Abstract
Age-induced loss of skeletal muscle mass and function, termed sarcopenia, may be the result of diminished response to anabolic stimulation. This review will explore the hypothesis that alterations in the expression of microRNA with aging contributes to reduced muscle plasticity resulting in impaired skeletal muscle adaptations to exercise-induced anabolic stimulation.
Keywords: myomiR, sarcopenia, exercise, anabolic resistance, miR
Summary for Table of Contents
Altered expression of microRNA to exercise is blunted with aging, contributing to diminished anabolic capacity of skeletal muscle and potentially sarcopenia.
Introduction
It is well established that mechanical load placed on skeletal muscle during resistance exercise activates anabolic intracellular signaling processes, increases the rate of muscle protein synthesis, and with repeated exposure (i.e., training) leads to gains in muscle mass (1). With aging, there is insufficient skeletal muscle plasticity, resulting in a blunted rate of skeletal muscle protein synthesis (i.e., anabolism) following acute exposure to potent anabolic stimuli, such as resistance exercise (2–4). Diminished anabolism with aging has been termed ‘anabolic resistance’ (5). Failure to preserve normal anabolic processes while proteolytic (i.e., breakdown) mechanisms are maintained or upregulated with aging results in development of sarcopenia; age-associated decline in skeletal muscle mass and function (5, 6). Over time, the progressive loss in muscle mass can compromise an individual’s quality of life, leading to loss of independence and diminished health-span.
To minimize declines in skeletal muscle mass and mobility with aging, an understanding of the underlying molecular processes regulating anabolism is crucial to determine potential therapeutic targets. Recently, our group (7) has identified dysregulation of microRNA (miRNA; small non-coding RNA, approximately 18–25 nucleotides in length) with aging as a potential mechanism governing the adaption within skeletal muscle in response to anabolic stimulation. Specifically, divergent responses in miRNA expression between younger and older individuals following a bout of resistance exercise showed impairment of anabolic signaling with aging (7). Additionally, we have reported that this blunted anabolic response to resistance exercise in skeletal muscle with aging can also be observed in discordant expressions of circulating (serum) miRNA (c-miRNA) profiles (8).
This review will provide a contemporary overview of the biogenesis and function of miRNA with specific focus on role of miRNA in governing biological processes involved in skeletal muscle anabolism and catabolism. Based on work from our laboratory, the purpose of this review is to examine the hypothesis that alterations in the expression of miRNA with aging contributes to reduced muscle plasticity resulting in impaired adaptations to exercise-induced anabolic stimulation. Additionally, we will explore the hypothesis that circulating miRNA can be used as a non-invasive marker reflective of age-related ‘anabolic resistance’ following resistance exercise.
microRNA Biogenesis and Function
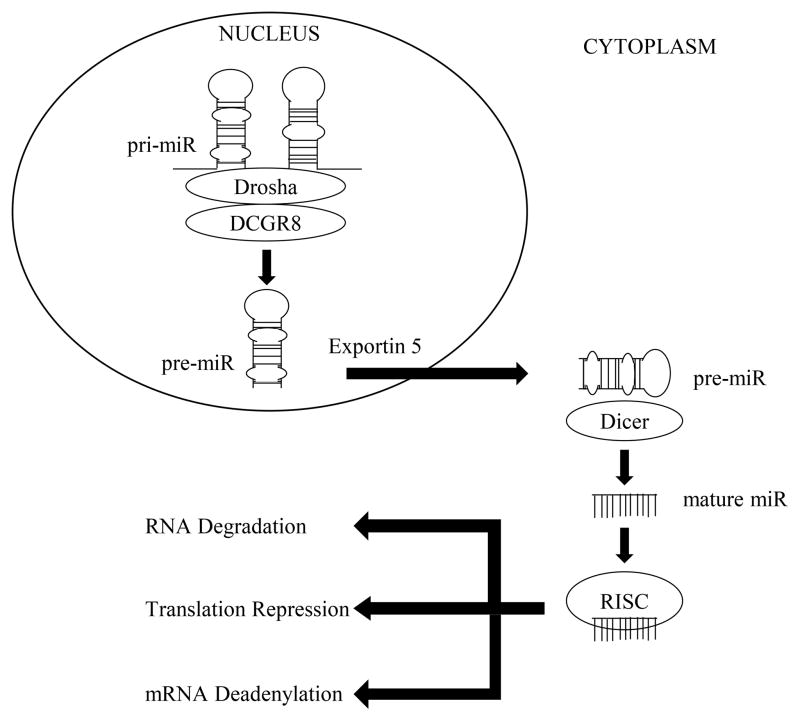
Biogenesis of miRNA begins in the nucleus of the cell (Figure 1), where it is processed and translocated into the cytoplasm to form a mature miRNA (9). The mature miRNA is bound by Argonaute, forming a protein complex called RNA-induced silencing complex (RISC) (10). This RISC complex allows miRNA to bind to target mRNA, resulting in post-transcriptional modifications that repress the translation of protein (11). Through this mechanism of negative inhibition miRNA regulate gene expression. miRNA-dependent gene regulation is a complex process, as one miRNA can regulate hundreds to thousands of genes (12). The ability for one miRNA to inhibit the expression of a large number of genes allows a single miRNA to repress several mRNA in a common biological pathway, resulting in robust regulation of an entire molecular process (13). Additionally, one gene can be targeted by multiple miRNA, resulting in cooperative/redundant regulation of a signal molecular process (12). Through these mechanisms of regulation, miRNA have a critical role in the development and maintenance of physiological process that determine muscle fiber number, type/phenotype, and mass/size (14).
Figure 1.
microRNA Biogenesis; Biogenesis of miRNA begins in the nucleus of the cell where RNA polymerase II transcribes primary-miRNA (pri-miRNA), consisting of thousands of nucleotides with stem-loop structures. The enzyme Drosha, a member of the ribonuclease (RNase) III superfamily of double-stranded RNA-specific endoribonuclease, together with DiGeorge syndrome critical region gene (DGCR8), cleaves the stem-loop structure of the pri-miRNA to form precursor miRNA (pre-miRNA). Conversion of pri-miRNA to pre-miRNA is a critical step, as it is site-specific, dictating the sequence of the mature miRNA. The pre-miRNA translocates out of the nucleus into the cytoplasm by small RNA transporter Exportin 5, which is a GTP dependent process. Once in the cytoplasm, pre-miRNA is processed by Dicer, another enzyme in the RNase III family, to form mature miRNA. One strand of the mature miRNA is bound by Argonaute, a protein that directly binds to miRNA, forming a protein complex called RNA-induced silencing complex (RISC), allowing miRNA to bind to target mRNA, resulting in post-transcriptional modifications that repress the translation of protein
microRNA Regulation of Skeletal Muscle Anabolism
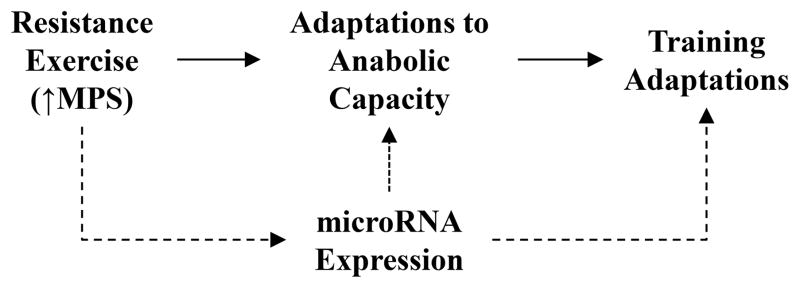
Skeletal muscle is highly enriched with specific microRNA (miR-1, miR-133a, miR-133b, miR-206, miR-208, miR-221, miR-222, miR-486, miR-499) that have together been termed myomiRs (14, 15). Though the exact mechanisms remain unclear, in young, healthy individuals, miRNA expression is acutely altered by anabolic stimulation (7, 16–18). In general, following resistance exercise there is a downregulation in miR-1, miR-133a, miR133b, and miR-206 expression (16, 19), while more metabolically demanding endurance exercise results in an upregulation or no change in expression of these miRNA (19–21). Divergent response of in miRNA expression to various exercise modes appears to be due, at least in part, to miRNA being sensitive to alteration in the rate of muscle protein synthesis, with miR-206 and miR-499 expression reported to be inversely associated with muscle protein synthesis during exercise (19). Given that miRNA function through negative inhibition, concurrent reductions in miRNA expression with increased muscle protein synthesis rates suggest a potential feed-forward mechanism, where acute anabolic stimulation downregulates the inhibition of miRNA to initiate training adaptions through enhanced translation of proteins regulating skeletal muscle anabolism (Figure 2).
Figure 2.
Schematic of hypothesized feed forward mechanism of elevated muscle protein synthesis (MPS) rates modulation microRNA expression to aid in regulation of training adaptions.
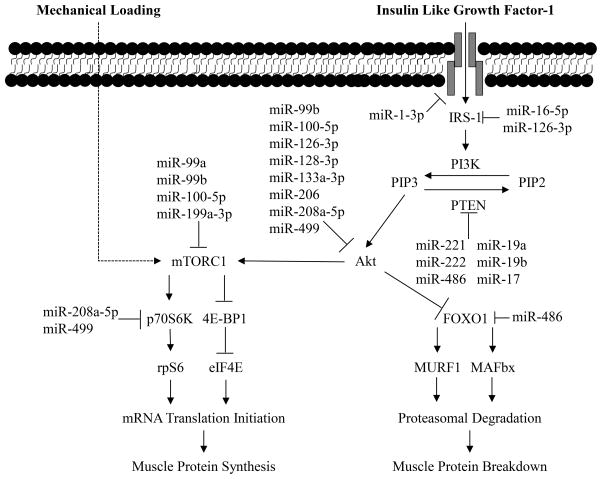
There is a growing body of evidence that miRNA have a significant impact on skeletal muscle growth, with several miRNA participating in the regulation of signaling proteins involved in muscle protein synthesis (mechanistic target of rapamycin; mTORC1) and breakdown (factor forkhead box O 1; FOXO1) signaling cascades (Figure 3). In muscle, miR-1, miR-133a-3p, and miR-199a-3p target IGF-1 and IGF-1R, blunting rates of protein synthesis (22, 23). During periods of muscle growth, induced by mechanical load, miR-1 and miR-133a expression is downregulated to allow for activation of mTORC1 signaling through IGF-1, resulting in increased rates of protein synthesis (24). In muscle atrophy, miR-199a-3p expression is increased (25), with this overexpression resulting in impairment of muscle hypertrophy, diminishing phosphorylation of Akt and mTORC1 (26). Additionally, miR-99a, miR-99b and miR-100-5p influence cellular growth by both directly and indirectly mediating translation of mTORC1. Specifically, increased expression of miR-99a and miR-99b inhibit transcription of mTOR, while miR-100-5p targets both Akt and mTOR, resulting in diminished total protein content and hypertrophy (27, 28). Furthermore, it is possible that miRNA play an important role in the shifting of intracellular signaling from catabolism to anabolism. Expression of the upstream inhibitor of Akt, phosphatase and tensin homolog (PTEN) is diminished by miR-221, miR-222, and miR-486 to promote cellular growth (29, 30). The miR-17~92 cluster, which contains 7 miRNA, may also participates in alterations in Akt-mTOR signaling. Similar to miR-221 and miR-222, miRNA in the miR-17~92 cluster (miR-17-5p, miR-19a-3p and miR-19b-3p) inhibit PTEN, promoting Akt-mTOR signaling (31). Increased Akt activity due to alterations in miRNA expression may not only promote synthesis, but diminish protein breakdown as well, resulting in a positive protein balance. Upregulation of miR-486 activates Akt and diminishes FOXO1 protein expression. Diminished FOXO1 protein content by miR-486 results in reductions in transcription of atrophy proteins MAFbx and MuRF1, potentially minimizing muscle protein breakdown (29). Additionally, miRNA, particularly miR-1, miR-133a, miR-133b, and miR-206, can further regulate skeletal muscle mass through control of transcription factors governing myogenesis/regeneration (15). Given that the majority of work in altered miRNA expression with aging has focus on anabolism, regulation of miRNA on myogenesis is outside the scope of this review. For a review of miRNA regulation of myogenesis please see Kirby et al. (32)
Figure 3.
Interaction between microRNA and intracellular signaling pathways regulating skeletal muscle protein synthesis and breakdown. Activation of mTORC1 triggers downstream signaling through p70 ribosomal S6 kinase (p70 S6K), ribosomal protein S6 (rpS6), eukaryotic elongation factor 2 kinase (eEF2), and eukaryotic initiation factor 4E-binding protein (4E-BP1), increasing mRNA translational efficiency and muscle protein synthesis. Muscle protein breakdown from ubiquitination results through the muscle-specific E3 class of ubiquitin ligases, atrogin-1/muscle atrophy F-box (MAFbx), and muscle RING finger-1 (MuRF1). Activity of atrogin-1/MAFbx and MuRF1 are regulated by the forkhead box O (FOXO) family of transcription factors, which when dephosphorylated translocate to the nucleus to mediate increased expression of these ubiquitin ligases. The ubiquinated proteins are transferred to the 26S proteasome for subsequent degradation.
Aging and Skeletal Muscle microRNA Expression
There have been a limited number of investigations examining the potential role of modulation in miRNA expression on age-associated declines in skeletal muscle anabolism (7, 16, 33). Following performance of knee extension exercise fixed at 70% of participant’s one repetition maximum (RM) and ingestion of 20 g EAA, expression of miR-1 was reduced in young participants, with no change observed in older individuals (16). A divergent response in miR-1 expression with aging likely indicates a lack of an anabolic response to the bout of resistance exercise. As miR-1 inhibits IGF-1 in skeletal muscle, reductions in miR-1 expression during periods of hypertrophy suggest potential activation of the mTORC1 pathway through IGF-1 signaling (22). In agreement with these findings, our laboratory (7) recently observed that following a single bout of resistance exercise, expression of 60 miRNA assessed in skeletal muscle were not altered in older participants, while younger participants experienced a significant reduction in the expression of 16 of these 60 miRNA. The absence of exercise-induced miRNA regulation with aging was accompanied by a blunted gene transcription response, and diminished activation of the mTORC1 signaling cascade compared to younger participants (3). Impairment of resistance exercise-induced alterations in skeletal muscle miRNA and mRNA expression, as well as diminished phosphorylation of mTORC1 signaling with aging suggests a potential link governing ‘anabolic resistance.’
To further examine whether altered miRNA expression was a potential mechanistic target for diminished muscle mass with aging, principal component analysis was conducted on miRNA with differing expression between younger and older to determine which miRNA distinguished aging (7). This analysis identified miR-126-3p as a potential target influencing the divergent anabolic response to resistance exercise with aging. To test the role of miR-126-3p on regulation of molecular pathways controlling skeletal muscle anabolism, in vitro analysis was performed, manipulating the expression of miR-126-3p through transfection of miR-126-3p inhibitor or mimetic for 24-hrs in myocytes and myotubes. Inhibition of miR-126-3p protein content of insulin receptor substrate 1 increased 50%, while FOXO1 decreased 25% compared to control myocytes. Additionally, when miR-126-3p was overexpressed, myogenic regulators MyoD and Myf5 were 60% and 50%, respectively, lower compared to controls. Following 30 min exposure to IGF-1 in myotubes upregulation of p-AktSer473 was greater in miR-126-3p inhibited myotubes compared to control. Similarly, a downstream target of mTORC1, p-rpS6Ser240/244 was activated to a greater extent in miR-126-3p inhibited myotubes compared to controls. Together, findings from in vivo and in vitro analysis identify miR-126-3p dysregulation with aging as a novel regulator suppressing skeletal muscle regeneration and growth following exercise-induced adaptions within skeletal muscle.
In agreement with findings from our laboratory, high-throughput analysis of 754 miRNA identified 26 miRNA that were differentially expressed with aging in response to resistance exercise, or a combination of the two (17). Top cellular functions of these miRNA were determined using Ingenuity Pathway Analysis. This bioinformatics analysis revealed that 6 (miR-99a-5p, miR-99b-5p, miR-100-5p, miR-149-3p, miR196b-5p, and miR-199a) of these 26 miRNA were validated to target proteins within the Akt-mTORC1 signaling cascade. As described above, members of the miR-99/100 family are of particular interest, as these miRNA directly target mTOR to suppress protein synthesis and anabolism (27, 28). Specifically, following acute resistance exercise miR-99b-5p and miR-100-5p expression were diminished in young but not old participants. Again, the lack of response in expression of miRNA associated with regulation of anabolic signaling proteins with aging in skeletal muscle indicate dysregulation in miRNA expression following acute anabolic stimulus may contribute to age-associated declines in skeletal muscle mass.
Circulating microRNA
While miRNA have been shown to function in the cell where they are transcribed, recently, it has been reported that an alternative fate exists, where rather than remaining in the cytoplasm of the cell, miRNA can be packaged and exported into the circulation (c-miRNA) (34). Presence of miRNA in the circulation can be the result of multiple mechanisms and transporters. Within the cytoplasm, membrane-derived vesicles (exosomes and microvesicles) can take up pre and mature miRNA, where they can then be released into the circulation to be transferred to recipient cells (34). In addition to exosomal and microvesicle transportation, c-miRNA are actively transported in RNA binding protein (Argonaute2), as well as high density lipoproteins (high density lipoprotein (HDL) and low density lipoprotein (LDL)) (35, 36). Furthermore, miRNA can be present in circulation passively in apoptotic bodies that have been shed by tissues (37). Once released into circulation, c-miRNA can be taken up by recipient cells to inhibit transcription of target genes (35). The mechanism involved in the uptake of exosome bound c-miRNA by recipient cells remains elusive, however, it has been suggested that miRNA may be removed from circulation by endocytosis, fusion to the plasma membrane, scavenger receptor uptake, or interaction at the cellular surface to alter intracellular signaling (38).
Though the field of c-miRNA research is relatively new, since 2011 over 30 manuscripts have been published reporting the influence of acute and chronic exercise on alterations in the expression of c-miRNA profiles, which have recently been described in a review by Sapp et al. (39). Together these manuscripts clearly show that c-miRNA profiles can be altered by a bout of exercise and/or training (39). Furthermore, while only a small number of studies have been conducted, modulation in c-miRNA expressions appears to be sensitive to exercise modality (40, 41) and fitness level/training mode (42, 43). As exercise has been well-established to alter physiological processes within multiple tissues, adaptions in c-miRNA expression in response to acute exercise stimulation and training status indicates that c-miRNA profiles may be reflective of the underlying physiological status at the cellular level. Additionally, as c-miRNA can participate in cell-to-cell communication (35), modulation of expression profiles may indicate a functional role in governing training adaptions. The application of c-miRNA as a non-invasive marker of skeletal muscle adaptions to exercise may be of particular interest in aging research, as attainment of muscle samples, especially in older frail individuals, can be difficult due to lower amounts of the tissue and high infiltration of intermuscular fat.
Influence of Aging on Circulating microRNA Expression Following Resistance Exercise
Recent findings from the our laboratory (8) reported that aging results in a divergent response in c-miRNA expression following a bout of resistance exercise. Of 90 c-miRNA assessed 25 c-miRNA were altered by aging and/or resistance exercise. Using principle component analysis to group c-miRNA, 10 c-miRNA (miR-19b-3p, miR-193-5p, miR-19a-3p, miR-106-5p, miR-20a-5p, miR-17-5p, miR143-3p, miR-26b-5p, miR-18a-5p, and miR-93-5p) were identified to explain the majority of the variance within the dataset. Following resistance exercise expression of all 10 c-miRNA were increased in younger participants, and decreased in older participants. Functional analysis was then performed assessing interactions between c-miRNA-to-mRNA expressions in skeletal muscle (7) using Ingenuity Pathway Analysis (IPA). Outcomes of IPA revealed an absence of an anabolic response to resistance exercise with aging, as markers of anabolic signaling, IGF-1 and mTOR, were identified as top canonical pathways in younger, but not older participants. Further strengthening the bioinformatics data from this investigation, positive associations were observed between the expressions of miR-19a-3p, miR-19b-3p, miR-20a-5p, miR-26b-5p, miR-143-3p, and miR-195-5p to the phosphorylation status (e.g., activity) of p-AktSer473 and p-p70S6KThr389. This finding suggests that increased expressions of these c-miRNA may be indicative of an anabolic response within skeletal muscle. Coupling state-of-the-art integrative analytics with findings from traditional bench-top techniques strengthen that c-miRNA can be used as noninvasive markers to predict adaptations reflective of molecular processes in skeletal muscle to acute resistance exercise with aging.
Interestingly, 7 out of the 10 c-miRNA (miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a, miR-93 and miR-106b) identified by our PCA results are members of the miR-17~92 cluster or extended miRNA families (31, 44). Clustering of miRNA indicate that they are generated from a primary transcript and have a large overlap in their sequences and thus function (44). As described in the “microRNA Regulation of Skeletal Muscle Mass” section, members of the miR-17~92 cluster have shown convergence of these miRNA on Akt-mTORC1 signaling within tissue (45). A main target of these miRNA is PTEN, an inhibitor of the PI3K-Akt pathway. Inhibition of PTEN can promote cellular survival and proliferation through increased activation of Akt-mTORC1 signaling (31). As a single miRNA can target hundreds of different genes, identification of divergent c-miRNA profiles following resistance exercise that are members of the same family of miRNA, thus sharing similar target genes, enhances the potential use of c-miRNA as potential predictive markers of resistance exercise-induced adaptions.
Future Direction of Research: Exosomal-miRNA as signal transducers in intercellular communication
Exosomes, small extracellular membrane vesicles with the size range of 40–100 nm that are formed by exocytosis of multivesicular endosomes, contribute to multiple aspects of physiology, metabolism and disease, including communication between cells (46). Exosome can be released from multiple cell types, including skeletal muscle, and contain proteins, lipids, DNA, and RNA and importantly miRNAs (47). Given that release of exosomes from cells would indicate an active process by which c-miRNA may participate in cell-to-cell communication, determining how select miRNAs are transported in exosomes to target organs could illuminate the function of altered c-miRNA profiles and improve our understanding and application of therapeutic approaches aimed at maintaining and/or improving muscle health.
Though presently not much is known regarding alterations in exosome derived c-miRNA in humans, cell culture and rodent models suggests that exosomes carrying specific miRNAs, such as miR-1, miR-21, miR-133, miR-182, and miR-206, are targeted to myocytes and modulate the physiology and pathology status of myocytes by altering gene expression (48, 49). We hypothesize that miRNA carried in exosomes have essential roles as signal transducers that trigger the adaptations of muscle and other organs such as the adipose tissue and liver in order to maintain homeostasis. Indeed, miRNAs shuttled between cells are shown to be preserved and mediated by microvesicles including exosomes, which are emerging as potent promoters of genetic transfer (50). Recent work had highlighted that extracellular vesicles are able to efficiently deliver their parental cell-derived molecular cargo to target cells, resulting in structural changes at the RNA, protein, or even the phenotypic level (34). These data provide evidence for the importance of understanding the role of exosomes and their cargo in adaptation of skeletal muscle with age, exercise and chronic disease. For these reasons, exosomes have recently gained major scientific interest as a therapeutic application for a drug delivery system. Conceivably, determining the essential miRNA cargo packaged in exosomes may reveal crucial miRNAs for targeting skeletal muscle in an attempt to mitigate sarcopenia.
Conclusion
In conclusion, findings from our laboratory (7) and others (16, 17) show that in response to resistance exercise, modulation in skeletal muscle miRNA expression is blunted with aging. Additionally, we have reported discrepancies in miRNA expression with aging are also present in circulation, with differences in c-miRNA profiles reflective of ‘anabolic resistance’ (8). Alterations in miRNA expression may be part of a feed forward mechanism stimulating skeletal muscle growth. Elevations in the rate of skeletal muscle protein synthesis following resistance exercise stimulate downregulations in specific microRNA to diminish translation inhibition of proteins within the mTORC1 signaling cascade (19). As such, discordant responses in skeletal muscle and circulating miRNA expression to resistance exercise may be a potential mechanism blunting skeletal muscle plasticity and ultimately resulting in age-associated declines in skeletal muscle mass and function (i.e., sarcopenia).
Key Points.
microRNA are small non-coding RNA that regulate skeletal muscle mass by targeting
In young healthy individuals skeletal muscle microRNA are inversely associated with rates of skeletal muscle protein synthesis following exercise.
Aging blunts response of microRNA expression to resistance exercise, resulting in impaired skeletal muscle adaptations to exercise-induced anabolic stimulation.
Circulating microRNA profiles reflect ‘anabolic resistance’ in skeletal muscle with aging.
Acknowledgments
We would like to acknowledge the research of other scientists that have contributed to the knowledge base in the field of skeletal muscle and circulating microRNA but could not be cited in this manuscript due to character count and citation limitations.
Footnotes
Conflicts of Interest: The authors report no conflicts of interest.
Disclosure of Funding: This material is based on the work supported by the U.S. Department of Agriculture (USDA), under agreement No. 58-1950-4-003. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the USDA. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. This work was also supported by the Boston Claude D. Pepper Center Older American Independence Centers (OAIC; 1P30AG031679). L.M.M is supported by T32 NIDDK training grant # 5T32DK062032-23. D.A.R. is supported by NIA K01 award # KAG047247A-A1.
References
- 1.Drummond MJ, Dreyer HC, Fry CS, Glynn EL, Rasmussen BB. Nutritional and contractile regulation of human skeletal muscle protein synthesis and mTORC1 signaling. J Appl Physiol (1985) 2009;106:1374–84. doi: 10.1152/japplphysiol.91397.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayhew DL, Kim JS, Cross JM, Ferrando AA, Bamman MM. Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. J Appl Physiol (1985) 2009;107:1655–62. doi: 10.1152/japplphysiol.91234.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivas DA, Morris EP, Haran PH, Pasha EP, da Morais MS, Dolnikowski GG, Phillips EM, Fielding RA. Increased ceramide content and NFkappaB signaling may contribute to the attenuation of anabolic signaling after resistance exercise in aged males. J Appl Physiol (1985) 2012;113:1727–36. doi: 10.1152/japplphysiol.00412.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fry CS, Drummond MJ, Glynn EL, Dickinson JM, Gundermann DM, Timmerman KL, Walker DK, Dhanani S, Volpi E, Rasmussen BB. Aging impairs contraction-induced human skeletal muscle mTORC1 signaling and protein synthesis. Skelet Muscle. 2011;1:11. doi: 10.1186/2044-5040-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haran PH, Rivas DA, Fielding RA. Role and potential mechanisms of anabolic resistance in sarcopenia. J Cachexia Sarcopenia Muscle. 2012;3:157–62. doi: 10.1007/s13539-012-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fielding RA, Vellas B, Evans WJ, Bhasin S, Morley JE, Newman AB, Abellan van Kan G, Andrieu S, Bauer J, Breuille D, et al. Sarcopenia: an undiagnosed condition in older adults. Current consensus definition: prevalence, etiology, and consequences. International working group on sarcopenia. J Am Med Dir Assoc. 2011;12:249–56. doi: 10.1016/j.jamda.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivas DA, Lessard SJ, Rice NP, Lustgarten MS, So K, Goodyear LJ, Parnell LD, Fielding RA. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J. 2014;28:4133–47. doi: 10.1096/fj.14-254490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margolis LM, Lessard SJ, Ezzyat Y, Fielding RA, Rivas DA. Circulating MicroRNA Are Predictive of Aging and Acute Adaptive Response to Resistance Exercise in Men. J Gerontol A Biol Sci Med Sci. 2016 doi: 10.1093/gerona/glw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 10.Schraivogel D, Meister G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem Sci. 2014;39:420–31. doi: 10.1016/j.tibs.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci U S A. 2008;105:8866–71. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–42. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy JJ. The MyomiR network in skeletal muscle plasticity. Exerc Sport Sci Rev. 2011;39:150–4. doi: 10.1097/JES.0b013e31821c01e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–6. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab. 2008;295:E1333–40. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zacharewicz E, Della Gatta P, Reynolds J, Garnham A, Crowley T, Russell AP, Lamon S. Identification of microRNAs linked to regulators of muscle protein synthesis and regeneration in young and old skeletal muscle. PLoS One. 2014;9:e114009. doi: 10.1371/journal.pone.0114009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camera DM, Ong JN, Coffey VG, Hawley JA. Selective Modulation of MicroRNA Expression with Protein Ingestion Following Concurrent Resistance and Endurance Exercise in Human Skeletal Muscle. Front Physiol. 2016;7:87. doi: 10.3389/fphys.2016.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolis LM, McClung HL, Murphy NE, Carrigan CT, Pasiakos SM. Skeletal Muscle myomiR Are Differentially Expressed by Endurance Exercise Mode and Combined Essential Amino Acid and Carbohydrate Supplementation. Front Physiol. 2017;8:182. doi: 10.3389/fphys.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell AP, Lamon S, Boon H, Wada S, Guller I, Brown EL, Chibalin AV, Zierath JR, Snow RJ, Stepto N, et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J Physiol. 2013;591:4637–53. doi: 10.1113/jphysiol.2013.255695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen S, Scheele C, Yfanti C, Akerstrom T, Nielsen AR, Pedersen BK, Laye MJ. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J Physiol. 2010;588:4029–37. doi: 10.1113/jphysiol.2010.189860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, Cimino V, De Marinis L, Frustaci A, Catalucci D, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377–85. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua Y, Zhang Y, Ren J. IGF-1 deficiency resists cardiac hypertrophy and myocardial contractile dysfunction: role of microRNA-1 and microRNA-133a. Journal of cellular and molecular medicine. 2012;16:83–95. doi: 10.1111/j.1582-4934.2011.01307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol (1985) 2007;102:306–13. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 25.Roberts TC, Blomberg KE, McClorey G, El Andaloussi S, Godfrey C, Betts C, Coursindel T, Gait MJ, Smith CI, Wood MJ. Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA transcriptome of the mdx mouse with implications for therapy. Mol Ther Nucleic Acids. 2012;1:e39. doi: 10.1038/mtna.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia L, Li YF, Wu GF, Song ZY, Lu HZ, Song CC, Zhang QL, Zhu JY, Yang GS, Shi XE. MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. Int J Mol Sci. 2014;15:296–308. doi: 10.3390/ijms15010296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei F, Liu Y, Guo Y, Xiang A, Wang G, Xue X, Lu Z. miR-99b-targeted mTOR induction contributes to irradiation resistance in pancreatic cancer. Mol Cancer. 2013;12:81. doi: 10.1186/1476-4598-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin Y, Tymen SD, Chen D, Fang ZJ, Zhao Y, Dragas D, Dai Y, Marucha PT, Zhou X. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS One. 2013;8:e64434. doi: 10.1371/journal.pone.0064434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Small EM, O’Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci U S A. 2010;107:4218–23. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 32.Kirby TJ, Chaillou T, McCarthy JJ. The role of microRNAs in skeletal muscle health and disease. Front Biosci. 2015;20:37–77. doi: 10.2741/4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drummond MJ, McCarthy JJ, Sinha M, Spratt HM, Volpi E, Esser KA, Rasmussen BB. Aging and microRNA expression in human skeletal muscle: a microarray and bioinformatics analysis. Physiol Genomics. 2011;43:595–603. doi: 10.1152/physiolgenomics.00148.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 35.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2(100):ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 38.Hackl M, Heilmeier U, Weilner S, Grillari J. Circulating microRNAs as novel biomarkers for bone diseases - Complex signatures for multifactorial diseases? Mol Cell Endocrinol. 2016;432:83–95. doi: 10.1016/j.mce.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 39.Sapp RM, Shill DD, Roth SM, Hagberg JM. Circulating microRNAs in acute and chronic exercise: more than mere biomarkers. J Appl Physiol (1985) 2017;122:702–17. doi: 10.1152/japplphysiol.00982.2016. [DOI] [PubMed] [Google Scholar]
- 40.Banzet S, Chennaoui M, Girard O, Racinais S, Drogou C, Chalabi H, Koulmann N. Changes in circulating microRNAs levels with exercise modality. J Appl Physiol (1985) 2013;115:1237–44. doi: 10.1152/japplphysiol.00075.2013. [DOI] [PubMed] [Google Scholar]
- 41.Uhlemann M, Mobius-Winkler S, Fikenzer S, Adam J, Redlich M, Mohlenkamp S, Hilberg T, Schuler GC, Adams V. Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur J Prev Cardiol. 2014;21:484–91. doi: 10.1177/2047487312467902. [DOI] [PubMed] [Google Scholar]
- 42.Wardle SL, Bailey ME, Kilikevicius A, Malkova D, Wilson RH, Venckunas T, Moran CN. Plasma microRNA levels differ between endurance and strength athletes. PLoS One. 2015;10:e0122107. doi: 10.1371/journal.pone.0122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bye A, Rosjo H, Aspenes ST, Condorelli G, Omland T, Wisloff U. Circulating microRNAs and aerobic fitness--the HUNT-Study. PLoS One. 2013;8(2):e57496. doi: 10.1371/journal.pone.0057496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Concepcion CP, Bonetti C, Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 2012;18(3):262–7. doi: 10.1097/PPO.0b013e318258b60a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranji N, Sadeghizadeh M, Shokrgozar MA, Bakhshandeh B, Karimipour M, Amanzadeh A, Azadmanesh K. MiR-17-92 cluster: an apoptosis inducer or proliferation enhancer. Mol Cell Biochem. 2013;380:229–38. doi: 10.1007/s11010-013-1678-7. [DOI] [PubMed] [Google Scholar]
- 46.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–89. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 47.Safdar A, Saleem A, Tarnopolsky MA. The potential of endurance exercise-derived exosomes to treat metabolic diseases. Nat Rev Endocrinol. 2016;12:504–17. doi: 10.1038/nrendo.2016.76. [DOI] [PubMed] [Google Scholar]
- 48.Matsuzaka Y, Tanihata J, Komaki H, Ishiyama A, Oya Y, Ruegg U, Takeda SI, Hashido K. Characterization and Functional Analysis of Extracellular Vesicles and Muscle-Abundant miRNAs (miR-1, miR-133a, and miR-206) in C2C12 Myocytes and mdx Mice. PLoS One. 2016;11:e0167811. doi: 10.1371/journal.pone.0167811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He WA, Calore F, Londhe P, Canella A, Guttridge DC, Croce CM. Microvesicles containing miRNAs promote muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci U S A. 2014;111:4525–9. doi: 10.1073/pnas.1402714111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Consalvi S, Sandona M, Saccone V. Epigenetic Reprogramming of Muscle Progenitors: Inspiration for Clinical Therapies. Stem Cells Int. 2016;2016:6093601. doi: 10.1155/2016/6093601. [DOI] [PMC free article] [PubMed] [Google Scholar]