Abstract
Previous work demonstrates that proteases present in human milk release hundreds of peptides derived from milk proteins. However, the question of whether human milk protein digestion begins within the mammary gland remains incompletely answered. The primary objective of this study was to determine whether proteolytic degradation of human milk proteins into peptides begins within the mammary gland. The secondary objectives were to determine which milk proteases participate in the proteolysis and to predict which released peptides have bioactivity. Lactating mothers (n = 4) expressed their milk directly into a mixture of antiproteases on ice followed by immediate freezing of the milk to limit post-expression protease activity. Samples were analyzed for their peptide profiles via mass spectrometry and database searching. Peptidomics-based protease prediction and bioactivity prediction were each performed with several different approaches. The findings demonstrate that human milk contains more than 1,100 unique peptides derived from milk protein hydrolysis within the mammary gland. These peptides derived from 42 milk proteins and included 306 potential bioactive peptides. Based on the peptidomics data, plasmin was predicted to be the milk protease most active in the hydrolysis of human milk proteins within the mammary gland. Milk proteases actively cleave milk proteins within the mammary gland, initiating the release of functional peptides. Thus, the directly breastfed infant receives partially pre-digested proteins and numerous bioactive peptides.
Keywords: Peptides, Human, Mother's milk, Casein, Whey
Introduction
Human milk has evolved to assist infants in digestion and development of the immune system and microbiome. Part of these functions may derive from mother’s milk proteases, which appear to initiate the digestion of milk proteins into functional peptides within the mammary gland. Previous work shows that milk contains hundreds of peptides derived from proteolysis of milk proteins [1–3]]. Moreover, a variety of proteases are known to be present in human milk [4, 5]. Our bioinformatic analyses predicted that these milk proteases were responsible for the cleavage of the peptides identified in human milk [6]. These enzymes were also shown to be present in their active forms within milk—including, from highest to lowest active concentration, carboxypeptidase B2, kallikrein, plasmin, elastase, thrombin and cytosol aminopeptidase [7]. Based on these observations, we hypothesized that proteases begin to break down milk proteins into peptides after milk is synthesized inside the mammary gland. Though previous peptidomic studies [1–3] took precautions to limit possible proteolysis after expression by freezing the milk as soon as possible, and by limiting thaw time and extraction time, these techniques did not fully control for post-expression proteolysis. In one study [1], heat-treatment of a single milk sample was used to control for post-expression proteolysis; however, proteases remain active until milk reaches the temperature needed to inactivate them, and plasmin, which is very heat-stable [8], may not have been fully inactivated after heating, which may have allowed for some post-expression proteolysis.
Therefore, the question of whether breast milk protein digestion begins in the mammary gland remained incompletely answered. Should proteolysis occur in the mammary gland, the biological importance of this proteolysis has yet to be determined, though it may aid in milk protein digestion in infants prior to complete gastrointestinal tract development or exert intra-mammary gland function.
In order to test the hypothesis that digestion of milk proteins occurs within the mammary gland, breast milk was pumped directly into a mixture of protease inhibitors on ice, mixed and then frozen on dry ice before immediate transfer to a −80 °C freezer to prevent any possible post-expression proteolysis. After peptide extraction, peptidomics analyses using an Orbitrap Fusion Lumos mass spectrometer were performed to determine if peptides were present in these samples, and thus, if they were produced via proteolysis within the mammary gland.
Materials and methods
Chemicals
HPLC-grade acetonitrile (ACN) was obtained from Fisher Scientific (Waltham, MA), trifluoroacetic acid (TFA) and HPLC-grade formic acid (FA) were obtained from EMD Millipore (Billerica, MA), and trichloroacetic acid was obtained from Sigma-Aldrich (St. Louis, MO).
Participants and samples
The study was approved by the Institutional Review Board at Oregon State University and informed consent was obtained from all participants. Duplicate human milk samples were collected from four mothers at 2–4 months of lactation; all mothers gave birth to term infants (Table 1). Exclusion criteria for the study included any signs of clinical mastitis and current antibiotic usage. All human milk samples were collected between 9:00 and 10:00 AM and approximately three hours after last milk expression. Prior to pumping, mothers thoroughly washed their hands and cleansed their nipples with a moist single-use paper towel. Breasts were pumped using a hospital-grade Medela Symphony electrical pump (Salem, Oregon, USA). All components that contacted the mother or her milk were single use. The milk was pumped into an 80-mL breastmilk collection container (6109S-100; Medela) prepared with protease inhibitor solution and held on ice. To make the protease inhibitor solution, one tablet of a protease inhibitor cocktail (cOmplete™ mini protease inhibitor cocktail tablets, 11836153001, Roche, Basel, Switzerland) was added to 1 mL of MilliQ water, according to the manufacturer's instructions. The exact components of the cocktail are proprietary and withheld; however, they inhibit a wide array of serine, cysteine and metalloproteases as well as calpains. Pumping was continued until the level of milk reached a premarked line for 10 mL. Immediately after this line had been reached, the mother stopped pumping and passed the milk bottle to the researchers. The milk was immediately mixed well by gentle shaking and divided into 1-mL aliquots in 1.5-mL Eppendorf tubes and placed on dry ice in a closed and isolated storage box, within one minute after expression. For each mother, a second milk sample was collected immediately after the first, following the above procedure. The samples were immediately transported on dry ice to storage in a −80 °C freezer (reaching the freezer within 30 min of expression) until further analysis. Milks were stored at −80 °C for a maximum of 3 weeks prior to peptidomic analyses.
Table 1.
Demographic information of mothers and their infants participating in this study
| Mothers (n) | 4 |
| Infant sex (male/female) | 1/3 |
| Gestational age (weeks) | 39 ± 2 |
| Lactation age (weeks) | 13.1 ± 4.3 |
| Mothers age at delivery (years) | 31 ± 4.2 |
| Number of pregnancies | 1.8 ± 0.5 |
| C-section | 1 |
| Vaginal birth | 3 |
Sample preparation
One milliliter aliquots of the eight milk samples (two samples from each of four mothers) were thawed on ice (approximately 30 min). Peptide extraction was performed according to the method described previously [1]. Briefly, milk fat was removed and the infranate was collected by pipette. Milk proteins were then precipitated from the skimmed milk by addition of trichloroacetic acid and 600 μL of the supernatant containing the peptides were collected.
Trichloroacetic acid, salts, oligosaccharides and lactose were removed from the peptide solution and peptides were extracted via C18 reverse-phase preparative chromatography 96-well plates (Glygen, Columbia, MD) according to the previous procedure [9]. The purified peptide solutions were frozen at −80 °C and lyophilized using a freeze dry system (Labconco FreeZone 4.5 L, Kansas City, MO). After drying, the samples were rehydrated in 40 μL of 0.1 % FA in water.
Liquid chromatography nano-electrospray ionization mass spectrometry
Liquid chromatography (LC) separation was performed on a Waters nanoAcquity UPLC (Waters Corporation, Milford, MA, USA) with a nanospray source. One microliter of each sample was loaded onto the column. Peptides were first loaded onto a 180 μm × 20 mm, 5-μm bead 2G nanoAcquity UPLC trap column (reverse phase) for enrichment and online desalting, and then onto a 100 μm × 100 mm, 1.7-μm bead Acquity UPLC Peptide BEH C18 column (Waters) for analytical separation. Peptides were eluted using a gradient of 0.1% FA in water (A) and 99.9% ACN, 0.1% FA (B) with a flow rate of 500 nL/min. The 120-min gradient consisted of 3–10% solvent B over 3 min, 10–30% solvent B over 99 min, 30–90% solvent B over 3 min, 90% solvent B for 4 min, 90–3% solvent B over 1 min, and finally held at 3% solvent B for 10 min. Each sample was followed by a 30-min column wash.
The mass spectrometry instrument used was a Thermo Scientific Orbitrap Fusion Lumos. Spectra were collected with positive-ionization mode and an electrospray voltage of 2,400 V. The MS scan range was 400–1500 m/z at a resolution of 120K. The automatic gain control target was set to 4.0 × 105, with a maximum injection time of 50 ms. The fragmentation mode was set to collision-induced dissociation and the collision energy was 35%. The MS cycle time was set to 3 s, with data-dependent analysis and automated precursor peak selection. Precursors were excluded (within 10 ppm mass error) after one fragmentation for 60 s. Precursors were selected for fragmentation based on the following criteria: most intense peaks, ion-intensity threshold 5.0 × 103 and charge state 2–7. Fragments were detected with the ion trap with an automatic scan range.
Spectra were analyzed by database searching in Thermo Proteome Discoverer (v2.1.0.81) using an in-house human milk protein sequence database. Potential modifications included phosphorylation of serine and threonine and oxidation of methionine. Only peptides identified with high confidence were included (P < 0.01), and peptide sequences with multiple modifications were grouped into a single peptide for counts. Counts measured the number of unique peptide sequences identified in a sample. Abundance measured the area under the curve of the eluted peak (ion intensity).
Data analysis
Identified peptides were examined for homology with literature-identified bioactive peptides using our recently created Milk Bioactive Peptide Database (MBPDB) [10]. The MBPDB is a comprehensive source for all milk bioactive peptides. The search was performed as a sequence search that searches for bioactive peptides matching the query peptide sequence. The similarity threshold was set to 80%, with the amino acid scoring matrix set to identity. “Get extra output” was selected to obtain the specific percentage similarity between the query sequence and the database sequence.
Peptides were mapped to the parent sequence of the major human milk proteins using an in-house tool (PepEx) [2], which can be accessed at http://mbpdb.nws.oregonstate.edu/pepex/. PepEx mapping provides a visual overview of the position of peptides released within the parent protein sequence by totaling the abundance or count of each amino acid from the peptidomic data. The protein hydrophobicity distribution was determined using the method described by Tanford [11]. The hydrophobicity was calculated as an average of seven amino acids and a linear weight variation with 25% relative weight of the amino acids at the window edge.
Excel was used to create tables of which amino acids were present in the P1 and P1’ positions of the N- and C-termini of each peptide identified. The cleavage specificity of known milk proteases were mapped to these amino acids for visual interpretation in the figures (based on the most abundant amino acids listed at P1 and P1’ for Merops proteases [12]): plasmin (MEROPS ID: S01.233), thrombin (MEROPS ID: S01.217), elastase (MEROPS ID: S01.131), kallikrein 6 (MEROPS ID: S01.236) and kallikrein 11 (MEROPS ID: S01.257). To our knowledge, of the known kallikreins, only kallikreins 6 and 11 have been identified in human milk [13].
Proteasix, an online tool, was used to predict enzymes involved in cleavage sites using a cleavage site database built from information on human proteases from the cleavage site specificity matrix within the Merops database [14]. Proteasix was used to predict which proteases were involved in peptide cleavage within the eight milk samples. The enzymes searched against were plasmin (PLG; P00747), neutrophil elastase (ELANE; P08246), thrombin (F2; P00734), kallikrein 6 (KLK 6; Q92876) and kallikrein 11 (KLK 11; Q9UBX7). Predicted and observed cleavages were combined in the result output.
Results
In total, 1,107 unique peptide sequences derived from 42 different proteins were identified in the eight human milk samples from four mothers, based on a database search against the human milk proteins with > 99.9% confidence. These peptides derived mostly from β-casein (32% of the total peptide abundance), osteopontin (22%), αs1-casein (10%), butyrophilin subfamily 1 member A1 (9%), polymeric immunoglobulin receptor (7%), perilipin 2 (4%), bile salt-activated lipase (4%), mucin 1 (3%) and fibrinogen α-chain (2%). Of the 1,107 peptides, 319 peptide sequences were identified in at least one of the two breast milk samples from all four mothers (Supplementary Table 1). The combined abundance of these 319 peptides accounted for 95 ± 5% of the total abundance of peptides. Another 317 peptides were found in only one of the mothers’ milks (40 in mother 1, 102 in mother 2, 94 in mother 3, and 81 in mother 4). These peptides made up <1% of total peptide abundance.
On average, 532 ± 38 peptides were identified in the eight milk samples. Identified peptides derived mainly from, in order of peptide count, β-casein (174 ± 13 sequences), osteopontin (115 ± 48), αs1-casein (59 ± 12), butyrophilin (50 ± 6), polymeric immunoglobulin receptor (43 ± 5), bile salt-activated lipase (19 ± 4), perilipin-2 (16 ± 7), mucin 1 (15 ± 2) and fibrinogen α-chain (10 ± 4) (Fig. 1A). Though κ-casein and lactoferrin are among the most abundant proteins in milk, ≤ 4 κ-casein-derived peptides (six total) and 0–1 lactoferrin-derived peptides (one total) were identified in each sample. Peptides from κ-casein and lactoferrin only accounted for less than 0.01% of the total peptide abundance (Fig. 1B). Both in terms of count and abundance, peptides found in all four mothers’ milks were greater than peptides found in only three, only two, or in just one mother across all major milk proteins. However, the percentage of total abundance of peptides found in all four (95 ± 5%) is higher than the percentage of total count of peptides found in all four (53 ± 6%).
Fig. 1.
Count (A) and abundance (B) of peptides derived from milk proteins and identified in human breast milk. Results are shown as means ± SD, n = 8. CASB, β-casein; OSTP, osteopontin; CASA1, αs1-casein; BT1A1, butyrophilin subfamily 1 member A1; PIGR, polymeric immunoglobulin receptor; CEL, bile salt-activated lipase; PLIN2, perilipin-2; MUC1, mucin-1; FIBA, fibrinogen α-chain; CASK, κ-casein; TRFL, lactoferrin
Enzyme cleavage
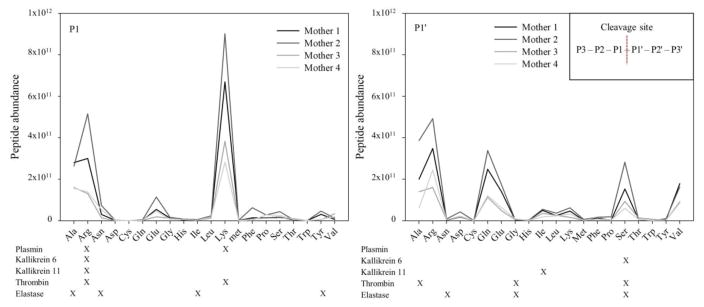
Each of the 532 ± 38 peptides identified in human milk samples have both an N- and C-terminus, resulting in potentially double the number of cleavage sites as peptides identified. However, of these peptides, 36 ± 7 and 39 ± 5 peptides had C- and N-termini, respectively, which matched the parent protein’s C- and N-termini and therefore did not count as a cleavage site. The final number of potential cleavage sites was therefore 990 ± 74. To identify which enzymes in human milk were responsible for cleaving the proteins in milk into peptides, we used several methods. First, we investigated the amino acids at the P1 and P1’ position of each cleavage site. The highest number of peptides had Lys or Arg in the P1 position (33.6%, Fig. 2), which accounted for 64% of the total abundance of peptides. This finding suggests that the majority of cleavages within the mammary gland are due to plasmin/kallikrein/thrombin (all of which can cleave after a Lys or Arg, and so cannot be distinguished based on their P1 cleavage specificity). The high count of Ser at P1’ matches with known specificities for elastase/thrombin/kallikrein 6. These results were conserved when examining the peptide abundances (Fig. 3) with few exceptions. Additionally, of the 1,107 peptides identified, 348 ± 56 peptides were identified that differed from another peptide by the removal of one N-terminal amino acid, and 328 ± 43 peptides that differed by the removal of one C-terminal amino acid (making up 74 ± 7 % of the total peptide abundance). These sets of peptides may result from exopeptidase activity.
Fig. 2.
Count of peptides derived from milk proteins and identified in human breast milk and distributed according to their cleavage site. Results are shown as means ± SD, n = 8
Fig. 3.
Abundance of peptides derived from milk proteins and identified in human breast milk and distributed according to their cleavage site. Results are shown as means ± SD, n = 8
For cleavage count (Fig. 2), all four mothers had highly similar numbers for each amino acid at both the P1 and P1’ positions. For cleavage abundance (Fig. 3), although the abundance of each amino acid at the P1 and P1’ positions were slightly different between the mothers, the overall trend of which amino acids were more abundant than others remained highly conserved.
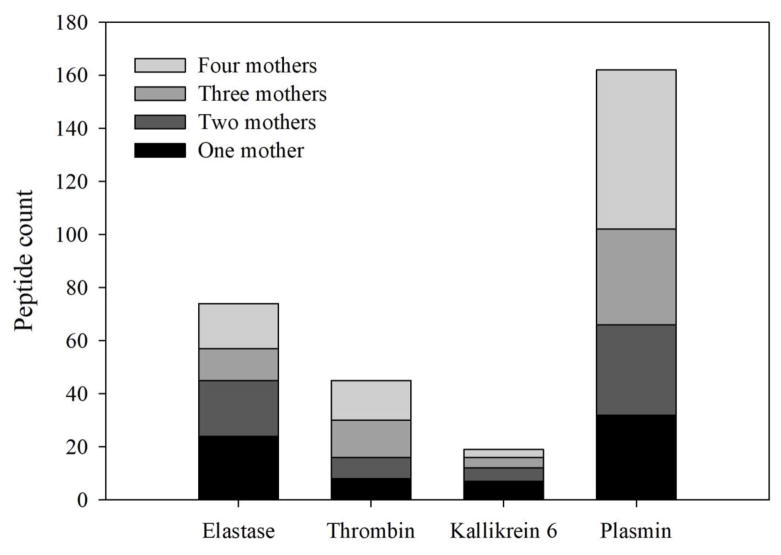
To extend the analysis of enzymes active in milk within the mammary gland, an online tool (Proteasix) was used to predict which enzyme could have caused the cleavage of milk proteins into the identified peptides. Proteasix assigned 162 ± 17 cleavage sites to one of the specified enzymes whereas 816 ± 50 cleavage sites were not assigned to an enzyme. Proteasix predicted that plasmin and elastase were most involved in cleaving the human milk proteins inside the mammary gland (Table 2). No cleavage sites were assigned to kallikrein 11. Next, we examined how many peptides assigned to a specific protease were found in all four mothers (Fig. 4). Overall, 32.7% of the predicted cleavages resulted in peptides that were identified in all four mothers (22.9% for elastase, 33.3% for thrombin, 15.8% for kallikrein 6, and 37% for plasmin).
Table 2.
Counts of cleavages by proteases predicted to be involved in protein cleavage of human breast milk proteins as estimated by Proteasix. Results are shown as means ± SD, n = 8
| Count of cleavages | |||||
|---|---|---|---|---|---|
| Plasmin | Elastase | Thrombin | Kallikrein 6 | Kallikrein 11 | Unassigned |
| 99.0 ± 16.1 | 30.9 ± 4.0 | 24.4 ± 3.5 | 8.3 ± 2.9 | 0.0 ± 0.0 | 816.1 ± 50.2 |
Fig. 4.
Count of peptides predicted by Proteasix to derive from specific protease activity
As described above, the cleavage site specificities of thrombin, plasmin and kallikrein 6 are overlapping and might not be distinguishable in all situations. On average, 18 ± 3 of the cleavage sites were assigned to two or more enzymes by Proteasix. The Proteasix results also identify which of the cleavage sites have been previously observed to occur with a specific protease and a specific protein precursor. Of the 163 ± 17 cleavages matched to the searched enzymes, 77 ± 18 had previously been observed; and of these known cleavages, 66 ± 16 were assigned to plasmin and 11 ± 2 were assigned to thrombin.
Peptide distribution
The PepEx peptide distribution analysis demonstrates that though varying in peptide count and abundance, proteases from all four mothers released peptides from similar regions across the sequence of αs1-casein, β-casein and osteopontin (Fig. 5 and Supplementary Fig. 1). This pattern was also apparent for polymeric immunoglobulin receptor, butyrophilin subfamily 1 member A1 and bile salt-activated lipase (Supplementary Fig. 2–4). The peptide abundance peaks and peptide count peaks align in all three figures. The regions that generated the highest abundance of peptides were osteopontin f(169–246), β-casein f(16–54) and αs1-casein f(16–51). These areas were 13.5 times, 11.4 times and 176 times higher in abundance than the peptides from the rest of their protein sequence, respectively. The position of the two major cleavage site amino acids, Arg and Lys, are also shown, along with the hydrophobicity distribution of αs1-casein, β-casein and osteopontin on their respective protein sequences. The cleaved Arg and Lys sites that released the largest abundance of peptides were found in osteopontin (Arg 152, Lys 154, Lys 159, Lys 187, Lys 225), β-casein (Arg 1, Lys 18, Lys 23, Lys 39) and αs1-casein (Arg 36). Consistently across all three proteins, the cleavage sites giving rise to the highest count of peptides and with the strongest ion intensities were in hydrophilic areas of the three proteins. The only exception was Lys 187 of osteopontin, which lies in a neutral part of the protein sequence.
Fig. 5.
Abundance of peptides identified in human breast milk from four mothers mapped on the sequence of β-casein (A), osteopontin (B) and αs1-casein (C). Results are shown as means, n = 2. Grey vertical lines represent Arg or Lys P1 cleavage sites. The hydrophobicity score is shown as a heat map. Green is hydrophobic, red is hydrophilic
Bioactive peptides
Searching the identified peptides against the MBPDB revealed 37 unique peptides in the milk samples that were highly homologous (≥ 80% sequence match) to previously identified bioactive peptides. Two of these peptides are identical to the known bioactive peptides β-casein f(185–211) QELLLNPTHQIYPVTQPLAPVHNPISV [15] and β-casein f(105–117) SPTIPFFDPQIPK [16], with antimicrobial and BALB/c3T3 (mouse fibroblast) cell-proliferation stimulatory effects, respectively. Multiple peptides homologous to these two were also identified in the milk samples (Table 3). We identified 21 other peptides highly homologous to β-casein f(185–211) and 5 to β-casein f(105–117). The high degree of sequence similarity suggests the possibility for antimicrobial and cell-proliferation-inducing activity within these 26 peptides. Of the remaining peptides, nine were derived from β-casein and are homologous with known ACE-inhibitory peptides, and one derived from lactoferrin and is homologous with a known antimicrobial peptide.
Table 3.
Bioactive peptides identified in the human breast milk samples by searching the Milk Bioactive Peptide Database (MBPDB), with a threshold value of 80%. n = 8
| Query peptide | Protein | Bioactive peptide | Function | Alignment |
|---|---|---|---|---|
| QELLLNPTHQIYPVTQPLAPVHNPISV | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 100.0 |
| NQELLLNPTHQIYPVTQPLAPVHNPISV | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 96.4 |
| ELLLNPTHQIYPVTQPLAPVHNPISV | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 96.3 |
| NQELLLNPTHQIYPVTQPLAPVHNPIS | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 96.3 |
| QELLLNPTHQIYPVTQPLAPVHNPIS | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 96.3 |
| LNQELLLNPTHQIYPVTQPLAPVHNPISV | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 93.1 |
| LNQELLLNPTHQIYPVTQPLAPVHNPIS | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 92.9 |
| ELLLNPTHQIYPVTQPLAPVHNPIS | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 92.6 |
| NQELLLNPTHQIYPVTQPLAPVHNPI | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 92.6 |
| LLNQELLLNPTHQIYPVTQPLAPVHNPISV | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 90.0 |
| LLNQELLLNPTHQIYPVTQPLAPVHNPIS | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 89.7 |
| LLNQELLLNPTHQIYPVTQPLAPVHNPI | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 89.3 |
| LLLNPTHQIYPVTQPLAPVHNPIS | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 88.9 |
| LLNPTHQIYPVTQPLAPVHNPISV | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 88.9 |
| NQELLLNPTHQIYPVTQPLAPVHNP | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 88.9 |
| LLNPTHQIYPVTQPLAPVHNPIS | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 85.2 |
| LNPTHQIYPVTQPLAPVHNPISV | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 85.2 |
| ALLLNQELLLNPTHQIYPVTQPLAPVHNPISV | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 84.4 |
| LLNQELLLNPTHQIYPVTQPLAPVH | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 81.5 |
| LNPTHQIYPVTQPLAPVHNPIS | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 81.5 |
| NPTHQIYPVTQPLAPVHNPISV | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 81.5 |
| NQELLLNPTHQIYPVTQPLAPVH | β-casein | QELLLNPTHQIYPVTQPLAPVHNPISV | Antimicrobial | 81.5 |
| SPTIPFFDPQIPK | β-casein | SPTIPFFDPQIPK | Stimulates cell proliferation | 100.0 |
| KSPTIPFFDPQIPK | β-casein | SPTIPFFDPQIPK | Stimulates cell proliferation | 92.9 |
| SPTIPFFDPQIPKL | β-casein | SPTIPFFDPQIPK | Stimulates cell proliferation | 92.9 |
| SPTIPFFDPQIP | β-casein | SPTIPFFDPQIPK | Stimulates cell proliferation | 92.3 |
| LKSPTIPFFDPQIPK | β-casein | SPTIPFFDPQIPK | Stimulates cell proliferation | 86.7 |
| SPTIPFFDPQIPKLTD | β-casein | SPTIPFFDPQIPK | Stimulates cell proliferation | 81.3 |
| DTVYTKGRVMP | β-casein | TVYTKGRVMP | ACE-inhibitory | 90.9 |
| KDTVYTKGRVMP | β-casein | TVYTKGRVMP | ACE-inhibitory | 83.3 |
| TVYTKGRVMPVL | β-casein | TVYTKGRVMP | ACE-inhibitory | 83.3 |
| PFFDPQIPK | β-casein | PFFDPQIP | ACE-inhibitory | 88.9 |
| FFDPQIPK | β-casein | PFFDPQIP | ACE-inhibitory | 87.5 |
| LRQAQEKFGKDKSPKFQL | Lactoferrin | WNLLRQAQEKFGKDKSPK | Antimicrobial | 83.3 |
| IYPSFQPQPLI | β-casein | YPSFQPQPLIYP | ACE-inhibitory | 83.3 |
| PSFQPQPLIYP | ACE-inhibitory | 81.8 | ||
| IYPSFQPQPLIYP | ACE-inhibitory | 84.6 | ||
| DKIYPSFQPQPL | β-casein | DKIYPSFQPQPLIYP | ACE-inhibitory | 80.0 |
| QDKIYPSFQPQPL | β-casein | DKIYPSFQPQPLIYP | ACE-inhibitory | 80.0 |
Discussion
Due to the measures taken to limit post-expression proteolytic activity (expressing into antiproteases held on ice followed by immediate freezing), the 1,107 milk protein-derived peptides found in these milk samples most likely represent the result of proteolytic activity within the mammary gland. However, the quantitative degree of this intra-mammary gland protein digestion was not examined and should be measured in future studies. Milk expression typically takes place every 3–5 hours at 2–4 months of lactation. In the time between milk expressions, milk proteins and enzymes exit the mammary epithelial acinar cells and are stored in lactiferous ducts for a sufficient incubation time to allow indigenous proteases to hydrolyze milk proteins. As most of the milk proteases are active at body temperature [17–19], storage of milk within the mammary gland provides favorable conditions for activity of milk proteases. Most milk proteases also function well at the neutral pH of breast milk (including plasmin, kallikrein, thrombin, elastase, carboxypeptidase B2 and cytosol aminopeptidase) [7]. Only cathepsin D is nonfunctional at milk’s pH and requires activation at a more acidic pH [7].
The identification of numerous milk protein-derived peptides within milk that was treated to prevent post-expression proteolysis in the present study matches well with the findings of the only previous study to address this question of whether milk proteases are active within the mammary gland. In the previous study [1], the determination of whether milk peptides were created within the mammary gland relied on a single sample of breast milk and controlled post-expression proteolysis by heat-treatment after milk was collected. The present study more carefully examined this question by analyzing two milk samples from each of four mothers with post-expression proteolysis prevented by immediate application of antiproteases, pumping on ice and immediate freezing. Therefore, the present study greatly substantiates the preliminary findings of the previous study, more strongly supporting that proteolysis does indeed occur within the mammary gland.
The present study identified a much larger count of unique sequences (1,107) than were identified in previous studies of human milk peptidomics. Previous studies included peptides with different modifications in their total count, which were not used herein. When including modified peptides as unique sequences, the total peptide count identified in the present study is 1,578, which is substantially higher than the 328 [1], 445 [3], 579 [2], and 649 peptides [20] identified in human milk in previous studies. The lactation stage of these previous studies range from 2 weeks to 8 weeks, which is slightly earlier than in the current study (13.1 ± 4.3). Although the peptide profile differentiates over lactation stage [21], the increased number of peptides identified in this study likely derives from the use of the Orbitrap Fusion, which allows for faster spectral acquisition than the Q-TOF used in earlier studies, allowing increased chances for peptide identification. The present study identified peptides derived from 42 human milk proteins in milk. The most abundant peptide parent proteins in this study (β-casein, αs1-casein, osteopontin, polymeric immunoglobulin receptor and butyrophilin) were also the most abundant in previous studies of peptidomics of human milk samples [1, 20, 2]. Additionally, the location of abundant peptides within the parent protein sequences aligns between the studies. The overall alignment of peptide results in this study with those of previous studies suggests that the peptides observed in those studies were generated via digestion within the mammary gland.
All four mothers’ milk samples had unique components of their peptide profiles. Many of the peptide sequences identified were absent in one or more of the mothers’ milk samples. This differential peptide release likely results from variations in the proteolytic activity of the native milk proteases between mothers. However, as mentioned above, the peptides that were shared between all four mothers accounted for more than 95% of the total peptide abundance. The peptides that were found in only one to three of the mothers’ milk thus represent a very small portion of the peptide component of human milk, indicating that protease activity is highly similar between mothers.
The results from the P1-P1’ Excel analysis, and to a lesser extent the Proteasix prediction, support this conclusion. The cleavage counts of each amino acid at the P1 and P1’ positions were conserved across the four mothers (Fig. 2), indicating similar rates of activity for all native proteases. The cleavage abundances of each amino acid were different between mothers (Fig. 3). However, the relative amounts of amino acid cleavage within each mother’s milk followed a similar trend for all four. Thus, the mothers have different degrees of protease activity, yet the comparative rates of activity of the different proteases is similar. From Proteasix, roughly one-third of all peptides predicted to result from specific enzyme activity were found in all four mothers (Fig. 4). Again, the overall proteolytic activity in the mammary gland is highly similar between mothers.
The combined examination of predicted cleavage sites in Excel and Proteasix suggests that plasmin is the most active enzyme in cleaving milk proteins into peptides. Plasmin, kallikrein and thrombin all have closely related P1 and P1’ cleavage specificity. Proteasix was used to distinguish among these enzymes as it uses more complex cleavage specificity by accounting for amino acids from P4 to P2’ and P4 to P4’, respectively. Results from Proteasix confirmed plasmin to be the most active enzyme, but it also found several cleavages that matched the specificity of thrombin and kallikrein 6. The distinction between thrombin and plasmin activity comes from the additional cleavage site specificities that thrombin has (Pro at P2, hydrophobic amino acids at P3 and P4, not Asp or Glu at P1’). In support of our in silico data, we previously measured the activity of proteases in human milk post-expression and found that plasmin, elastase, kallikrein and thrombin were all active [7]. However, measuring enzyme activity post-expression does not fully represent what occurs inside the mammary gland. Using the peptidomics data of the milk samples to predict for enzyme activity based on released peptides might be the current best approach to circumvent this limitation.
In bovine milk, indigenous plasmin activity has been shown to hydrolyze αs1-, αs2- and β-casein but not κ-casein during storage [22]. Similarly, in our study, we found that human milk proteases hydrolyzed both β-casein and αs1-casein, whereas little digestion of κ-casein was identified.
Milk also contains cytosol aminopeptidase and carboxypeptidase B2, which can sequentially release amino acids from the N- and C-termini, respectively. The activities of carboxypeptidase B2 and cytosol aminopeptidase are harder to determine by cleavage site prediction, but our previous study has confirmed their activity in human milk post-expression [7]. Carboxypeptidase B2 cleaves Lys and Arg residues off the C-terminus [23] and may account for some of the cleavages with a Arg or Lys at the P1’ position (Figure 2). Cytosol aminopeptidase preferentially cleaves Leu from the N-terminus and has activity for all other amino acids as well [24]. These enzymes were not available for inclusion in the Proteasix analyses and could account for much of the unassigned cleavages in these analyses. Many of the identified peptides corresponded to a set of peptides with sequential removal of amino acids from the N- and C-termini, which indicates exopeptidase activity in the human mammary gland.
Besides proteases, milk also contains a wide array of protease inhibitors, including (from highest to lowest abundance) α-1-antitrypsin, antithrombin III, α-1-antichymotrypsin, α-2-antiplasmin and plasma serine protease inhibitor [7]. In this study, no peptides were identified from these inhibitors. This finding implies either that these protease inhibitors are not degraded by milk proteases within the mammary gland, or that the concentrations of their released peptides are too low to be detected. The fact that some proteolysis occurs in the mammary gland despite the presence of numerous protease inhibitors suggests that the activity of protease inhibitors is not as high as the activity of proteases.
The regions of peptide release in the mammary gland are highly conserved across all four mothers (Fig. 4 & 5) in terms of peptide abundance and count. For milk from all four mothers, the majority of peptide release came from the regions αs1-casein f(16–51), β-casein f(16–54) and osteopontin f(169–246). The near identical release of peptides from these regions suggests that there may be a physiological role for peptide release. Indeed, a large number of bioactive peptides were predicted to derive from these regions.
Only two peptides matched the exact sequence of known bioactive peptides; however, 35 sequences had ≥ 80% homology to known bioactive peptides. These two bioactive peptides were also identified in previous studies on peptides in human milk [1, 6]. Though this number of homologous bioactive peptides is small compared with the overall number of peptides identified, it is not unexpected. These bioactive peptides may exert their action inside the mammary gland or within the infant. However, the effect of these peptides has only been confirmed using in vitro studies, and for the infant to benefit from them, they would need to survive further digestion from the infant digestive enzymes. Survival may be enhanced by post-translational glycosylation of milk proteins which can release peptides more resistant to proteolytic digestion [25, 26]. Determining whether site-selective glycosylation improves the survivability of peptides with bioactive function necessitates further research.
Human milk proteins as a source of bioactive peptides have been studied far less than bovine proteins [10]. The fewer number of identified peptides limits the number of results the homology search can produce, as the search relies on comparisons with previously identified bioactive peptides. Additionally, most studies that identified bioactive peptides in human milk only did so after incubation of milk with additional proteolytic enzymes, thus they did not examine the peptides naturally released in the milk [27, 28]. The overlap in peptides identified from these types of studies and the present one may be minimal. Most of the identified bioactive peptides from human milk derive from either β-casein or lactoferrin. However, in this study, we were unable to find significant proteolysis of lactoferrin by indigenous milk proteases in the mammary gland, as only one peptide was identified from this protein in milk from only one of the mothers. However, this peptide matched closely to a known antimicrobial peptide. Lactoferrin digestion and release of lactoferrin-derived bioactive peptide begins in the infant stomach [20]. Interestingly, to date, no bioactive peptides have been identified from osteopontin despite it being one of the largest contributors of peptides in milk.
Overall, these findings clearly support for the first time that milk protease digestion of milk proteins begins within the mammary gland. Therefore, the directly breastfed infant receives partially digested proteins and an array of bioactive peptides within milk. We do not yet know how significant the contribution of this early digestion is to the infant’s overall protein digestion. Future research could further investigate whether lactation age, gestational age, mother age and number of pregnancies affect protein digestion inside the mammary gland.
Supplementary Material
Supplemental Table 1 All identified peptides from the mother’s milk samples. Shared indicates whether the peptide was found in all four mothers. “Single mother” lists the mother’s milk sample the peptide was found in if only found in one of the four mothers. Homology displays the percent homology of the peptide with the Milk Bioactive Peptide Database
Supplemental Fig. 1 Count of peptides identified in human breast milk from four mothers mapped on the sequence of β-casein (A), osteopontin (B) and αs1-casein (C). Results are shown as means, n = 2. Grey vertical lines represent Arg or Lys P1 cleavage sites. The hydrophobicity score is shown as a heat map. Green is hydrophobic, red is hydrophilic
Supplemental Fig. 2 Count (A) and abundance (B) of peptides identified in human breast milk from four mothers mapped on the sequence of polymeric immunoglobulin receptor. Results are shown as means, n = 2
Supplemental Fig. 3 Count (A) and abundance (B) of peptides identified in human breast milk from four mothers mapped on the sequence of butyrophilin subfamily 1 member A1. Results are shown as means, n = 2
Supplemental Fig. 4 Count (A) and abundance (B) of peptides identified in human breast milk from four mothers mapped on the sequence of bile salt-activated lipase. Results are shown as means, n = 2
Acknowledgments
The authors thank Melinda Spooner for assisting in collecting the human milk samples and Cora J. Dillard for editing the manuscript. SDN and DCD planned the study. SDN collected the samples. SDN and RB conducted the experiments and data analysis. SDN, RB and DCD prepared the manuscript. All authors read and approved the final manuscript. The authors acknowledge the Mass Spectrometry Center at Oregon State University, which is supported in part by the National Institute of Health grant S10OD020111 (CSM).
Funding Supported by the K99/R00 Pathway to Independence Career Award, Eunice Kennedy Shriver Institute of Child Health & Development of the National Institutes of Health (R00HD079561) (DCD).
Footnotes
Compliance with Ethical Standards
Ethical Approval This present study was approved and carried out in accordance with the guidelines from the Institutional Review Board at Oregon State University. All mothers gave their informed consent.
Conflict of Interest The authors declare that they have no conflict of interest.
References
- 1.Dallas DC, Guerrero A, Khaldi N, Castillo PA, Martin WF, Smilowitz JT, et al. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res. 2013;12(5):2295–304. doi: 10.1021/pr400212z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerrero A, Dallas DC, Contreras S, Chee S, Parker EA, Sun X, et al. Mechanistic peptidomics: factors that dictate the specificity on the formation of endogenous peptides in human milk. Mol Cell Proteomics. 2014;13(12):3343–51. doi: 10.1074/mcp.M113.036194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dallas DC, Smink CJ, Robinson RC, Tian T, Guerrero A, Parker EA, et al. Endogenous human milk peptide release is greater after preterm birth than term birth. J Nutr. 2015;145(3):425–33. doi: 10.3945/jn.114.203646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dallas DC, Murray NM, Gan J. Proteolytic systems in milk: perspectives on the evolutionary function within the mammary gland and the infant. J Mammary Gland Biol Neoplasia. 2015;20(0):133–147. doi: 10.1007/s10911-015-9334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dallas DC, German JB. Enzymes in human milk. Nestle Nutr Inst Workshop Ser. 2017;88:129–36. doi: 10.1159/000455250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khaldi N, Vijayakumar V, Dallas DC, Guerrero A, Wickramasinghe S, Smilowitz JT, et al. Predicting the important enzyme players in human breast milk digestion. J Agric Food Chem. 2014;62(29):7225–32. doi: 10.1021/jf405601e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demers-Mathieu V, Nielsen SD, Underwood MA, Borghese R, Dallas DC. Analysis of milk from mothers who delivered prematurely reveals few changes in proteases and protease inhibitors across gestational age at birth and infant postnatal age. J Nutr. 2017;146(6):1152–9. doi: 10.3945/jn.116.244798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prado BM, Ismail B, Ramos O, Hayes KD. Thermal stability of plasminogen activators and plasminogen activation in heated milk. Int Dairy J. 2007;17(9):1028–33. [Google Scholar]
- 9.Dallas DC, Guerrero A, Parker EA, Garay LA, Bhandari A, Lebrilla CB, et al. Peptidomic profile of milk of holstein cows at peak lactation. J Agric Food Chem. 2014;62(1):58–65. doi: 10.1021/jf4040964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen SD, Beverly RL, Qu Y, Dallas DC. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017;232:673–82. doi: 10.1016/j.foodchem.2017.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanford C. Contribution of hydrophobic interactions to the stability of the globular conformation of proteins. J Am Chem Soc. 1962;84(22):4240–7. [Google Scholar]
- 12.Klein J, Eales J, Zürbig P, Vlahou A, Mischak H, Stevens R. Proteasix: A tool for automated and large-scale prediction of proteases involved in naturally occurring peptide generation. Proteomics. 2013;13(7):1077–82. doi: 10.1002/pmic.201200493. [DOI] [PubMed] [Google Scholar]
- 13.Palmer DJ, Kelly VC, Smit A-M, Kuy S, Knight CG, Cooper GJ. Human colostrum: identification of minor proteins in the aqueous phase by proteomics. Proteomics. 2006;6(7):2208–16. doi: 10.1002/pmic.200500558. [DOI] [PubMed] [Google Scholar]
- 14.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2011;40(D1):D343–D50. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minervini F, Algaron F, Rizzello CG, Fox PF, Monnet V, Gobbetti M. Angiotensin I-converting-enzyme-inhibitory and antibacterial peptides from Lactobacillus helveticus PR4 proteinase-hydrolyzed caseins of milk from six species. Appl Environ Microbiol. 2003;69(9):5297–305. doi: 10.1128/AEM.69.9.5297-5305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azuma N, Nagaune S-I, Ishino Y, Mori H, Kaminogawa S, Yamauchi K. DNA-synthesis stimulating peptides from human β-casein. Agric Biol Chem. 1989;53(10):2631–34. [Google Scholar]
- 17.Derechin M. The assay of human plasminogen with casein as substrate. Biochem J. 1961;78:443–48. doi: 10.1042/bj0780443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Draper AM, Zeece MG. Thermal stability of cathepsin D. J Food Sci. 1989;54(6):1651–2. [Google Scholar]
- 19.Levison PR, Tomalin G. Studies on the temperature-dependent autoinhibition of human plasma kallikrein I. Biochem J. 1982;205(3):529–34. doi: 10.1042/bj2050529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dallas DC, Guerrero A, Khaldi N, Borghese R, Bhandari A, Underwood MA, et al. A peptidomic analysis of human milk digestion in the infant stomach reveals protein-specific degradation patterns. J Nutr. 2014;144(6):815–20. doi: 10.3945/jn.113.185793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dingess KA, de Waard M, Boeren S, Vervoort J, Lambers TT, van Goudoever JB, et al. Human milk peptides differentiate between the preterm and term infant and across varying lactational stages. Food & Function. 2017;8(10):3769–82. doi: 10.1039/c7fo00539c. [DOI] [PubMed] [Google Scholar]
- 22.Rauh VM, Johansen LB, Ipsen R, Paulsson M, Larsen LB, Hammershøj M. Plasmin activity in UHT milk: relationship between proteolysis, age gelation, and bitterness. J Agric Food Chem. 2014;62(28):6852–60. doi: 10.1021/jf502088u. [DOI] [PubMed] [Google Scholar]
- 23.Mao SS, Cooper CM, Wood T, Shafer JA, Gardell SJ. Characterization of plasmin-mediated activation of plasma procarboxypeptidase B. Modulation by glycosaminoglycans. J Biol Chem. 1999;274(49):35046–52. doi: 10.1074/jbc.274.49.35046. [DOI] [PubMed] [Google Scholar]
- 24.Burley SK, David PR, Taylor A, Lipscomb WN. Molecular structure of leucine aminopeptidase at 2.7-A resolution. Proc Natl Acad Sci U S A. 1990;87(17):6878–82. doi: 10.1073/pnas.87.17.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell D, Oldham NJ, Davis BG. Site-selective chemical protein glycosylation protects from autolysis and proteolytic degradation. Carbohydr Res. 2009;344(12):1508–14. doi: 10.1016/j.carres.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 26.Ihara S, Miyoshi E, Nakahara S, Sakiyama H, Ihara H, Akinaga A, et al. Addition of β1–6 GlcNAc branching to the oligosaccharide attached to Asn 772 in the serine protease domain of matriptase plays a pivotal role in its stability and resistance against trypsin. Glycobiology. 2004;14(2):139–46. doi: 10.1093/glycob/cwh013. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez-Ledesma B, Quiros A, Amigo L, Recio I. Identification of bioactive peptides after digestion of human milk and infant formula with pepsin and pancreatin. Int Dairy J. 2007;17(1):42–9. [Google Scholar]
- 28.Tsopmo A, Romanowski A, Banda L, Lavoie JC, Jenssen H, Friel JK. Novel anti-oxidative peptides from enzymatic digestion of human milk. Food Chem. 2011;126(3):1138–43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 All identified peptides from the mother’s milk samples. Shared indicates whether the peptide was found in all four mothers. “Single mother” lists the mother’s milk sample the peptide was found in if only found in one of the four mothers. Homology displays the percent homology of the peptide with the Milk Bioactive Peptide Database
Supplemental Fig. 1 Count of peptides identified in human breast milk from four mothers mapped on the sequence of β-casein (A), osteopontin (B) and αs1-casein (C). Results are shown as means, n = 2. Grey vertical lines represent Arg or Lys P1 cleavage sites. The hydrophobicity score is shown as a heat map. Green is hydrophobic, red is hydrophilic
Supplemental Fig. 2 Count (A) and abundance (B) of peptides identified in human breast milk from four mothers mapped on the sequence of polymeric immunoglobulin receptor. Results are shown as means, n = 2
Supplemental Fig. 3 Count (A) and abundance (B) of peptides identified in human breast milk from four mothers mapped on the sequence of butyrophilin subfamily 1 member A1. Results are shown as means, n = 2
Supplemental Fig. 4 Count (A) and abundance (B) of peptides identified in human breast milk from four mothers mapped on the sequence of bile salt-activated lipase. Results are shown as means, n = 2