Abstract
The cell cycle checkpoint proteins ataxia-telangiectasia-mutated-and-Rad3-related kinase (ATR) and its major downstream effector checkpoint kinase 1 (CHK1) prevent the entry of cells with damaged or incompletely replicated DNA into mitosis when the cells are challenged by DNA damaging agents, such as radiation therapy (RT) or chemotherapeutic drugs, that are the major modalities to treat cancer. This regulation is particularly evident in cells with a defective G1 checkpoint, a common feature of cancer cells, due to p53 mutations. In addition, ATR and/or CHK1 suppress replication stress (RS) by inhibiting excess origin firing, particularly in cells with activated oncogenes. Those functions of ATR/CHK1 make them ideal therapeutic targets. ATR/CHK1 inhibitors have been developed and are currently used either as single agents or paired with radiotherapy or a variety of genotoxic chemotherapies in preclinical and clinical studies. Here, we review the status of the development of ATR and CHK1 inhibitors. We also discuss the potential mechanisms by which ATR and CHK1 inhibition induces cell killing in the presence or absence of exogenous DNA damaging agents, such as RT and chemotherapeutic agents. Lastly, we discuss synthetic lethality interactions between the inhibition of ATR/CHK1 and defects in other DNA damage response (DDR) pathways/genes.
Keywords: ATR, CHK1, DNA damage response, Cell cycle checkpoints, DNA replication stress, Synthetic lethality
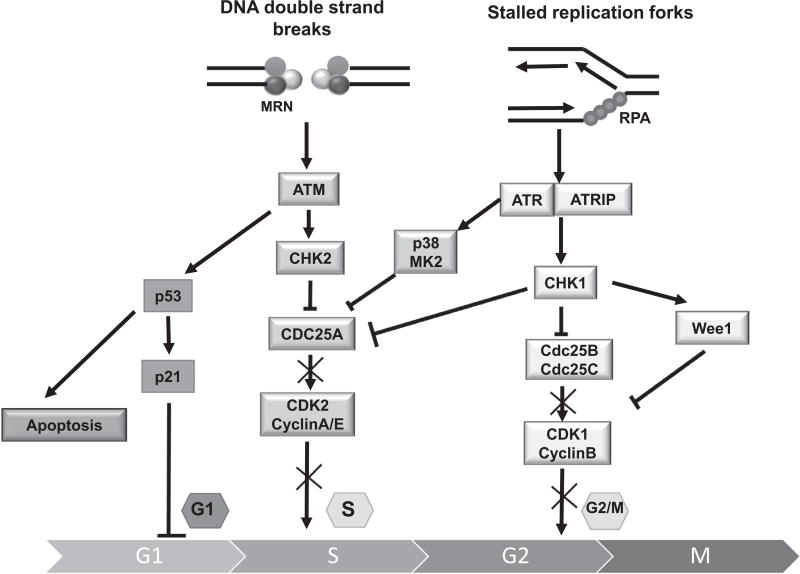
Human genomic DNA is threatened by a variety of exogenous and endogenous agents, such as ultraviolet radiation (UV), ionizing radiation (IR) and chemotherapeutic drugs [1], reactive oxygen species, lipid peroxidation products and DNA replication stress (RS; defined as any situation leading to instability of replication forks) [2,3]. Defending the integrity of the genome is the DNA damage response (DDR), an extensive network of pathways which detects and signals DNA damage for subsequent processing. The two major components of DDR are (a) the activation of cell cycle checkpoints, which is thought to allow cells time to repair their damaged DNA before moving to the next stage of the cell cycle and (b) facilitation of direct DNA repair. When DNA double-strand breaks (DSBs), the most dangerous type of DNA damage, are generated, the MRE11/NBS1/RAD5 complex activates the ataxia telangiectasia mutated (ATM) /checkpoint kinase 2 (CHK2) pathway to promote S-phase cell cycle arrest and the p53-associated G1/S-phase checkpoint [4]. When single-strand DNA (ssDNA) is created at sites of DNA damage or stressed replication forks, RPA-coated ssDNA activates ATR and its binding partner ATRIP (Fig. 1) [3,5,6]. The serine/threonine kinase CHK1 is the key downstream regulator of the ATR response and is phosphorylated by ATR on Ser-317 and Ser-345 [7]. Activated CHK1 triggers the intra-S- and G2/M-phase checkpoints [8–11]. Inhibitory phosphorylation by CHK1 of the phosphatase CDC25A and its subsequent proteasomal degradation leads to a decrease in CDK2 activity in S-phase [12]. CHK1-mediated phosphorylation of CDC25B at Ser-323 and CDC25C at Ser-216 also promotes their degradation, which abolishes the activation of CDK1/cyclin B kinases, thereby causing G2/M arrest [13,14]. The tyrosine kinase Wee1 is a dual-specificity kinase that can phosphorylate CDK1 at Tyr15 and inhibit CDK1 kinase activity [15]. In yeast and Xenopus egg extracts [16,17], CHK1 could activate Wee1 to halt the G2/M transition and prevent cells with damaged DNA from entering mitosis. However, it is not clear if a similar regulation exists in mammalian cells. There is crosstalk between the ATM/CHK2 and ATR/CHK1 pathways, and they share many substrates [18]. However, they cannot compensate for the loss of each other [19,20]. Most cancer cells have dysregulated G1 checkpoints, making them dependent on S and G2 checkpoints, which are activated by ATR/CHK1 signaling.
Fig. 1.
Overview of the DNA damage-induced checkpoint pathways. The ATM/CHK2 and ATR/CHK1 pathways are activated by DNA double-strand breaks or by DNA single-strand breaks and replication stress, respectively. Cell cycle checkpoints are induced primarily through p53, CHK2, CHK1 and p38/MK2, which are phosphorylated by ATM and ATR. Activated p53 leads to G1-phase arrest and induces apoptosis. The phosphorylation of CDC25 by CHK2 and CHK1 abolishes the activation of CDKs, thus stopping cell cycle progression either in S-phase or at the boundary of G2/M. Cancer cells deficient in p53 due to mutation or deletion lack the G1 checkpoint and are more dependent on the intra-S and G2/M checkpoints.
In addition to their regulation of cell cycle checkpoints, ATR/CHK1 have multiple additional roles in protecting cells from DNA damage, including the stabilization of stalled replication forks, the control of replication origin firing at specialized start sites, and the promotion of homologous recombination (HR). During replication stress, ATR/CHK1 stabilizes stalled replication forks to prevent their collapse and DSB formation and helps to restart stalled replication forks [18,21,22]. In addition, ATR/CHK1 suppresses replication initiation, particularly in cells expressing activated oncogenes [23] (Fig. 2, see discussion below). These properties of the ATR/CHK1 pathway have made it an attractive target for therapeutic intervention. ATR/CHK1 signaling regulates HR, a major pathway required for repair of DSBs/collapsed replication forks. The role of ATR/CHK1 in promoting HR, particular gene-conversion-mediated HR, has been well established [9,24–30]. It is believed that ATR/CHK1 promotes HR via regulation of HR factors, such as facilitation of phosphorylation of RAD51 [26], FANCE [29] and FANCD2 [30]. In addition, CHK1 recruits the HR proteins BRCA2 and RAD51 [9,26] to DNA damage foci. In most of the studies demonstrating a role for ATR/CHK1 in HR repair, DR-GFP, an HR reporter designed to detect gene conversion non-crossover recombination was used [24–28,31]. Interestingly, we demonstrated that ATR depletion increased HR, as detected by sister chromatid exchange [27], which is mediated by recombination during replication in vertebrates [32] and is a reflection of crossover recombination. Thus, ATR/CHK1 may have a different impact on the various subtypes of HR.
Fig. 2.
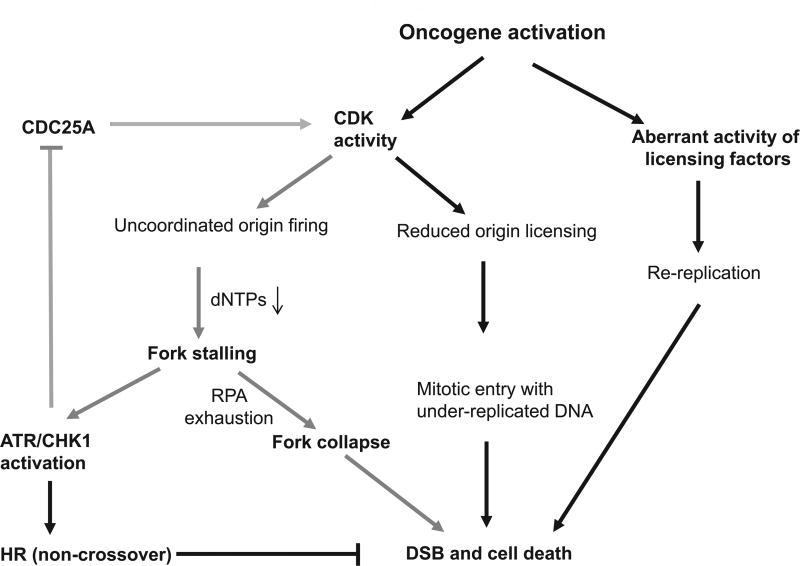
ATR/CHK1 suppresses oncogene-induced RS. Oncogene activation causes replication stress via uncoordinated origin firing, a reduction in licensed replication origins and inappropriate re-licensing of newly replicated DNA. Increased CDK activity leads to not only dysregulated replication initiation but also a deceased number of licensed replication origins, leading to under-replicated DNA. ATR/CHK1 signaling inhibits oncogene-induced replication stress via CDKs. Although it has been demonstrated that ATR/CHK1 suppresses replication stress via inhibition of CDK-induced replication initiation, it is not clear if ATR/CHK1 suppress CDK-mediated regulation of the number of licensed replication origins. CDK signaling suppressed by ATR/CHK1 is presented by gray arrows.
Inhibitors of both ATR and CHK1 have been developed and are being tested in clinical trials. However, the development and clinical application of CHK1 inhibitors are more advanced than that of ATR inhibitors for several reasons. First, it was thought that inhibition of ATR might affect normal cells extensively in vivo, because ATR has a broader biological function than CHK1 [33,34]. Second, it is difficult to purify active ATR protein for the development of ATR inhibitors. ATR is a large protein with 2644 amino acids that makes compound screening in kinase assays challenging. Thus, an enzymatic analysis is more difficult to perform on the upstream ATR than on the downstream CHK1 [35]. Third, the crystal structure of ATR has not been reported, and hence there is no clue as to possible allosteric binding regions for drug design [36]. Thus, results from CHK1 inhibitor-related studies are far richer than from ATR inhibitors. Nevertheless, there has been some recent progress in the development of small molecule ATR inhibitors. In this review, we first discuss the chemo- and radio-sensitization activities of ATR/CHK1 inhibitors, with a comparison of mechanisms by which ATR/CHK1 inhibition acts as a chemosensitizer and radiosensitizer. All the inhibitors discussed in this review are listed in Table 1. In addition, we also discuss recent new discoveries that ATR and CHK1 inhibitors can be used as single agents. Last, we summarize the additional opportunities for new use of ATR/CHK1 inhibitors based on synthetic lethal interactions. The aims of this review are to emphasize the promise of ATR and CHK1 inhibitors for cancer therapy and that the need to exploit biomarkers to guide the use of these agents.
Table 1.
ATR/CHK1 inhibitors discussed in this review.
| Agents | Targets | Preclinical research |
Treatment | Clinical trials | Detail | References |
|---|---|---|---|---|---|---|
| UCN-01 | CHK1 | cellular/xenograft | Cisplatin | NCT00045513 | Phase I/II combined with fludarabine phosphate | [40–44,62,136– 138,146,203,217] |
| CHK2 | Doxorubicin | NCT00006464 | Phase I combined with cisplatin | |||
| CDK1 | HDAC inhibitor | NCT00004059 | Phase I combined with fluorouracil | |||
| CDK2 | NCT00030888 | Phase II single | ||||
| MK2 | NCT00072267 | Phase II combined with topotecan hydrochloride | ||||
| PKC | NCT00019838 | Phase I combined with fludarabine phosphate | ||||
| NCT00003289 | Phase I single | |||||
| NCT00042861 | Phase I combined with multiple chemo drugs | |||||
| NCT00047242 | Phase I combined with irinotecan hydrochloride | |||||
| NCT00045500 | Phase I combined with prednisone | |||||
| NCT00045747 | Phase II combined with fluorouracil | |||||
| NCT00045175 | Phase I combined with topotecan hydrochloride | |||||
| NCT00001444 | Phase I single | |||||
| NCT00072189 | Phase II single | |||||
| NCT00012194 | Phase I combined with cisplatin | |||||
| NCT00301938 | Phase I combined with perifosine | |||||
| NCT00098956 | Phase II combined with topotecan hydrochloride | |||||
| NCT00031681 | Phase I combined with irinotecan hydrochloride | |||||
| NCT00039403 | Phase I combined with gemcitabine | |||||
| NCT00004263 | Phase I combined with cytarabine | |||||
| NCT00082017 | Phase II single | |||||
| NCT00036777 | Phase I combined with carboplatin | |||||
| XL844 | CHK1 | cellular/xenograft | Gemcitabine | NCT00475917 | Phase I single/combined with gemcitabine | [45] |
| CHK2 | NCT00234481 | Phase I single | ||||
| VEGFR-2 | ||||||
| VEGFR-3 | ||||||
| CBP501 | CHK1 | cellular/xenograft | CDDP | NCT00700336 | Phase I/II combined with pemetrexed and cisplatin | [46–48] |
| CHK2 | Bleomycin | NCT00551512 | Phase I combined with cisplatin | |||
| MAPKAP-K2 | Cisplatin | NCT00942825 | Phase II combined with cisplatin and pemetrexed | |||
| c-Tak1 | Pemetrexed | |||||
| PLK1 | ||||||
| AZD7762 | CHK1 | Cellular/xenograft | Nocodazole | NCT00937664 | Phase I single/combined with gemcitabine | [49–59,65– 68,139,196,210,211] |
| CHK2 | Camptothecin | NCT00413686 | Phase I single/combined with gemcitabine | |||
| Gemcitabine | NCT00473616 | Phase I single/combined with irinotecan | ||||
| Irinotecan | ||||||
| Gemcitabine | ||||||
| Radiation | ||||||
| Cisplatin | ||||||
| Temozolomide | ||||||
| HDAC inhibitor | ||||||
| LY2603618 | CHK1 | Cellular/xenograft | Gemcitabine | NCT00415636 | Phase I combined with pemetrexed | [69–74] |
| Pemetrexed | NCT00839332 | Phase I/II combined with gemcitabine | ||||
| Cisplatin | NCT00988858 | Phase II combined with pemetrexed | ||||
| NCT01296568 | Phase I single | |||||
| NCT01139775 | Phase I/II combined with gemcitabine or pemetrexed | |||||
| NCT01341457 | Phase I combined with gemcitabine | |||||
| NCT01358968 | Phase I single | |||||
| MK-8776 | CHK1 | Cellular/xenograft | Antimetabolites | NCT01870596 | Phase II combined with cytarabine | [75–83,135,140,189,196] |
| Gemcitabine | NCT00779584 | Phase I/II combined with gemcitabine | ||||
| Radiation | NCT00907517 | Phase I combined with cytarabine | ||||
| Pemetrexed | NCT01521299 | Phase I combined with hydroxyurea | ||||
| Cytarabine | ||||||
| PF-00477736 | CHK1 | Cellular/xenograft | Gemcitabine | NCT00437203 | Phase I combined with gemcitabine | [84–88,153,188] |
| Carboplatin | ||||||
| Docetaxel | ||||||
| Radioimmunotherapy | ||||||
| Radiation | ||||||
| LY2606368 | CHK1 | Cellular/xenograft | BMN673 | NCT01115790 | Phase I single | [89,90] |
| CHK2 | NCT02873975 | Phase II single | ||||
| NCT02203513 | Phase II single | |||||
| NCT02808650 | Phase I single | |||||
| NCT02514603 | Phase I single | |||||
| NCT02860780 | Phase I combined with ralimetinib | |||||
| NCT02555644 | Phase I combined with radiation and multiple chemo drugs | |||||
| NCT02735980 | Phase II single | |||||
| NCT02124148 | Phase I combined with multiple chemo drugs | |||||
| NCT02778126 | Phase I single | |||||
| NCT02649764 | Phase I combined with fludarabine and cytarabine | |||||
| 2e | CHK1 | Cellular/xenograft | Doxorubicin | [91] | ||
| Camptothecin | ||||||
| CCT244747 | CHK1 | Cellular | Radiation | [92–94] | ||
| Gemcitabine | ||||||
| CHIR-124 | CHK1 | Cellular/xenograft | Radiation | [95–97] | ||
| Top1 inhibitor | ||||||
| HDAC inhibitor | ||||||
| GNE-783 | CHK1 | Cellular | Temozolomide | [98] | ||
| GNE-900 | CHK1 | Cellular/xenograft | Gemcitabine | [98] | ||
| CPT-11 | ||||||
| Temozolomide | ||||||
| PD-321852 | CHK1 | Cellular | Gemcitabine | [99] | ||
| Wee1 | ||||||
| PD-407824 | CHK1 | Cellular | Gemcitabine | [100] | ||
| Wee1 | ||||||
| SAR-020106 | CHK1 | Cellular/xenograft | Radiation | [101–104] | ||
| Radiochemo | ||||||
| SB-218078 | CHK1 | Cellular | Doxorubicin | [62,105–107] | ||
| HSP inhibitor | ||||||
| S1181 | CHK1 | Cellular | Gemcitabine | [108] | ||
| V158411 | CHK1 | Cellular/xenograft | Irinotecan | [109] | ||
| CHK2 | ||||||
| CH-1 | CHK1 Aurora A | Cellular | Hypoxia | [181] | ||
| AR323 | CHK1 | Cellular | [154] | |||
| AR678 | CHK1 | Cellular | [154] | |||
| AR458323 | CHK1 | Cellular | MK-1775 | [187] | ||
| Schisandrin B | ATR | Cellular | UV | [110,111] | ||
| NU6027 | CDK2 | Cellular | PARP inhibitor | [112,113,205] | ||
| CDK1 | Hydroxyurea | |||||
| DNA-PK | Cisplatin | |||||
| ATR | ||||||
| NVP-BEZ235 | PI3K | Cellular | [114] | |||
| MTOR | ||||||
| ATR | ||||||
| ATM | ||||||
| DNA-PKcs | ||||||
| Torin 2 | MTOR | Cellular | [115] | |||
| ATR | ||||||
| ATM | ||||||
| DNA-PKcs | ||||||
| ETP-46464 | MTOR | Cellular | [114] | |||
| ATR | ||||||
| ATM | ||||||
| DNA-PKcs | ||||||
| PI3K | ||||||
| Compound | ATR | Cellular | Cisplatin | [116] | ||
| 45 | ||||||
| VE-821 | ATR | Cellular/xenograft | Radiation | [53,117– 125,158,192,199,205] | ||
| Cisplatin | ||||||
| Topotecan | ||||||
| Veliparib | ||||||
| VE-822 (VX-970) | ATR | Cellular/xenograft | Radiation | NCT02157792 | Phase I combined with multiple chemo drugs | [124,126–128] |
| Cisplatin | NCT02627443 | Phase II combined with multiple chemo drugs | ||||
| NCT02567409 | Phase II combined with multiple chemo drugs | |||||
| NCT02487095 | Phase I/II combined with Topotecan | |||||
| NCT02595892 | Phase II combined with Gemcitabine | |||||
| NCT02589522 | Phase I combined with radiotherapy | |||||
| NCT02567422 | Phase I combined with Cisplatin | |||||
| NCT02595931 | Phase I combined with Irinotecan | |||||
| NCT02723864 | Phase I combined with Veliparib and Cisplatin | |||||
| AZ20 | ATR | Cellular | [129] | |||
| AZD6738 | ATR | Cellular/xenograft | Radiation | NCT02264678 | Phase I/Ib combined with multiple chemo drugs | [130–133] |
| Cisplatin | NCT02223923 | Phase I single/combined with radiotherapy | ||||
| Top1 inhibitor | NCT01955668 | Phase I single | ||||
| NCT02630199 | Phase I combined with Paclitaxel |
Clinical trial data are from https://clinicaltrials.gov/.
ATR/CHK1 inhibitors and chemo- and radio-sensitization
The initial idea is that ATR/CHK1 inhibitors would enhance killing of tumor cells by cytotoxic drugs or by radiotherapy through blocking cell cycle checkpoints, especially in p53-deficient cells [11,37,38]. Thus, the early studies focused on the activities of ATR/CHK1 inhibitors in chemo- and radio-sensitization. It has been reported that the inhibition or knockdown of ATR and/or CHK1 renders cells sensitive to a wide variety of DNA-damaging agents, including IR, antimetabolites (hydroxyurea, gemcitabine, cytarabine, 5-fluorouracil, gemcitabine), DNA cross-linking agents (cisplatin), topoisomerase I and II poisons (topotecan, SN-38, etoposide, doxorubicin) and alkylating agents (methyl-methane sulfonate). A recent review has summarized this broad panel of sensitized agents [39]. We will discuss selected combinations in this review.
CHK1 inhibitors
UCN-01 (7-hydroxystaurosporine) is the first CHK1 inhibitor with broad-spectrum efficacy against the protein kinase C family [40]. However, further application of UCN-01 has been hindered by its lack of specificity and by its long half-life, due to its binding to alpha acidic-glycoprotein that leads to hyperglycemia [41–44]. XL844 is a potent ATP-competitive inhibitor of CHK1, CHK2, VEGFR-2 and VEGFR-3. XL844 in combination with gemcitabine blocks CDC25A degradation, abrogates the S-phase checkpoints and increases DNA damage. Thus, XL844 potentiates gemcitabine activity in vitro and in xenografts [45]. However, clinical evaluation of XL844 was prematurely terminated (NCT00475917 and NCT00234481) for unknown reasons. CBP501 inhibits kinases that phosphorylate Ser-216 of CDC25C, including MAPKAP-K2, C-TAK1, and CHK1 [46]. Although a phase I study demonstrated that CBP501 is well tolerated in patients with advanced solid tumors, both as monotherapy and in combination with cisplatin [47], in a randomized phase II trial the addition of CBP501 failed to improve the efficacy of pemetrexed/cisplatin treatment in patients with advanced malignant pleural mesothelioma [48]. Because of the nonspecific targeting of UCN-01, XL844 and CBP501, their clinical applications have been limited.
Subsequently, more potent and specific CHK1 inhibitors have been developed. Most are ATP-competitive but with varying potencies and specificities. As a potent CHK1/2 dual inhibitor, AZD7762 is an ATP-competitive drug which suppresses CHK1-mediated phosphorylation of CDC25C with an IC50 of 5 nmol/L [49]. It has been suggested that AZD7762 treatment impairs CHK1 autophosphorylation (S296 CHK1), stabilizes CDC25A and potentiates the ATR/ATM-mediated CHK1 phosphorylation (S345 CHK1) [50]. Thus, it has been proposed that increased pS296 CHK1 and/or pS345 CHK1 can serve as a predictive biomarker of CHK1 inhibitor sensitivity [51,52]. However, a recent study demonstrated that p-CHK1 may not be a readout for ATR/CHK1 inhibition in some cancer cells and that the enhanced CDC25A expression is strongly associated with ATR/CHK1 inhibition [53]. Interestingly, AZD7762 sensitizes pancreatic cancer cells and tumors to gemcitabine, in association with induction of pS345 CHK1, with a strict schedule in which gemcitabine is administered concurrently with or before AZD7762 [52]. Although AZD7762 is a dual inhibitor that potently inhibits both CHK1 and CHK2 [49,54], its function mainly depends on the suppression of CHK1, because neither siRNA-mediated CHK2 knockdown [55,56] nor a CHK2-specific inhibitor [57] led to the sensitization to DNA damaging agents by AZD7762. AZD7762 in combination with chemotherapeutic drugs decreases tumor growth, especially cancers with p53 mutations or deletion [37,49,57–59]. Also, AZD7762 increased IR-induced apoptosis in cancer cells with compromised p53 [50,57,60,61]. However, effects of CHK1 inhibitors independent of p53 were also observed, e.g., UCN-01 or SB-218078 on cell survival [62,63]. Similarly, short-term cell viability, as well as the long-term ability to form colonies under CHK1 depletion, was similar in isogenic cell lines regardless of the status of p53, although abrogation of the DNA-damage-induced cell cycle arrest by CHK1 inactivation was p53 dependent [64]. These results indicate that p53 is not the only determinant. Indeed, CHK1 inhibition by AZD7762 preferentially kills HR-deficient tumor cells, because HR-deficient cells rely more heavily on ATR/CHK1 for survival [65]. Interestingly, AZD7762 sensitizes pancreatic cells to IR via inhibition of the G2/M checkpoint and HR, regardless of p53 status [50]. In support of this result, we recently demonstrated that AZD7762 was more effective in targeting radioresistant breast cancer cells that are proficient in HR independent of p53 status, although CHK1 inhibition abrogated the IR-induced G2/M-phase checkpoint [66]. Therefore, p53 may not be the sole determinant of the efficacy of ATR/CHK1 inhibitors, and CHK1 signaling driving cell cycle checkpoints may not be the only mechanism contributing to cell killing. Unfortunately, the clinical trial of AZD7762 was terminated due to cardiac toxicity [67] and multiple adverse effects [68].
LY2603618 is the first selective and potent CHK1 inhibitor in which replacement of the piperidine moiety with a morpholine guarantees the potent inhibition of CHK1 but reduces the risk of cardiac toxicity [69]. In preclinical studies, LY2603618 increased the in vitro potency of gemcitabine in p53-mutant HT29 cells but not in p53-wild type HCT116 cells, and it abrogated the G2/M checkpoints triggered by treatment with gemcitabine in xenograft models [69,70]. A Phase I clinical trial demonstrated that LY2603618 administered approximately 1 day after pemetrexed showed acceptable safety and pharmacokinetic profiles [71]. Two out of 14 non-small-cell lung cancer (NSCLC) patients achieved a confirmed partial response after treatment with a combination of LY2603618, pemetrexed and cisplatin [70]. LY2603618 in combination with gemcitabine showed acceptable safety and tolerability in Japanese patients with solid tumors [72]. One out of 50 patients with NSCLC achieved a partial response in a Phase I study of LY2603618 in combination with gemcitabine [73]. Unfortunately, p53 status appeared to be not important for LY2603618 efficacy when combined with pemetrexed in a Phase II evaluation of patients with advanced or metastatic NSCLC [74]. Thus, the clinical studies of LY2603818 did not produce promising results.
MK-8776 (SCH 900776) is a selective CHK1 inhibitor that induces DSBs and cell death when combined with the DNA antimetabolites hydroxyurea, gemcitabine or pemetrexed in xenograft models [75–79]. In acute myelogenous leukemia cells, MK-8776 abolished the cytarabine-induced S-phase checkpoint and increased Ara-C (cytarabine; cytosine arabinoside)-induced apoptosis [80]. Moreover, MK-8776 radiosensitized p53-defective NSCLC and HNSCC tumors by abrogation of G2/M arrest and by inhibition of DSB repair [81]. A phase I trial suggested that MK-8776 is well tolerated both as a single agent and in combination with gemcitabine in patients with refractory acute leukemias [82]. Two out of 30 patients with advanced solid tumors showed a partial response when treated with MK-8776 in combination with gemcitabine [83]. A phase II clinical trial using Ara-C with and without MK-8776 in patients with refractory acute leukemia (NCT01870596) has been completed. However, no results have yet been reported.
PF-00477736 is a potent, selective ATP-competitive CHK1 inhibitor that abrogated cell cycle arrest induced by gemcitabine and carboplatin in multiple p53-defective cancer cell lines in vitro [84]. In xenografts, PF-00477736 augmented the antitumor activity of gemcitabine and docetaxel by inducing checkpoint abrogation and mitotic catastrophe [84,85]. PF-00477736 also potentiated the anti-proliferative effect of gemcitabine against both parental and cancer stem cell-like NSCLC populations [86]. The combination of PF-00477736 and gemcitabine enhanced 177Lu-anti-EGFR-directed radioimmunotherapy against pancreatic ductal adenocarcinoma both in vitro and in vivo [87]. In addition, PF-0047736 sensitized HPV-positive HNSCC cell lines to IR [88]. Unfortunately, a Phase I trial of PF-00477736 (NCT00437203) in combination with gemcitabine in patients with advanced solid tumors was terminated due to business reasons.
LY2606368 (Prexasertib) is a CHK1/CHK2 dual inhibitor that preferentially binds to and inhibits CHK1 in vitro [89]. Treatment of cancer cells with LY2606368 resulted in DNA damage, premature entry into mitosis and cell death [89]. Similar results were obtained in xenograft tumor models where tumor growth was significantly inhibited after LY2606368 treatment [89]. Several clinical trials of LY2606368 have been initiated, but only one Phase I clinical trial to determine biosafety and dose for LY2606368 as monotherapy has been completed [90], and no data have yet been published.
New CHK1 inhibitors are still in preclinical development. 2e and CCT244747 are potent and selective CHK1 inhibitors that abrogated G2 and S checkpoint arrest caused by doxorubicin and camptothecin and increased cytotoxicity of several anticancer drugs both in vivo and in vitro [91–93]. CCT244747 radiosensitized p53-deficient head-and-neck squamous cell carcinoma (HNSCC) cells to paclitaxel-based chemoradiotherapy [94]. CHIR-124 is a selective, quinolone-based CHK1 inhibitor that is structurally unrelated to other known CHK1 inhibitors [95]. CHIR-124 interacted synergistically with topoisomerase poisons in p53-mutant solid tumor cell lines and in an orthotopic breast cancer xenograft model [95]. Treatment with CHIR-124 potentiated the efficacy of IR, histone deacetylase inhibitors and gemcitabine in inhibiting cancer proliferation [96,97]. A comparison study of two structurally related CHK1 inhibitors revealed that GNE-783 was more effective at sensitizing cells to antimetabolite-based DNA-damaging agents, whereas GNE-900 preferentially enhanced the activity of gemcitabine [98]. Treatment with two CHK1/Wee1 dual inhibitors, PD-321852 or PD-407824, led to sensitization of pancreatic cancer cells to gemcitabine [99,100]. SAR-020106 is a potent and highly selective isoquinoline CHK1 inhibitor [101,102] that augmented the efficacies of irinotecan and gemcitabine in human colon carcinoma xenograft models [101] and sensitized p53-deficient cancer cells to IR [103,104]. An indolocarbazole inhibitor of CHK1, SB-218078, sensitized p53 mutant cancer lines to IR, topotecan or quinacrine by abolishing the G2 checkpoint [105–107]. Two newly developed CHK1 inhibitors, S1181 and the CHK1/CHK2 dual inhibitor V158411, potentiated p53-defective cancer cells to different chemotherapeutic drugs [108,109].
In summary, despite the promising preclinical studies of CHK1 inhibitors in chemo- and radio-sensitization, results from clinical trials have been less impressive. In addition, independence of p53 has been observed in both preclinical studies and in clinical trials. Thus, other new factors/biomarkers affecting the efficacy of CHK1 inhibitors need to be identified. The most important break-through to be made in cancer treatment with these inhibitors may be to establish inhibitor-specific patient stratification criteria, since a lower dose of a CHK1 inhibitor is expected to be effective if the appropriate patients were selected. In addition, a new generation of CHK1 inhibitors with reduced toxicity needs to be developed in the future.
ATR inhibitors
The natural product Schisandrin B is the first ATR-specific inhibitor that reduced the phosphorylation of CHK1 by inhibiting ATR protein kinase activity following UV treatment of lung cancer cells [110,111]. Nevertheless, application of this drug is limited, due to the high dose that is required (30 µM for cellular assays) [110]. More recently, NU6027, NVP-BEZ235, torin 2, and ETP-46464 were reported to be more potent ATR inhibitors that sensitized tumor cells to a variety of genotoxic chemotherapies, but they also inhibited other kinases, such as CDK2, PIK3, mTOR, and ATM [112–115]. Compound 45 is a novel ATR inhibitor that showed potent activity against ATM-defective HCT116 cells and spared normal HFL1 cells [116]. It also synergistically enhanced the killing effect of IR and cisplatin in multiple cell lines [116].
VE-821 is a potent and selective ATR inhibitor that was discovered in a high-throughput screen [117]. It inhibited the phosphorylation of CHK1 at Ser345 and sensitized cancer cells to IR [117–121] or chemotherapeutic drugs, including gemcitabine [117,122], cisplatin [53,123], topotecan [53], indotecan [124] and camptothecin [124]. Thus, decreased pCHK1 Ser345 can be used as a biomarker for ATR inhibition [125]. The chemosensitization effect of VE-821 was further confirmed in p53-defective, ATM-defective or inhibited cells [120,122]. The novel inhibitor VE-822 (VX970) that is developed by Vertex is a close analog of VE-821 but has optimized solubility and pharmacokinetic properties. This inhibitor selectively inhibits ATR kinase activity. Targeting ATR in vivo with VE-822 resulted in selective sensitization of pancreatic tumors to IR [126] and augmented the in vivo anti-tumor response to the topoisomerase I inhibitor irinotecan without additional toxicity to normal cells [124]. VE-822 also potentiates the effect of cisplatin in NSCLC cells in vitro and in human lung tumor xenografts [127]. In a study of esophageal cancer, VE-822 improved response of cells to cisplatin, carboplatin or radiation and increased tumor growth delay from radiotherapy in vivo [128]. In this study, the radiation experiments were also carried out under hypoxic conditions with the same conclusion. Given its minimal toxicity, VE-822 is the first ATR-specific inhibitor entering clinical trials. Nine phase I/II trials of VE-822 in combination with radiotherapy or chemotherapy are currently ongoing.
AZ20 is another selective and potent inhibitor of ATR that inhibited the proliferation of LoVo colorectal adenocarcinoma cells in vitro as a single agent [129]. AZD6738, an analog of AZ20, is the second ATR inhibitor in clinical trials. The solubility, bioavailability and pharmacokinetic properties of AZD6738 are better than those of AZ20. These advantages of AZD6738 make it suitable for oral dosing [130]. AZD6738 potentiated cisplatin effects against ATM-deficient NSCLC in vivo [131]. A recent study suggested radiosensitization by the ATR inhibitor AZD6738 through generation of acentric micronuclei. The induction of micronuclei was significantly more prominent for AZD6738 in comparison to CHK1 inhibition at isotoxic doses [132]. Furthermore, AZD6738 sensitized p53- or ATM-defective primary chronic lymphocytic leukemia (CLL) cells to chemotherapy and ibrutinib both in in vitro and in vivo assays [133]. However, the selectivity of ATR inactivation might not be restricted to p53-defective cells. ATR silencing sensitized HeLa (p53-defective) and U2OS (p53-wild-type) cells to topoisomerase I poisons [134]. Radiosensitization by AZD6738 to clinically relevant doses of fractionated radiation was demonstrated in vitro using a 3D tumor spheroid model. The radiosensitization by AZD6738 is independent of both p53 and BRCA2 status [132]. Thus, p53 status may not be the sole determinant of the outcome of ATR inhibitors. Three phase I clinical trials of AZD6738 in combination with radiotherapy or chemotherapy are currently recruiting patients (Table 1). Compared to studies related of CHK1 inhibitors, investigations of ATR inhibition are still in an early stage. It is hoped that results from the current clinical trials will validate those from preclinical studies. We look forward to the development of new improved ATR inhibitors in the future.
The mechanisms by which ATR/CHK1 inhibitors sensitize cells to chemotherapy and radiotherapy
ATR/CHK1 signaling regulates cell cycle checkpoints, replication fork stability, replication origin firing and HR. It remains unknown whether it is the disruption of one or all of these pathways by ATR/CHK1 inhibition that results in the potentiation of the response to radiotherapy or chemotherapeutic drugs. The mechanisms of chemosensitization and radiosensitization by the various ATR/CHK1 inhibitors may overlap but with differences, given the multiple mechanisms by which the majority of cancer chemotherapeutic agents work; for example, both antimetabolites and cytotoxic drugs inhibit DNA synthesis in S-phase either by inhibiting production of the necessary deoxyribonucleotide precursors or by directly damaging DNA, but IR causes DNA damage, particularly DSBs, throughout all stages of the cell cycle. ATR/CHK1 signaling is primarily activated by RPA-coated ssDNA in S-phase cells during replication stress, a situation typically caused by most chemotherapeutic drugs, whereas DSBs induced by IR predominately activate ATM. The most dramatic sensitization has been observed when CHK1 inhibitors are combined with hydroxyurea or gemcitabine, two antimetabolites that inhibit ribonucleotide reductase, thereby starving cells of deoxyribonucleotides and leading to stalling of replication fork progression. For instance, a 100-fold decrease in the IC50 for hydroxyurea has been reported upon addition of the CHK1 inhibitor MK-8776 [76,135]. The sensitization to gemcitabine in vitro was about 10-fold with other CHK1 inhibitors [45,49,84,101,135]. CHK1 inhibitors do not cause cell cycle delay when combined with hydroxyurea or gemcitabine, because cell cycle arrest induced by antimetabolites does not require checkpoint activation, as cells cannot complete DNA replication without dNTPs. Thus, the effects of ATR/CHK1 inhibitors on S-phase checkpoints may not be important to their ability to sensitize cells to antimetabolites. It is generally believed that the antimetabolites alone result in the accumulation of stalled replication forks. In the absence of CHK1, the stalled forks are not protected by DNA2 or SMARCAL1 and would become susceptible to MUA81 cleavage and increased cytotoxicity by causing fork collapse/DSBs [135]. The detailed mechanism has been discussed in a recent review [135].
Of note, CHK1 inhibitors do not appear to sensitize cells to all antimetabolites. For instance, no sensitization activity of MK-8776 was observed when combined with 5-FU [76], a drug that inhibits thymidylate synthase, thus depleting cells of thymidine and stalling replication fork progression. Therefore, stabilization of replication forks by CHK1 may not be the sole mechanism explaining the sensitization by CHK1 inhibition when combined with antimetabolites. The work from Engelke et al. has demonstrated that HR is involved in radiosensitization by CHK1 inhibition, because HR-proficient cells, MiaPaCa-2, BxPC-3, and AsPC-1, were sensitized to gemcitabine by MK8776, whereas HR-deficient Capan-1 cells were only minimally sensitized [77]. Another study suggested that CHK1 inhibition sensitizes cells to gemcitabine and/or hydroxyurea by abolishing G2/M arrest and inhibition of HR [50], although disruption of replication fork stability is thought to be critical for sensitization to antimetabolites (discussed above). In addition, it has been demonstrated that interference with cell cycle progression may be important for the CHK1 inhibitor UCN-01-induced cell killing when combined with cytotoxic drugs, the topoisomerase I inhibitor camptothecin/SN38 [136,137] or the crosslinking agent cisplatin [138]. ATR/CHK1 inhibitors potentiate the response to cisplatin by decreasing the drug-induced G2/M arrest and leading to polyploidy and subsequent mitotic catastrophe [47,48,53,70,116,123,127,131]. Therefore, ATR/CHK1 inhibition sensitizes the response to many chemotherapeutic drugs that act through different mechanisms; the sensitization mechanisms may overlap, depending on the mode of action of the chemotherapeutic drugs.
In contrast to the extensive sensitization of chemotherapeutic drugs by ATR/CHK1 inhibitors, more modest radiosensitization is observed; however, variations in experimental conditions, such as cell lines, types of drugs, testing models, experimental assays, endpoints and p53 status, may contribute to differences. A recent study evaluated the effect of UCN-01 and MK-8776 on cell viability after exposure to IR using a clonogenic survival assay. The sensitizer enhancement ratios (SERs), which is defined as the ratio of radiation doses resulting in 10% survival (LD90 doses) under non-sensitized vs. sensitized conditions, for UCN-01 and MK-8776, were found to be 1.13 and 1.22 in EMT6 cells, and 1.07 and 1.39 in HeLa cells, respectively. Although AZD7762 was a very effective radiosensitizer in both in vitro and in vivo assays, MK8776 produced less radiosensitization [50,60,77]; radiosensitization by AZD7762 may be in part attributable to its activity against CHK2 or some other targets. Others have found that inhibition of CHK2 radiosensitizes cancer cells [139]. Clonogenic assays showed radiosensitization by AZD6738 in both p53 wild-type and p53 mutant cells with SERs from 1.43 to 2.27 at the 37% survival condition [132]. In pancreatic cancer cells, a low dose of AZD7762 reduced cell survival with an SER of 1.5 at the LD90 [50].
It is generally accepted that the mechanisms of ATR/CHK1 inhibition-mediated radiosensitization are related to abrogation of the G2 checkpoint, particularly in p53-deficient cells [50,57,60, 61,81,88,96,103–105]. In the accepted model, p53 triggers cell-cycle arrest in response to genotoxic stress: normal cells arrest in G1 (via p53), which is thought to allow time for DNA repair, whereas, p53-deficient tumor cells rely on CHK1 to arrest cell-cycle progression in the S- and G2-phases. Cells deficient in p53 undergo mitotic catastrophe and apoptosis when the S and G2 checkpoints are abrogated by inhibition of ATR or CHK1 [37,40,44,81,133]. Although HR may also be important for radiosensitization by ATR/CHK1 inhibition [28], it was also found that radiosensitization by the CHK1 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of HR in a p53-independent manner [50], and the ATR inhibitor AZD6738 worked similarly in a manner independent of p53 and the HR protein BRCA2 [132]. Interestingly, HR may not be involved in radiosensitization when a CHK1 inhibitor is applied post-IR, as evidenced by a recent study demonstrating that CHK1 inhibition by MK-8776 had no influence on the radiation-induced DSB levels or repair kinetics if the drug was applied immediately after irradiation [140], a finding that differs from a previous study in which MK-8776 was applied prior to IR [81]. Thus, some mechanisms of radiosensitization and chemo-sensitization by ATR/CHK1 inhibition are shared, but they can also be different. These mechanisms are far more sophisticated than we discussed. A combination of ATR/CHK1 inhibitors with a variety of chemotherapeutic drugs has been discussed in a recent review [39].
Given the fact that the combination of radiation with chemotherapy is the most effective therapy for some types of cancer, such as unresectable pancreatic cancer, the role of CHK1 inhibitors on chemoradiation also has been studied. The work of Engelke et al. suggested that CHK1 inhibition by AZD7762 could sensitize pancreatic cancer cells and tumors to combinations of gemcitabine and radiation, with inhibition of HR repair as an important underlying mechanism of this sensitization [50]. A similar result was obtained with a second inhibitor MK8776 [78]. These results supported clinical investigation of CHK1 inhibitors with gemcitabine-radiation in locally advanced pancreatic cancer patients [77]. The same study also demonstrated that MK8776 combined with gemcitabine or gemcitabine-radiation produced a significantly greater increase in pCHK1 (S345) in tumors relative to small intestine. Thus, pCHK1 (S345) may be a useful biomarker of the inhibition of CHK1 and sensitization activity as well as for CHK1 inhibition and DNA damage. However, correlation of pCHK1 (S345) status with clinical response needs to be validated.
In summary, ATR/CHK1 inhibition can sensitize cells to a broad array of DNA damaging agents, although the mechanisms of sensitization may differ with different treatment modalities. It is possible that biomarkers to predict radiosensitization and chemosensitization may differ although they overlap. Thus, it will be important to identify biomarkers which predict the superiority of one modality over others. The future challenge is to identify the patient populations that are sensitive to particular treatment regimens.
Applications of ATR/CHK1 inhibitors as single agents
Recent studies suggest that inactivation of ATR or CHK1 could target cancer cells as single agents. Results exploring this strategy are starting to emerge.
Targeting cancer cells with ATR and CHK1 inhibitors, used as single agents
Despite their well-established role in cell cycle regulation following DNA damage, the essential function of CHK1/ATR in this response is poorly understood. The initial thoughts that ATR and CHK1 were tumor suppressors are countered by several recent studies in different models demonstrating that ATR/CHK1 signaling may promote tumor progression. An extra allele of CHK1 limits oncogene (RAS/E1A)-induced RS and DSB formation, and therefore promotes transformation [141]. In addition, suppression of ATR function in oncogenic KRAS-expressing cells synergistically increases genomic instability and DNA damage-induced cell death [142]. In support of the idea that ATR/CHK1 is indispensable for tumor growth, amplification of ATR or CHK1 is frequently observed in genomically unstable cancers, including squamous cell lung carcinoma, ovarian cancer and head-and-neck cancer [65]. In addition, expression of ATR or CHK1 is increased in triple-negative breast cancer (TNBC) [143], neuroblastoma [144], T-cell acute lymphoblastic leukemia [145], acute myeloid leukemia [146] and hepatocellular carcinoma [147]. The oncogenes MYC and E2F1 upregulate CHK1 mRNA and protein expression both in vitro and in vivo [148,149]. Thus, compared to normal cells, cancer cells that carry oncogene-induced RS rely more heavily on ATR/CHK1 for survival. The reliance on ATR/CHK1 signaling for growth of tumor cells provides a therapeutic window for ATR/CHK1 inhibitors as single agents. Thus, the value of CHK1 inhibitors, and perhaps ATR inhibitors, as single agents has been recognized.
In a comprehensive loss-of-function screen of the protein kinome for neuroblastoma, CHK1 was identified as a candidate therapeutic target [144]. Oncogene expression is a major cause of RS [150,151]. In support of the hypothesis that inhibition of the ATR-CHK1 pathway is antineoplastic when targeted to cancers with high levels of RS [51,141,152–154], the CHK1 inhibitor UCN-01 decreased the clonogenicity of AML samples with high levels of RS and complex karyotypes [146]. The CHK1 inhibitor CCT244747 showed marked antitumor activity as a monotherapeutic agent in a MYCN-driven neuroblastoma [92]. As single agents, V158411, PF-477736 and AZD7762 potently inhibited the proliferation of TNBC [51]. In MYC-overexpressing B-cell lymphoma cells, CHK1 inhibition by siRNA or the inhibitor Chekin led to increased cell death both in vitro and in vivo via caspase-dependent apoptosis pathways [148]. CHK1 inhibitors, as single agents, specifically target a subtype of cancer cells expressing Cyclin E and MYC [152]. We also reported that a CHK1 inhibitor has antitumor activity in radioresistant breast cancer cells via enhancing RS, perhaps caused by the oncogenes CDC25A/Myc/-Src/H-ras/E2F1 [66].
Treatment with the CHK1 inhibitor PF-00477736 increased DNA damage and DDR signaling and led to caspase-dependent apoptosis and cell death in Eµ-myc lymphoma cells [153]. The two CHK1-specific inhibitors AR323 and AR678 were shown to be selectively cytotoxic to melanoma cell lines with endogenous RS [154]. In addition, a hypomorphic ATR pathway was synthetic lethal in the combination with oncogenic RAS expression in cultured cells [142] or in oncogenic RAS-driven murine tumors in vivo [155]. In the murine ATR-Seckel syndrome model, a decrease in ATR expression completely prevented the development of Myc-induced lymphomas or pancreatic tumors, which had high levels of RS [152]. A cell-based screen identified ATR inhibitors with synthetic lethal properties for cancer cells overexpressing cyclin E [114]. Pharmacologic inhibition of ATR led to chromosome breakage under conditions that stall replication forks. ATR inhibition is particularly toxic for p53-deficient cells, this toxicity being exacerbated by overexpression of cyclin E, a condition causing RS. In this study, the compound NVP-BEZ235, a dual PI3K/mTOR inhibitor, was found to be very potent against ATM, ATR and DNA-PKcs [114]. In addition, lack of p53 exacerbated RS induced by the ATR deficiency and augmented the detrimental effect [156]. Combined deletion of p53 and ATR in the mouse hindered tissue regeneration, induced high levels of DNA damage and led to synthetic lethality [157]. Furthermore, ATR inhibition by VE-821 in U2OS cells was shown to rewire cellular signaling networks induced by RS and activate DSB repair driven by ATM [158]. A study of LY2606368 in patients with solid tumors with RS or HR deficiency (NCT02873975) is ongoing. This study is testing if the CHK1 inhibitor is a possible treatment for advanced solid tumors that harbor genetic alterations in the HR pathway or with genetic alterations that indicate RS. Eight clinical trials using CHK1 inhibitors as single agents have reported results [47,83,159–163], while three clinical trials (NCT01564251, NCT01115790 and NCT02203513) are still recruiting patients. However, all of these trials focus on finding the maximum tolerated dose or the tolerance of the tested drugs. None of the clinical trials of ATR inhibitors as single agents to treat cancers has been completed at the time of preparation of this manuscript.
ATR/CHK1 inhibitors target cancer cells, as single agents, by regulating RS
DNA replication is a tightly regulated process. In late M- and early G1-phases of the cell cycle when CDK activity is low, replication is initiated by the association of origin recognition complex proteins with DNA replication origins, followed by the loading of CDT1 and CDC6 proteins [164]. Then, the six minichromosome maintenance (MCM) proteins that are the core components of the replicative DNA helicase are recruited to the origins to complete the pre-replication complexes. Activation of DBF4-dependent kinase and CDKs subsequently trigger the formation of functional bidirectional replication forks by recruiting replication initiation factor CDC45, GINS complex, RPA and DNA polymerases [164]. There is a regulation mechanism to ensure that all of the licensed replication origins do not fire simultaneously. Although how oncogenes cause RS is not fully understood, uncoordinated replication initiation, reduced licensing and re-usage of origins could be involved [165–167] (Fig. 2). For instance, overexpression of the oncogene cyclin E causes increased replication initiation [167] and also reduces the number of licensed origins by impairing prereplication complex assembly [165,166]. In that model of oncogene-induced replication initiation/firing, activation of oncogenes results in increased origin firing due to increased CDK activity, which in turn depletes the dNTP pool, as too many ongoing replication forks consume the limited dNTP pool. Replication stalling produces massively elongated ssDNA regions that are protected by RPA coating. With extended RS time and a limited supply of RPA, uncoated ssDNA causes DSB formation [168,169] (Fig. 2). CHK1/ATR suppresses oncogene-induced RS [51,141,152–154,170,171] by controlling replication initiation (Fig. 2). In the presence of replication stalling, activated CHK1/ATR phosphorylates CDC25A and promotes its degradation. ATR/CHK1-mediated loss of CDC25A activity suspends CDKs, such as CDK2, in an inactive phosphorylated state, blocking initiation of DNA replication origins (Fig. 2). The resulting inhibition of CDK2 activity prevents accumulation of the initiation factor CDC45 at unfired origins. Without CHK1 to regulate CDC25A, CDK2 activity is increased, leading to uncoordinated and excessive DNA replication initiation [172] and consequently the slowing and stalling of replication forks, due to an insufficient supply of replication factors [173–175]. Although ATR/CHK1 suppress replication stress via inhibition of CDK-induced replication initiation, it is not clear if ATR/CHK1 also suppresses CDK-mediated regulation on the licensed replication origins. In addition, the promotion of HR by ATR/CHK1 may contribute to its suppression of RS. Thus, the inhibition of ATR/CHK1 enhances RS via multiple mechanisms.
In support of the hypothesis that the conditions causing RS likely result in dependence on ATR/CHK1, hypoxia, a common biological source of RS frequently found in the majority of solid tumors [176], activates ATR/CHK1 signaling [177]. Severe levels of hypoxia (<0.1% O2) induce DDR and RS that activates both ATR and ATM signaling for survival [178–180]. Thus, it is not surprising to find that inducible knockdown of ATR and siRNA knockdown of CHK1 lead to increased DNA damage and decreased survival of hypoxic cells [177]. Cazares-Korner et al. suggested that the CHK1/Aurora A dual inhibitor CH-1 specifically decreases the viability of cancer cell lines under hypoxic conditions [181]. A unique aspect of this study is the development of a bioreductive prodrug of CH-01, which functions specifically in hypoxic conditions. A significant loss of viability of cancer cells exposed to hypoxia was observed when CH-01 is present [181]. The mechanism by which ATR/CHK1 inhibition targets hypoxic cells may be related to a defect in HR, since HR-defective cells are more reliant on ATR/CHK1 signaling for survival [65], and decreased HR in hypoxic cancer cells was observed to result from down-regulation of Rad51 and other proteins [182–185]. Thus, targeting hypoxic cells through targeting DDR is a promising strategy for cancer therapy [180]. Specifically, inhibition of ATR/CHK1 may be a promising therapy for tumors with high levels of hypoxia-induced RS, which needs to be further explored. Interestingly, a very recent study from Foskolou et al. suggested that ribonucleotide reductase (RNR), the only enzyme capable of de novo synthesis of dNTPs, requires subunit switching in hypoxia to maintain DNA replication. RNR consists of two homodimeric subunits, RRM1/RRM2 and RRM1/RRM2B. RRM1/RRM2B is favored over RRM1/RRM2 in order to preserve ongoing replication and avoid the accumulation of DNA damage [186]. Thus, this study raises the possibility of targeting tumors with hypoxia by targeting RRM2B.
Synthetic lethality between inhibition of ATR/CHK1 signaling and inhibition of other DDR pathways/genes
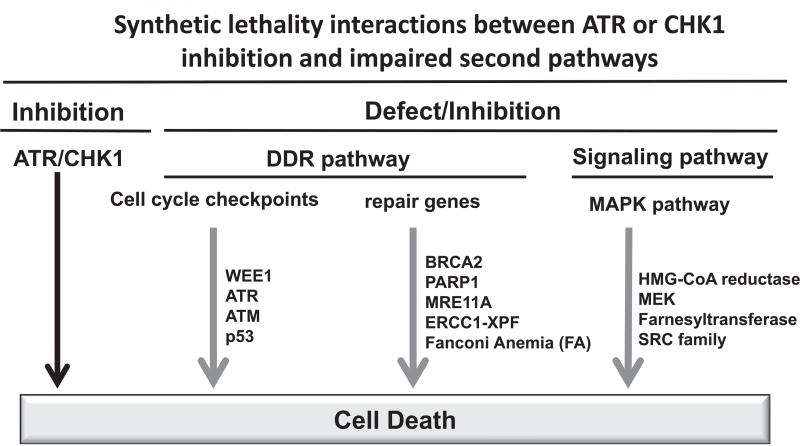
Synthetic lethality has attracted much attention from cancer biologists recently, as it provides a new angle for cancer therapy. It is a genetic concept describing any situation in which a combination of deficiencies in the expression of two genes leads to cell death, whereas a deficiency in only one of these genes does not. The potential synthetic lethality interactions with ATR/CHK1 are listed in Fig. 3. The synergistic cytotoxic effect of combined inhibition of WEE1 and CHK1 has been observed in preclinical research both in vitro and in vivo [187–193]. These two proteins have complementary but distinct roles. Both WEE1 and CHK1 suppress the activity of CDK and subsequent origin firing, but CHK1 has additional targets, not shared with WEE1, to promote fork progression and stability [194], providing a rationale for combination. Double knockdown of WEE1 and CHK1 by siRNA did not increase γ-H2AX levels more than those of single knockdowns; however, most of the cells lacking WEE1 and CHK1 underwent apoptosis [195]. Inhibition of WEE1 by siRNA or MK-1775, the only WEE1 inhibitor in clinical trial, potentiates the anti-cancer efficacy of the CHK1 inhibitor AR458323 and increases the level of apoptosis in prostate and lung cancer cell lines [187]. In a high-throughput synthetic lethality screen, WEE1 was also identified as a target that sensitized human ovarian cancer OVCAR-5 cells to the CHK1 inhibitor PF-0077736 [188]. The proposed mechanism for the synergistic effect is that inhibition of these two kinases interrupts DNA replication, induces DNA damage, allows the entry of unrepaired DNA into mitosis and results in apoptosis [188]. A similar synergistic effect was found between the CHK1 inhibitor MK-8776 and MK-1775 both in vitro and in vivo [189–191]. The CHK1/2 inhibitor AZD7762 synergized with MK-1775 in malignant melanoma cell lines both in vitro (2D and 3D cultures) and in xenograft models [196]. Consistent with the results with CHK1 inhibitors, ATR inhibition has a synthetic lethality with WEE1 inhibition in U2OS cells [197]. The ATR inhibitor VE-821, as a single agent, had no effect on the cell cycle of HeLa cells, but premature mitosis was induced when combined with sublethal concentrations of the WEE1 inhibitor MK-1775 [192]. In addition, two siRNA screens to identify synthetic lethal genes toward ATR inhibition suggested that knockdown of CHK1 is synthetic lethal to ATR inhibition [197,198]. Most interestingly, a combination of inhibitors of CHK1 (AZD77762) and ATR (VE-821), which are thought to be in the same pathway, led to DNA damage accumulation and subsequent cell death [199]. It was found that either the ATR or the CHK1 inhibitor alone leads to mitotic catastrophe, while the combination results in extensive DNA damage that likely leads to replication catastrophe [200]. It is proposed that cancer cells with disrupted CHK1 rely more heavily on ATR for survival, because the DNA damage caused by CHK1 inhibition activates ATR [199]. Of course, other mechanisms may also be involved; for instance, there is an ATR-dependent, CHK1-independent, intra-S-phase checkpoint that suppresses origin firing [200]. Interestingly, inhibition of CHK1 had no effect on B-cell transformation, but specific inhibition of ATR dramatically decreased the transformation efficiency of EBV [201]. Furthermore, ATR and CHK1 may not act linearly in the kinase cascade pathway and have distinct functions [199,202]. All the findings above warrant further investigation in clinical trials.
Fig. 3.
The potential synthetic lethality interactions with ATR/CHK1 inhibitions. See text for description. The defect in the second pathway is presented by gray arrows.
ATR or CHK1 inhibitor also undergoes synthetic lethality with a defect in other genes that are involved in DDR. The ATR inhibitor AZD6738 was reported to have single-agent anti-tumor activity in ATM-deficient xenograft models [130], and induced synthetic lethality in p53- or ATM-defective CLL cells both in vitro and in vivo [133]. The authors proposed that in p53- or ATM-defective CLL cells, inhibition of ATR signaling by AZD6738 can cause an accumulation of unrepaired DNA damage, which would be carried through into mitosis because of defective cell cycle checkpoints, resulting in cell death by mitotic catastrophe. In addition, a synthetic lethality screen revealed that the sensitivity to an ATR inhibitor treatment was enhanced in mantle cell lymphoma with ATM Loss-of-Function. The mechanism of this synthetic lethality is that cancers with ATM mutations have impaired DSB repair pathways and depend on compensatory repair mechanisms for survival [197]. Therefore, impairing ATR activity may selectively sensitize cancer cells to killing.
The CHK1 inhibitor UCN-01 suppressed the growth of pancreatic cancer cell lines with deficient BRCA2 [203]. CHK1 inhibition by siRNA knockdown or UCN-01 was more effective in cell lines deficient in Fanconi’s Anemia (FA) DNA repair pathway [204]. In addition, the replication factor C (RFC)-related protein RAD17 was identified to be synthetic lethal to multiple CHK1 inhibitors [193]. Similar results were also found with the ATR inhibitor VE-821 [122]. ATR inhibition by NU6027 or VE-821 was shown to be synthetic lethal with XRCC1 loss in multiple ovarian cancer lines [205]. Loss of the structure-specific endonuclease ERCC1-XPF (ERCC4), which is important for nucleotide excision repair, was synthetic lethal with the ATR inhibitor VE-821 or the CHK1 inhibitor AZD7762 [198]. The synthetic lethality between inhibition of ATR and loss of MRE11A was also confirmed [129]. In a synthetic lethality screen directed against 288 DNA-repair genes using DLD1 colorectal cancer cells, POLD1 was identified to have synthetic lethality with ATR and CHK1 inhibitors [206]. However, those synthetic lethalities described above have not yet been confirmed in clinical trials.
Inhibition of ATR/CHK1 also sensitizes cells to PARP inhibition. Both ATR- and CHK1-deficient cells were sensitive to the PARP inhibitor KU0058684 in an RNAi screen [207]. Multiple types of breast cancers were effectively killed by a combination of PARP inhibitors and CHK1 inhibitors both in vitro and in vivo [61,208]. Similar results were also found in prostate [209] and p53-mutant pancreatic cancer cells [38]. Another study demonstrated that the CHK1/2 dual inhibitor AZD7762 synergized with the MEK1/2 inhibitor AZD6244 by inducing massive apoptosis of myeloma cells [210,211]. The ATR inhibitor NU6027 was synthetic lethal with the PARP inhibitor PF-01367338 in breast and ovarian cancer cells [113]. It was found that NU6027 impaired G2/M arrest and HR, which could explain the increasing sensitivity to DNA-damaging agents and PARP inhibitors, providing proof-of-concept data for clinical development of ATR inhibitors [113]. The ATR inhibitor VE-821 and the CHK1 inhibitor MK-8776 further sensitized HR-deficient ovarian cancer cells to the PARP inhibitor veliparib (ABT-888) [53]. The synergy between an ATR inhibitor and a PARP inhibitor was observed in more than 100 cell lines with p53 mutations [212]. Although synthetic lethality between ATR/CHK1 inhibition and a defect in various other factors involved in DDR might have a variety of explanations; most mechanisms are related to the fact that a defect in DDR factors, such as WEE1, PARP, HR proteins or replication proteins, results in increased RS which activates ATR/CHK1 signaling; thus in the presence of DDR defects that can cause RS, cells rely more heavily on ATR/CHK1 signaling for survival.
Last, ATR/CHK1 inhibition also has a synthetic lethal interaction with inhibitors targeting the mitogen-activated protein kinase (MAPK) signaling pathway. CHK1 inhibition was synergistically lethal with HMG-CoA reductase inhibitors [213], MEK and farnesyl-transferase inhibitors [214] and SRC family inhibitors [215,216]. A combination of the nonspecific CHK1 inhibitor UCN-01 and a MAPK inhibitor led to mitochondrial dysfunction and apoptosis in human leukemia cells [217]. A similar result was reported in human multiple myeloma cells [218]. It is believed that the MAPK pathway is activated following exposure of leukemic cells to UCN-01. Interruption of this process by MEK inhibition triggers perturbations in cell cycle regulatory pathways and several signaling cascades that culminate in mitochondrial injury, caspase activation and subsequent apoptosis. Thus, a combination of UCN-01 with MEK inhibitors may represent a novel anti-leukemic strategy.
Altogether, inhibition of ATR/CHK1 signaling had greater antitumor efficacy when combined with other inhibitors targeting DDR pathways or signaling pathways. Thus, identification of new synthetic lethal combinations will significantly improve the outcome of ATR/CHK1-associated cancer therapy.
Conclusions and future prospects
Extensive preclinical data support the clinical application of ATR/CHK1 inhibitors for targeted cancer therapy with or without combination with radiotherapy and chemotherapy. However, compelling results from clinical trials are lacking for most combinations. The clinical efficacy of ATR/CHK1 inhibitors as single agents remains unknown. One of the keys to improving the efficacy of ATR/CHK1 inhibitors is to identify the appropriate patient population to target and to develop more highly specific ATR/CHK1 inhibitors. New synthetic lethality interactions between ATR/CHK1 inhibitors and other inhibitors targeting DDR pathways need to be identified. The rapid progress of high throughput synthetic lethality screening and next generation sequencing should dramatically improve the ability to verify unknown targets. However, only a small portion of potential synthetic lethality situations has been confirmed. In addition, development of new generations of pharmacological inhibitors with improved bioavailability and substrate specificity are also critical for clinical implementation. Lastly, successful application of ATR/CHK1 inhibitors in clinic trials will also rely on optimized schedules to increase their efficacy and on the identification of biomarkers that are suitable for these agents. The results from ongoing and future clinical trials will help to elucidate the role of ATR/CHK1 inhibitors in cancer therapeutics.
Acknowledgments
The authors apologize to colleagues whose work was not cited because of space limitations or ignorance. Research in the authors’ laboratory is supported by a grant R01CA154625 from the National Cancer Institute, seed grants from the Case Comprehensive Cancer Center and VeloSano Bike to Cure Foundation, a start-up fund from Department of Radiation Oncology, Case Western Reserve University School of Medicine to J. Zhang and the Case Comprehensive Cancer Center support grant (P30 CA043703).
Abbreviations
- RT
radiation therapy
- ATR
ataxia-telangiectasia-mutated-and-Rad3-related kinase
- ATM
ataxia-telangiectasia mutated
- CHK1
checkpoint kinase 1
- CHK2
checkpoint kinase 2
- IR
ionizing radiation
- DDR
DNA damage response
- RS
replication stress
- DSBs
DNA double-strand breaks
- ssDNA
single-strand DNA
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 2.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–85. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 3.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–58. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 5.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 6.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–6. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Vidanes G, Lin YC, Mori S, Siede W. Characterization of a Saccharomyces cerevisiae homologue of Schizosaccharomyces pombe Chk1 involved in DNA-damage-induced M-phase arrest. Mol General Genetics: MGG. 2000;262:1132–46. doi: 10.1007/pl00008656. [DOI] [PubMed] [Google Scholar]
- 8.Tse AN, Carvajal R, Schwartz GK. Targeting checkpoint kinase 1 in cancer therapeutics. Clin Cancer Res. 2007;13:1955–60. doi: 10.1158/1078-0432.CCR-06-2793. [DOI] [PubMed] [Google Scholar]
- 9.Bahassi EM, Ovesen JL, Riesenberg AL, Bernstein WZ, Hasty PE, Stambrook PJ. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene. 2008;27:3977–85. doi: 10.1038/onc.2008.17. [DOI] [PubMed] [Google Scholar]
- 10.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–83. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busino L, Chiesa M, Draetta GF, Donzelli M. Cdc25A phosphatase: combinatorial phosphorylation, ubiquitylation and proteolysis. Oncogene. 2004;23:2050–6. doi: 10.1038/sj.onc.1207394. [DOI] [PubMed] [Google Scholar]
- 13.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–5. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 14.Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, et al. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A Cancer. Cell. 2003;3:247–58. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe N, Broome M, Hunter T. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 1995;14:1878–91. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–54. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Kumagai A, Dunphy WG. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol Biol Cell. 2001;12:551–63. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 19.Zaugg K, Su YW, Reilly PT, Moolani Y, Cheung CC, Hakem R, et al. Cross-talk between Chk1 and Chk2 in double-mutant thymocytes. Proc Natl Acad Sci U S A. 2007;104:3805–10. doi: 10.1073/pnas.0611584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol. 2014;6:442–57. doi: 10.1093/jmcb/mju045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, et al. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–23. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortez D. Preventing replication fork collapse to maintain genome integrity. DNA Repair. 2015;32:149–57. doi: 10.1016/j.dnarep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecona E, Fernandez-Capetillo O. Replication stress and cancer: it takes two to tango. Exp Cell Res. 2014;329:26–34. doi: 10.1016/j.yexcr.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–43. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- 25.Brown AD, Sager BW, Gorthi A, Tonapi SS, Brown EJ, Bishop AJ. ATR suppresses endogenous DNA damage and allows completion of homologous recombination repair. PLoS One. 2014;9:e91222. doi: 10.1371/journal.pone.0091222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 27.Feng Z, Zhang J. A dual role of BRCA1 in two distinct homologous recombination mediated repair in response to replication arrest. Nucl Acids Res. 2012;40:726–38. doi: 10.1093/nar/gkr748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–43. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D’Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007;27:3098–108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhi G, Wilson JB, Chen X, Krause DS, Xiao Y, Jones NJ, et al. Fanconi anemia complementation group FANCD2 protein serine 331 phosphorylation is important for fanconi anemia pathway function and BRCA2 interaction. Cancer Res. 2009;69:8775–83. doi: 10.1158/0008-5472.CAN-09-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–8. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–9. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 34.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol: CB. 2000;10:479–82. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 35.Fang B. Development of synthetic lethality anticancer therapeutics. J Med Chem. 2014;57:7859–73. doi: 10.1021/jm500415t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fokas E, Prevo R, Hammond EM, Brunner TB, McKenna WG, Muschel RJ. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treatment Rev. 2014;40:109–17. doi: 10.1016/j.ctrv.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Investig. 2012;122:1541–52. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vance S, Liu E, Zhao L, Parsels JD, Parsels LA, Brown JL, et al. Selective radiosensitization of p53 mutant pancreatic cancer cells by combined inhibition of Chk1 and PARP1. Cell Cycle. 2011;10:4321–9. doi: 10.4161/cc.10.24.18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rundle S, Bradbury A, Drew Y, Curtin NJ. Targeting the ATR-CHK1 Axis in Cancer Therapy. Cancers. 2017:9. doi: 10.3390/cancers9050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunch RT, Eastman A. Enhancement of cisplatin-induced cytotoxicity by 7-hydroxystaurosporine (UCN-01), a new G2-checkpoint inhibitor. Clin Cancer Res. 1996;2:791–7. [PubMed] [Google Scholar]
- 41.Fuse E, Tanii H, Kurata N, Kobayashi H, Shimada Y, Tamura T, et al. Unpredicted clinical pharmacology of UCN-01 caused by specific binding to human alpha1-acid glycoprotein. Cancer Res. 1998;58:3248–53. [PubMed] [Google Scholar]
- 42.Fuse E, Hashimoto A, Sato N, Tanii H, Kuwabara T, Kobayashi S, et al. Physiological modeling of altered pharmacokinetics of a novel anticancer drug, UCN-01 (7-hydroxystaurosporine), caused by slow dissociation of UCN-01 from human alpha1-acid glycoprotein. Pharmaceut Res. 2000;17:553–64. doi: 10.1023/a:1007512832006. [DOI] [PubMed] [Google Scholar]
- 43.Dent P, Tang Y, Yacoub A, Dai Y, Fisher PB, Grant S. CHK1 inhibitors in combination chemotherapy: thinking beyond the cell cycle. Mol Intervent. 2011;11:133–40. doi: 10.1124/mi.11.2.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O’Connor PM, et al. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275:5600–5. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 45.Matthews DJ, Yakes FM, Chen J, Tadano M, Bornheim L, Clary DO, et al. Pharmacological abrogation of S-phase checkpoint enhances the anti-tumor activity of gemcitabine in vivo. Cell Cycle. 2007;6:104–10. doi: 10.4161/cc.6.1.3699. [DOI] [PubMed] [Google Scholar]
- 46.Sha SK, Sato T, Kobayashi H, Ishigaki M, Yamamoto S, Sato H, et al. Cell cycle phenotype-based optimization of G2-abrogating peptides yields CBP501 with a unique mechanism of action at the G2 checkpoint. Mol Cancer Therapeut. 2007;6:147–53. doi: 10.1158/1535-7163.MCT-06-0371. [DOI] [PubMed] [Google Scholar]
- 47.Shapiro GI, Tibes R, Gordon MS, Wong BY, Eder JP, Borad MJ, et al. Phase I studies of CBP501, a G2 checkpoint abrogator, as monotherapy and in combination with cisplatin in patients with advanced solid tumors. Clin Cancer Res. 2011;17:3431–42. doi: 10.1158/1078-0432.CCR-10-2345. [DOI] [PubMed] [Google Scholar]
- 48.Krug LM, Wozniak AJ, Kindler HL, Feld R, Koczywas M, Morero JL, et al. Randomized phase II trial of pemetrexed/cisplatin with or without CBP501 in patients with advanced malignant pleural mesothelioma. Lung cancer. 2014;85:429–34. doi: 10.1016/j.lungcan.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Zabludoff SD, Deng C, Grondine MR, Sheehy AM, Ashwell S, Caleb BL, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Therapeut. 2008;7:2955–66. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 50.Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70:4972–81. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bryant C, Rawlinson R, Massey AJ. Chk1 inhibition as a novel therapeutic strategy for treating triple-negative breast and ovarian cancers. BMC cancer. 2014;14:570. doi: 10.1186/1471-2407-14-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsels LA, Qian Y, Tanska DM, Gross M, Zhao L, Hassan MC, et al. Assessment of chk1 phosphorylation as a pharmacodynamic biomarker of chk1 inhibition. Clin Cancer Res. 2011;17:3706–15. doi: 10.1158/1078-0432.CCR-10-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huntoon CJ, Flatten KS, Wahner Hendrickson AE, Huehls AM, Sutor SL, Kaufmann SH, et al. ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 2013;73:3683–91. doi: 10.1158/0008-5472.CAN-13-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oza V, Ashwell S, Almeida L, Brassil P, Breed J, Deng C, et al. Discovery of checkpoint kinase inhibitor (S)-5-(3-fluorophenyl)-N-(piperidin-3-yl)-3-ureidothiophene-2-carboxamide (AZD7762) by structure-based design and optimization of thiophenecarboxamide ureas. J Med Chem. 2012;55:5130–42. doi: 10.1021/jm300025r. [DOI] [PubMed] [Google Scholar]
- 55.McNeely S, Conti C, Sheikh T, Patel H, Zabludoff S, Pommier Y, et al. Chk1 inhibition after replicative stress activates a double strand break response mediated by ATM and DNA-dependent protein kinase. Cell Cycle. 2010;9:995–1004. doi: 10.4161/cc.9.5.10935. [DOI] [PubMed] [Google Scholar]
- 56.Karnitz LM, Flatten KS, Wagner JM, Loegering D, Hackbarth JS, Arlander SJ, et al. Gemcitabine-induced activation of checkpoint signaling pathways that affect tumor cell survival. Mol Pharmacol. 2005;68:1636–44. doi: 10.1124/mol.105.012716. [DOI] [PubMed] [Google Scholar]
- 57.Ma Z, Yao G, Zhou B, Fan Y, Gao S, Feng X. The Chk1 inhibitor AZD7762 sensitises p53 mutant breast cancer cells to radiation in vitro and in vivo. Molecular medicine reports. 2012;6:897–903. doi: 10.3892/mmr.2012.999. [DOI] [PubMed] [Google Scholar]
- 58.Gadhikar MA, Sciuto MR, Alves MV, Pickering CR, Osman AA, Neskey DM, et al. Chk1/2 inhibition overcomes the cisplatin resistance of head and neck cancer cells secondary to the loss of functional p53. Mol Cancer Therapeut. 2013;12:1860–73. doi: 10.1158/1535-7163.MCT-13-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landau HJ, McNeely SC, Nair JS, Comenzo RL, Asai T, Friedman H, et al. The checkpoint kinase inhibitor AZD7762 potentiates chemotherapy-induced apoptosis of p53-mutated multiple myeloma cells. Mol Cancer Therapeut. 2012;11:1781–8. doi: 10.1158/1535-7163.MCT-11-0949. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell JB, Choudhuri R, Fabre K, Sowers AL, Citrin D, Zabludoff SD, et al. In vitro and in vivo radiation sensitization of human tumor cells by a novel checkpoint kinase inhibitor, AZD7762. Clin Cancer Res. 2010;16:2076–84. doi: 10.1158/1078-0432.CCR-09-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang Y, Hamed HA, Poklepovic A, Dai Y, Grant S, Dent P. Poly(ADP-ribose) polymerase 1 modulates the lethality of CHK1 inhibitors in mammary tumors. Mol Pharmacol. 2012;82:322–32. doi: 10.1124/mol.112.078907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirose Y, Berger MS, Pieper RO. Abrogation of the Chk1-mediated G(2) checkpoint pathway potentiates temozolomide-induced toxicity in a p53-independent manner in human glioblastoma cells. Cancer Res. 2001;61:5843–9. [PubMed] [Google Scholar]
- 63.Tse AN, Schwartz GK. Potentiation of cytotoxicity of topoisomerase i poison by concurrent and sequential treatment with the checkpoint inhibitor UCN-01 involves disparate mechanisms resulting in either p53-independent clonogenic suppression or p53-dependent mitotic catastrophe. Cancer Res. 2004;64:6635–44. doi: 10.1158/0008-5472.CAN-04-0841. [DOI] [PubMed] [Google Scholar]
- 64.Zenvirt S, Kravchenko-Balasha N, Levitzki A. Status of p53 in human cancer cells does not predict efficacy of CHK1 kinase inhibitors combined with chemotherapeutic agents. Oncogene. 2010;29:6149–59. doi: 10.1038/onc.2010.343. [DOI] [PubMed] [Google Scholar]
- 65.Krajewska M, Fehrmann RS, Schoonen PM, Labib S, de Vries EG, Franke L, et al. ATR inhibition preferentially targets homologous recombination-deficient tumor cells. Oncogene. 2015;34:3474–81. doi: 10.1038/onc.2014.276. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Lai J, Du Z, Gao J, Yang S, Gorityala S, et al. Targeting radioresistant breast cancer cells by single agent CHK1 inhibitor via enhancing replication stress. Oncotarget. 2016;7:34688–702. doi: 10.18632/oncotarget.9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sausville E, Lorusso P, Carducci M, Carter J, Quinn MF, Malburg L, et al. Phase I dose-escalation study of AZD7762, a checkpoint kinase inhibitor, in combination with gemcitabine in US patients with advanced solid tumors. Cancer Chemother Pharmacol. 2014;73:539–49. doi: 10.1007/s00280-014-2380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seto T, Esaki T, Hirai F, Arita S, Nosaki K, Makiyama A, et al. Phase I, dose-escalation study of AZD7762 alone and in combination with gemcitabine in Japanese patients with advanced solid tumours. Cancer Chemother Pharmacol. 2013;72:619–27. doi: 10.1007/s00280-013-2234-6. [DOI] [PubMed] [Google Scholar]
- 69.King C, Diaz H, Barnard D, Barda D, Clawson D, Blosser W, et al. Characterization and preclinical development of LY2603618: a selective and potent Chk1 inhibitor. Investig New Drugs. 2014;32:213–26. doi: 10.1007/s10637-013-0036-7. [DOI] [PubMed] [Google Scholar]
- 70.Calvo E, Chen VJ, Marshall M, Ohnmacht U, Hynes SM, Kumm E, et al. Preclinical analyses and phase I evaluation of LY2603618 administered in combination with pemetrexed and cisplatin in patients with advanced cancer. Investig New Drugs. 2014;32:955–68. doi: 10.1007/s10637-014-0114-5. [DOI] [PubMed] [Google Scholar]
- 71.Weiss GJ, Donehower RC, Iyengar T, Ramanathan RK, Lewandowski K, Westin E, et al. Phase I dose-escalation study to examine the safety and tolerability of LY2603618, a checkpoint 1 kinase inhibitor, administered 1 day after pemetrexed 500 mg/m(2) every 21 days in patients with cancer. Investig New Drugs. 2013;31:136–44. doi: 10.1007/s10637-012-9815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doi T, Yoshino T, Shitara K, Matsubara N, Fuse N, Naito Y, et al. Phase I study of LY2603618, a CHK1 inhibitor, in combination with gemcitabine in Japanese patients with solid tumors. Anti-cancer Drugs. 2015;26:1043–53. doi: 10.1097/CAD.0000000000000278. [DOI] [PubMed] [Google Scholar]
- 73.Calvo E, Braiteh F, Von Hoff D, McWilliams R, Becerra C, Galsky MD, et al. Phase I Study of CHK1 Inhibitor LY2603618 in Combination with Gemcitabine in Patients with Solid Tumors. Oncology. 2016;91:251–60. doi: 10.1159/000448621. [DOI] [PubMed] [Google Scholar]
- 74.Scagliotti G, Kang JH, Smith D, Rosenberg R, Park K, Kim SW, et al. Phase II evaluation of LY2603618, a first-generation CHK1 inhibitor, in combination with pemetrexed in patients with advanced or metastatic non-small cell lung cancer. Investig New Drugs. 2016;34:625–35. doi: 10.1007/s10637-016-0368-1. [DOI] [PubMed] [Google Scholar]
- 75.Guzi TJ, Paruch K, Dwyer MP, Labroli M, Shanahan F, Davis N, et al. Targeting the replication checkpoint using SCH 900776, a potent and functionally selective CHK1 inhibitor identified via high content screening. Mol Cancer Therapeut. 2011;10:591–602. doi: 10.1158/1535-7163.MCT-10-0928. [DOI] [PubMed] [Google Scholar]
- 76.Montano R, Chung I, Garner KM, Parry D, Eastman A. Preclinical development of the novel Chk1 inhibitor SCH900776 in combination with DNA-damaging agents and antimetabolites. Mol Cancer Therapeut. 2012;11:427–38. doi: 10.1158/1535-7163.MCT-11-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Engelke CG, Parsels LA, Qian Y, Zhang Q, Karnak D, Robertson JR, et al. Sensitization of pancreatic cancer to chemoradiation by the Chk1 inhibitor MK8776. Clin Cancer Res. 2013;19:4412–21. doi: 10.1158/1078-0432.CCR-12-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Montano R, Thompson R, Chung I, Hou H, Khan N, Eastman A. Sensitization of human cancer cells to gemcitabine by the Chk1 inhibitor MK-8776: cell cycle perturbation and impact of administration schedule in vitro and in vivo. BMC Cancer. 2013;13:604. doi: 10.1186/1471-2407-13-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grabauskiene S, Bergeron EJ, Chen G, Chang AC, Lin J, Thomas DG, et al. CHK1 levels correlate with sensitization to pemetrexed by CHK1 inhibitors in non-small cell lung cancer cells. Lung Cancer. 2013;82:477–84. doi: 10.1016/j.lungcan.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schenk EL, Koh BD, Flatten KS, Peterson KL, Parry D, Hess AD, et al. Effects of selective checkpoint kinase 1 inhibition on cytarabine cytotoxicity in acute myelogenous leukemia cells in vitro. Clin Cancer Res. 2012;18:5364–73. doi: 10.1158/1078-0432.CCR-12-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bridges KA, Chen X, Liu H, Rock C, Buchholz TA, Shumway SD, et al. MK-8776, a novel chk1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Oncotarget. 2016 doi: 10.18632/oncotarget.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karp JE, Thomas BM, Greer JM, Sorge C, Gore SD, Pratz KW, et al. Phase I and pharmacologic trial of cytosine arabinoside with the selective checkpoint 1 inhibitor Sch 900776 in refractory acute leukemias. Clin Cancer Res. 2012;18:6723–31. doi: 10.1158/1078-0432.CCR-12-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Daud AI, Ashworth MT, Strosberg J, Goldman JW, Mendelson D, Springett G, et al. Phase I dose-escalation trial of checkpoint kinase 1 inhibitor MK-8776 as monotherapy and in combination with gemcitabine in patients with advanced solid tumors. J Clin Oncol. 2015;33:1060–6. doi: 10.1200/JCO.2014.57.5027. [DOI] [PubMed] [Google Scholar]
- 84.Blasina A, Hallin J, Chen E, Arango ME, Kraynov E, Register J, et al. Breaching the DNA damage checkpoint via PF-00477736, a novel small-molecule inhibitor of checkpoint kinase 1. Mol Cancer Therapeut. 2008;7:2394–404. doi: 10.1158/1535-7163.MCT-07-2391. [DOI] [PubMed] [Google Scholar]
- 85.Zhang C, Yan Z, Painter CL, Zhang Q, Chen E, Arango ME, et al. PF-00477736 mediates checkpoint kinase 1 signaling pathway and potentiates docetaxel-induced efficacy in xenografts. Clin Cancer Res. 2009;15:4630–40. doi: 10.1158/1078-0432.CCR-08-3272. [DOI] [PubMed] [Google Scholar]
- 86.Fang DD, Cao J, Jani JP, Tsaparikos K, Blasina A, Kornmann J, et al. Combined gemcitabine and CHK1 inhibitor treatment induces apoptosis resistance in cancer stem cell-like cells enriched with tumor spheroids from a non-small cell lung cancer cell line. Front Med. 2013;7:462–76. doi: 10.1007/s11684-013-0270-6. [DOI] [PubMed] [Google Scholar]
- 87.Al-Ejeh F, Pajic M, Shi W, Kalimutho M, Miranda M, Nagrial AM, et al. Gemcitabine and CHK1 inhibition potentiate EGFR-directed radioimmunotherapy against pancreatic ductal adenocarcinoma. Clin Cancer Res. 2014;20:3187–97. doi: 10.1158/1078-0432.CCR-14-0048. [DOI] [PubMed] [Google Scholar]
- 88.Busch CJ, Kriegs M, Laban S, Tribius S, Knecht R, Petersen C, et al. HPV-positive HNSCC cell lines but not primary human fibroblasts are radiosensitized by the inhibition of Chk1. Radiother Oncol. 2013;108:495–9. doi: 10.1016/j.radonc.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 89.King C, Diaz HB, McNeely S, Barnard D, Dempsey J, Blosser W, et al. LY2606368 Causes replication catastrophe and antitumor effects through CHK1-dependent mechanisms. Mol Cancer Therapeut. 2015;14:2004–13. doi: 10.1158/1535-7163.MCT-14-1037. [DOI] [PubMed] [Google Scholar]
- 90.Hong D, Infante J, Janku F, Jones S, Nguyen LM, Burris H, et al. Phase I study of LY2606368, a checkpoint kinase 1 inhibitor, in patients with advanced cancer. J Clin Oncol. 2016;34:1764–71. doi: 10.1200/JCO.2015.64.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang GT, Li G, Mantei RA, Chen Z, Kovar P, Gu W, et al. 1-(5-Chloro-2-alkoxyphenyl)-3-(5-cyanopyrazin-2-yl)ureas [correction of cyanopyrazi] as potent and selective inhibitors of Chk1 kinase: synthesis, preliminary SAR, and biological activities. J Med Chem. 2005;48:3118–21. doi: 10.1021/jm048989d. [DOI] [PubMed] [Google Scholar]
- 92.Walton MI, Eve PD, Hayes A, Valenti MR, De Haven Brandon AK, Box G, et al. CCT244747 is a novel potent and selective CHK1 inhibitor with oral efficacy alone and in combination with genotoxic anticancer drugs. Clin Cancer Res. 2012;18:5650–61. doi: 10.1158/1078-0432.CCR-12-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lainchbury M, Matthews TP, McHardy T, Boxall KJ, Walton MI, Eve PD, et al. Discovery of 3-alkoxyamino-5-(pyridin-2-ylamino)pyrazine-2-carbonitriles as selective, orally bioavailable CHK1 inhibitors. J Med Chem. 2012;55:10229–40. doi: 10.1021/jm3012933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barker HE, Patel R, McLaughlin M, Schick U, Zaidi S, Nutting CM, et al. CHK1 inhibition radiosensitizes head and neck cancers to paclitaxel-based chemoradiotherapy. Mol Cancer Therapeut. 2016;15:2042–54. doi: 10.1158/1535-7163.MCT-15-0998. [DOI] [PubMed] [Google Scholar]
- 95.Tse AN, Rendahl KG, Sheikh T, Cheema H, Aardalen K, Embry M, et al. CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clin Cancer Res. 2007;13:591–602. doi: 10.1158/1078-0432.CCR-06-1424. [DOI] [PubMed] [Google Scholar]
- 96.Tao Y, Leteur C, Yang C, Zhang P, Castedo M, Pierre A, et al. Radiosensitization by Chir-124, a selective CHK1 inhibitor: effects of p53 and cell cycle checkpoints. Cell Cycle. 2009;8:1196–205. doi: 10.4161/cc.8.8.8203. [DOI] [PubMed] [Google Scholar]
- 97.Lee JH, Choy ML, Ngo L, Venta-Perez G, Marks PA. Role of checkpoint kinase 1 (Chk1) in the mechanisms of resistance to histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2011;108:19629–34. doi: 10.1073/pnas.1117544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xiao Y, Ramiscal J, Kowanetz K, Del Nagro C, Malek S, Evangelista M, et al. Identification of preferred chemotherapeutics for combining with a CHK1 inhibitor. Mol Cancer Therapeut. 2013;12:2285–95. doi: 10.1158/1535-7163.MCT-13-0404. [DOI] [PubMed] [Google Scholar]
- 99.Azorsa DO, Gonzales IM, Basu GD, Choudhary A, Arora S, Bisanz KM, et al. Synthetic lethal RNAi screening identifies sensitizing targets for gemcitabine therapy in pancreatic cancer. J Transl Med. 2009;7:43. doi: 10.1186/1479-5876-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parsels LA, Morgan MA, Tanska DM, Parsels JD, Palmer BD, Booth RJ, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Therap. 2009;8:45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walton MI, Eve PD, Hayes A, Valenti M, De Haven Brandon A, Box G, et al. The preclinical pharmacology and therapeutic activity of the novel CHK1 inhibitor SAR-020106. Mol Cancer Therap. 2010;9:89–100. doi: 10.1158/1535-7163.MCT-09-0938. [DOI] [PubMed] [Google Scholar]
- 102.Reader JC, Matthews TP, Klair S, Cheung KM, Scanlon J, Proisy N, et al. Structure-guided evolution of potent and selective CHK1 inhibitors through scaffold morphing. J Med Chem. 2011;54:8328–42. doi: 10.1021/jm2007326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Borst GR, McLaughlin M, Kyula JN, Neijenhuis S, Khan A, Good J, et al. Targeted radiosensitization by the Chk1 inhibitor SAR-020106. Int J Radiat Oncol Biol Phys. 2013;85:1110–8. doi: 10.1016/j.ijrobp.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 104.Touchefeu Y, Khan AA, Borst G, Zaidi SH, McLaughlin M, Roulstone V, et al. Optimising measles virus-guided radiovirotherapy with external beam radiotherapy and specific checkpoint kinase 1 inhibition. Radiother Oncol. 2013;108:24–31. doi: 10.1016/j.radonc.2013.05.036. [DOI] [PubMed] [Google Scholar]
- 105.Jackson JR, Gilmartin A, Imburgia C, Winkler JD, Marshall LA, Roshak A. An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res. 2000;60:566–72. [PubMed] [Google Scholar]
- 106.Robles AI, Wright MH, Gandhi B, Feis SS, Hanigan CL, Wiestner A, et al. Schedule-dependent synergy between the heat shock protein 90 inhibitor 17-(dimethylaminoethylamino)-17-demethoxygeldanamycin and doxorubicin restores apoptosis to p53-mutant lymphoma cell lines. Clin Cancer Res. 2006;12:6547–56. doi: 10.1158/1078-0432.CCR-06-1178. [DOI] [PubMed] [Google Scholar]
- 107.Preet R, Siddharth S, Satapathy SR, Das S, Nayak A, Das D, et al. Chk1 inhibitor synergizes quinacrine mediated apoptosis in breast cancer cells by compromising the base excision repair cascade. Biochem Pharmacol. 2016;105:23–33. doi: 10.1016/j.bcp.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 108.Koh SB, Courtin A, Boyce RJ, Boyle RG, Richards FM, Jodrell DI. CHK1 inhibition synergizes with gemcitabine initially by destabilizing the DNA replication apparatus. Cancer Res. 2015;75:3583–95. doi: 10.1158/0008-5472.CAN-14-3347. [DOI] [PubMed] [Google Scholar]
- 109.Massey AJ, Stokes S, Browne H, Foloppe N, Fiumana A, Scrace S, et al. Identification of novel, in vivo active Chk1 inhibitors utilizing structure guided drug design. Oncotarget. 2015;6:35797–812. doi: 10.18632/oncotarget.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nishida H, Tatewaki N, Nakajima Y, Magara T, Ko KM, Hamamori Y, et al. Inhibition of ATR protein kinase activity by schisandrin B in DNA damage response. Nucl Acids Res. 2009;37:5678–89. doi: 10.1093/nar/gkp593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tatewaki N, Nishida H, Yoshida M, Ando H, Kondo S, Sakamaki T, et al. Differential effect of schisandrin B stereoisomers on ATR-mediated DNA damage checkpoint signaling. J Pharmacol Sci. 2013;122:138–48. doi: 10.1254/jphs.13048fp. [DOI] [PubMed] [Google Scholar]
- 112.Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Therap. 2015;149:124–38. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 113.Peasland A, Wang LZ, Rowling E, Kyle S, Chen T, Hopkins A, et al. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer. 2011;105:372–81. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–7. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu Q, Wang J, Kang SA, Thoreen CC, Hur W, Ahmed T, et al. Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h] [1,6]naphthyridin-2(1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. J Med Chem. 2011;54:1473–80. doi: 10.1021/jm101520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Charrier JD, Durrant SJ, Golec JM, Kay DP, Knegtel RM, MacCormick S, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem. 2011;54:2320–30. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 117.Prevo R, Fokas E, Reaper PM, Charlton PA, Pollard JR, McKenna WG, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Therap. 2012;13:1072–81. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pires IM, Olcina MM, Anbalagan S, Pollard JR, Reaper PM, Charlton PA, et al. Targeting radiation-resistant hypoxic tumour cells through ATR inhibition. Br J Cancer. 2012;107:291–9. doi: 10.1038/bjc.2012.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vavrova J, Zarybnicka L, Lukasova E, Rezacova M, Novotna E, Sinkorova Z, et al. Inhibition of ATR kinase with the selective inhibitor VE-821 results in radiosensitization of cells of promyelocytic leukaemia (HL-60) Radiat Environ Biophys. 2013;52:471–9. doi: 10.1007/s00411-013-0486-5. [DOI] [PubMed] [Google Scholar]
- 120.Salovska B, Fabrik I, Durisova K, Link M, Vavrova J, Rezacova M, et al. Radiosensitization of human leukemic HL-60 cells by ATR kinase inhibitor (VE-821): phosphoproteomic analysis. Int J Mol Sci. 2014;15:12007–26. doi: 10.3390/ijms150712007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fujisawa H, Nakajima NI, Sunada S, Lee Y, Hirakawa H, Yajima H, et al. VE-821, an ATR inhibitor, causes radiosensitization in human tumor cells irradiated with high LET radiation. Radiat Oncol. 2015;10:175. doi: 10.1186/s13014-015-0464-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–30. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 123.Mohni KN, Thompson PS, Luzwick JW, Glick GG, Pendleton CS, Lehmann BD, et al. A synthetic lethal screen identifies DNA repair pathways that sensitize cancer cells to combined ATR inhibition and cisplatin treatments. PLoS One. 2015;10:e0125482. doi: 10.1371/journal.pone.0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Josse R, Martin SE, Guha R, Ormanoglu P, Pfister TD, Reaper PM, et al. ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase i inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 2014;74:6968–79. doi: 10.1158/0008-5472.CAN-13-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen T, Middleton FK, Falcon S, Reaper PM, Pollard JR, Curtin NJ. Development of pharmacodynamic biomarkers for ATR inhibitors. Mol Oncol. 2015;9:463–72. doi: 10.1016/j.molonc.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fokas E, Prevo R, Pollard JR, Reaper PM, Charlton PA, Cornelissen B, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hall AB, Newsome D, Wang Y, Boucher DM, Eustace B, Gu Y, et al. Potentiation of tumor responses to DNA damaging therapy by the selective ATR inhibitor VX-970. Oncotarget. 2014;5:5674–85. doi: 10.18632/oncotarget.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leszczynska KB, Dobrynin G, Leslie RE, Ient J, Boumelha AJ, Senra JM, et al. Preclinical testing of an Atr inhibitor demonstrates improved response to standard therapies for esophageal cancer. Radiother Oncol. 2016;121:232–8. doi: 10.1016/j.radonc.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Foote KM, Blades K, Cronin A, Fillery S, Guichard SS, Hassall L, et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl] pyrimidin-2-y l}-1H–indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J Med Chem. 2013;56:2125–38. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- 130.Jones CD, Blades K, Foote KM, Guichard SM, Jewsbury PJ, McGuire T, et al. Abstract 2348: discovery of AZD6738, a potent and selective inhibitor with the potential to test the clinical efficacy of ATR kinase inhibition in cancer patients. Cancer Res. 2013;73:2348. [Google Scholar]
- 131.Vendetti FP, Lau A, Schamus S, Conrads TP, O’Connor MJ, Bakkenist CJ. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget. 2015;6:44289–305. doi: 10.18632/oncotarget.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dillon MT, Barker HE, Pedersen M, Hafsi H, Bhide SA, Newbold KL, et al. Radiosensitization by the ATR Inhibitor AZD6738 through Generation of Acentric Micronuclei. Mol Cancer Therap. 2017;16:25–34. doi: 10.1158/1535-7163.MCT-16-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kwok M, Davies N, Agathanggelou A, Smith E, Oldreive C, Petermann E, et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood. 2016;127:582–95. doi: 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- 134.Flatten K, Dai NT, Vroman BT, Loegering D, Erlichman C, Karnitz LM, et al. The role of checkpoint kinase 1 in sensitivity to topoisomerase I poisons. J Biol Chem. 2005;280:14349–55. doi: 10.1074/jbc.M411890200. [DOI] [PubMed] [Google Scholar]
- 135.Thompson R, Eastman A. The cancer therapeutic potential of Chk1 inhibitors: how mechanistic studies impact on clinical trial design. Br J Clin Pharmacol. 2013;76:358–69. doi: 10.1111/bcp.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kohn EA, Ruth ND, Brown MK, Livingstone M, Eastman A. Abrogation of the S phase DNA damage checkpoint results in S phase progression or premature mitosis depending on the concentration of 7-hydroxystaurosporine and the kinetics of Cdc25C activation. J Biol Chem. 2002;277:26553–64. doi: 10.1074/jbc.M202040200. [DOI] [PubMed] [Google Scholar]
- 137.Shao RG, Cao CX, Shimizu T, O’Connor PM, Kohn KW, Pommier Y. Abrogation of an S-phase checkpoint and potentiation of camptothecin cytotoxicity by 7-hydroxystaurosporine (UCN-01) in human cancer cell lines, possibly influenced by p53 function. Cancer Res. 1997;57:4029–35. [PubMed] [Google Scholar]
- 138.Lee SI, Brown MK, Eastman A. Comparison of the efficacy of 7-hydroxystaurosporine (UCN-01) and other staurosporine analogs to abrogate cisplatin-induced cell cycle arrest in human breast cancer cell lines. Biochem Pharmacol. 1999;58:1713–21. doi: 10.1016/s0006-2952(99)00258-0. [DOI] [PubMed] [Google Scholar]
- 139.Jobson AG, Lountos GT, Lorenzi PL, Llamas J, Connelly J, Cerna D, et al. Cellular inhibition of checkpoint kinase 2 (Chk2) and potentiation of camptothecins and radiation by the novel Chk2 inhibitor PV1019 [7-nitro-1H–indole-2-carboxylic acid {4-[1-(guanidinohydrazone)-ethyl]-phenyl}-amide] J Pharmacol Exp Therapeut. 2009;331:816–26. doi: 10.1124/jpet.109.154997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suzuki M, Yamamori T, Bo T, Sakai Y, Inanami O. MK-8776, a novel Chk1 inhibitor, exhibits an improved radiosensitizing effect compared to UCN-01 by exacerbating radiation-induced aberrant mitosis. Transl Oncol. 2017;10:491–500. doi: 10.1016/j.tranon.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lopez-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J Exp Med. 2012;209:455–61. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70:9693–702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Verlinden L, VandenBempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, et al. The E2F–regulated gene Chk1 is highly expressed in triple-negative estrogen receptor /progesterone receptor /HER-2 breast carcinomas. Cancer Res. 2007;67:6574–81. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- 144.Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci U S A. 2011;108:3336–41. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sarmento LM, Povoa V, Nascimento R, Real G, Antunes I, Martins LR, et al. CHK1 overexpression in T-cell acute lymphoblastic leukemia is essential for proliferation and survival by preventing excessive replication stress. Oncogene. 2015;34:2978–90. doi: 10.1038/onc.2014.248. [DOI] [PubMed] [Google Scholar]
- 146.Cavelier C, Didier C, Prade N, Mansat-De Mas V, Manenti S, Recher C, et al. Constitutive activation of the DNA damage signaling pathway in acute myeloid leukemia with complex karyotype: potential importance for checkpoint targeting therapy. Cancer Res. 2009;69:8652–61. doi: 10.1158/0008-5472.CAN-09-0939. [DOI] [PubMed] [Google Scholar]
- 147.Hong J, Hu K, Yuan Y, Sang Y, Bu Q, Chen G, et al. CHK1 targets spleen tyrosine kinase (L) for proteolysis in hepatocellular carcinoma. J Clin Investig. 2012;122:2165–75. doi: 10.1172/JCI61380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hoglund A, Nilsson LM, Muralidharan SV, Hasvold LA, Merta P, Rudelius M, et al. Therapeutic implications for the induced levels of Chk1 in Mycexpressing cancer cells. Clin Cancer Res. 2011;17:7067–79. doi: 10.1158/1078-0432.CCR-11-1198. [DOI] [PubMed] [Google Scholar]
- 149.Carrassa L, Broggini M, Vikhanskaya F, Damia G. Characterization of the 5’flanking region of the human Chk1 gene: identification of E2F1 functional sites. Cell Cycle. 2003;2:604–9. [PubMed] [Google Scholar]
- 150.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 151.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 152.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–5. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ferrao PT, Bukczynska EP, Johnstone RW, McArthur GA. Efficacy of CHK inhibitors as single agents in MYC-driven lymphoma cells. Oncogene. 2012;31:1661–72. doi: 10.1038/onc.2011.358. [DOI] [PubMed] [Google Scholar]
- 154.Brooks K, Oakes V, Edwards B, Ranall M, Leo P, Pavey S, et al. A potent Chk1 inhibitor is selectively cytotoxic in melanomas with high levels of replicative stress. Oncogene. 2013;32:788–96. doi: 10.1038/onc.2012.72. [DOI] [PubMed] [Google Scholar]
- 155.Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Investig. 2012;122:241–52. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–8. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ruzankina Y, Schoppy DW, Asare A, Clark CE, Vonderheide RH, Brown EJ. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet. 2009;41:1144–9. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wagner SA, Oehler H, Voigt A, Dalic D, Freiwald A, Serve H, et al. ATR inhibition rewires cellular signaling networks induced by replication stress. Proteomics. 2016;16:402–16. doi: 10.1002/pmic.201500172. [DOI] [PubMed] [Google Scholar]
- 159.Wickremsinhe ER, Hynes SM, Palmieri MD, Mitchell MI, Abraham TL, Rehmel JF, et al. Disposition and metabolism of LY2603618, a Chk-1 inhibitor following intravenous administration in patients with advanced and/or metastatic solid tumors. Xenobiotica; The Fate of Foreign Compounds in Biological Systems. 2014;44:827–41. doi: 10.3109/00498254.2014.900589. [DOI] [PubMed] [Google Scholar]
- 160.Dees EC, Baker SD, O’Reilly S, Rudek MA, Davidson SB, Aylesworth C, et al. A phase I and pharmacokinetic study of short infusions of UCN-01 in patients with refractory solid tumors. Clin Cancer Res. 2005;11:664–71. [PubMed] [Google Scholar]
- 161.Sausville EA, Arbuck SG, Messmann R, Headlee D, Bauer KS, Lush RM, et al. Phase I trial of 72-hour continuous infusion UCN-01 in patients with refractory neoplasms. J Clin Oncol. 2001;19:2319–33. doi: 10.1200/JCO.2001.19.8.2319. [DOI] [PubMed] [Google Scholar]
- 162.Rini BI, Weinberg V, Shaw V, Scott J, Bok R, Park JW, et al. Time to disease progression to evaluate a novel protein kinase C inhibitor, UCN-01, in renal cell carcinoma. Cancer. 2004;101:90–5. doi: 10.1002/cncr.20313. [DOI] [PubMed] [Google Scholar]
- 163.Li T, Christensen SD, Frankel PH, Margolin KA, Agarwala SS, Luu T, et al. A phase II study of cell cycle inhibitor UCN-01 in patients with metastatic melanoma: a California Cancer Consortium trial. Investig New Drugs. 2012;30:741–8. doi: 10.1007/s10637-010-9562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Ann Rev Genet. 2007;41:237–80. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Boyer AS, Walter D, Sorensen CS. DNA replication and cancer: From dysfunctional replication origin activities to therapeutic opportunities. Sem Cancer Biol. 2016;37–38:16–25. doi: 10.1016/j.semcancer.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 166.Ekholm-Reed S, Mendez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, et al. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013;32:3744–53. doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- 168.Puigvert JC, Sanjiv K, Helleday T. Targeting DNA repair, DNA metabolism and replication stress as anti-cancer strategies. FEBS J. 2016;283:232–45. doi: 10.1111/febs.13574. [DOI] [PubMed] [Google Scholar]
- 169.Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 170.Abdullah MF, Hoffmann ER, Cotton VE, Borts RH. A role for the MutL homologue MLH2 in controlling heteroduplex formation and in regulating between two different crossover pathways in budding yeast. Cytogenet Genome Res. 2004;107:180–90. doi: 10.1159/000080596. [DOI] [PubMed] [Google Scholar]
- 171.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–62. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J. 2012;443:13–26. doi: 10.1042/BJ20112100. [DOI] [PubMed] [Google Scholar]
- 173.Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, Caldecott KW. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–26. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A. 2010;107:16090–5. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Scorah J, McGowan CH. Claspin and Chk1 regulate replication fork stability by different mechanisms. Cell Cycle. 2009;8:1036–43. doi: 10.4161/cc.8.7.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 177.Hammond EM, Dorie MJ, Giaccia AJ. Inhibition of ATR leads to increased sensitivity to hypoxia/reoxygenation. Cancer Res. 2004;64:6556–62. doi: 10.1158/0008-5472.CAN-04-1520. [DOI] [PubMed] [Google Scholar]
- 178.Hammond EM, Green SL, Giaccia AJ. Comparison of hypoxia-induced replication arrest with hydroxyurea and aphidicolin-induced arrest. Mutat Res. 2003;532:205–13. doi: 10.1016/j.mrfmmm.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 179.Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol. 2002;22:1834–43. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Olcina M, Lecane PS, Hammond EM. Targeting hypoxic cells through the DNA damage response. Clin Cancer Res. 2010;16:5624–9. doi: 10.1158/1078-0432.CCR-10-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cazares-Korner C, Pires IM, Swallow ID, Grayer SC, O’Connor LJ, Olcina MM, et al. CH-01 is a hypoxia-activated prodrug that sensitizes cells to hypoxia/reoxygenation through inhibition of Chk1 and Aurora A. ACS Chem Biol. 2013;8:1451–9. doi: 10.1021/cb4001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, et al. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–18. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chan N, Koritzinsky M, Zhao H, Bindra R, Glazer PM, Powell S, et al. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res. 2008;68:605–14. doi: 10.1158/0008-5472.CAN-07-5472. [DOI] [PubMed] [Google Scholar]
- 184.Chan N, Pires IM, Bencokova Z, Coackley C, Luoto KR, Bhogal N, et al. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res. 2010;70:8045–54. doi: 10.1158/0008-5472.CAN-10-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Scanlon SE, Glazer PM. Multifaceted control of DNA repair pathways by the hypoxic tumor microenvironment. DNA Repair. 2015;32:180–9. doi: 10.1016/j.dnarep.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Foskolou IP, Jorgensen C, Leszczynska KB, Olcina MM, Tarhonskaya H, Haisma B, et al. Ribonucleotide reductase requires subunit switching in hypoxia to maintain DNA replication. Mol Cell. 2017;66:e9. doi: 10.1016/j.molcel.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Davies KD, Cable PL, Garrus JE, Sullivan FX, von Carlowitz I, Huerou YL, et al. Chk1 inhibition and Wee1 inhibition combine synergistically to impede cellular proliferation. Cancer Biol Therap. 2011;12:788–96. doi: 10.4161/cbt.12.9.17673. [DOI] [PubMed] [Google Scholar]
- 188.Carrassa L, Chila R, Lupi M, Ricci F, Celenza C, Mazzoletti M, et al. Combined inhibition of Chk1 and Wee1: in vitro synergistic effect translates to tumor growth inhibition in vivo. Cell Cycle. 2012;11:2507–17. doi: 10.4161/cc.20899. [DOI] [PubMed] [Google Scholar]
- 189.Guertin AD, Martin MM, Roberts B, Hurd M, Qu X, Miselis NR, et al. Unique functions of CHK1 and WEE1 underlie synergistic anti-tumor activity upon pharmacologic inhibition. Cancer Cell Int. 2012;12:45. doi: 10.1186/1475-2867-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Russell MR, Levin K, Rader J, Belcastro L, Li Y, Martinez D, et al. Combination therapy targeting the Chk1 and Wee1 kinases shows therapeutic efficacy in neuroblastoma. Cancer Res. 2013;73:776–84. doi: 10.1158/0008-5472.CAN-12-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chila R, Basana A, Lupi M, Guffanti F, Gaudio E, Rinaldi A, et al. Combined inhibition of Chk1 and Wee1 as a new therapeutic strategy for mantle cell lymphoma. Oncotarget. 2015;6:3394–408. doi: 10.18632/oncotarget.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mak JP, Man WY, Ma HT, Poon RY. Pharmacological targeting the ATR-CHK1-WEE1 axis involves balancing cell growth stimulation and apoptosis. Oncotarget. 2014;5:10546–57. doi: 10.18632/oncotarget.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Shen JP, Srivas R, Gross A, Li J, Jaehnig EJ, Sun SM, et al. Chemogenetic profiling identifies RAD17 as synthetically lethal with checkpoint kinase inhibition. Oncotarget. 2015;6:35755–69. doi: 10.18632/oncotarget.5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Beck H, Nahse-Kumpf V, Larsen MS, O’Hanlon KA, Patzke S, Holmberg C, et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol. 2012;32:4226–36. doi: 10.1128/MCB.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Dominguez-Kelly R, Martin Y, Koundrioukoff S, Tanenbaum ME, Smits VA, Medema RH, et al. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol. 2011;194:567–79. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Magnussen GI, Emilsen E, Giller Fleten K, Engesaeter B, Nahse-Kumpf V, Fjaer R, et al. Combined inhibition of the cell cycle related proteins Wee1 and Chk1/2 induces synergistic anti-cancer effect in melanoma. BMC Cancer. 2015;15:462. doi: 10.1186/s12885-015-1474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Menezes DL, Holt J, Tang Y, Feng J, Barsanti P, Pan Y, et al. A synthetic lethal screen reveals enhanced sensitivity to ATR inhibitor treatment in mantle cell lymphoma with ATM loss-of-function. Mol Cancer Res: MCR. 2015;13:120–9. doi: 10.1158/1541-7786.MCR-14-0240. [DOI] [PubMed] [Google Scholar]
- 198.Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014;74:2835–45. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sanjiv K, Hagenkort A, Calderon-Montano JM, Koolmeister T, Reaper PM, Mortusewicz O, et al. Cancer-specific synthetic lethality between ATR and CHK1 kinase activities. Cell Reports. 2016;14:298–309. doi: 10.1016/j.celrep.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Luciani MG, Oehlmann M, Blow JJ. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J Cell Sci. 2004;117:6019–30. doi: 10.1242/jcs.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mordasini V, Ueda S, Aslandogmus R, Berger C, Gysin C, Huhn D, et al. Activation of ATR-Chk1 pathway facilitates EBV-mediated transformation of primary tonsillar B-cells. Oncotarget. 2017;8:6461–74. doi: 10.18632/oncotarget.14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S Phase. Mol Cell. 2015;59:1011–24. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hattori H, Skoulidis F, Russell P, Venkitaraman AR. Context dependence of checkpoint kinase 1 as a therapeutic target for pancreatic cancers deficient in the BRCA2 tumor suppressor. Mol Cancer Therap. 2011;10:670–8. doi: 10.1158/1535-7163.MCT-10-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chen CC, Kennedy RD, Sidi S, Look AT, D’Andrea A. CHK1 inhibition as a strategy for targeting Fanconi Anemia (FA) DNA repair pathway deficient tumors. Mol Cancer. 2009;8:24. doi: 10.1186/1476-4598-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sultana R, Abdel-Fatah T, Perry C, Moseley P, Albarakti N, Mohan V, et al. Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibition is synthetically lethal in XRCC1 deficient ovarian cancer cells. PLoS One. 2013;8:e57098. doi: 10.1371/journal.pone.0057098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hocke S, Guo Y, Job A, Orth M, Ziesch A, Lauber K, et al. A synthetic lethal screen identifies ATR-inhibition as a novel therapeutic approach for POLD1-deficient cancers. Oncotarget. 2016;7:7080–95. doi: 10.18632/oncotarget.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–15. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 208.Booth L, Cruickshanks N, Ridder T, Dai Y, Grant S, Dent P. PARP and CHK inhibitors interact to cause DNA damage and cell death in mammary carcinoma cells. Cancer Biol Therap. 2013;14:458–65. doi: 10.4161/cbt.24424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Li L, Chang W, Yang G, Ren C, Park S, Karantanos T, et al. Targeting poly(ADP-ribose) polymerase and the c-Myb-regulated DNA damage response pathway in castration-resistant prostate cancer. Sci Signal. 2014;7:ra47. doi: 10.1126/scisignal.2005070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pei XY, Dai Y, Youssefian LE, Chen S, Bodie WW, Takabatake Y, et al. Cytokinetically quiescent (G0/G1) human multiple myeloma cells are susceptible to simultaneous inhibition of Chk1 and MEK1/2. Blood. 2011;118:5189–200. doi: 10.1182/blood-2011-02-339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pei X-Y, Dai Y, Chen S, Bodie W, Kramer L, Dent P, et al. The MEK1/2 inhibitor AZD6244 (ARRY-142886) interacts synergistically with the novel Chk1 inhibitor AZD7762 to induce apoptosis in human multiple myeloma cells. Cancer Res. 2014;68 LB-103-LB- [Google Scholar]
- 212.Pollard J, Reaper P, Peek A, Hughes S, Djeha H, Cummings S, et al. Abstract 3711: Pre-clinical combinations of ATR and PARP inhibitors: Defining target patient populations and dose schedule. Cancer Res. 2016;76:3711. [Google Scholar]
- 213.Dai Y, Khanna P, Chen S, Pei XY, Dent P, Grant S. Statins synergistically potentiate 7-hydroxystaurosporine (UCN-01) lethality in human leukemia and myeloma cells by disrupting Ras farnesylation and activation. Blood. 2007;109:4415–23. doi: 10.1182/blood-2006-09-047076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dai Y, Chen S, Pei XY, Almenara JA, Kramer LB, Venditti CA, et al. Interruption of the Ras/MEK/ERK signaling cascade enhances Chk1 inhibitor-induced DNA damage in vitro and in vivo in human multiple myeloma cells. Blood. 2008;112:2439–49. doi: 10.1182/blood-2008-05-159392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Mitchell C, Hamed HA, Cruickshanks N, Tang Y, Bareford MD, Hubbard N, et al. Simultaneous exposure of transformed cells to SRC family inhibitors and CHK1 inhibitors causes cell death. Cancer Biol Therap. 2011;12:215–28. doi: 10.4161/cbt.12.3.16218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tang Y, Dai Y, Grant S, Dent P. Enhancing CHK1 inhibitor lethality in glioblastoma. Cancer Biol Therap. 2012;13:379–88. doi: 10.4161/cbt.19240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dai Y, Yu C, Singh V, Tang L, Wang Z, McInistry R, et al. Pharmacological inhibitors of the mitogen-activated protein kinase (MAPK) kinase/MAPK cascade interact synergistically with UCN-01 to induce mitochondrial dysfunction and apoptosis in human leukemia cells. Cancer Res. 2001;61:5106–15. [PubMed] [Google Scholar]
- 218.Pei XY, Dai Y, Tenorio S, Lu J, Harada H, Dent P, et al. MEK1/2 inhibitors potentiate UCN-01 lethality in human multiple myeloma cells through a Bim-dependent mechanism. Blood. 2007;110:2092–101. doi: 10.1182/blood-2007-04-083204. [DOI] [PMC free article] [PubMed] [Google Scholar]