Summary
Invasive fungal infections are accompanied by high mortality rates that range up to 90%. At present, only three different compound classes are available for use in the clinic, and these often suffer from low bioavailability, toxicity and drug resistance. These issues emphasize an urgent need for novel antifungal agents. Herein, we report the identification of chemically versatile benzamide and picolinamide scaffolds with antifungal properties. Chemogenomic profiling and biochemical assays with purified protein identified Sec14p, the major phosphatidylinositol/phosphatidylcholine transfer protein in Saccharomyces cerevisiae, as the sole essential target for these compounds. A functional variomics screen identified resistance-conferring residues that localized to the lipid-binding pocket of Sec14p. Determination of the X-ray co-crystal structure of a Sec14p-compound complex confirmed binding in this cavity and rationalized both the resistance conferring-residues and the observed structure-activity relationships. Taken together, these findings open new avenues for rational compound optimization and development of novel antifungal agents.
Keywords: Sec14p, lipid transfer protein, chemogenomics, target identification, functional variomics, co-crystal, antifungal, benzamide, picolinamide
eTOC blurb
Rising mortality rates and a limited armamentarium ask for novel, effective antifungal agents. Pries et al. have identified benz- and picolinamide derived compounds that target the fungal lipid transfer protein Sec14. Intersecting structure-activity relationship data, high-resolution genetics data and the first Sec14p crystal structure in complex with an inhibitor, pave the way for rational evaluation of this compound/target pair for its antifungal potential.
Introduction
Invasive fungal infections (IFIs) are associated with high morbidity and mortality rates (Schmiedel and Zimmerli, 2016). Moreover, the incidence of fungal infections is rapidly escalating as the numbers of premature infants, patients receiving immunosuppressive therapy, and patients afflicted with AIDS, neoplastic disease and advanced age increase (Pfaller et al., 2006). Although the incidence of IFIs is rising, the launch of initiatives aimed at antifungal drug discovery is lagging (Schmiedel and Zimmerli, 2016). Only three compound classes (i.e. azoles, polyenes, and echinocandins) are currently in therapeutic use (Prasad et al., 2016; Roemer and Krysan, 2014), and these act on the fungal plasma membrane, its biosynthetic pathways and cell wall components, respectively. However, these drugs are only modestly effective in reducing the high mortality rates associated with IFIs because of: (i) restrictions in route of administration, (ii) limited spectrum of activity, (iii) poor bioavailability in target tissues, (iv) significant toxicities that result from undesirable drug interactions, and (v) the emergence of drug resistant pathogens (Brown et al., 2012). All of these factors highlight the acute demand for the discovery and development of novel classes of small molecule inhibitors (SMI) against a wider range of targets.
Genome-wide fitness profiling approaches using the surrogate model fungus Saccharomyces cerevisiae are proving themselves as powerful tools for identifying novel antifungal agents and developing hypotheses for their cellular target (Hoepfner et al., 2012; Richie et al., 2013). Haploinsufficiency profiling (HIP) identifies candidate pathways for direct inhibition by the compound as genes are identified in which one functional copy, compared to two, confers hypersensitivity to inhibition by that compound. In homozygous profiling (HOP), both gene copies are deleted so that information on synthetic lethal gene interactions and compensating pathways is accessed (Giaever et al., 1999; Hoon et al., 2008; Lee et al., 2014; Lum et al., 2004; Parsons et al., 2006; Roemer et al., 2011). Using the HIP-HOP approach, we identified compounds with benzamide and picolinamide scaffolds as inhibitors of Sec14p - the major phosphatidylinositol transfer protein (PITP) in S. cerevisiae whose activity is essential for cell viability (Bankaitis et al., 1990; Bankaitis et al., 1989). A battery of genetic and biochemical assays corroborated the target hypothesis. Finally, we report the first crystal structure for a Sec14p::SMI complex - thus providing a detailed mechanism of inhibition. Taken together, these findings outline a path for rationale synthesis of the next generation of small molecule Sec14p inhibitors with optimized antifungal properties.
Results
Chemogenomic profiling
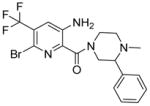
Screening of the Novartis compound archive for small molecule inhibitors (SMI) with antifungal activity (Richie et al., 2013) identified the picolinamide-containing compound 1 (Table 1) as novel inhibitor of S. cerevisiae growth with an IC50 value of 13.5 μM. Lack of potent cytotoxicity against mammalian HCT116 cells (Table 1) led us to initiate follow up studies on this chemotype. To identify the target protein of compound 1 haploinsufficiency profiling (HIP) and homozygous profiling (HOP) experiments were performed. For data analysis, the strain sensitivity was plotted against the z-score, which relates the sensitivity score of a strain in the compound profile to the variability in sensitivity of that strain across all the >3000 compounds tested in the dataset (Hoepfner et al., 2014).
Table 1. Structures and IC50 values for active compounds.
IC50 values were determined in a cell based growth assay with the heterozygous SEC14 deletion strain against a wildtype control, with a luminescent cell viability assay on HCT116 cells and in an in vitro lipid transfer assay, as indicated. “nd” indicates “not determined”.
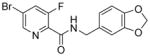
| Cpd | Structure | IC50 [μM] S. cerevisiae wildtype | fold IC50 shift S. cerevisiae wildtype/wildtypeΔ8 | IC50 [μM] S. cerevisiae SEC14/sec14 | IC50 [μM] HCT116 | IC50 [μM] lipid transfer |
|---|---|---|---|---|---|---|
| 1 |
|
13.5 | 13.8x | 5.9 | 30.4 | nd |
| 2 |
|
28.5 | 2.7x | 9.4 | >100 | 6.7 |
| 3 |
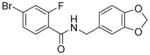
|
6.6 | 2.8x | 1.7 | >100 | 0.7 |
| 4 |
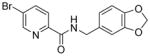
|
134.9 | 2.3x | 99.4 | >100 | 29.4 |
| 5 |
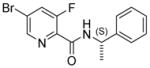
|
42.9 | 2.7x | 32.6 | >100 | 30.7 |
| 6 |
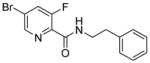
|
>200 | nd | >200 | >100 | 129 |
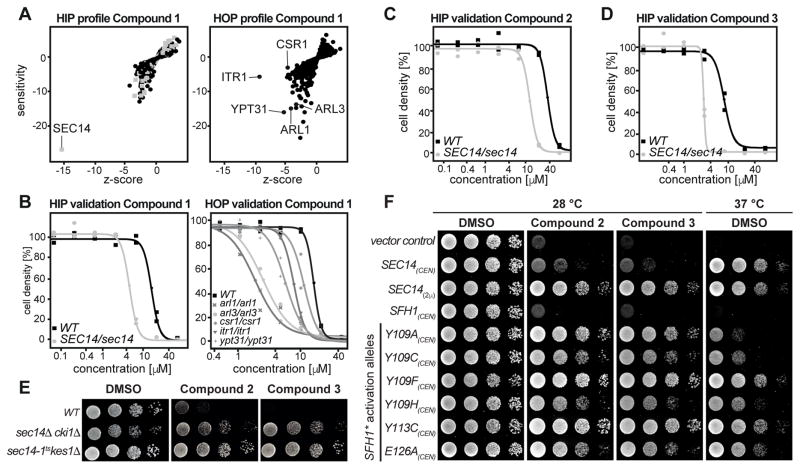
Execution of the HIP-HOP assay at a sub-lethal compound concentration (20 μM) identified a single strain to be significantly hypersensitive to compound 1: the strain carrying a heterozygous deletion for SEC14 (Figure 1A). SEC14 encodes an essential phosphatidylinositol (PtdIns)/phosphatidylcholine (PtdCho) transfer protein (PITP) that plays a crucial role in protein transport from the trans-Golgi network (TGN) and endosomal system. As such, Sec14p function is essential for yeast cell viability (Bankaitis et al., 1990). HOP (Figure 1A) identified strains deleted for ITR1(Nikawa et al., 1991), encoding a myo-inositol transporter, YPT31 (Benli et al., 1996), encoding a Rab GTPase essential for Golgi function, ARL1 (Lee et al., 1997) and ARL3 (Huang et al., 1999) encoding two ARF-like GTPases that are involved in TGN/endosomal membrane traffic and SFH2/CSR1 (Li et al., 2000), a non-classical PITP, to be synthetic lethal. The significance of the HOP screen gene set was that it recapitulated independent screens that previously revealed genetic interactions between SEC14 and ARL1, ARL3, YPT31 and CSR1/SFH2 (Fairn et al., 2007; Mousley et al., 2008). Hypersensitivities of the strains identified in the genome-wide, pooled experiment, were validated by recording individual growth curves with the specific single strains picked from the HIP and HOP collection (Figure 1B). Taken together, the data from the genome-wide fitness profiling suggested that compound 1 exerts its antifungal action through Sec14p.
Figure 1.
Chemogenomic profiling and hypersensitivity validation. A) Calculated profiles of the chemogenomic profiling experiment. HIP outlines hits directly affected by the test compound. HOP identifies synthetic interactions with the target. Essential genes are depicted by grey boxes, non-essential genes by black dots. B) Single strain validation of hits from the chemogenomic profiling experiments as recorded in duplicates. C, D) Confirmation of hypersensitivity of the Sec14/sec14 HIP strain against compound 2 and 3 as recorded in duplicates. E) The wildtype and two “bypass Sec14p” strains were spotted on rich medium (YPD) supplemented with DMSO or 20 μM of compound 2 and 3 and incubated at 30 °C for 48 hours. F) Transformants of the temperature-sensitive sec14-1ts yeast strain harboring centromeric (CEN) plasmids carrying either SFH1 or the designated SFH1 activation alleles (SFH1*) were spotted on minimal media supplemented with 120 μM of compound 2, 30 μM of compound 3 or DMSO as indicated and incubated at permissive (left) or restrictive (right) temperature. Transformants harboring YCplac33 (empty vector), or SEC14 expressed from a centromeric (CEN) or a multi-copy plasmid (2μ) served as controls.
Chemical derivatization
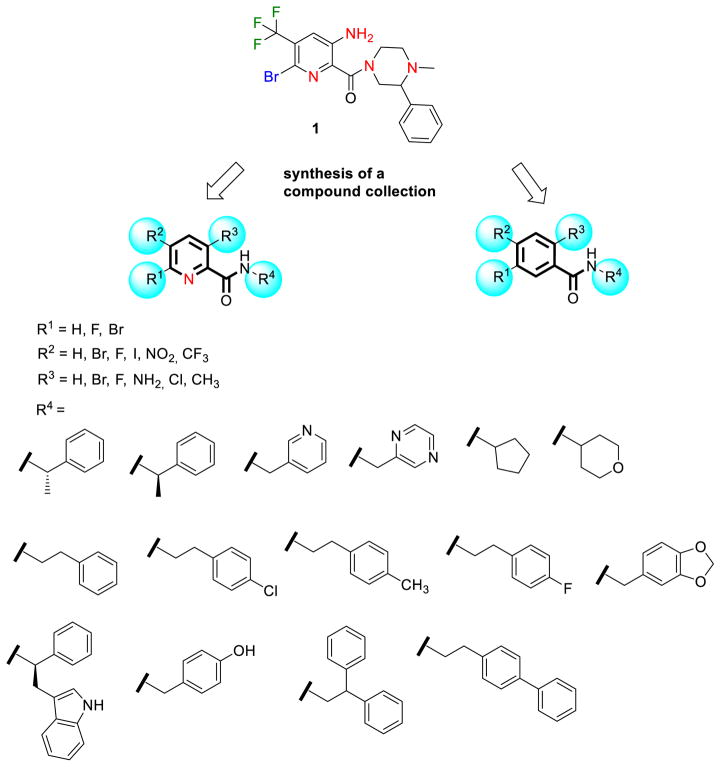
Sec14p is an attractive antifungal target as it has been shown to be druggable (Filipuzzi et al., 2016; Nile et al., 2014), it executes essential functions in several pathogenic fungi and it is required for efficient secretion of pathogenicity factors (Chayakulkeeree et al., 2011). We therefore sought to delineate structure-activity relationships by producing and testing a small compound collection generated by coupling different carboxylic acids and amines via established chemical synthesis methods (El-Faham and Albericio, 2010; El-Faham et al., 2009) (Figure 2, supplementary Data S1). All derivatives were tested for their activity against wildtype S. cerevisiae cells (supplementary Table S1). To ensure that the SMIs still acted at the level of Sec14p, hypersensitivity against the heterozygous SEC14 deletion strain, as observed in the HIP assay for compound 1, was verified. For compounds 2, 3 (Figure 1C, D), 4 and 5 (Table 1) decreased fitness of the SEC14/sec14Δ heterozygote was observed relative to the isogenic wildtype. Of this SMI series, the benzamide compound 3 was the most potent with an IC50 of 6.6 μM. These data indicated that the nitrogen of the picolinamide moiety was not essential for the Sec14p-targeted inhibition of these compounds. Compound 3 however had much reduced solubility in pure aqueous solutions at pH 7.4 (supplementary Table S2), thereby hampering subsequent testing by requiring careful handling when diluting into a variety of fungal and mammalian culture media. Substituting the benzodioxole with benzene, biphenyl, fluorobenzene, pyridine or pyrazine functional groups either reduced potency or ablated inhibitory activity entirely (supplementary Table S1). Similarly, exchange of bromine and fluorine on the benzamide/picolinamide moiety was also incompatible with inhibitor activity. One desirable property of the bioactive derivatives was that all active compounds of the collection showed reduced cytotoxicity for HCT116 cells compared to compound 1 (Table 1).
Figure 2.
Synthesis of a collection of benz- and picolinamides for structure-activity relationship studies.
Validation of Sec14p as cellular target
Several independent approaches were taken to establish Sec14p as the direct target of the most bioactive benzamide (compound 3) and picolinamides (compounds 1, 2, 4, 5, 6). First, we exploited the fact that the otherwise essential Sec14p activity is rendered dispensable in mutant cells deleted for structural genes of the cytidine diphosphate-choline pathway (e.g. CKI1 (Cleves et al., 1991)) or the structural gene for the oxysterol binding protein homolog Kes1p (Fang et al., 1996). In these “bypass Sec14p” mutants, a functional and Sec14p-independent balance of lipid metabolism and PtdIns-4-phosphate signaling for exocytic membrane trafficking from the yeast TGN/endosomal system is restored (Cleves et al., 1991; Fang et al., 1996; Li et al., 2002). It was previously shown that the growth inhibitory activity of highly specific Sec14p-targeted SMIs is alleviated in genetic backgrounds carrying loss-of-function cki1 or kes1 alleles (Nile et al., 2014). Thus, the cki1 and kes1 “bypass Sec14p” mutants were spotted on agar plates supplemented with compound 2 and 3 to a final concentration of 20 μM (Figure 1E). Indeed, growth of the “bypass Sec14p” mutants was indifferent to challenge by these compounds.
To further investigate target specificity, we assessed the inhibitory effects of compound 2 and 3 on Sfh1p, the closest Sec14p homolog (Schaaf et al., 2011) in S. cerevisiae. Sfh1p is a non-essential protein and therefore cannot be identified as a drug target by HIP profiling. In these experiments, we adopted the previously described strategy of assessing the inhibitory effects of compound 2 and 3 on Sfh1p activation mutants that were identified by a directed evolution approach (Khan et al., 2016; Nile et al., 2014) and for which single residue substitutions endow this pseudo-Sec14p with robust Sec14p-like PITP activities in vivo and in vitro (Schaaf et al., 2011). All SFH1 activation alleles (SFH1*) expressed from a single-copy plasmid endowed resistance to both compounds 2 and 3 – even when the SMI was used at concentrations sufficiently high (120 and 30 μM, respectively) that even an additional copy of wildtype SEC14 failed to restore cell growth (Figure 1F). Rather, the SFH1*-associated SMI-resistance phenotype resembled that of yeast expressing a multi-copy episomal SEC14 expression plasmid or yeast carrying cki1 or kes1 ‘bypass Sec14p’ mutations. Taken together, these results strongly suggest that Sec14p is the sole essential target for these compounds in the yeast cell.
SMIs inhibit Sec14p lipid transfer activity in vitro
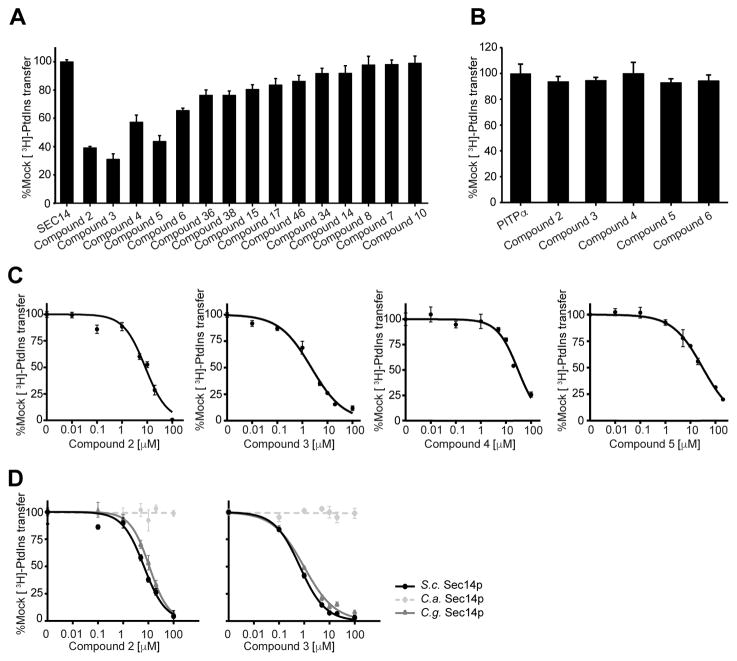
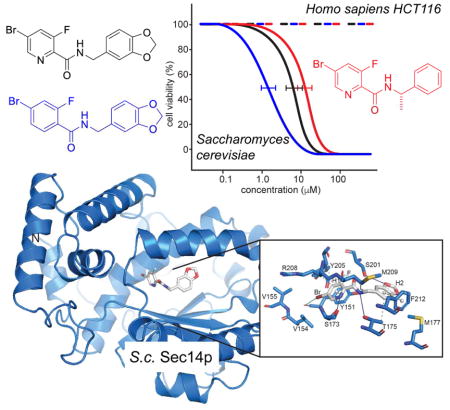
Based on the knowledge gained from the cell-based growth inhibition tests, a subset of 15 compounds was chosen for direct analysis of their inhibitory effect on Sec14p-mediated phospholipid transfer in a purified in vitro system (Khan et al., 2016; Nile et al., 2014; Schaaf et al., 2008) (Figure 3A). To exclude the possibility that compound activity was due to non-specific membrane-active effects, the selected scaffolds were tested in parallel in phospholipid transfer assays reconstituted with the structurally unrelated mammalian PtdIns/PtdCho transfer protein PITPα (Figure 3B). Recombinant Sec14p was pre-incubated with acceptor membranes and compounds for 30 min at 37 °C, the assay was initiated by the addition of radiolabeled rat liver microsomes, and the reaction was terminated after incubation of the assay for 30 min at 37 °C. The [3H]-PtdIns transfer activities in the presence of test compounds were normalized to DMSO (mock) controls. Compounds 2, 3, 4, 5 and 6 showed clear inhibition of PtdIns-transfer mediated by Sec14p whereas no such inhibitory effects were observed when PtdIns-transfer was catalyzed by the mammalian PITPα. Furthermore, IC50 values were determined for active compounds in subsequent titration experiments (Figure 3C, Table 1). In agreement with the growth inhibition data, compound 3 again scored as the most potent inhibitor of Sec14p PtdIns-transfer activity with an IC50 of 0.7 μM (IC50 = 7 μM in S. cerevisiae). To assess if compound 2 and 3 also inhibited the Sec14p of pathogenic species we recorded IC50 curves as described above on the purified porteins of C. albicans and C. glabrata. In this experiment both compounds scored similar activity on S. cerevisiae and C. glabrata but did not inhibit PtdIns-transfer by the C. albicans protein in the tested concentration range (Figure 3D). Activity on at least one other important fungal pathogen and the absence of any measurable inhibition of PtdIns-transfer by the mammalian PITPα not only indicated that the Sec14p-directed inhibitory effect of compound 3 was fungal specific (and not the result of non-specific membrane-active effect), but it further emphasized the potential of this chemotype as antifungal lead.
Figure 3.
Analysis of lipid transfer inhibition with recombinant Sec14p. A) Fifteen compounds were tested for inhibition of Sec14p-mediated [3H]-PtdIns transfer at a fixed concentration of 20 μM SMI (see Materials and Methods). Values represent mean ± s.e.m. of triplicate assay determinations from two independent experiments. Total radiolabel inputs per assay ranged from 8356–9604 c.p.m., backgrounds from 436–489 c.p.m. and the transfer efficiency from 14–16% of total input [3H]-PtdIns. Activities were normalized against the mock condition set at 100%. B) The five small molecules that inhibited Sec14p [3H]-PtdIns transfer by > 30% in the end-point assays in (a) were tested for inhibition of the structurally unrelated mammalian PITPα. [3H]-PtdIns input for these assays ranged from 10703–11202 c.p.m., assay background from 403–415 c.p.m., and [3H]-PtdIns transfer efficiencies ranged from 10%-11% of total input radiolabel. Values represent mean + s.e.m. of triplicate assay determinations from two independent experiments. C) IC50 values for Sec14p-active compounds. Small molecule inhibitors of Sec14p were titrated into PtdIns-transfer assays to determine IC50 values. [3H]-PtdIns input into these assays varied from 9649 - 11034 c.p.m, backgrounds ranged from 243–648 c.p.m., and transfer efficiencies as functions of total [3H]-PtdIns input ranged from 12–18%. D) IC50 curves for compounds 2 and 3 tested against Sec14p of S. cerevisisae, C. albicans and C.glabrata. IC50 values represent a 95% confidence interval from two independent experiments, with triplicate determinations for each data point
Single amino acid substitutions render Sec14p resistant to inhibition by compound 2 and 3
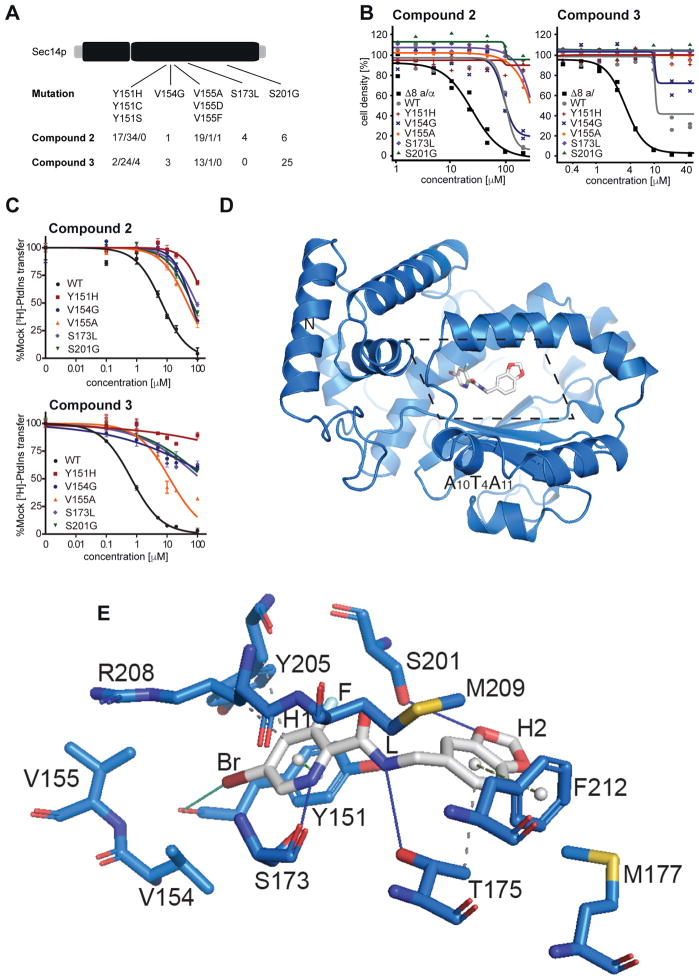
The genetic and biochemical data strongly supported Sec14p as the sole essential cellular target of this SMI series. To obtain additional resolution for how this chemotype engages its target, and to identify amino acids mediating interaction and selectivity for these compounds, a functional variomics screen was deployed (Huang et al., 2013; Pries et al., 2016). This assay exploited a library of plasmid-encoded SEC14 genes mutagenized by error-prone polymerase chain reaction (PCR) and screens for genetically dominant resistance of respective yeast transformants against the compound of interest. 5 × 106 BY4743 wild-type cells transformed with library DNA previously identified to be of a complexity above 2 × 105 (Huang et al., 2013) were plated onto 20 cm plates with synthetic defined media lacking uracil and containing growth inhibitory concentrations of compound 2 (250 μM) or compound 3 (100 μM). Distinct colonies appeared after 72 hours and 96 fast-growing colonies were selected from both SMI-resistance screens. The plasmid-encoded SEC14 genes were then subjected to nucleotide sequence analysis and single nucleotide polymorphisms (SNPs) were identified. For both compounds, single amino acid substitutions involving Tyr151, Val154, Val155, and Ser201 were recovered. Among the 96 clones picked against each compound, the frequencies of mutations identified with unambiguous sequences were as follows: Tyr151 substitutions were identified 51 and 30 times in compound 2 and compound 3 selections respectively, Val154 was recovered 1 and 3 times, Val155 21 and 14 times, and Ser201 6 and 25 times, respectively. Furthermore, the compound 2 resistance screen identified a missense substitution of Ser173 to Leu in four resistant clones (Figure 4A). Genotype-phenotype linkage of the most prominent SNPs, was verified by reintroduction of the plasmid encoded mutant SEC14 genes into the BY4743 8 MATa/α strain, that is deleted for eight genes involved in drug resistance. Resistance phenotypes were subsequently assessed by dose-response experiments (Figure 4B). Integration of an additional SEC14 wildtype allele already yielded a 5- fold IC50 shift with compound 2 and 3-fold IC50 shift with compound 3 compared to wildtype cells. However, with the sole exception of SEC14V154G, all mutant alleles tested endowed naïve yeast cells with significantly increased, or effectively complete, resistance to SMI challenge.
Figure 4.
Structural and functional analysis of Sec14p-compound interactions. A) SNPs and mutation frequency as identified by functional variomics for both tested compounds. B) Validation of identified mutations by integrative transformation of indicated SEC14 allele into cells. C) Validation of identified mutations by in vitro testing on recombinant protein. D) Overview of the structure of Sec14p (as cartoon, in marine) in complex with compound 2 (in sticks representation, in grey). The section displayed in detail in panel e is outlined by dotted lines. e) Detailed view (rotated 45° along the x-axis for clarity) of the binding pocket of Sec14p bound to compound 2 as identified by co-crystallization at 2.6 Å resolution. Interacting residues and relevant secondary structure elements of Sec14p are labeled; side chains are colored in marine and shown as sticks. H- and halogen-bonding are visualized as solid lines in blue and green, respectively while hydrophobic interactions are shown as grey dashed lines. p-p stackings are depicted as green dashed lines (light green for parallel stacking; smudge green for perpendicular stacking) with aromatic ring centers as grey spheres. Functional groups of compound 2 are indicated.
Next, these Sec14p variants were purified as recombinant proteins from E. coli and [3H]-PtdIns-transfer activities were measured for each in SMI titrations in vitro. All of these mutant Sec14ps exhibited substantially increased resistance to compound 2 and compound 3-mediated inhibition of [3H]-PtdIns-transfer relative to wildtype Sec14p. With regard to compound 2, all mutant proteins exhibited IC50s that were some 7- to 30-fold higher than that of wild-type Sec14p (Figure 4C, supplementary Table S3). Moreover, all mutant proteins but one were essentially indifferent to challenge with compound 3. That exception, Sec14pV155A, exhibited a more modest 17-fold increase in IC50 relative to wild-type Sec14p (Figure 4C, supplementary Table S3). In general, the IC50s for each mutant protein were reflective of the levels of SMI resistance for the corresponding mutant yeast strains as measured by cell growth. These data unambiguously validate the identified resistance conferring mutations by directly measuring PtdIns activities and demonstrating that the magnitude of intrinsic Sec14p resistance scaled proportionately to the SMI-resistance of the corresponding yeast strain.
Mode of ligand binding
To identify the mechanism of inhibition, Sec14p was co-crystallized with picolinamide compound 2 and the crystal structure of the complex was determined at 2.6 Å resolution. Compound 2 was prioritized for increased potency over compound 1 and better solubility than compound 3 (supplementary Table S2). The structure was solved by Molecular Replacement (MR) using Sec14p (pdb id: 1aua (Sha et al., 1998a)) as input model in PHASER (Mccoy et al., 2007). The model was rebuilt and refined using PHENIX and COOT (Adams et al., 2010; Emsley and Cowtan, 2004). The final model exhibited an Rfactor of 19.9% and an Rfree of 22.5% with good stereochemistry (supplementary Table S4). The asymmetric unit (ASU) contained one molecule and Sec14p residues from 4 to 303 were identified in the final model. A discrete portion of the electron density in the 2mFo-DFc map (contoured at 1 sigma) revealed the presence of the ligand (supplementary Figure S1). As expected, the protein exhibited a typical Sec14p fold consisting of a globular structure with an N-terminal tripod motif and a C-terminal lobe that included a large hydrophobic pocket of sufficient volume to accommodate a single phospholipid molecule (Schaaf et al., 2008; Sha et al., 1998a) (Figure 4D, supplementary Figure S2). In the structure, Sec14p showed an open conformation with the helical gate (helix A10T4A11 (Bankaitis et al., 2010; Ryan et al., 2007; Schaaf et al., 2008)) displaced, as in the previously described detergent-bound Sec14p (Sha et al., 1998b). Superposition with a known structure of Sec14p in the open conformation showed that the two structures were very similar overall with root mean square deviation (rmsd) of 0.336 Å over 1917 atoms (pdb id.: 1aua (Sha et al., 1998a)) (supplementary Figure S2).
In the crystallized complex, compound 2 occupied the lipid-binding pocket of Sec14p where it was deeply buried in the cavity away from the solvent-exposed Sec14p surface (Figure 4D, E). The buried surface area of the ligand was 440.32 Å2. The bromo-pyridine group (H1) of the picolinamide moiety pointed toward the core of the cavity while the 1,3-benzodioxole ring (H2) was oriented towards solvent. The bromine was in van der Waals contact with Val154 and Val155 (range from 3.7–4.2 Å) and interacted with the carboxyl group of Tyr151 (3.51 Å). The pyridine ring engaged in a π-π stacking interaction with Tyr151 on one side, and in a hydrogen-bond (H-bond) interaction between the amine and the carboxyl of Ser173 on the other. On this side, the Arg208 side chain and backbone atoms engaged in van der Waals interactions and further stabilized SMI binding. The fluorine atom interacted with Ser201 and Tyr151 and was in long-range interactions with Tyr205. The linker (L) that connected rings H1 and H2 of compound 2 was stabilized by interactions with Ser201 and Tyr151 as well as with Met209 and Thr175. The 1,3-benzodioxole ring was positioned for H-bond interaction with Ser201 and also engaged in van der Waals interactions with Phe212, Thr175 and Met177 on the other side. These structural data were fully consistent with the results of the functional variomics screen described above that independently identified missense substitutions for Tyr151, Val154, Val155, Ser201, and Ser173 as incompatible with SMI binding.
Structure-activity relationship analysis
Structure-activity relationship (SAR) data assigned the relative importance of individual functional groups for the benz- and picolinamide analogs to their activities as Sec14p inhibitors. In that regard, an obligatory requirement for the para-Br on the phenyl ring (H1) and the H1 ring itself were notable - as evidenced by compound 2–6 showing inhibition of Sec14p PtdIns-transfer activity while compounds 7–11 were ineffective inhibitors (supplemental Table S1). The crystal and in silico docking data rationalized these obligate requirements (Figure 4D, E, supplementary Figure S3, S4). The hydrophobic planar ring system (H1) was critical for inhibitor activity as it: (i) acted as a scaffold to properly position the hydrophobic para-Br group for a functionally essential interaction with V154V155 and the backbone carboxyl of Tyr151, and (ii) provided a planar ring system for stacking interactions within the hydrophobic sub-pocket composed of residues Y151, V154, V155 and R208. Those latter stacking interactions lent significant stability to scaffold binding. The fluorine on Group R3 at the meta position of planar ring H1 contributed additional hydrophobicity (compounds 2, 3, 5, 6). Polar substitutions (compounds 29, 34, 38) and incorporation of larger moieties (e.g. CH3, Cl, Br; see compounds 7, 11, 36) at that position were not well tolerated. Similarly, a halogen substitution on the R1 group (compounds 9, 10, 11) also failed to enhance the inhibitory activities of these compounds.
Nitrogen substitution in the hydrophobic planar ring H1 resulted in decreased activity, while inhibitory activity was enhanced when H1 was a hydrophobic phenyl ring system. The carboxyl oxygen (A) of the carboxamide group was required for the activity of the compounds as it favorably contributed to the polar amphipathic microenvironment of the PtdCho binding site. Similarly, the carboxamide amide nitrogen contributed to polarity of the molecule and engaged residue Thr175 in polar and/or H-bond interactions.
The methylene group linker (L) that connects the carboxamide group and planar ring H1 with the proximal hydrophobic planar ring system (H2) also contributed to the activity of the picolinamide analogs. The SAR indicated the methylene group was well tolerated and oriented the benzodioxole planar ring H2 in a conformation favorable for docking to the amphipathic region of the binding cavity. However, extending the linker region (L), or substitution of methyl or phenyl group on the linker, was incompatible with inhibition of Sec14p activity. Several modifications of the hydrophobic planar ring system H2 were investigated and affect the potencies of active picolinamides. Alterations in size of that ring system, or modification of the ring system with polar substitutions, was also predicted to compromise SMI incorporation into the Sec14p hydrophobic cleft with the result that inhibitory activity was ablated.
Structure-based optimization of the benzamide/picolinamide series
Based on the Sec14p::compound 2 co-crystal structure, a medicinal chemistry transformation approach was applied to design novel chemical structures by structure-based transformation and optimization of existing ligands. Van der Waals interaction maps of the Sec14p lipid binding pocket were generated to identify steric incompatibility boundaries. For this purpose, we also generated an extended set of strains with mutations in the binding pocket (supplementary Figure S6). These analyses suggest the availability of space between the respective C-terminal section of β-strand B3 and the A9 helix that might be productively exploited in alternative derivatizations of the benzodioxole planar ring H2 in further compound optimizations. Electrostatic maps and contact preference statistics were also created/applied to identify preferred energetically favorable locations for: (i) hydrophobic entities, and (ii) hydrogen bond donors and acceptors for potential ligand substitutions (Figure 4E, supplementary Figure S4, S5). This model was used to screen fragment substitutions with filters based on drug-like properties and favorable pharmacophoric features for optimal binding (see Materials and Methods). Out of total ~41,000 fragments screened, 582 fragments were retrieved and the top 15 were ranked and prioritized based on their estimated binding affinities (supplemental Figure S5). The novel designed compounds are devoid of chemically reactive groups, fullfill Lipinski’s rule of five for bioavailable drug-like (Lipinski et al., 2001), and are predicted to exhibit higher Sec14p binding affinities relative to compound 2. This strategy represents a prototypic approach comprising all structural and genetic data collected in this study for the design of second-generation, improved Sec14p SMIs as candidates for synthesis.
Importance of the VV motif in SMI sensitivity
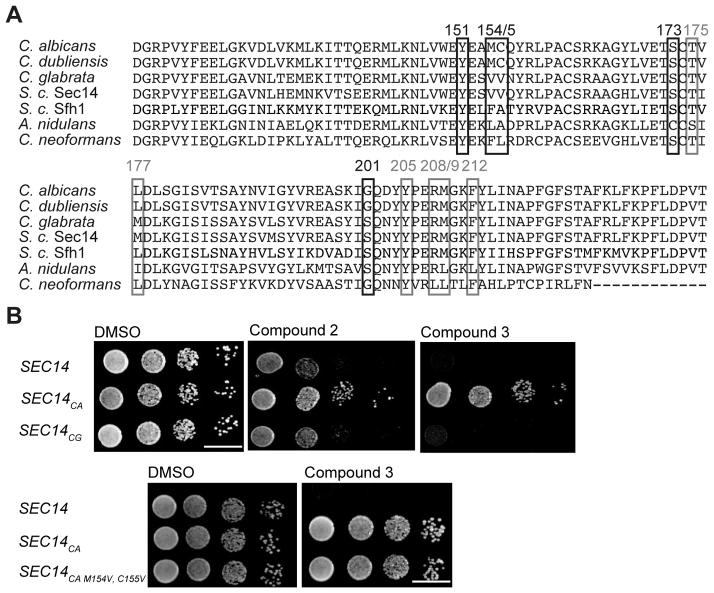
Residues Val154 and Val155 constitute what is termed the VV-motif, and it was previously recognized that: (i) this motif is a reliable predictor of sensitivity of fungal Sec14ps to NPPMs, and (ii) that this motif is not conserved between the Sec14ps of S. cerevisiae and other pathogenic fungi (Khan et al., 2016) (Figure 5A). This observation took on added significance given that the Sec14ps of S. cerevisiae and Candida glabrata both naturally harbor a VV motiv and the [3H]-PtdIns transfer activities of both proteins were sensitive to inhibition by compounds 2 and 3 in vitro (IC50s of 6.7 μM and 10.9 μM for compound 2, and 0.7 μM vs 1.0 μM for compound 3, respectively – Figure 3D). In contrast, the Candida albicans Sec14p, which is divergent at these positions was completely resistant to inhibition by both compounds 2 and 3 in vitro - even at concentrations that approached saturation in aqueous solution (IC50s > 100 μM for each compound). The co-crystal data obtained in this study confirmed the VV motif to be involved in interactions with bioactive picolinamides and benzamides. Thus, we tested whether reconstitution of the VV motif sensitized otherwise resistant Sec14ps to these SMI chemotypes. To that end, Sec14p-deficient S. cerevisiae were reconstituted with the Sec14p of C. albicans and C. glabrata by the previously described strategy (Khan et al., 2016), and those strains were spotted on rich medium (YPD) containing 20 μM inhibitor. Only the strain expressing C. glabrata Sec14p (naturally harbors the VV motif) was sensitive to SMI challenge whereas the strain reconstituted with C. albicans Sec14p was resistant to SMI challenge (Figure 5B). To further investigate whether reconstitution of the VV motif in a Sec14p that naturally lacks it sensitized the protein to SMI treatment, the VV-motif was transplaced into the C. albicans Sec14p. However, unlike the case with NPPMs (Khan et al., 2016), transplacement of the VV-motif into Sec14CA (M154V, C155V double mutant) was insufficient to render the strain sensitive to challenge with compound 3 (Figure 5B). Why this is so remains an important and outstanding conundrum.
Figure 5.
Sec14p sequence comparison and investigations into the VV motif. A) Protein sequence alignment of Saccharomyces cerevisiae Sec14p and Sfh1p and Sec14p of the pathogenic fungi Candida albicans, Candida dubliniensis, Candida glabrata, Aspergillus nidulans and Cryptococcus neoformans. Resistance-conferring amino acids identified by the functional variomics screen are marked with black boxes, sites predicted by co-crystal structure to be involved in compound interactions with grey boxes. B) A wildtype strain and isogenic derivatives expressing physiological levels of C. albicans and C. glabrata Sec14 PITPs were spotted on rich medium (YPD) agar medium supplemented with the indicated compounds and incubated at 30 °C for 48 hours. Transplacement of the VV-motif into Sec14pCA (the M154V, C155V double mutant) did not render this PITP sensitive to compound 3 (lower panel).
Antifungal activity
The initial aim of this study was to identify novel antifungal compounds. Identification of Sec14p as cellular target of the tested compounds by the methodologies described above motivated prioritization of the two optimized compounds 2 and 3 for further antifungal testing. As Sec14ps play an important role in pathogenicity and virulence of pathogenic fungi (Chayakulkeeree et al., 2011), it was of interest to determine whether compounds 2 and 3 exerted activity beyond the S. cerevisiae PITP. Thus, the inhibitory effects of compound 2 and compound 3 were tested in vitro following the Clinical Laboratory Standards Institute guidelines against four diverse and clinically relevant pathogens (Wayne, 2008a, b). These included two dimorphic fungi of the Candida genus (C. albicans and C. glabrata), one filamentous fungus (Aspergillus brasiliensis) and, as representative of the Basidiomycota, the yeast Cryptococcus neoformans. Posaconazole, a triazole compound, was used as positive control. For compound 2, a minimal inhibitory concentration (MIC) of 100 μM was measured for C. albicans and C. glabrata, and an MIC of 50 μM for Aspergillus brasiliensis. Compound 3 inhibited growth of C. glabrata with a MIC of 50 μM, but residual growth was observed at higher concentrations (likely due to compound precipitation at higher dose, see supplementary Table S2). No growth inhibition was observed for C. neoformans (Table 2).
Table 2. Growth inhibitory concentrations against pathogenic fungi.
Minimal inhibitory concentrations on solid medium have been determined following the Clinical Laboratory Standards Institute reference in triplicates.
| S. cerevisiae BY4743 | S. cerevisiae BY4743Δ8 |
Candida albicans ATCC 10231 |
Candida glabrata ATCC 2001 |
Cryptococcus neoformans DSM 70219 |
Aspergillus brasiliensis ATCC 16404 |
|
|---|---|---|---|---|---|---|
| Minimal inhibitory concentration [μM] | ||||||
| Posaconazole | <0.3 | <0.3 | <0.3 | 3.125 | <0.3 | <0.3 |
| Compound 2 | 200 | 100 | 100 | 100 | >200 | 50 |
| Compound 3 | >200 | 50 | >200 | 50 | >200 | >200 |
Discussion
This report describes the identification of a series of picolinamide and benzamide-containing compounds as a novel class of small molecule inhibitors of the S. cerevisiae PtdIns/PtdCho transfer protein Sec14p. Although there are two previous reports of Sec14 inhibitors (Filipuzzi et al., 2016; Nile et al., 2014), this study presents the first Sec14p::SMI co-crystal structure. This structure, rationalized both the functional variomics and SAR results of more than 40 generated analogues, supports strong structural fungal selectivity over the mammalian homolog and represents a quantum leap for rational design of the next generation of improved Sec14p inhibitors. The picolinamides and benzamides share some similarity with related inhibitors reported before by Nile et al. 2014. However, they also differ considerable from these by the substituent patterns and the heteroatom in the aromatic acid part and by different amine substituents than reported before thus opening up the chemical landscape for the design of novel Sec14p SMIs.
Sec14p is the major PITP of the budding yeast S. cerevisiae and potentiates the efficient production of phosphoinositides, in particular PtdIns(4)P, whose signaling is specifically channeled to protein trafficking in the yeast TGN-endosomal system (Bankaitis et al., 1990) and for biogenesis of secretory vesicles from the TGN (Phillips et al., 2006). The sequence homology of Sec14p from S. cerevisiae with that of pathogenic fungi ranges from 45 to 86 %, but amino acids that line the lipid binding cavity surface are highly conserved. Although the role of Sec14p in other fungal species is less well characterized, it is reported to be involved in yeast dimorphism, sporulation and in sustaining mycelial growth, all prerequisites for the infectivity and pathogenicity of fungal pathogens (Chayakulkeeree et al., 2011; Lopez et al., 1994; Monteoliva et al., 1996; Phillips et al., 2006; Rudge et al., 2004). Compared with the in vitro potency measured on some fungal proteins, the recorded potencies on the tested pathogenic fungi were relatively low. But the used cell-based assay only recpitulated mitotic growth but not any of the other functional aspects of Sec14p inhibition stated above. While the presented compounds need further evaluation to qualify as clinical candidates, the analytical platform and structural data identify a path for application of rational medicinal chemistry approaches.
The fact that, at least in S. cerevisiae, SMI-sensitivity are overcome by loss of function in any one of one of seven different genes (“bypass Sec14p” mutations (Bankaitis et al., 1990; Cleves et al., 1989; Fang et al., 1996)), or by single amino acid substitutions in Sec14p itself, would seem to reduce enthusiasm for Sec14p as attractive antifungal target. However, ‘bypass Sec14p’ mutations derange lipid metabolism and are accompanied by pleiotropic effects that generally reduce fitness of the organism – particularly under stress conditions (Cleves et al., 1989; Mousley et al., 2012). Sec14p amino acid substitutions that result in SMI-resistance influence key residues required for optimal protein function and therefore are also likely to be deleterious under conditions of high Sec14p demand as in pathogenic settings.
Specificity of target is always a primary issue in drug design. The presented benz- and picolinamides, while toxic to fungi, were not cytotoxic to mammalian cells and failed to inhibit the structurally unrelated mammalian PtdIns-transfer proteins in vitro. The benz- and picolinamides reported here, target the PtdCho headgroup coordination substructure of the Sec14p lipid binding pocket. As none of the mammalian SEC14L proteins exhibit this substructure, it is highly unlikely these activities will be targeted. Thus, the benz- and picolinamide compounds described herein represent priviledged scaffolds that exhibit exquisite specificity among fungal Sec14-like PtdIns/PtdCho-transfer proteins.
In summary, the structural and functional data gained by the experimental approach in this study, together with the experimental tools and data from previous studies (Filipuzzi et al., 2016; Khan et al., 2016; Nile et al., 2014), now provide the scientific community a robust roadmap to the design, synthesis, and validation of the next generation of Sec14p inhibitors aimed at treating severe fungal infections.
Significance
Due to increasing numbers of elderly or immunocompromised patients severe antifungal infections are on the rise and the mortality numbers are unacceptably high. Existing treatments suffer from emerging drug resistance against established pathogens and lack of efficacy as the spectrum of pathogenic fungi identified in patients today is much broader than a few decades ago. Despite this, the current pharmaceutical drug discovery pipeline lists less than a handful of compounds in clinical phases that modulate novel targets. This report describes benz- and picolinamide compounds with versatile and robust chemistry that exert antifungal activity against the model organism Saccharomyces cerevisiae but also against pathogenic Candida and Aspergillus species. Using a combination of genetic methodologies, the fungal lipid-transfer protein Sec14 was identified as the target of these compounds. Importantly, the tested benz- and picolinamides, while toxic to fungi, were not cytotoxic to mammalian cells and failed to inhibit the structurally diverged mammalian lipid-transfer proteins in vitro, further emphasizing their antifungal potential. A potent compound of the series was successfully co-crystallized with Sec14p yielding the first high-resolution structural dataset for a phosphatidylinositol-transfer protein in complex with an inhibitor. The compound-protein data obtained by the structural approach was in full agreement with a series of genetic point mutants and the structure-activity data derived from 48 compound derivatives. Combined, this dataset paves the way and provides the protocols for rational optimization of benz- and picolinamides as antifungal Sec14 inhibitors.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents can be directed to and will be fulfilled by the Lead Contact, Dominic Hoepfner (dominic.hoepfner@novartis.com). For simplicity, if compounds are requested they can also directly be requested from the Waldmann lab (Herbert.Waldmann@mpi-dortmund.mpg.de) or genetic reagents listed in the key resource table can be requested from the indicated labs (Bankaitis lab: vytas@medicine.tamhsc.edu, Schaaf lab: gabriel.schaaf@uni-bonn.de). Signing of a material transfer agreement (MTA) will be required for reagents originating from Novartis. The MTA will restrict use of provided materials to research purposes in pre-clinical context and will exclude any administration to humans. Special conditions may apply if reagents are requested for experiments on human embryonic stem cells or for in vivo experiments.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| E.c. BL21 | New England BioLabs | Cat# C2527I |
| E.c. BL21- CodonPlus (DE3)-RIL | Agilent Technologies | #230245 |
| Fungal Strains | ||
| S.c. BY4743 | OpenBiosyste ms | Cat# YSC1050 |
| S.c. BY4743Δ8 | Dominic Hoepfner’s lab | BY4743Δ8 |
| S.c heterozygous, genome-wide deletion collection | OpenBiosyste ms | Cat# YSC1055 |
| S.c homozygous, genome-wide deletion collection | OpenBiosyste ms | Cat# YSC1056 |
| S.c. CTY1-1A | Bankaitis’ lab | CTY1-1A |
| S.c. CTY159 | Bankaitis’ lab | CTY159 |
| S.c. CTY303 | Bankaitis’ lab | CTY303 |
| C.albicans | American Type Culture Collection (ATCC) | Cat# 10231 |
| C.glabrata | American Type Culture Collection (ATCC) | Cat# 2001 |
| A.brasiliensis | American Type Culture Collection (ATCC) | Cat# 16404 |
| C.neoformans | Royal Botanic Gardens, Kew | Cat# DSM70219 |
| Mammalian Cell Lines | ||
| Human colorectoral carcinoma cell line HCT116 | American Type Culture Collection (ATCC) | Cat# CCL-247 |
| Recombinant DNA | ||
| Plasmid: SEC14CG | Bankaitis’ lab | pDK247 |
| Plasmid: SEC14CA | Bankaitis’ lab | pDK9 |
| Plasmid: SEC14CAM154V, C154V | Bankaitis’ lab | pDK265 |
| Plasmid: pDR195-SEC14 | Bankaitis’ lab | pDR195 |
| Plasmid: YCplac33-SFH1 | Schaaf’s lab | N/A |
| Plasmid: YCplac33- SFH1* | Schaaf’s lab | N/A |
| Plasmid: YCplac33- SEC14 | Schaaf’s lab | N/A |
| Plasmid: BYInt | Hoepfner’s lab | pBYInt_LEU |
| Chemicals and Recombinant Proteins | ||
| Compounds 1- 48 | Waldman’s lab | Compound number as listed in Table S1 |
| His8-Sec14 | Schaaf’s lab | N/A |
| His8-Sec14Y151H | Bankaitis’ lab | N/A |
| His8-Sec14V154G | Bankaitis’ lab | N/A |
| His8-Sec14V155A | Bankaitis’ lab | N/A |
| His8-Sec14S173L | Bankaitis’ lab | N/A |
| His8-Sec14S201G | Bankaitis’ lab | N/A |
| Software and Algorithms | ||
| Tibco Spotfire | TIBCO Software Inc. | http://www.spotfire.tibco.com |
| GOLD Suite | The Cambridge Crystallographic Data Centre (CCDC) | https://www.ccdc.cam.ac.uk |
| Pymol | Schrödigner | https://pymol.org |
| XDS | Kabsch et al., Max-Plack Institute, Heidelberg | http://xds.mpimf-heidelberg.mpg.de |
| Phaser | McCoy et al., University of Cambridge | http://www.ccp4.ac.uk/html/phaser.html |
| Coot | Emsley et al., University of Cambridge | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| MOE | Chemical Computing Group | https://www.chemcomp.com/ |
| Critical Commercial Assays | ||
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat# G7573 |
| Deposited Data | ||
| S.c. Sec14::compound 2 co-crystal structure | Protein databank | PDB 6F0E (http://www.rcsb.org/pdb/explore/explore.do?structureId=6F0E) |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Microbial strain description, culture conditions and plasmids construction
Strains BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0), BY4743Δ8 (as above but with snq2Δ::KanMX4/snq2Δ::KanMX4, pdr3Δ::KanMX4/pdr3Δ::KanMX4, pdr5Δ::KanMX4/pdr5Δ::KanMX4, pdr1Δ::NatMX4/pdr1Δ::NatMX4, yap1Δ::NatMX4/yap1Δ::NatMX4, pdr2Δ/pdr2Δ, yrm1Δ/yrm1Δ, yor1Δ/yor1Δ), CTY182 (MATa ura3-52 lys2-801 his3 -200) and its isogenic derivatives CTY1-1A (MATa ura3-52 lys2-801 his3 -200 sec14-1ts), CTY159 (MATa ura3-52 lys2-801 3his3-200 sec14-1ts kes1 Δ), and CTY303 (MATa ura3-52 lys2-801 his3Δ-200 sec14Δ cki1Δ::HIS3)(Cleves et al., 1991; Fang et al., 1996; Li et al., 2002). Gene replacement constructs were used to integrate SEC14CG, SEC14CA and SEC14CAM154V, C154V expression cassettes into the LEU2 locus as described by Khan et. al.(Khan et al., 2016). Yeast strains were grown on YPD (yeast extract 1%, peptone 2%, dextrose 2%) plates or in liquid medium. If plasmid selection was necessary, the strains were grown on synthetic complete medium lacking uracil. Compound 1 was obtained from the Novartis compound store. All other reagents, chemicals, and buffer salts were purchased from Sigma-Aldrich Chemicals (St. Louis, MO), Fluka (Buchs, Switzerland), Alfa Aesar (Karlsruhe, Germany) and Acros Organics (Geel, Belgium). Centromeric YCplac33(URA3) plasmids carrying SEC14, SFH1 and sfh1 activation alleles under control of a SEC14 promoter fragment(Schaaf et al., 2011) as well as episomal high-copy (2μ) plasmid pDR195-SEC14 in which the SEC14 ORF (without its intron) is expressed under control of a strong plasma membrane ATPase (PMA1) promoter fragment(Schaaf et al., 2008) were described previously. Individual mutations were introduced in YCplac33-SEC14 by site-directed mutagenesis (QuickChangeTM, Stratagene).
METHOD DETAILS
Compound synthesis
Unless otherwise noted, all commercially available compounds were used as provided without further purifications. Chemicals and solvents were purchased from the companies Sigma Aldrich, Alfa Aesar and Acros Organics. Dry solvents (e.g. dimethylformamide (DMF)) were used as commercially available. Solvents for preparative HPLC (acetonitrile HPLC grade) were used as commercially available. Analytical thin-layer chromatography (TLC) was performed on Merck silica gel aluminum plates with F-254 indicator. Compounds were visualized by irradiation with UV light or potassium permanganate staining. Solvent mixtures are understood as volume/volume. Extended information on compound synthesis and analytical methods can be found in supplemental Data S1.
Compound analysis by NMR
1H-NMR and 13C-NMR were recorded on a Mercury VX400 (Varian) (400 MHz), Bruker DRX400 (400 MHz), Bruker DRX500 (500 MHz), INOVA500 (500 MHz) and INOVA600 (600 MHz) using CDCl3, MeOD, CD2Cl2, DMSO as solvent. Data are reported in the following order: chemical shift (δ) values are reported in ppm with the solvent resonance as internal standard (CDCl3: δ = 7.26 ppm for 1H, δ = 77.16 ppm for 13C, MeOD: δ = 3.34 ppm for 1H, δ = 49.86 ppm for 13C; CD2Cl2: δ = 5.32 ppm for 1H, δ = 54.00 ppm for 13C DMSO: δ = 2.54 ppm for 1H, δ = 40.45 ppm for 13C); multiplicities are indicated br s (broad singlet), s (singlet), d (doublet), dd (double doublet), t (triplet), q (quartet) m (multiplet); coupling constants (J) are given in Hertz (Hz). Extended information on compound synthesis and analytical methods can be found in supplemental Data S1.
Compound analysis by LC-MS
LC-MS measurements were performed on a LCQ Advantage ESI from Agilent 1100 series. As precolumn, a VP 50/21 Nucleodur C18 Gravity 5 μm was used and as main column a 125/4 Nucleodur C18 Gravity 3 μm. The flow rate was 1 mL per min. Eluent A: H2O + 0.1% HCOOH; Eluent B: Acetonitril + 0.1% HCOOH. The parameters used for the different methods (named A, B, C, D) are listed below:
Method A
Used Gradient: 0–1 min: 90% A/10% B; 1–10 min: 0% A/100% B; 10–12 min: 0% A/100% B; 12–12.1 min: 90% A/10% B; 12.1–15 min: 90% A/10% B. Furthermore, LC-MS measurements were performed on a LCQ Fleet from Thermo Ultimate 3000 series. As precolumn a VP 50/21 Nucleodur C18 Gravity 5 μm was used and as main column a 50/2 Nucleodur C18 Gravity 1.8 μm. The flow rate was 0.4 mL per min. Eluent A: H2O + 0.1% HCOOH; Eluent B: Acetonitril + 0.1% HCOOH.
Method B
Used Gradient: 0–0.5 min: 90% A/10% B; 0.5–7.5 min: 5% A/95% B; 7.5–9.0 min: 5% A/95% B; 9.0–11 min: 90 % A/10% B. High resolution mass spectra were recorded on a LTQ Orbitrap mass spectrometer coupled to an Accela HPLC-System (HPLC column: Hypersyl GOLD, 50 mm × 1 mm, particle size 1.9 μm, ionization method: electron spray ionization). GC-MS measurements were done on an Agilent Technologies 7890A GC System. It was used a 5975C inert Xl MSD with Triple-Axis Detector.
Method C
DB_100_S: flow rate: 20 ml per min, 1 min hold at 100 °C, during 10 min increase to 300 °C, at 11 min 300 °C for 5 min.
The preparative HPLC purifications were carried out on an Agilent HPLC (1100 series) with LC/MSD VL (ESI-MS) mass detector with parallel UV-detection. A reversed-phase C18 column (Nucleodur C18, diameter 10 mm, Macherey & Nagel) was used.
Method D: flow rate 6.0 mL/min, (A = acetonitrile + 0.1 % trifluoroacetic acid (TFA), B = water + 0.1 % TFA). Gradient: 1–25 min: 10 % A, 90 % B; 26–28 min: 100 % A, 0 % B; 28.10–30 min: 10 % A, 90 % B. Optical activities were measured with a Schmidt + Haensch Polartronic HH8 polarimeter. Chemical yields refer to pure isolated substances. Melting points were measured on the apparature B-450 instrument from Büchi. Up open capillaries were used.
The amine (1.0 eq) was dissolved in dimethylformamide (DMF) (1 mL) at room temperature. In a separate flask, the carboxylic acid (1.0 eq) was dissolved in DMF (1 mL) and 1-cyano-2-ethoxy-2-oxoethylidenaminooxy)dimethylamino-morpholino-carbenium hexafluorophosphate (COMU) (1.5 eq.), ethyl (hydroxyimino)-cyanoacetate (Oxyma) (1.5 eq) were added. After addition of N,N-diisopropylethylamine (DIPEA) (4.5 eq.) to the mixture, the flask was shaken for 30–60 s and the mixture was added dropwise to the solution of the amine and stirred at room temperature for 18 h. The reaction mixture was diluted with 1 M HCl (50 mL) and extracted with ethylacetate (3 × 50 mL). The combined organic phases were washed with sat. NaHCO3 (1 × 50 mL), with sat. NaCl (1 × 50 ml), dried with magnesium sulfate and concentrated in vacuo. The crude residue was purified by preparative HPLC (method D). Aqueous solubility of compounds was determined by spectrophotometrical measurement of the kinetic solubility of a 500 μM compound solution in aqueous buffer at pH 7.4 compared to a solution in the organic solvent acetonitrile after 90 minutes of vigorous shaking at room temperature. Permeability through artificial membranes (PAMPA) was performed at an initial concentration of 500 μM of the compound in the donor compartment. After an incubation period of 20 hours, absorption of the receiver wells was measured by spectrophotometry and permeation was calculated by normalization of the compound flux across a blank filter. Extended information on chemical synthesis, analytical data and NMR spectra are collected in supplementary Data S1.
Chemogenomic profiling (HIP-HOP)
Potency determination
The growth-inhibitory potency of compounds was determined using wildtype S. cerevisiae BY4743 as described by Pierce et al.(Pierce et al., 2007). OD600 values of exponentially growing cultures in rich medium were recorded with a robotic system. Twelve-point serial dilutions of the tested compounds were assayed in 96-well plates with a reaction volume of 150 μl, start OD600 was 0.05. Solutions containing dimethyl sulfoxide (DMSO) were normalized to 2%. IC50 values were calculated using logistic regression curve fits generated by TIBCO Spotfire v.6.5.3 (TIBCO Software Inc.).
HIP assay execution
The HIP assay was performed in 24 well plates (Greiner 662102), with 1600 μl/well YPD. Experimental compounds were tested at n=2 within the same plate at or close to their IC30 concentration. Each plate contained two no drug controls, one positive control (Benomyl, CMB991), 10 experimental compounds in duplicates and one contamination control that received no cells. A standard experiment was 4 plates/40 experimental compounds processed robotically on a BiomekFX (Beckman Coulter) without human intervention. YPD/compound filled wells were inoculated with ~250 yeast cells/strain (100 μl of a 1.5 OD600/ml culture) from an overnight log phase pre-culture to start the experiment. The plates were pipetted with a standard 96 pipettor head by providing tip boxes pre-configured with a special tip pattern. Plates were incubated for 16 hours in a robotic shaking incubator at 30°C/550 RPM allowing for ~5 doublings. ~250 yeast cells/strain (120 μl of a 1.2 OD600/ml culture) were subsequently transferred into a pre-configured 24 well plate that was stored in a robotic plate reservoir at 4°C until 30 minutes prior to its use where it was pre-warmed to 30°C. Once inoculated the new plate was incubated at 30°C/550 RPM to allow the next 5 yeast generations (generation 6–10) and the plate containing the first 5 doubling cultures was stored at 4°C. This procedure was repeated 2x more until the final plate containing the yeast with ~20 generations were stored at 4°C. Detailed information about the robotic setup have been published previously (Hoepfner et al., 2014).
HOP assay execution
The HOP assay was performed similar to the HIP experiment but the duration was reduced to ~5 doublings and no dilutions were necessary. Before the experiment, aliquots of the HOP pool were thawed and recovered for 3 hours in YPD. The robotic system inoculated the wells prefilled with YPD and compound at the onset of the experiment with ~320 yeast cells/strain (110 μl of a 1.5 OD600/ml culture) from an overnight log phase pre-culture. Plates were incubated for 16 hours in a robotic shaking incubator at 30°C/550RPM allowing for ~5 doublings and where then stored at 4°C. .HIP, HOP, and microarray analysis was performed as described previously(Hoepfner et al., 2014; Pierce et al., 2007). Sensitivity was computed as the median absolute deviation logarithmic (MADL) score for each compound/concentration combination. Z-scores are based on a robust parametric estimation of gene variability from >3000 different profiles and were computed as described in detail in Hoepfner et al. (Hoepfner et al., 2014).
gDNA extraction, TAG amplification, and hybridization
An aliquot of 5 OD600 units of yeast/well from the HIP and HOP experiments were arrayed in 96 well plates, spun and the supernatant discarded. gDNA extraction was performed using the ChargeSwitch kit (Invitrogen #18000) in a partially automated process. 150 ul/well of Zymolyase buffer (2 U Zymolyase, 50 ng RNase A, in 20 mM DTT and 20 mM Tris pH 7.5) were added and the cells were incubated at 37°C/700 RPM for 45 minutes. 300 μl/well lysis buffer (L18, Invitrogen) was added and the plate incubated at 56°C/700 RPM for 30 minutes. 200 μl/well ice cold precipitation buffer (N2, Invitrogen) was added and the precipitate pelleted by centrifugation at 4°C. The supernatant was saved to a new deep well plate (AB-0932, Abgene) suitable to lock on the Invitrogen MagnaRack magnet and 40 μl/well of pre-dispersed ChargeSwitch magnetic beads were added. The plate was incubated at room temperature for 5 minutes followed by incubation on the MagnaRack for 5 minutes to pellet the beads. All supernatant was carefully aspirated. The deep well plate was removed from the MagnaRack and 500 ul/well wash buffer (W12, Invitrogen) were added and mixed to disperse the beads. The washing was repeated three times and the wash buffer completely removed. Finally, 70 μl of elution buffer (E6, Invitrogen) were added. The beads were resuspended by mixing and the plate incubated for 10 min at RT. The beads were pelleted for one last time by incubation on the magnet for 5 minutes and the supernatant (containing the gDNA) was transferred to a new plate. The TAG PCR amplification and GenFlex Tag16K v2 hybridization protocol was used as described (Pierce et al., 2006).
Processing of TAG16K v2 data
The raw probe intensity values of the CEL are summarized and normalized to tag intensities as described (Pierce et al., 2006). Tags with low intensity values in control samples are removed by computing an intensity value threshold based on the comparison of the correlation between the logarithmic intensity ratios for uptags and downtags across different intensity ranges. The tag intensities are then averaged to obtain a strain intensity value. In order to measure the relative abundance of each strain with respect to the averages of the control samples we compute MAD logarithmic (MADL) scores for each compound/concentration combination. If we denote the logarithm of the ratio of the average intensity of the compound samples over the average intensity of the control samples as rL, then the MADL score is given as (rL – med(rL))/MAD(rL) where the median and MAD are computed over all strains in one sample. MADL scores can be viewed as robustly computed experiment-wise z-scores. We also compute the t-test p-value, p, between the two replicates for a compound and the four to eight control replicates as a measure of the variability of the compound and control sample intensities across the experiment and. The final (adjusted) score aL is decreased for highly variable strains and computed as:
Then, we compute gene-wise z-scores (across all experiments) which are based on a robust parametric estimation of gene variability allowing for up to 15% outliers. To do this we consider the adjusted MADL scores (a1, …, an) of a strain over n experiments. Usually, the z-score transformation of score ai is defined as zi = (ai - ā)/σ where ā is the mean and σ the standard deviation of the values ai. However, due to the special nature of the HIP-HOP data it is advantageous to introduce a number of changes to the computation of the adjusted MADL z-score transformation. Since an (adjusted) MADL score of zero indicates a relative growth rate of a strain in the compound treated sample that is equal to the relative growth rate of the strain in the untreated control, we set ā = 0, for all strains. What remains is to estimate σ as a measure of the variability of the strain which is described in the following.
In order to avoid biasing the computation of the standard deviation of a strain profile we allow at most five entries of the same compound/concentration combination in the data set. The samples of the sixth or greater occurance are discarded. We normalize the positive and negative scores separately; that is, we treat the positive and the negative scores as the halves of two separate distributions (artificially creating two perfectly symmetric distributions in this way). In the following we only consider the negative scores. Let the profile of the negative scores of a gene be (s1, …, sn) where we assume that the scores are sorted in ascending order. We assume that the scores follow a normal distribution N(μ,σ). Note that since the distribution is perfectly symmetric around 0, μ = 0; moreover, for each i, the expectation of the score si is the i/(2n+1)-quantile qi of the standard normal distribution N(0,1) times σ. In other words, si/qi is an estimator σ̂i for σ, for each i. These estimators are computed only for the indices i in the interval [0.15n, 0.85n], that is, we consider only the middle 70% of the data to estimate σ. We assume that the scores si are drawn for a normal distribution only if the standard deviation of the σ̂i is at most 0.05 times the mean σ of σ̂i; in this case, the estimators σ̂i are considered to be consistent and σ a good estimate of σ. If the standard deviation is larger, we abandon the parametric approach and just set σ to be the standard deviation of sample (s1, …, sn, –s1, …, –sn). The estimate σ of σ obtained from the single-array data is used to compute the normalized z-score transformation (a1/σ, …, an/σ) of the adjusted MADL scores (a1, …, an).
Growth curves
HIP-HOP profiles were validated by picking the individual strains from the HIP or HOP collections (OpenBiosystems, Cat # YSC1056 and YSC1055) and testing log-phase cultures in 96-well microtiter plates in YPD medium with serial dilutions of the compound. The assay volume was 150 μl/well, start OD600 was 0.05, DMSO was normalized to 2%. Curves were calculated by taking the 13–14 h OD600 measurements and applying a logistic regression curve fit in TIBCO Spotfire v6.5.3. Strain HO/YDL228C was used as the wildtype reference.
For determination of cell viability in HCT116 cells 1500 cells/well were seeded in a 384 well plate and incubated for 72 hours with serial dilutions of the compound. The readout was done with CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Expression and purification of recombinant proteins
Recombinant S. cerevisiae, C. glabrata and C. albicans Sec14 proteins were purified as previously described (Khan et al., 2016; Schaaf et al., 2008). The murine PITPα structural gene was subcloned into pET28b(+) as an Nco1-SacI PCR fragment encoding an N-terminal octa-histidine epitope tag appended to the PITPα open reading frame. The construct was transformed in E. coli BL21 (DE3) cells and cultured in LB plus antibiotics at 37°C until an OD600 of 0.6 was reached. Protein production was induced by adding IPTG to a final concentration of 60μM at which time the culture was shifted to 16°C and incubated overnight with shaking. Cells were pelleted, resuspended in buffer A (50 mM NaH2PO4 300 mM NaCl pH 7.8) supplemented with PMSF and 2-mercaptoethanol (1 mM each, final concentration). Cells were disrupted by two successive passages through a French Press (10,000 p.s.i.), and crude lysates clarified by two successive rounds of centrifugation at 2800 g and 27,000 g for 30 minutes each. Clarified lysates were incubated with Co-TALON metal affinity beads for 3 hours at 4°C followed by exhaustive washing with buffer A. Bound proteins were eluted in a 20–200 mM imidazole gradient incremented in 1 ml steps of 20, 40, 60, 80, 100, 150, and 200 mM imidazole in buffer A and evaluated by SDS-PAGE using BSA mass standards to estimate PITPα yield.
Lipid transfer assays
Compounds of interest were maintained as 20 mM stock solutions in DMSO and stored at room temperature in the dark. In vitro sensitivities of Sec14p to the compounds was investigated as described by Nile et al.(Nile et al., 2014). Briefly, protein was incubated with DMSO/compound, as applicable, in the presence of liposomes. Compound concentrations were fixed at 20 μM throughout the experiments. Total [3H]-PtdIns input per assay ranged from 8356–9604 c.p.m., background transfer from 436–489 c.p.m., and transfer efficiencies from 14–16%. Protein concentrations were clamped at 287 nM (~10 μg per assay).
For IC50 determinations, % [3H]PtdIns transfer (normalized to mock) was plotted as a function of log10 of the compound concentration. The IC50 was determined using GraphPad Prism Version 6 software. IC50 values are determined at 95% confidence level from at least two independent experiments, each done in triplicate. [3H]-PtdIns input for Sec14, its mutant proteins, Sec14CA and Sec14CG, ranged from 7111–11517 c.p.m. per assay; background ranged between 212–510 c.p.m. per assay. The transfer efficiencies are as follows: Sec14 (25–30%), Sec14CA (39–43%), Sec14CG (30–34%), Sec14Y151H (32–35%), Sec14V154G (15–18%), Sec14V155A (26–31%), Sec14S201G (40–43%) and Sec14S173L (25–27%). Protein was clamped at 10 μg per assay throughout these experiments.
Functional variomics
A functional variomics screen to isolate SMI-resistant Sec14p variants was performed (Huang et al., 2013; Pries et al., 2016). 6 × 107 BY4743 yeast transformants harboring the SEC14 variomic library (the SEC14 ORF amplified by error-prone PCR and cloned under control of the endogenous promoter into a low-copy expression plasmid, at a pre-determined complexity above 2 × 105 primary alleles, (Huang et al., 2013)) were plated onto a 15 cm petridish containing uracil-free synthetic complete agar supplemented with either 250 μM of compound 2 or 100 μM of compound 3. The plates were incubated for four days at 30 °C. Ninety-six resistant colonies were picked for each condition, the plasmids purified using the Wizard SV 96 plasmid DNA purification system (A2250, Promega,). The inserts were amplified by PCR Platinum Hot Start PCR Master Mix (13000012, Invitrogen) using the oligonucleotides 5′CTGTTGGGAAGGGCGATC3′ and 5′CCAGGCTTTACACTTTATGCT3′ respectively and performing the reaction as specified in Invitrogen’s manual. Single nucleotide polymorphisms (SNPs) were then idenetified by Sanger sequencing.
Mutant validation
To confirm the SNPs identified with the functional variomics screen, the isolated plasmids were cut with SacI and BamHI, ligated with a pBYInt-URA plasmid and transformed into BY4743 8 MATa/α strain, that is deleted for eight genes involved in drug resistance (efflux pumps SNQ2, PDR5, YOR1; transcription factors PDR1, PDR2, PDR3, YAP1, YRM1, detailed genotype is described in the Fungal Strains section above). This sensitized strain was chosen to open up the observable resistance window in the dose-response validation as testing of high compound concentrations were limited by solubility (Supplemental Table S2). Growth curves were recorded over 24 hours as described in the growth curve section described above. In vitro validation with recombinant protein was performed as described above.
Protein expression, purification, and crystallization
Octahistidine-tagged Sec14p was purified from BL21-CodonPlus (DE3)-RIL cells (Agilent Technolgies) as described for Sfh1(Schaaf et al., 2006) with minor modifications. Protein expression was induced with 60 μM isopropyl-β-D-thiogalactoside at 16°C for 20 h, prior to harvesting cells and extracting protein in a modified lysis buffer (300 mM NaCl, 25 mM sodium phosphate pH 7.5 and 5 mM β-mercaptoethanol). After purification with a Ni-NTA affinity resin (Macherey-Nagel), the protein was subjected to size exclusion chromatography (Superdex 75 16/600 column, GE Healthcare) at a flow rate of 1 mL/min in modified lysis buffer. Fractions of the second peak were pooled and concentrated to 5 mg ml−1. Initial crystallization screens were carried out manually by sampling an array of circa 1500 conditions that represent variations on crystallization conditions established for the Sec14 homolog Sfh1 (Schaaf et al., 2006; Schaaf et al., 2008). In this initial screen, His8-Sec14p was supplemented with 1 volume % compound 2 (30 mM in DMSO), and a sitting-drop geometry was employed in which drops consisted of 1 μL protein/compound solution and 1 μl well solution. After an incubation time of ca. 1 week at room temperature, crystals appeared in one condition where the solution consisted of 129,5 mM sodium acetate, 64,8 mM TRIS, 4,6 % (w/v) PEG 4000, and 11.9 % (v/v) glycerol adjusted to pH 7.0 (by acetic acid).
Crystallography
For data collection, crystals were transferred to a cryo solution (129.5 mM sodium acetate, 64.8 mM TRIS, 10 % (w/v) PEG 4000, 20 % (v/v) glycerol, pH 7.0) and flash frozen in liquid nitrogen. Diffraction data were collected at the Beamline PXII of the Swiss Light Source. Data scaling and processing was performed with XDS(Kabsch, 1993). The structure was solved by Molecular Replacement with PHASER(Mccoy et al., 2007) using the structure of Sec14p prior to removal of the detergent coordinates (pdb id.: 1aua (Sha et al., 1998a)). Cycles of model building and restrained refinement were carried out in COOT and PHENIX(Adams et al., 2010; Emsley and Cowtan, 2004). Ligand restraints were generated with Jligand(Lebedev et al., 2012). Protein-ligand interaction was analysed with PLIP(Salentin et al., 2015). Protein structure figures were generated using PyMOL (http://www.pymol.org). The coordinates and structure factors have been deposited in the RSCB PDB database with ID code 6F0E.
In silico docking
Computational docking was carried out using the genetic algorithm-based ligand docking program GOLD 5.2.1 that exhaustively explores ligand conformations and provides limited flexibility of protein side chains with hydroxyl groups by reorienting hydrogen bond donor and acceptor groups. For computational docking, the Sec14p::compound 2 crystal structure was used. The binding site was defined by taking cognate ligand in the crystal structure as reference center and defining a 10 Å boundary around it using the GOLD cavity detection algorithm. GOLD docking was carried out without constraint to explore all possible solutions. To explore all possible binding modes, docking was carried out with early termination turned off. All other parameters were as the defaults. Compounds of interest were docked and scored using CHEMPLP scoring function within GOLD as it gives the highest success rates for pose prediction and virtual screening experiments against diverse validation test sets.
Structure-based design and MedChem transformation
Structure-based optimization and MedChem Transformation of the picolinamide series was carried out using MOE software (https://www.chemcomp.com/). The Sec14p::compound 2 crystal structure was used as starting structure for optimization. Binding pocket within the Sec14p Lipid Binding Domain was mapped in an area 6Å around the co-crystallized ligand. Compound 2 was used as lead template for transformation and optimization. Van der Waals interaction surface were generated near the lead template to determine accessible space for R group substitutions. Ligand R-vectors were generated on the lead template to show positions at which the heavy atom could be substituted without introducing > 2 kcal/mol of Van der Waals clash energy.
To ascertain biochemically favorable regions for substitutions, Electrostatic Feature Maps were generated that predict the electrostatically preferred locations of hydrophobic, H-bond acceptor and H-bond donor sites. Similarly, probabilistic receptor preference maps were calculated to predict non-bonded contact preferences - i.e. the preferred locations of hydrophobic and hydrophilic ligand atoms. A fragment screen was generated and drug-like filters were used such that total molecular weight of compound does not exceed 500 daltons, total polar surface area (TPSA) descriptor between 40 and 140 with no reactive group substitutions. These parameters were imposed on an MOE fragment and linkers database screen for R-group substitutions. Considering the amphipathic nature of the sub-pocket, a pharmacophoric feature was included in the query that would position the fragment for interaction with Glu124. Binding poses of compounds retrieved from the screen were refined by energy minimization within the pocket, and their binding affinities estimated using the GBVI/WSA dG scoring function.
Antifungal testing
Fungal strains used for testing were as follows: Candida albicans (ATCC 10231), Candida glabrata (ATCC 2001), Cryptococcus neoformans (DSM 70219) Aspergillus brasiliensis (ATCC 16404). Antifungal susceptibility testing was performed in triplicates according to the Clinical and Laboratory Standards Institute (CLSI) guidelines for broth microdilution M27-A3 and M38-A2 (Wayne, 2008a, b). By performing 1:2 serial dilutions, 10 concentrations in a range between 200 and 0.3 μM were tested for growth inhibitory activity on solid medium in 8 cm petridishes prepared with 15 ml Mueller-Hinton agar medium (2.0 g7/l beef extract, 17.5 g/l casein hydrolysate, 1.5 g/l starch, 17 g/l agar), with pH adjusted to pH 7.2, in triplicates. 1 μl spore inoculi were spotted manually onto the plates and absence of growth was scored visually after 72 hours incubation at 30°C to determine the minimal inhibitory concentration.
QUANTIFICATION AND STATISTICAL ANALYSIS
Chemogenomic profiles were acquired as independent duplicates and then averaged and computed against 8 no drug controls as outlined in the corresponding section above. The statistical analysis performed in described in detail in the chemogenomic profiling section. Fungal growth curves were acquired as duplicates, biochemical inhibition assays were conducted in triplicates, cytotoxicity assays were conducted in triplicates. Curves were calculated using the logistic regression curve fit function of TIBCO Spotfire (TIBCO Software Inc.) or Prism (GraphPad Inc.). MIC experiments were determined in triplicates and lack of colony formation scored visually.
DATA AND SOFTWARE AVAILABILITY
Used software packages are listed in the corresponding experimental sections and the key resources table. The co-crystal structures have been uploaded to RCSB protein data bank and can be found and retrieved under the code 6F0E or the following link: http://www.rcsb.org/pdb/explore/explore.do?structureId=6F0E.
Supplementary Material
Highlights.
Identification of selective, antifungal activity of benz- and picolinamide compounds
Target identification of the lipid transfer protein Sec14p by multiple methods
Selective inhibition of fungal but not mammalian Sec14 lipid transfer proteins
First Sec14p-inhibitor co-crystal structure solved
Acknowledgments
We would like to thank Jürg Eichenberger and Nicole Hartmann for processing and Sven Schuierer for analyzing the HIP-HOP microarray data. Furthermore, we thank David Estoppey for performing the cell viability assay on HCT116 cells, Dr. Matthias Baumann for conducting compound solubility and permeability assays and Eva M. Winklbauer for cloning support. We also thank the staff at the Swiss Light Source synchrotron for assistance during data collection, and the Laboratory for Molecular Simulation and High Performance Research Computing at Texas A&M University for providing software, support and computer time. DK, AT and VB were supported by grants GM44530 and BE-0017 to V.B. from the National Institutes of Health and Robert A. Welch Foundation, respectively. P.J., A.L.K., M.F., and G.S. were supported by the Deutsche Forschungsgemeinschaft (Emmy Noether grant SCHA 1274/2-1, SFB 1101/TP A05 and research grant SCHA 1274/4-1 to G.S.) and P.J. also by the Studienstiftung des deutschen Volkes. F.B. and Z.H. received support from the Max Planck Gesellschaft, the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013), ERC grant agreement n° 310957 and the Deutsche Forschungsgemeinschaft (BO3588/2-1 to F.B.).
Footnotes
Author Contributions
V.P., D.K., P.J., I.F., T.A., R.R., S.Z., designed and performed genetic experiments. C.N., S.T., designed, synthesized and analyzed compounds, P.J., A.L.K., M.F. conducted crystallization experiments, Z.H., F.B., determined and analyzed the structure, A.T., F.P. generated docking poses, A.T. developed the SAR, conducted the MD simulations, structure-based design, and MedChem Transformation of new analogs. D.K. purified Sec14p and the various mutant derivative proteins designed and performed the in vitro lipid transfer experiments. V.P., F.B., G.S., V.B., H.W., and D.H. conceived the study, analyzed data, prepared figures and wrote the manuscript.
Competing financial interests
The authors with the affiliation Novartis Institutes for BioMedical research are employees of Novartis Pharma AG and may own stock in the company. All other authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. The Journal of cell biology. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Mousley CJ, Schaaf G. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci. 2010;35:150–160. doi: 10.1016/j.tibs.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benli M, Döring F, Robinson DG, Yang X, Gallwitz D. Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast. The EMBO Journal. 1996;15:6460–6475. [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden Killers: Human Fungal Infections. Science Translational Medicine. 2012;4:165rv113–165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Chayakulkeeree M, Johnston SA, Oei JB, Lev S, Williamson PR, Wilson CF, Zuo X, Leal AL, Vainstein MH, Meyer W, et al. SEC14 is a specific requirement for secretion of phospholipase B1 and pathogenicity of Cryptococcus neoformans. Molecular Microbiology. 2011;80:1088–1101. doi: 10.1111/j.1365-2958.2011.07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves AE, McGee TP, Whitters EA, Champlon KM, Altken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. The Journal of cell biology. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Faham A, Albericio F. COMU: a third generation of uronium-type coupling reagents. J Pept Sci. 2010;16:6–9. doi: 10.1002/psc.1204. [DOI] [PubMed] [Google Scholar]
- El-Faham A, Subiros Funosas R, Prohens R, Albericio F. COMU: a safer and more effective replacement for benzotriazole-based uronium coupling reagents. Chemistry. 2009;15:9404–9416. doi: 10.1002/chem.200900615. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fairn GD, Curwin AJ, Stefan CJ, McMaster CR. The oxysterol binding protein Kes1p regulates Golgi apparatus phosphatidylinositol-4-phosphate function. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15352–15357. doi: 10.1073/pnas.0705571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MK, Bankaitis VA. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. The EMBO Journal. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- Filipuzzi I, Cotesta S, Perruccio F, Knapp B, Fu Y, Studer C, Pries V, Riedl R, Helliwell SB, Petrovic KT, et al. High-Resolution Genetics Identifies the Lipid Transfer Protein Sec14p as Target for Antifungal Ergolines. PLoS Genet. 2016;12:e1006374. doi: 10.1371/journal.pgen.1006374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nature genetics. 1999;21:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- Hoepfner D, Helliwell SB, Sadlish H, Schuierer S, Filipuzzi I, Brachat S, Bhullar B, Plikat U, Abraham Y, Altorfer M, et al. High-resolution chemical dissection of a model eukaryote reveals targets, pathways and gene functions. Microbiological Research. 2014;169:107–120. doi: 10.1016/j.micres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Hoepfner D, Karkare S, Helliwell SB, Pfeifer M, Trunzer M, De Bonnechose S, Zimmerlin A, Tao J, Richie D, Hofmann A, et al. An Integrated Approach for Identification and Target Validation of Antifungal Compounds Active against Erg11p. Antimicrobial Agents and Chemotherapy. 2012;56:4233–4240. doi: 10.1128/AAC.06332-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon S, Smith AM, Wallace IM, Suresh S, Miranda M, Fung E, Proctor M, Shokat KM, Zhang C, Davis RW, et al. An integrated platform of genomic assays reveals small-molecule bioactivities. Nature chemical biology. 2008;4:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- Huang CF, Buu LM, Yu WL, Lee FJS. Characterization of a Novel ADP-ribosylation Factor-like Protein (yARL3) in Saccharomyces cerevisiae. Journal of Biological Chemistry. 1999;274:3819–3827. doi: 10.1074/jbc.274.6.3819. [DOI] [PubMed] [Google Scholar]
- Huang Z, Chen K, Zhang J, Li Y, Wang H, Cui D, Tang J, Liu Y, Shi X, Li W, et al. A functional variomics tool for discovering drug-resistance genes and drug targets. Cell Rep. 2013;3:577–585. doi: 10.1016/j.celrep.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- Khan D, McGrath KR, Dorosheva O, Bankaitis VA, Tripathi A. Structural elements that govern Sec14-like PITP sensitivities to potent small molecule inhibitors. Journal of Lipid Research. 2016;57:650–662. doi: 10.1194/jlr.M066381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, Murshudov GN. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr D. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, St Onge RP, Proctor MJ, Wallace IM, Nile AH, Spagnuolo PA, Jitkova Y, Gronda M, Wu Y, Kim MK, et al. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science. 2014;344:208–211. doi: 10.1126/science.1250217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FJS, Huang CF, Yu WL, Buu LM, Lin CY, Huang MC, Moss J, Vaughan M. Characterization of an ADP-ribosylation Factor-like 1 Protein inSaccharomyces cerevisiae. Journal of Biological Chemistry. 1997;272:30998–31005. doi: 10.1074/jbc.272.49.30998. [DOI] [PubMed] [Google Scholar]
- Li X, Rivas MP, Fang M, Marchena J, Mehrotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA. Analysis of oxysterol binding protein homologue Kes1p function in regulation of 23 Sec14p-dependent protein transport from the yeast Golgi complex. The Journal of Cell Biology. 2002;157:63–78. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Routt SM, Xie Z, Cui X, Fang M, Kearns MA, Bard M, Kirsch DR, Bankaitis VA. Identification of a Novel Family of Nonclassic Yeast Phosphatidylinositol Transfer Proteins Whose Function Modulates Phospholipase D Activity and Sec14p-independent Cell Growth. Molecular Biology of the Cell. 2000;11:1989–2005. doi: 10.1091/mbc.11.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Lopez MC, Nicaud JM, Skinner HB, Vergnolle C, Kader JC, Bankaitis VA, Gaillardin C. A phosphatidylinositol/phosphatidylcholine transfer protein is required for differentiation of the dimorphic yeast Yarrowia lipolytica from the yeast to the mycelial form. The Journal of cell biology. 1994;125:113–127. doi: 10.1083/jcb.125.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteoliva L, Sánchez M, Pla J, Gil C, Nombela C. Cloning of Candida albicans SEC14 gene homologue coding for a putative essential function. Yeast. 1996;12:1097–1105. doi: 10.1002/(SICI)1097-0061(19960915)12:11%3C1097::AID-YEA990%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Mousley CJ, Tyeryar K, Ile KE, Schaaf G, Brost RL, Boone C, Guan X, Wenk MR, Bankaitis VA. Trans-Golgi network and endosome dynamics connect ceramide homeostasis with regulation of the unfolded protein response and TOR signaling in yeast. Molecular biology of the cell. 2008;19:4785–4803. doi: 10.1091/mbc.E08-04-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousley CJ, Yuan P, Gaur NA, Trettin KD, Nile AH, Deminoff SJ, Dewar BJ, Wolpert M, Macdonald JM, Herman PK, et al. A sterol-binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell. 2012;148:702–715. doi: 10.1016/j.cell.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Tsukagoshi Y, Yamashita S. Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. Journal of Biological Chemistry. 1991;266:11184–11191. [PubMed] [Google Scholar]
- Nile AH, Tripathi A, Yuan P, Mousley CJ, Suresh S, Wallace IM, Shah SD, Pohlhaus DT, Temple B, Nislow C, et al. PITPs as targets for selectively interfering with phosphoinositide signaling in cells. Nat Chem Biol. 2014;10:76–84. doi: 10.1038/nchembio.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Pappas PG, Wingard JR. Invasive Fungal Pathogens: Current Epidemiological Trends. Clinical Infectious Diseases. 2006;43:S3–S14. [Google Scholar]
- Phillips SE, Vincent P, Rizzieri KE, Schaaf G, Bankaitis VA, Gaucher EA. The Diverse Biological Functions of Phosphatidylinositol Transfer Proteins in Eukaryotes. Critical Reviews in Biochemistry and Molecular Biology. 2006;41:21–49. doi: 10.1080/10409230500519573. [DOI] [PubMed] [Google Scholar]
- Pierce SE, Davis RW, Nislow C, Giaever G. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat Protocols. 2007;2:2958–2974. doi: 10.1038/nprot.2007.427. [DOI] [PubMed] [Google Scholar]
- Pierce SE, Fung EL, Jaramillo DF, Chu AM, Davis RW, Nislow C, Giaever G. A unique and universal molecular barcode array. Nat Methods. 2006;3:601–603. doi: 10.1038/nmeth905. [DOI] [PubMed] [Google Scholar]
- Prasad R, Shah AH, Rawal MK. Antifungals: Mechanism of Action and Drug Resistance. In: Ramos J, Sychrová H, Kschischo M, editors. Yeast Membrane Transport. Cham: Springer International Publishing; 2016. pp. 327–349. [Google Scholar]
- Pries V, Cotesta S, Riedl R, Aust T, Schuierer S, Tao J, Filipuzzi I, Hoepfner D. Advantages and Challenges of Phenotypic Screens: The Identification of Two Novel Antifungal Geranylgeranyltransferase I Inhibitors. Journal of Biomolecular Screening. 2016;21:306–315. doi: 10.1177/1087057115610488. [DOI] [PubMed] [Google Scholar]
- Richie DL, Thompson KV, Studer C, Prindle VC, Aust T, Riedl R, Estoppey D, Tao J, Sexton JA, Zabawa T, et al. Identification and Evaluation of Novel Acetolactate Synthase Inhibitors as Antifungal Agents. Antimicrobial Agents and Chemotherapy. 2013;57:2272–2280. doi: 10.1128/AAC.01809-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Krysan DJ. Antifungal Drug Development: Challenges, Unmet Clinical Needs, and New Approaches. Cold Spring Harbor Perspectives in Medicine. 2014;4 doi: 10.1101/cshperspect.a019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Xu D, Singh SB, Parish CA, Harris G, Wang H, Davies JE, Bills GF. Confronting the challenges of natural product-based antifungal discovery. Chemistry & biology. 2011;18:148–164. doi: 10.1016/j.chembiol.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Rudge SA, Sciorra VA, Iwamoto M, Zhou C, Strahl T, Morris AJ, Thorner J, Engebrecht J. Roles of Phosphoinositides and of Spo14p (phospholipase D)-generated Phosphatidic Acid during Yeast Sporulation. Molecular Biology of the Cell. 2004;15:207–218. doi: 10.1091/mbc.E03-04-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MM, Temple BRS, Phillips SE, Bankaitis VA. Conformational dynamics of the major yeast phosphatidylinositol transfer protein Sec14p: Insight into the mechanisms of phospholipid exchange and diseases of Secl4p-like protein deficiencies. Molecular biology of the cell. 2007;18:1928–1942. doi: 10.1091/mbc.E06-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic acids research. 2015;43:W443–447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Betts L, Garrett TA, Raetz CRH, Bankaitis VA. Crystallization and preliminary X-ray diffraction analysis of phospholipid-bound Sfh1p, a member of the Saccharomyces cerevisiae Sec14p-like phosphatidylinositol transfer protein family. Acta Crystallogr F. 2006;62:1156–1160. doi: 10.1107/S1744309106041728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Dynowski M, Mousley CJ, Shah SD, Yuan P, Winklbauer EM, de Campos MKF, Trettin K, Quinones MC, Smirnova TI, et al. Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution. Molecular Biology of the Cell. 2011;22:892–905. doi: 10.1091/mbc.E10-11-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CRH, Redinbo MR, et al. Functional Anatomy of Phospholipid Binding and Regulation of Phosphoinositide Homeostasis by Proteins of the Sec14 Superfamily. Molecular Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel Y, Zimmerli S. Common invasive fungal diseases: an overview of invasive candidiasis, aspergillosis, cryptococcosis, and Pneumocystis pneumonia. Swiss Med Wkly. 2016;146:w14281. doi: 10.4414/smw.2016.14281. [DOI] [PubMed] [Google Scholar]
- Sha B, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature. 1998a;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- Sha BD, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature. 1998b;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- Wayne P. CLSI document MA38-A2. 2008a. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard - 2nd ed. In Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard - 2nd ed CLSI document MA38-A2. [Google Scholar]
- Wayne P. CLSI document M27-A3. 2008b. Reference method for broth dilution antifungal susceptibility testing of yeast; approved standard - 3rd ed. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.