Significance
Next-generation sequencing (NGS) has allowed the comprehensive study of the genome and transcriptome. However, a similarly broad, highly multiplexed, and inexpensive method for proteomics using NGS remains elusive. Here, we describe a phage display-based method using preselected antibodies that are genetically encoded and capable of simultaneous profiling of hundreds of cell-surface targets on cells in culture or singly at low cost and without the need for chemical conjugation to purified antibodies. We use the method to identify cell-surface proteins that change in cancer cells, some of which are coordinately regulated and could lead to new biomarkers and cancer targets.
Keywords: phage display, NGS, cell surface proteomics, biomarkers, leukemia
Abstract
Human cells express thousands of different surface proteins that can be used for cell classification, or to distinguish healthy and disease conditions. A method capable of profiling a substantial fraction of the surface proteome simultaneously and inexpensively would enable more accurate and complete classification of cell states. We present a highly multiplexed and quantitative surface proteomic method using genetically barcoded antibodies called phage-antibody next-generation sequencing (PhaNGS). Using 144 preselected antibodies displayed on filamentous phage (Fab-phage) against 44 receptor targets, we assess changes in B cell surface proteins after the development of drug resistance in a patient with acute lymphoblastic leukemia (ALL) and in adaptation to oncogene expression in a Myc-inducible Burkitt lymphoma model. We further show PhaNGS can be applied at the single-cell level. Our results reveal that a common set of proteins including FLT3, NCR3LG1, and ROR1 dominate the response to similar oncogenic perturbations in B cells. Linking high-affinity, selective, genetically encoded binders to NGS enables direct and highly multiplexed protein detection, comparable to RNA-sequencing for mRNA. PhaNGS has the potential to profile a substantial fraction of the surface proteome simultaneously and inexpensively to enable more accurate and complete classification of cell states.
Next-generation sequencing (NGS) has revolutionized our ability to sensitively and broadly detect and quantify DNA and RNA sequences, even at the single-cell level (1, 2). Although RNA-sequencing (RNAseq) is sensitive and highly multiplexed, mRNA levels do not necessarily correlate with protein abundance (3). Here we describe an approach for multiplexed detection of membrane proteins on cells using genetically barcoded antibody-phage.
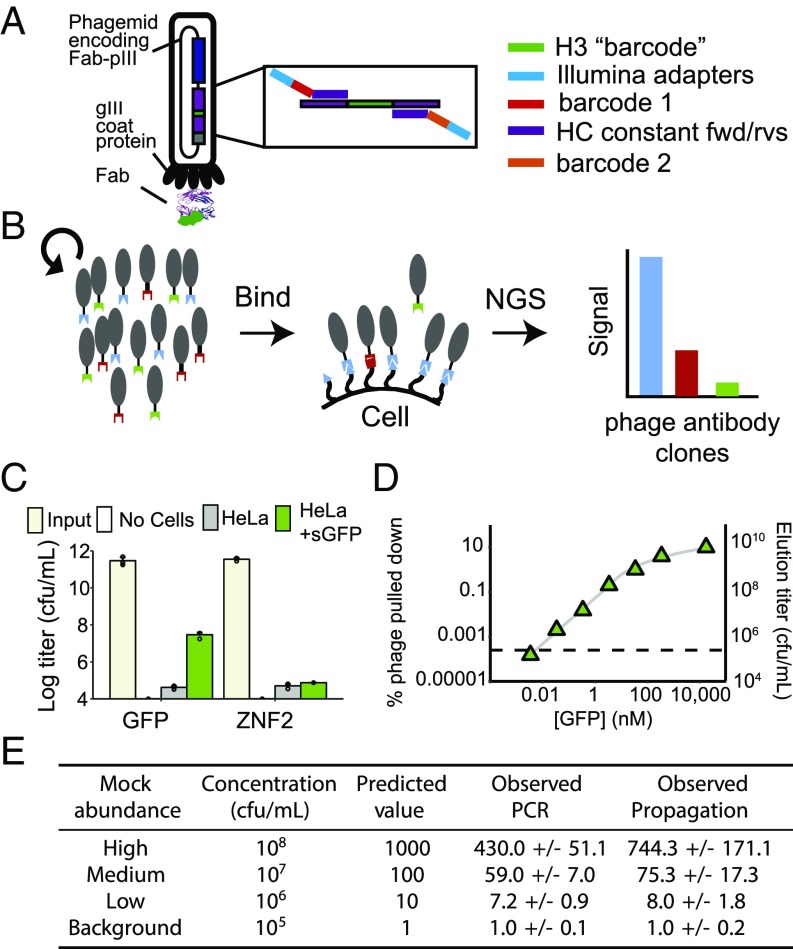
Key to the technology we call phage-antibody NGS (PhaNGS) is a collection of defined fragment antibodies (Fabs) previously selected to bind specific targets of interest using high-throughput phage display (SI Appendix, Fig. S1) (4–6). Each Fab-phage was selected from a large synthetic Fab library (∼1010 unique sequences) built from a stable, VH3-V1-based Trastuzumab scaffold (7) and validated for affinity and selectivity. The selected Fabs are genetically encoded and displayed on a phage particle that packages its specific Fab gene within (Fig. 1A). Taking inspiration from previous work that linked deep sequencing and phage libraries (8), we reasoned that the individual Fab-phage can be distinguished and quantified by NGS of the highly variable complementarity determining region (CDR) H3 loop, which represents both the major binding determinant of the encoded Fab and a unique DNA barcode. The fixed scaffold in which the CDR is embedded allows the use of a common set of primers flanking the H3 region for amplification and sequencing (SI Appendix, Fig. S2) (9). We hypothesized that a pool of preselected Fab-phage specific for a defined set of extracellular proteins would bind their cognate proteins on cells at levels in proportion to receptor abundance and allow for target quantification by NGS (Fig. 1B). The displayed Fab is attached to the phage on the opposite end from its antigen binding site and is known to retain virtually the same affinity for its target as when expressed as soluble Fab (SI Appendix, Fig. S3) (10).
Fig. 1.
Design and validation of the PhaNGS method. (A) The Fab is fused to the gene III coat protein on the M13 phagemid within the viral particle. Each phage antibody is preselected for binding to specific proteins and can be uniquely identified by the sequence of its CDR H3 (green bar). (B) Diagram of the PhaNGS method, which consists of three steps: (i) A collection of phage antibodies is assembled that bind to specific targets of interest; (ii) the library is bound to a cell sample, and nonbinding phage antibodies are washed away; and (iii) the bound phage antibodies are propagated, amplified, and subjected to next-generation DNA sequencing to quantify the retained phage antibodies. (C) One antitarget (GFP.01, n = 3) and one control phage (ZNF2.01, n = 3) were profiled against a HeLa line stably overexpressing GFP tethered to the cell surface (green bar), along with its parental line (gray bar) and a no-cell control. Error bars show SD of three replicates. Input titer for GFP/ZNF2 phage was measured at 3 × 1011/4 × 1011 cfu/mL. The phage titers after binding and washing were 3 × 107/8 × 104 cfu/mL for HeLaGFP, 4 × 104/5 × 104 for the HeLa cell control, and undetectable for the no-cell control. (D) To investigate the limit of detection of PhaNGS, a single, strong phage (Kd = ∼0.5 nM at 2 × 1011 cfu/mL) was panned against varying concentrations of immobilized GFP (green triangles) or BSA alone (dashed line), acid eluted, and titered. Results are shown below the graph. A total of 3.4 pM of GFP was indistinguishable from BSA, however 34 pM appeared to be above the detection limit, which we approximate here at a titer of ∼106 cfu/mL phage at around 10 pM GFP. To obtain a value for the fraction of infective phage actually expressing a Fab, the same data are displayed as fraction of phage pulled down. We see the fraction level off at around 0.1, suggesting that 10% of GFP.01 phage actually display a Fab. (E) A mock phage mixture was assembled from four different Fab-phage clones with concentrations ranging from 105−8 cfu/mL, a 4-log range expected to match the range present on cells (14) and expected phage titers from above, to examine the accuracy and dynamic range of the PhaNGS method. Propagating the phage mixture to saturation in culture followed by PCR was more accurate than direct PCR without propagation.
Results
Validation of the PhaNGS Method in Model Experiments.
To assess the feasibility of the approach, we used simple phage titering to measure binding of an anti-GFP Fab-phage (GFP-phage) to HeLa cells engineered to express a membrane-anchored GFP or parental (Fig. 1C). After binding and washing, we found 400 times more phage on the GFP-expressing cells than control cells. A control Fab-phage directed to the intracellular transcription factor ZNF2 showed similar low-titer binding to either cell line.
To assess the sensitivity of detection of GFP-phage, we immobilized varying concentrations of GFP on beads ranging from about 2 pM to 20 M. Eluted phage titers were linear over a 4-log range and showed detectable signal over background (defined as BSA binding) with as little as 30 pM of immobilized GFP (Fig. 1D). We also observed that the fractional recovery of GFP-phage peaked at 0.1, implying that about 10% of total phage particles for this clone display a Fab. This is consistent with earlier estimates of Fab display in monovalent phage format (11).
We next investigated the most efficient means of amplifying phage DNA for NGS over a 4-log range of concentration, either by direct PCR or by first propagating the Fab-phage in Escherichia coli to saturation followed by PCR (Fig. 1E). We found that propagation and PCR more closely matched the expected results than PCR alone, most likely because fewer rounds of PCR introduce less amplification bias (12). Bradbury and coworkers have shown that polyclonal pools of scFv-phage have similar replicative rates independent of the scFv (13). We tested this hypothesis for a pool of 155 Fab-phage clones with roughly equal starting titers and found little replicative advantage in the pool based on NGS counts (SI Appendix, Fig. S4). Indeed, after multiple rounds of propagation from 105 cfu/mL to 1011 cfu/mL, we saw only two clones with an obvious replicative advantage over the other Fab-phage in the pool.
Assembly and Validation of the PhaNGS Library.
We then assembled a larger pool of Fab-phages, which grew to encompass 179 clones preselected for high affinity and selectivity to bind 59 purified ectodomain targets broadly associated with cancer and immunology (SI Appendix, Fig. S5, and Dataset S1). We decided to include multiple Fab-phage per target to provide independent measures and to test if certain epitopes were more responsive than others. In the pool, we had an average of four unique Fab-phage per target with a range of one Fab-phage for some targets to up to 13 for others. The Fab-phage pool also includes ZNF2- and GFP-phage background controls, against which the raw NGS counts obtained from antisurface protein Fab-phages can be normalized to determine signal and define nonspecific binding (see Dataset S3).
To test this assembled PhaNGS library, we first profiled HEK293T cells, a standard human expression host derived from human embryonic kidney cells (SI Appendix, Fig. S6). We found that most Fab-phage in the pool showed comparable signal to the nonspecific controls, while about a dozen targets gave significant signal over background, including the tyrosine kinase receptors EGFR, EPHA4, FGFR4, FLT3, INSR, ROR1, and TYRO3. To determine how overexpression affects Fab-phage signal strength, we generated a tetracycline (Tet)-inducible construct for CDCP1, one of the targets basally expressed at background (SI Appendix, File S2). Upon induction with Tet, the NGS signal for each of three CDCP1-phages in the pool increased dramatically to between 50- and 80-fold above background, with small SDs between replicates. In other control experiments with GFP-phages of known affinity, we found that the NGS signals vary in rough proportion to affinity (SI Appendix, Fig. S7). Thus, we expect signal variation between unique Fab-phages to the same target is due to affinity differences.
PhaNGS Profiling of Diagnosis-Relapse Samples in Leukemia.
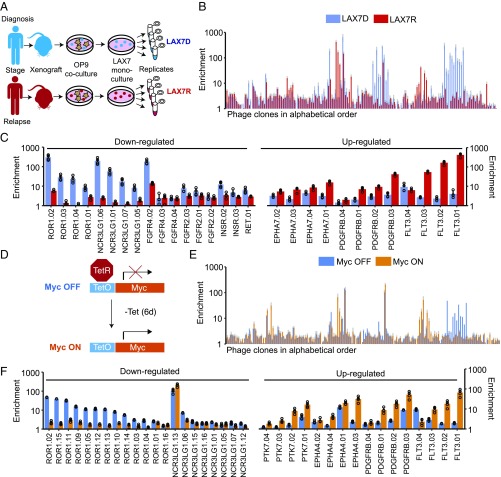
We next applied the pool of Fab-phages to profile B cells from an ALL patient before and after chemotherapy. Samples were obtained from the bone marrow of the patient at diagnosis (LAX7D) and after the development of resistance (LAX7R) following a standard 3-wk chemotherapy regimen (see Fig. 2A and Materials and Methods for sample details) (15). PhaNGS profiles on LAX7D and LAX7R were performed in quadruplicate and showed a 1,000-fold signal range (Fig. 2B). Out of the 144 antitarget phages used in this experiment, 125 showed significant signal above background in either the LAX7D or LAX7R conditions. These phages fell into several groups: About one-third of the Fab-phages showed little change in NGS signal between the LAX7D and LAX7R cells, suggesting little net change in target expression. The remaining clones fell into two groups (Fig. 2C): one where the target protein was high at diagnosis and decreased up to 10-fold in relapse (such as for ROR1 and NCR3LG1), and another where the target was low at diagnosis and increased up to 100-fold at relapse (such as for FLT3 and PDGFRB). FLT3 has previously been observed to be overexpressed and/or mutated in ALL and acute myeloid leukemia (AML) (16, 17). ROR1 also represents a major target of interest in ALL, chronic lymphocytic leukemia (CLL), and other leukemias (18–20). Despite the variation in NGS signal between Fab-phage against the same target, the ratio of the average fold-change between the LAX7D and LAX7R states for a family of antibodies was consistent, providing additional confidence in the changes observed (SI Appendix, Fig. S8A). The variation in relative NGS signal for antibodies to the same targets is likely due to signal suppression for the weakly bound antibodies, since signal-to-noise is less.
Fig. 2.
PhaNGS profiling of surface proteomes at diagnosis and relapse in a patient with acute lymphoblastic leukemia (ALL). (A) Bone marrow samples obtained from a patient diagnosed with ALL (Ph-negative) at diagnosis (LAX7D) and relapse (LAX7R) were grown as xenografts into immune-compromised NOD/SCIDc–/– mice, cocultured with OP9 cells, and later frozen as monoculture stocks. Samples were thawed and expanded in culture 1 wk before the PhaNGS profile experiment. Both cell populations were positive for CD10, CD19, and CD45. The LAX7R resistance sample possessed a KRASG12V mutation not detected at diagnosis. (B) PhaNGS profiles for 144 different Fab-phages (SI Appendix, Datasets S1 and S3) directed to 44 different membrane targets were allowed to bind to LAX7D (blue) or LAX7R (red) cells. The average value from four replicates, with SD (gray bars), is shown. (C) Targets are shown that were down- or up-regulated from LAX7D to LAX7R. (D) Experimental scheme for the P493-6 cell line in MycOFF and MycON conditions. Myc was repressed for 48 h with the addition of Tet (100 ng/mL, twice per day). The MycOFF state was harvested, Tet was washed out, and the cells recovered for 6 d before the MycON condition was harvested. (E) The extended bar chart displays the results of the PhaNGS profiling for the MycOFF to MycON experiment (blue and orange bars, respectively). The average value from four replicates, with SD (gray bars), is shown. (F) Targets are presented that were down- or up-regulated when transitioning from MycOFF to MycON.
We also performed RNAseq on the LAX7D and LAX7R samples and identified 11 transcripts for which targets were also present in the PhaNGS dataset (SI Appendix, Fig. S9). There was a weak correlation in the fold-change for the common targets (Pearson correlation coefficient, R = 0.17), possibly reflecting differences in receptor translation, trafficking, and stability. Such discrepancies between protein and RNA levels for mammalian cytosolic proteins have been reported and highlight the need for direct cell-surface protein quantitation (3).
PhaNGS Profiling of Myc-Induced Surface Proteomic Changes.
Oncogenes are known to induce significant changes in gene protein expression. Myc shows especially strong perturbation in expression profiles (21). We wished to use PhaNGS to explore how Myc expression alters the expression of surface targets in our library. We used a model B cell line, P493-6, that has been used to mimic Burkitt lymphoma (22). In these cells, Myc is expressed at high levels but can be repressed by addition of Tet. We cultured these cells, then repressed Myc expression by treating with Tet for 2 d to generate the OFF state (Fig. 2D). The repression of Myc stalled cell growth without inducing apoptosis and led to subtle changes in morphology, as reported (23). Cells were allowed to recover in the absence of Tet for 6 d to re-express Myc, during which time cell growth and morphology returned to that of the ON state. PhaNGS profiles were then generated for cells from the OFF and ON states (Fig. 2E). The general profile for the P493-6 B cells looks remarkably similar to that for the patient-derived ALL cells (Fig. 2B). Myc re-expression lead to down-regulation of NCR3LG1 (for most clones) and ROR1 and up-regulation of FLT3, PDGFRB, and PTK7 (Fig. 2F), the same pattern of expression changes seen between the diagnosis and relapse ALL cells (Fig. 2C and SI Appendix, Fig. S10). The fact that many of the same proteins exhibit similar changes in both the LAX7 and P493-6 experiments suggests that among these B cell subtypes, particular groups of receptors dominate the response to these perturbations, as has been seen for EFG stimulation in other cell types (24). It is noteworthy that the PhaNGS profile for the HEK293T cells is considerably different from the profile seen for the B cells, which suggests PhaNGS can be used as a sensitive probe of cell type.
Comparison of the PhaNGS Method to Mass Spectrometry and Flow Cytometry.
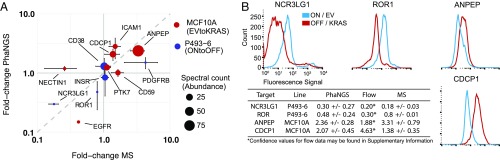
We next compared fold-change values observed for PhaNGS targets to those collected using more traditional protein detection methods. We used SILAC-based cell-surface capture (CSC) mass spectrometry to profile the fold-changes in cell-surface proteins in P493-6 cells induced by Myc repression or in MCF10A cells induced by KRASG12V and compared 12 common targets to those observed in PhaNGS experiments on the same cells (Fig. 3A). We found a good correlation (R = 0.66) despite the sparse overlap from the small target set in the PhaNGS pool and detection of mostly abundant glycoproteins in the CSC experiments. We also expressed and purified two Fabs identified from the PhaNGS experiments that were highly responsive in the Myc-inducible experiment (NCR3LG1 and ROR1) and two that were induced in the KRASG12V-transformed MCF10 cells (ANPEP and CDCP1). All four targets showed the same directional change and roughly the same fold-change by flow cytometry and PhaNGS (Fig. 3B).
Fig. 3.
Comparison of PhaNGS to established proteomic methods. (A) Comparison of fold-changes in surface expression of indicated surface proteins pre– and post–2-d suppression of Myc in P493-6 B cells or for empty vector to KRASG12V transformation of MCF10A cells as assessed by PhaNGS and Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) mass spectrometry. Dot size indicates spectral counts for mass spectrometry experiments to imply the abundance of each protein. Identity (y = x, gray dashed line) is shown as a benchmark for perfect agreement between MS and PhaNGS. R = 0.66 (regression line not pictured, y = 0.98x0.62). Where applicable, error bars for PhaNGS fold-change represent SD derived from unique Fab-phages against the same target. (B) Flow data corresponding to mass spectrometry vs. PhaNGS data for ANPEP, CDCP1, NCR3LG1, and ROR1. The fold-change values observed in the flow cytometry experiments closely match those observed for PhaNGS.
Single-Cell PhaNGS.
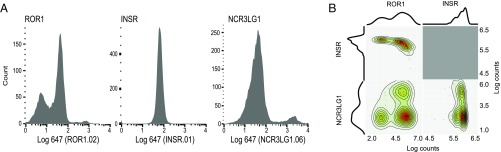
The PhaNGS profiles to this point have been on populations containing 1–10 × 106 cells. Bulk measurements of cancer cell populations can neglect to identify important nuances in cellular heterogeneity revealed by single-cell analysis (25). To extend the reach of the method, we sought to develop single-cell PhaNGS (SI Appendix, Fig. S11A). Here, Fab-phages were added to a population of cells, and then individual cells were flow-sorted into wells and each was profiled by NGS. We tested the single-cell method on HEK293T cells overexpressing cell surface-anchored GFP cells using GFP, CD55, CDCP1, and GHR-Fab phage (SI Appendix, Fig. S10B). For each of the 62 cells tested, we only observed a signal for the GFP-Fab-phage, as expected. We then tested single-cell PhaNGS on P493-6 cells against three targets we found via flow cytometry to exhibit dissimilar distributions of protein abundance (Fig. 4A). Our single-cell PhaNGS experiment found that for 84 single cells, the distributions observed by flow were closely recapitulated by PhaNGS (Fig. 4B).
Fig. 4.
Single-cell PhaNGS on P493-6 cells. (A) Flow cytometry histograms on a population of P493-6 MycON cells (n = 12,000), using purified Fabs for ROR1 (clone ROR1.02, Left), insulin receptor (clone INSR.01, Middle), and NCR3LG1 (clone NCR3LG1.06, Right). Log fluorescence values are indicated on the x axis (anti–FLAG-APC). ROR1 shows bimodal expression with a small low-signal peak and large high-signal peak, INSR shows unimodal expression, and NCR3LG1 shows bimodal expression with a large low-signal peak and small high-signal peak. (B) Results from single-cell PhaNGS using the corresponding ROR1, INSR, and NCR3LG1 Fab-phage antibodies on 84 individual P493-6 cells match observations from flow cytometry.
Discussion
We believe that PhaNGS represents an important advance in barcoded antibody technology. This initial PhaNGS demonstration using ∼150 antibodies is to our knowledge the largest simultaneous use of defined antibodies yet published, surpassing the theoretical limit for fluorophores using immunofluorescence (26), mass tags using CyTOF (27–30), or what has thus far been achieved using oligonucleotide-based barcodes (31). Moreover, we estimate a fixed cost of about $2 per replicate for each PhaNGS profile because the Fab-phages are genetically encoded and renewable (SI Appendix, Table S1). This compares with expensive and perishable antibody reagents needed for probe conjugation using other technologies. Although the PhaNGS experiments presented here were conducted with fewer than 200 unique Fab-phages, we estimate that PhaNGS could profile a target multiplicity of tens of thousands based on experiments using dilute input libraries.
Given the growing interest in the development of renewable antibodies (32) and the ability to industrialize their selection (5), we believe the PhaNGS technology will undergo tremendous growth in the size of the probe library in the next few years. Moreover, the application of microfluidic technology to the method will increase its use for single-cell analysis and could allow for simultaneous RNAseq-PhaNGS experiments. The method may also be amenable to alternative display systems such as ribosome display (33). Having identified Fab-phage hits, one can easily transition the recombinant antibody into a multitude of detection or bioengineered formats such as antibody–drug conjugates (ADCs), bispecific T-cell engagers (BiTEs), or chimeric antigen receptor (CAR) T cells. We believe the PhaNGS method will have general utility to profile how cell-surface proteins change in health and disease. Such data will be useful for identifying new combinatorial biomarkers and drug targets.
Materials and Methods
Patient Samples.
Patient-derived leukemia samples were collected with informed consent from all participants according to NCI/Cancer Therapy Evaluation Program-approved protocol ECOG E2993T5 and studied with approval of the Institutional Review Boards of the University of California San Francisco (UCSF). The samples were taken as bone marrow biopsies, blast content >80%.
Preparing Phage Input Libraries.
The PhaNGS input pool was constructed as follows: 5 L of each clone was transferred to respective wells of a 96-well round-bottom plate (Corning). Then, 100 L log phase XL-1 blue cells (Agilent, OD600 = 0.6–0.7) were added to each well before the plate was covered in a gas-permeable film (Diversified Biotech) and placed at 37 °C for 20 min. We then transferred 100 L of infected cells to respective wells of a 96-well deep-well plate (Corning Axygen) containing 400 L per well 2xYT broth with 100 g/mL carbenicillin and 1010 cfu/mL KO7 helper phage (NEB).
The deep-well plate was covered in a gas-permeable film and shaken at 1000 rpm and 37 °C for 18–24 h in an Infors HT shaker. Plates were spun down at 4,000 g for 15 min at room temperature, and the supernatant was consolidated into 50 mL tubes before adding 0.02% sodium azide and storing at 4 °C. This method leads to approximately equal quantities of each clone from a propagated supernatant (roughly 1011 cfu/mL total).
Panning Phage on Cells.
Cells were washed once (to remove media, DMSO) by spinning the cells down at 300 g for 5 min at 4 °C, pouring off the supernatant into liquid waste, resuspending in 1 mL cold PBS, spinning down, and decanting again. The final drops during decanting were removed by inverting and dabbing the tube on a paper towel. The washed cell pellet was then resuspended in 1 mL of the input phage mixture prepared above. The tube was end-over-end rotated for 20 min at 4 °C before spinning down and decanting as above. Cells were then washed four times with PBS, transferring to fresh 2 mL Eppendorf tubes, and inverting to coat the walls each time. To elute cell-bound phage, the pellet was resuspended in 900 L of 0.1 M acetic acid, pH 2–3, allowed to sit for 5 min, spun down, and 800 L of the acid eluent was transferred to a 1.5 mL Eppendorf tube containing 100 L 1 M Tris, pH 7.5, to neutralize. The neutralized solution was propagated as described above for the input pool or by flask (see SI Appendix). After propagation for 16 to 18 h, 50 L of propagated phage were transferred to a 96-well PCR plate and boiled.
For single-cell experiments, instead of elution, single cells were sorted by forward and side scatter (or fluorescence) into each well of a 96-well plate containing 50 L of 2xYT broth, then propagated via addition of 100 L log phase XL-1 culture.
Amplification and Purification.
The H3 “barcode” of each phagemid was amplified using boiled propagate template and flanking primers using Phusion polymerase (NEB). See Dataset S4 for primer design. The complete mix was then thermocycled for 12 cycles. Those samples showing bands by agarose gel were combined, gel purified, and submitted to a sequencing facility for analysis on a HiSeq4000 (Illumina) with a custom sequencing primer (order as shown): TGAGGACACTGCCGTCTATTATTGTGCTCGC (Tm = 67 °C, GC% = 52).
Mass Spectrometry.
Cell samples were generated by SILAC as described previously (34). Mass spectrometry work-up was performed as described previously (35). Samples were run on a Q-Exactive Mass Spectrometer (ThermoScientific Inc.). Data were analyzed using MaxQuant (36). Median SILAC ratio values were used to determine fold-change values between conditions.
Code Availability.
See https://github.com/sbpollock/PhaNGS-counting for scripts and details on how to convert “.fastq.gz” sequencing files into counts. See Dataset S3 for details on how these counts are interpreted.
Statistics.
Error bars represent SD, which was calculated using Excel’s STDEV.P function (see Dataset S3). Student’s t test was performed using Excel’s T.TEST function (two-tailed, homoscedastic).
Data Archival.
The sequencing data that support the findings of this study are available in the Gene Expression Omnibus (GEO) with the identifier GSE102712 for PhaNGS data and GSE102301 for RNAseq data. All other data supporting the findings of this study are available within the SI Appendix, Dataset S3.
Supplementary Material
Acknowledgments
We thank J. Diaz for HeLaGFP cells, I. Lui for help in preparing single-cell samples for sequencing, and S. Fodor and Z. Hill for constructive comments on the manuscript. This work used the Center for Advanced Technology at UCSF. This work also used the Vincent J. Coates Genomics Sequencing Laboratory at University of California, Berkeley, supported by NIH Instrumentation Grant S10 OD018174. This work was supported by NIH Grants R01CA191018 and P41CA196276 and the Harry and Dianna Hind Professorship in Pharmaceutical Sciences (to J.A.W.) as well as Canadian Institutes of Health Research Grants 365646 (to S.S. and J.M.) and 342551 (to J.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequencing data that support the findings of this study are available in the Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo [accession nos. GSE102712 (for PhaNGS data) and GSE102301 (for RNA-sequencing data)]. All other data supporting the findings of this study are available in the SI Appendix, Dataset S3.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721899115/-/DCSupplemental.
References
- 1.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Fan HC, Fu GK, Fodor SP. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 3.Pascal LE, et al. Correlation of mRNA and protein levels: Cell type-specific gene expression of cluster designation antigens in the prostate. BMC Genomics. 2008;9:246. doi: 10.1186/1471-2164-9-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang H, et al. Selection of recombinant anti-sh3 domain antibodies by high-throughput phage display. Protein Sci. 2015;24:1890–1900. doi: 10.1002/pro.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornsby M, et al. A high through-put platform for recombinant antibodies to folded proteins. Mol Cell Proteomics. 2015;14:2833–2847. doi: 10.1074/mcp.O115.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrard LJ, Yang M, O’Connell MP, Kelley RF, Henner DJ. Fab assembly and enrichment in a monovalent phage display system. Biotechnology (NY) 1991;9:1373–1377. doi: 10.1038/nbt1291-1373. [DOI] [PubMed] [Google Scholar]
- 7.Ewert S, Huber T, Honegger A, Pluckthun A. Biophysical properties of human antibody variable domains. J Mol Biol. 2003;325:531–553. doi: 10.1016/s0022-2836(02)01237-8. [DOI] [PubMed] [Google Scholar]
- 8.Matochko WL, et al. Deep sequencing analysis of phage libraries using Illumina platform. Methods. 2012;58:47–55. doi: 10.1016/j.ymeth.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Persson H, et al. Cdr-h3 diversity is not required for antigen recognition by synthetic antibodies. J Mol Biol. 2013;425:803–811. doi: 10.1016/j.jmb.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sidhu SS, Fairbrother WJ, Deshayes K. Exploring protein-protein interactions with phage display. Chembiochem. 2003;4:14–25. doi: 10.1002/cbic.200390008. [DOI] [PubMed] [Google Scholar]
- 11.Bass S, Greene R, Wells JA. Hormone phage: An enrichment method for variant proteins with altered binding properties. Proteins. 1990;8:309–314. doi: 10.1002/prot.340080405. [DOI] [PubMed] [Google Scholar]
- 12.Aird D, et al. Analyzing and minimizing pcr amplification bias in illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrara F, et al. Recombinant renewable polyclonal antibodies. MAbs. 2015;7:32–41. doi: 10.4161/19420862.2015.989047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumbers–The database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38:D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shojaee S, et al. Erk negative feedback control enables pre-b cell transformation and represents a therapeutic target in acute lymphoblastic leukemia. Cancer Cell. 2015;28:114–128. doi: 10.1016/j.ccell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kindler T, Lipka DB, Fischer T. Flt3 as a therapeutic target in aml: Still challenging after all these years. Blood. 2010;116:5089–5102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 17.Wellmann S, et al. Flt3 mutations in childhood acute lymphoblastic leukemia at first relapse. Leukemia. 2005;19:467–468. doi: 10.1038/sj.leu.2403655. [DOI] [PubMed] [Google Scholar]
- 18.Dave H, et al. Restricted cell surface expression of receptor tyrosine kinase ror1 in pediatric b-lineage acute lymphoblastic leukemia suggests targetability with therapeutic monoclonal antibodies. PLoS One. 2012;7:e52655. doi: 10.1371/journal.pone.0052655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mummery A, Narendran A, Lee KY. Targeting epigenetics through histone deacetylase inhibitors in acute lymphoblastic leukemia. Curr Cancer Drug Targets. 2011;11:882–893. doi: 10.2174/156800911796798922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daneshmanesh AH, et al. Monoclonal antibodies against ror1 induce apoptosis of chronic lymphocytic leukemia (cll) cells. Leukemia. 2012;26:1348–1355. doi: 10.1038/leu.2011.362. [DOI] [PubMed] [Google Scholar]
- 21.Dang CV, et al. The c-myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Pajic A, et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int J Cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Mezquita P, Parghi SS, Brandvold KA, Ruddell A. Myc regulates vegf production in b cells by stimulating initiation of vegf mRNA translation. Oncogene. 2004;24:889–901. doi: 10.1038/sj.onc.1208251. [DOI] [PubMed] [Google Scholar]
- 24.Itzhak DN, Tyanova S, Cox J, Borner GH. Global, quantitative and dynamic mapping of protein subcellular localization. Elife. 2016;5:e16950. doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 26.Black CB, Duensing TD, Trinkle LS, Dunlay RT. Cell-based screening using high-throughput flow cytometry. Assay Drug Dev Technol. 2011;9:13–20. doi: 10.1089/adt.2010.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandura DR, et al. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 28.Di Palma S, Bodenmiller B. Unraveling cell populations in tumors by single-cell mass cytometry. Curr Opin Biotechnol. 2015;31:122–129. doi: 10.1016/j.copbio.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Spitzer MH, Nolan GP. Mass cytometry: Single cells, many features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ullal AV, et al. Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci Transl Med. 2014;6:219ra9. doi: 10.1126/scitranslmed.3007361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradbury A, Pluckthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015;518:27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 33.Zahnd C, Amstutz P, Pluckthun A. Ribosome display: Selecting and evolving proteins in vitro that specifically bind to a target. Nat Meth. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 34.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, silac, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 35.Wollscheid B, et al. Mass-spectrometric identification and relative quantification of n-linked cell surface glycoproteins. Nat Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox J, Mann M. Maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.