free virus in the plasma of treated patients was analyzed as previously described (12). Briefly, 6-mL aliquots of plasma were thawed and ultracentrifuged at 170,000 × g for 30 min at 4 °C. Pelleted virus was resuspended in 400 μL of PBS (Invitrogen) and lysed, and the RNA was extracted via a silica bead-based RNA isolation protocol implemented on an EZ1 Biorobot (Qiagen). The RNA was eluted in 60 μL of elution buffer and subsequently treated with amplification grade DNase I (Invitrogen) according to the manufacturer’s instructions. To amplify the C2–V4 region of the env gene from RNA isolated from free plasma virus, the RNA was subjected to a one-step RT-PCR assay using a SuperScript III RT/Platinum Taq High-Fidelity DNA polymerase one-step RT-PCR kit (Invitrogen) followed by a nested PCR assay, using Platinum Taq High-Fidelity DNA polymerase, and 2.5 μL of the outer reaction as a template. Control reactions were carried out for all experimental amplifications, including a no-RT control to rule out DNA contamination and a no-template control. Primers for the outer and nested reactions were as follows: (outer forward) 5′-CTGTTAAATGGCAGTCTAGC-3′, (outer reverse) 5′-CACTTC TCCAATTGTCCCTCA-3′, (nested forward) 5′-ACAATGCTAAAACCATAATAGT-3′, and (nested reverse) 5′-CATACATTGCTTTTCCTACT-3′. PCR conditions for the one-step RT-PCR assay were as follows: reverse transcription at 50 °C for 30 min and denaturation at 94 °C for 3 min, followed by 40 cycles at 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 1 min. The PCR conditions for the nested reaction were as follows: denaturation at 94 °C for 3 min, followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 1 min. Products of the nested reaction were separated on 1% agarose gels. Bands of appropriate size were excised, and the corresponding amplicons were eluted using QIAquick Gel Extraction Kits. Isolated amplicons were subsequently cloned using a PCR2.1 TOPO cloning vector (Invitrogen), and at least six clones were sequenced from each PCR assay using an ABI Prism 3700 DNA analyzer (Applied Biosystems). Only sequences that could be shown to be derived from independent templates were analyzed. Phylogenetic analysis by neighbor joining was performed on previously published (12, 50) and novel sequences as discussed below. Novel sequences have been deposited in Genbank. Accession nos.: GQ256402–GQ256627, GQ261350–GQ261724, DQ391282–DQ391351, KF878519–KF878571, KF878848–KF878929, MG680752–MG680913, and MG751469–MG751764.
Phylogenetic Analysis.
A consensus sequence for each sample was generated with forward and reverse sequences using default assembly parameters on CodonCode Aligner software (CodonCode Corporation). Each consensus sequence was aligned with an HXB2 reference HIV-1 sequence from the Los Alamos National Laboratory HIV sequence database using BioEdit software. Aligned sequences were trimmed to the same length for phylogenetic tree generation. Genetic distances were calculated, and neighbor-joining trees (51) were generated using a maximum composite likelihood algorithm and default parameters using MEGA7 software (Molecular Evolutionary Genetics Analysis Program) (52). The conclusions were not sensitive to the method of tree generation, and neighbor-joining trees are shown in the figures.
Statistical Analysis of Clones.
A statistical test was designed to see if changes in the frequency distribution of clones carrying sequence-identical, replication-competent virus were significantly different between time points or if they could be attributed to sampling error alone. Under this null model, samples from two different time points are drawn from the same underlying distribution. Physiologically, this corresponds to a scenario in which the clonal composition of the latent reservoir does not change over time. To construct a null distribution of data for this model, data from two time points were aggregated and repartitioned randomly, while all margins were fixed [implemented using the R package vegan (53)]. To test whether the observed data supported a more extreme difference in proportions than the repartitioned data, the likelihood ratio test statistic was calculated for the observed data and the distribution of repartitioned data under a multinomial model. The likelihood ratio test statistic T was derived by finding the maximum likelihood parameter estimate vector for a multinomial model with the combined clone size data (n1+2) and a multinomial model with separated data (n1, n2), and then calculating their respective likelihoods:
The P values corresponding to the proportion of random partitions with statistic T were at least as large as that of the true observation. This test is equivalent to a Fisher’s exact test when the permuted matrices are ordered by the above statistic.
Acknowledgments
This work was supported by the NIH Martin Delaney I4C (Grant UM1 AI126603), Beat-HIV (Grant UM1 AI126620), and Delaney AIDS Research Enterprise (Grant UM1 AI12661) programs, and by the Howard Hughes Medical Institute and the Bill and Melinda Gates Foundation (Grant OPP1115715).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GQ256402–GQ256627, GQ261350–GQ261724, DQ391282–DQ391351, KF878519–KF878571, KF878848–KF878929, MG680752–MG680913, and MG751469–MG751764).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720665115/-/DCSupplemental.
References
- 1.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, et al. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitney JB, et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 7.Strain MC, et al. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: Intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci USA. 2003;100:4819–4824. doi: 10.1073/pnas.0736332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crooks AM, et al. Precise quantitation of the latent HIV-1 reservoir: Implications for eradication strategies. J Infect Dis. 2015;212:1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deeks SG. HIV: Shock and kill. Nature. 2012;487:439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 10.Archin NM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tobin NH, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: Expression of archival virus and replication of virus. J Virol. 2005;79:9625–9634. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey JR, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chomont N, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simonetti FR, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA. 2016;113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenzi JC, et al. Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc Natl Acad Sci USA. 2016;113:E7908–E7916. doi: 10.1073/pnas.1617789113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosmane NN, et al. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J Exp Med. 2017;214:959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bui JK, et al. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 2017;13:e1006283. doi: 10.1371/journal.ppat.1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scripture-Adams DD, Brooks DG, Korin YD, Zack JA. Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J Virol. 2002;76:13077–13082. doi: 10.1128/JVI.76.24.13077-13082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang FX, et al. IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART. J Clin Invest. 2005;115:128–137. doi: 10.1172/JCI22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho DD, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 23.Wei X, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 24.Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 2011;7:e1002288. doi: 10.1371/journal.ppat.1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dornadula G, et al. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282:1627–1632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 26.Havlir DV, et al. Productive infection maintains a dynamic steady state of residual viremia in human immunodeficiency virus type 1-infected persons treated with suppressive antiretroviral therapy for five years. J Virol. 2003;77:11212–11219. doi: 10.1128/JVI.77.20.11212-11219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer S, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mens H, et al. Amplifying and quantifying HIV-1 RNA in HIV infected individuals with viral loads below the limit of detection by standard clinical assays. J Vis Exp. 2011:2960. doi: 10.3791/2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermankova M, et al. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA. 2001;286:196–207. doi: 10.1001/jama.286.2.196. [DOI] [PubMed] [Google Scholar]
- 30.Persaud D, Zhou Y, Siliciano JM, Siliciano RF. Latency in human immunodeficiency virus type 1 infection: No easy answers. J Virol. 2003;77:1659–1665. doi: 10.1128/JVI.77.3.1659-1665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kieffer TL, et al. Genotypic analysis of HIV-1 drug resistance at the limit of detection: Virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis. 2004;189:1452–1465. doi: 10.1086/382488. [DOI] [PubMed] [Google Scholar]
- 32.Nettles RE, et al. Intermittent HIV-1 viremia (blips) and drug resistance in patients receiving HAART. JAMA. 2005;293:817–829. doi: 10.1001/jama.293.7.817. [DOI] [PubMed] [Google Scholar]
- 33.Gandhi RT, et al. AIDS Clinical Trials Group A5244 team The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: A randomized controlled trial. PLoS Med. 2010;7:e1000321. doi: 10.1371/journal.pmed.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besson GJ, McMahon D, Maldarelli F, Mellors JW. Short-course raltegravir intensification does not increase 2 long terminal repeat episomal HIV-1 DNA in patients on effective antiretroviral therapy. Clin Infect Dis. 2012;54:451–453. doi: 10.1093/cid/cir721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinoso JB, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci USA. 2009;106:9403–9408. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohn LB, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160:420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruner KM, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seddon B, Tomlinson P, Zamoyska R. Interleukin 7 and T cell receptor signals regulate homeostasis of CD4 memory cells. Nat Immunol. 2003;4:680–686. doi: 10.1038/ni946. [DOI] [PubMed] [Google Scholar]
- 40.Sprent J, Surh CD. Normal T cell homeostasis: The conversion of naive cells into memory-phenotype cells. Nat Immunol. 2011;12:478–484. doi: 10.1038/ni.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 42.Laird GM, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskey SB, Pohlmeyer CW, Bruner KM, Siliciano RF. Evaluating clonal expansion of HIV-infected cells: Optimization of PCR strategies to predict clonality. PLoS Pathog. 2016;12:e1005689. doi: 10.1371/journal.ppat.1005689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollack RA, et al. Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe. 2017;21:494–506.e4. doi: 10.1016/j.chom.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nickle DC, et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol. 2003;77:5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rethi B, et al. Loss of IL-7Ralpha is associated with CD4 T-cell depletion, high interleukin-7 levels and CD28 down-regulation in HIV infected patients. AIDS. 2005;19:2077–2086. doi: 10.1097/01.aids.0000189848.75699.0f. [DOI] [PubMed] [Google Scholar]
- 47.Honeycutt JB, et al. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med. 2017;23:638–643. doi: 10.1038/nm.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avalos CR, et al. Brain macrophages in simian immunodeficiency virus-infected, antiretroviral-suppressed macaques: A functional latent reservoir. MBio. 2017;8:e01186-17. doi: 10.1128/mBio.01186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenbloom DI, et al. Designing and interpreting limiting dilution assays: General principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect Dis. 2015;2:ofv123. doi: 10.1093/ofid/ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brennan TP, et al. Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source for residual viremia in patients on antiretroviral therapy. J Virol. 2009;83:8470–8481. doi: 10.1128/JVI.02568-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oksanen J, et al. (2017) Package ‘vegan’. Community ecology package, version, 2:9.