Abstract
HIV-1 Gag protein assembles into 100- to 120-nm diameter particles in mammalian cells. Recombinant HIV-1 Gag protein assembles in a fully defined system in vitro into particles that are only 25–30 nm in diameter and that differ significantly in other respects from authentic particles. However, particles with the size and other properties of authentic virions were obtained in vitro by addition of inositol phosphates or phosphatidylinsitol phosphates to the assembly system. Thus, the interactions between HIV-1 Gag protein molecules are altered by binding of inositol derivatives; this binding is apparently essential for normal HIV-1 particle assembly. This requirement is not seen in a deleted Gag protein lacking residues 16–99 within the matrix domain.
Keywords: retrovirus assembly, phosphatidylinsitol phosphates
The principal structural component from which a retrovirus particle is formed is the viral Gag protein. Spherical particles are assembled as arrays of radially arranged Gag monomers; the particles are coated with a membrane that is derived from the plasma membrane of the host cell as they are released by budding from the cell surface. After the particle is released, Gag is cleaved by the viral protease (PR) into the proteins of the infectious particle; this series of cleavage events is accompanied by profound morphological changes and is termed “maturation” of the particle. The cleavage products always include, from N to C terminus, matrix (MA), capsid (CA), and nucleocapsid (1).
Expression of the Gag protein is sufficient for assembly of virus-like particles (VLPs) in mammalian cells. Therefore, this protein would appear to be capable of the protein–protein interactions required for particle formation. This inference has generally been confirmed by in vitro assembly studies with recombinant retroviral Gag proteins; thus, Rous sarcoma virus (RSV) (2), Mason–Pfizer monkey virus (MPMV) (3), and Moloney murine leukemia virus (MoMuLV) (S.C. and A.R., unpublished data) Gag proteins purified from bacteria can assemble, under simple, fully defined conditions, into particles that closely resemble authentic immature virions. These standard conditions are, in general, a moderate ionic strength (0.1 M NaCl), neutral pH, and the addition of ≈4 μg of single-stranded DNA or RNA per 100 μg of Gag protein.
We have described the results of similar experiments using recombinant Gag protein of HIV-1 (4). (The recombinant HIV-1 Gag protein used in these experiments has a deletion of the C-terminal p6 domain, and also lacks the N-terminal myristoyl modification found on Gag protein that is produced in eukaryotic cells.) In contrast to the results with Gag proteins from other retroviruses, standard incubation conditions were not sufficient for the normal assembly of HIV-1 Gag protein. Although the protein formed spherical particles in these assembly reactions, the radius of curvature of these spheres was far smaller than in authentic HIV-1 virions, so that the particles were only 25–30 nm, rather than ≈100 nm, in diameter (4). We also noted that the addition of mammalian cell lysates to this system could eliminate this defect, resulting in the formation of VLPs of the correct size (4).
We now report that these particles also differ in several other respects from authentic HIV-1 virions. Cell lysates prevent these defects in addition to correcting the radius of curvature of the particles. Fractionation of the lysates has shown that the ability to alter the viral assembly process can be attributed to inositol pentakisphosphate (IP5). A number of inositol phosphate derivatives, including a phosphatidylinositol 3,4,5-trisphosphate (PIP3) analog, also correct the assembly process. We also show that a Gag protein lacking residues 16–99 in its matrix domain has lost the requirement for inositol phosphates in assembly in vitro.
Materials and Methods
Purification of HIV-1 Gag Protein and Assembly of Particles.
HIV-1 Gag protein was purified from bacteria as described (4). Gag proteins lacking residues 16–99 of MA (5, 6) (originally designated ΔMA-CA-NC-SP2, but referred to here as Δ16–99) were purified by the same procedure. Both of these proteins lack the p6 domain and also differ from Gag protein produced in eukaryotic cells by the absence of N-terminal myristoylation.
Particles were assembled (1 mg/ml protein, 0.04 mg/ml yeast tRNA) in 100 μl of buffer A (20 mM Tris, pH 7.5/0.1 M NaCl/10 mM DTT) with the indicated additions (5 μl of reticulocyte lysate or 2 μM IP5) for 1 h at room temperature.
Salt and RNase Treatments.
Particles were assembled in 100 μl of buffer B (buffer A + 1% Nonidet P-40; Roche Molecular Biochemicals). They were treated with 100 μg of RNase A (Roche) for 30 min or with 0.5 M NaCl for 20 min at room temperature, and were then centrifuged for 30 min at 21,000 × g in an Eppendorf 5417R microcentrifuge. Pellets and supernatants were analyzed after SDS/PAGE, either by Coomassie blue staining or by immunoblotting with sera directed against the viral CA protein. Immature MoMuLV or HIV-1 virions from cultured cells (each having an active-site mutation to inactivate PR) were incubated in buffer B for 1 h before treatment with RNase or NaCl and analysis as described above.
Protease Treatments.
After assembly in buffer B, particles were treated with 5 μg of HIV-1 PR for 1 h or 1 μg of trypsin (Sigma) for 20 min at 37°C. Immature virions from cell culture were incubated in buffer B for 1 h before exposure to proteases. Samples were then analyzed by immunoblotting as described above.
Fractionation of Reticulocyte Lysates.
Reticulocyte lysates (7) were fractionated by cation exchange chromatography (Q Sepharose, Amersham Pharmacia) and eluted with a gradient of 0.05–1 M ammonium bicarbonate. Fractions possessing activity in the salt-resistance assay were lyophilized, dissolved in water, and precipitated with 80% ethanol. The pellet was dissolved in 0.1 M ammonium bicarbonate and fractionated by gel filtration chromatography on a Superdex-30 (Amersham Pharmacia) column. Active fractions were pooled and precipitated again with 80% ethanol, and the pellet was dissolved in water before analysis.
Mass Spectrometry.
Samples were treated with 1 M HCl to replace sodium ions. They were then analyzed by positive-ion electrospray mass spectrometry using an Agilent Technologies mass selective detector over the range of 200–800 atomic mass units.
Electron Microscopy.
Particles were examined by negative-stain electron microscopy with 2% uranyl acetate on Formvar–carbon-coated grids.
Results
Effects of Mammalian Cell Lysate on Assembly of Particles.
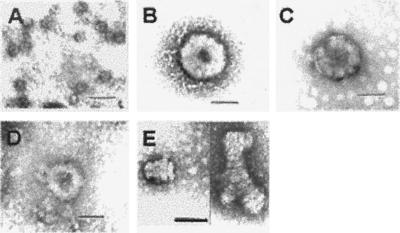
We reported that recombinant HIV-1 Gag protein assembles in vitro into particles that are far smaller than authentic HIV-1 virions, with a diameter of 25–30 nm rather than ≈100 nm (4). We tested the possibility that a factor in mammalian cells is responsible for the assembly of full-size particles by adding rabbit reticulocyte lysate to the standard assembly reaction. The effect of the lysate on the size and morphology of particles is shown in Fig. 1. Particles assembled in the presence of lysate (Fig. 1B) closely resembled authentic, immature virions (Fig. 1C), whereas particles assembled in the standard assembly reaction were far smaller (Fig. 1A). Examination of thin sections of the particles shown in Fig. 1B confirmed that they were hollow spheres (data not shown).
Figure 1.
Electron microscopy of negatively stained particles assembled in vitro and of authentic, immature HIV-1 virion. (A) Particles assembled in buffer A. (B) VLP assembled in buffer A plus 5% rabbit reticulocyte lysate. (C) Immature HIV-1 virion. (D) VLP assembled in buffer A plus 2 μM IP5. (E) “Stacked plates” assembled in buffer A plus 2 μM IP6. (Scale bars, 100 nm.)
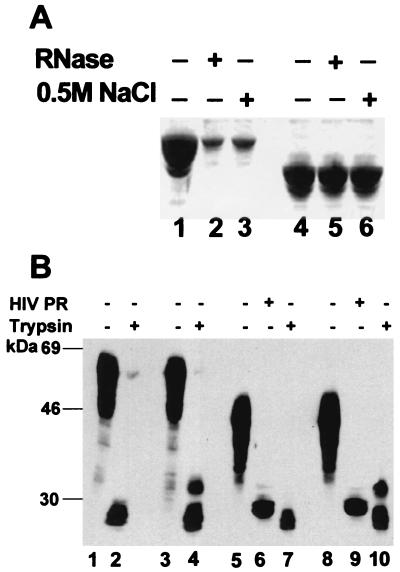
We performed other tests to further characterize the effects of cellular lysate on particle assembly. As reported (4), particles assembled in buffer alone were solubilized by treatment with either 0.5 M NaCl or RNase (if the nucleic acid used in the assembly was RNA) (Fig. 2A, lanes 2 and 3). However, particles formed in the presence of reticulocyte lysate remained pelletable after either of these treatments (Fig. 2B, lanes 2 and 3). Fig. 2D (lanes 6–9) shows that authentic immature HIV-1 particles (whose membranes had been removed with Nonidet P-40) were also resistant to these treatments. Interestingly, the resistance to these treatments was not seen in immature MoMuLV particles (Fig. 2D, lanes 6–9) (8), suggesting that it reflects a specific property of HIV-1 assembly.
Figure 2.
Nuclease and salt resistance of particles assembled in vitro and of authentic, immature particles. Particles assembled in buffer B (A), buffer B plus 5% rabbit reticulocyte lysate (B), or buffer B plus 2 μM IP5 (C), were treated with RNase A (lanes 2) or NaCl (lanes 3). Gag proteins in particles pelleted after treatment were examined by SDS/PAGE and Coomassie blue staining. We estimate that >90% of the Gag protein was solubilized in A (lanes 2 and 3), whereas >90% remained pelletable in B and C (lanes 2 and 3). (D) Immature MoMuLV and HIV-1 virions produced in mammalian cells were analyzed separately (lanes 1 and 2, respectively) or were mixed together (lanes 3–9) and treated with RNase A (lanes 6 and 7) or NaCl (lanes 8 and 9). They were also digested with HIV-1 PR to confirm that the lipid envelope was removed by the detergent (lane 3), or sedimented without RNase or NaCl treatment to confirm that the particles are stable in buffer B (lanes 4 and 5). Gag proteins in the samples in D were detected by immunoblotting with antibodies against the CA proteins of both HIV-1 and MoMuLV. We estimate that >90% of the HIV-1 Gag protein was still pelletable after NaCl or RNase treatment. P, pellet; S, supernatant.
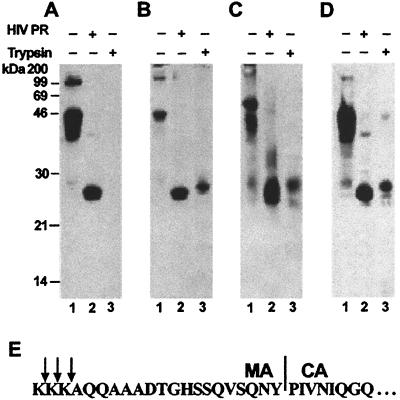
Another way of probing protein conformations in the particles is to test for resistance to proteolytic enzymes (9, 10). We compared the trypsin and PR sensitivities of the small particles formed under standard assay conditions and the larger VLPs assembled in the presence of lysate. Digestion of the Gag protein in particles was monitored by immunoblotting with anti-CA antibody. We found that CA protein was released by digestion of particles with HIV PR, whether or not they had been assembled in the presence of lysate (Fig. 3 A and B, lanes 2). Release of CA was also seen when detergent-treated, authentic immature particles were cleaved with HIV PR (Fig. 3C, lane 2). However, although particles assembled in buffer were completely susceptible to digestion with trypsin (Fig. 3A, lane 3), the addition of lysate to the assembly reactions led to the appearance of a trypsin-resistant band of 26 kDa (Fig. 3B, lane 3). The same trypsin-resistant fragment was also seen on digestion of the authentic immature HIV-1 particles (Fig. 3C, lane 3). N-terminal sequencing of this band showed that it begins within the C-terminal 21 residues of the MA domain (Fig. 3E).
Figure 3.
Susceptibility of particles to proteolytic digestion. Particles assembled in buffer B (A), particles assembled in buffer B plus 5% rabbit reticulocyte lysate (B), immature HIV-1 virions (C), or particles assembled in buffer B plus 2 μM IP5 (D) were digested with HIV-1 PR (lanes 2) or with trypsin (lanes 3). Digestion products were examined by SDS/PAGE and immunoblotting with antibodies against HIV-1 CA protein. (E) N-terminal sequence of the trypsin-resistant fragment. The ≈26-kDa band in lane 3 was a mixture of molecules with zero, one, or two N-terminal lysines. The first lysine is residue 112 of Gag.
Identification of Active Factor in Reticulocyte Lysates.
Rabbit reticulocyte lysates (7) were fractionated to purify the material(s) responsible for these alterations in particle assembly in vitro. The ability to make particles resistant to high-NaCl conditions (as in Fig. 2B) was used as an assay for screening active fractions during the purification. Ultimately, a highly purified fraction was obtained: starting with 2 liters of lysate, we obtained a fraction containing only 1 mg (total dry weight) of material. One microgram of this material was sufficient to completely alter the assembly properties of 1 mg of Gag protein (data not shown).
Activity in the purified fraction was lost after treatment with alkaline phosphatase, suggesting that a phosphorylated compound was responsible for the activity. Treatment with acetic anhydride followed by analysis with gas chromatography/electron impact ionization mass spectrometry identified myo-inositol hexaacetate in the phosphatase-treated sample, but not in the untreated sample, indicating that the source of the myo-inositol was an inositol phosphate (IP) (data not shown). Electrospray mass spectrometry analysis detected ions at m/z 501 and 523, representing inositol tetrakisphosphate (IP4) with 0 or 1 sodium ions; m/z 581, 603, and 625, representing IP5 with 0, 1, or 2 sodium ions; and m/z 661, 683, 705, 727, 749, and 771, representing inositol hexakisphosphate (IP6) with 0 to 5 sodium ions (data not shown).
Using these analytical results as guidance, we obtained a number of purified compounds from commercial sources and tested them individually by using the salt-resistance assay (Table 1). Two compounds, IP5 and IP6, had activity quantitatively equivalent to that of the active fraction purified from rabbit reticulocytes and had significantly higher activity than any of the other compounds. IP5 could also induce the formation of 100-nm particles (Fig. 1D) that were, like authentic, immature particles, resistant to RNase (Fig. 2C, lane 2) as well as to 0.5 M NaCl (Fig. 2C, lane 3). Digestion of these VLPs with trypsin also yielded the trypsin-resistant fragment (Fig. 3D, lane 3) characteristic of authentic particles. Thus, in all respects, the ability of the lysates to “correct” the assembly process in vitro could be completely accounted for by the presence of IP5 in these lysates. Addition of only one IP5 molecule per 10 Gag molecules was sufficient to render the VLPs completely salt-resistant.
Table 1.
Activities of compounds in NaCl-resistance assay
| Compound | Conc., mM | Compound/HIV Gag |
|---|---|---|
| Phosphoglycerates | ||
| 2-Phosphoglycerate | >2 | >100 |
| 2,3-Diphosphoglycerate | 1 | 50 |
| Polyphosphates | ||
| Tripolyphosphate | >1 | >50 |
| Tetrapolyphosphate | 0.1 | 5 |
| Sodium phosphate glass* | 0.035 | 2 |
| Inositol phosphates | ||
| IP3 | 0.54 | 27 |
| IP4 | 0.08 | 4 |
| IP5 | 0.002 | 0.1 |
| IP6 | 0.002 | 0.1 |
| Inositol sulfates | ||
| IS6 | >1 | >50 |
| Phosphatidylinositol phosphates | ||
| Dibutyryl PIP3 | 0.057 | 3 |
A number of compounds with multiple phosphate groups are active in inducing formation of NaCl-resistant VLPs. VLPs were assembled in the presence of the indicated compounds (Sigma or Calbiochem), and activity was quantitated using the salt-resistance asay. The minimum concentration of the compound required to produce 100% resistance and the molar ratio of each compound to Gag at that concentration are shown. Where multiple species with the same number of phosphates are listed (e.g., IP3), all the species gave similar results. IP3, D-myo-inositol 1,3,5-triphosphate and D-myo-inositol 3,4,5-triphosphate. IP4, D-myo-inositol 1,3,4,5-tetrakisphosphate, D-myo-inositol 2,3,4,6-tetrakisphosphate, and D-myo-inositol 3,4,5,6-tetrakisphosphate. IP5, D-myo-inositol 1,3,4,5,6-pentakisphosphate. IP6, D-myo-inositol hexakisphosphate. IS6, inositol hexasulfate. Dibutyryl PIP3, a derivative of PIP3 (phosphatidylinositol 3,4,5-trisphosphate) with a truncated (four-carbon) diacylglycerol tail.
Average chain length, 18.
To explore structure–activity relationships, we also tested a number of other compounds for activity in the assembly assays. Several compounds with multiple phosphate groups exhibited some activity in the NaCl-resistance assay, including inorganic polyphosphate (Table 1). Surprisingly, though IP6 (frequently termed “phytic acid”) was as active as IP5 in the RNase, NaCl, and trypsin assays, it did not induce the formation of 100-nm spherical VLPs. Rather, large, flat plates, which tended to stack on each other, were formed (shown in Fig. 1E); evidently, the additional phosphate group in this molecule interferes with the curvature of the assembled complex. Inositol hexasulfate, in which the phosphates had been replaced with sulfates, had no effect in the assembly assays, suggesting that phosphate groups are specifically required for activity. Another inositol derivative, dibutyryl PIP3, a water-soluble analog of PIP3, was also positive in all assays, although its specific activity in the NaCl-resistance assay, like that of the closely related compound IP4, was significantly lower than IP5 (Table 1 and data not shown).
Identification of a Region in Gag Essential for the Effect of IPs.
A report has shown that recombinant HIV-1 Gag protein (Δ16–99) lacking residues 16–99 (the majority of the 132-residue MA domain) assembles into full-size VLPs in vitro (6). Indeed, these VLPs were shown to have a morphology very similar to that of authentic immature virions (6). We compared the particles formed by our Gag protein and those formed by Δ16–99 (both lacking the p6 domain) in parallel assembly reactions that were not supplemented with IPs. These experiments confirmed that the Δ16–99 mutant forms full-size particles (data not shown), whereas the protein with an intact MA domain forms small particles in the absence of added cofactor.
We also tested the resistance of the VLPs formed from the Δ16–99 Gag protein to 0.5 M NaCl and to RNase digestion. As shown in Fig. 4A, these VLPs, unlike the particles assembled from our Gag protein (lanes 2 and 3), are fully resistant to these treatments (lanes 5 and 6), even when no cofactor is added. However, they only exhibited the ≈26-kDa trypsin-resistant band in the presence of IP5 (Fig. 4B). Thus, the Δ16–99 protein assembles into VLPs resembling authentic immature virions in most respects tested, despite the lack of IPs or any other cofactor in the assembly reaction.
Figure 4.
Properties of particles assembled from Δ16–99 Gag protein. (A) Resistance to RNase and 0.5 M NaCl. Particles assembled from Gag (lanes 1–3) or Δ16–99 Gag (lanes 4–6) were treated with RNase (lanes 2 and 5) or 0.5 M NaCl (lanes 3 and 6). Proteins in particles pelleted after treatment were examined by SDS/PAGE and Coomassie blue staining. We estimate that >90% of the Δ16–99 Gag protein, but ≤20% of the Gag protein, remained pelletable after the treatments. (B) Susceptibility to proteolytic digestion. Particles assembled from Gag (lanes 1–4) or Δ16–99 Gag (lanes 5–10) were digested with HIV-1 PR (lanes 6 and 9) or with trypsin (lanes 2, 4, 7, and 10) after assembly in buffer B with (lanes 3–4 and 8–10) or without (lanes 1, 2, and 5–7) 2 μM IP5. Digestion products were examined as in Fig. 3.
Discussion
We have shown here that particles assembled in vitro from recombinant HIV-1 Gag protein differ in a number of respects from authentic, immature HIV-1 particles: (i) they are far smaller than authentic particles, (ii) they can be dissociated by treatment with RNase or 0.5 M NaCl, and (iii) the CA region in the assembled particles is completely susceptible to trypsin digestion. Addition of a mammalian cell lysate leads to the assembly of particles that resemble the authentic particles in each of these respects. Because IPs were present in the active fractions purified from these lysates, and because commercially obtained, pure IPs can quantitatively replace the lysates in each of the assembly assays, the results strongly suggest that IPs modulate the normal process of HIV-1 particle assembly in mammalian cells.
Our data show that the molecular interactions between HIV-1 Gag molecules are altered by IPs. Preliminary results also demonstrate that our recombinant HIV-1 Gag protein binds IP6 (S.C. and A.R., unpublished data). There is already a wealth of structural information on HIV-1 proteins (11); it will be of great interest to determine how the structures of these proteins are affected by IP binding.
Our results also show that the region of the MA domain between residues 16 and 99 is essential for the effects described here, because Δ16–99 Gag protein assembles into 100-nm particles that are resistant to NaCl and RNase without the addition of cofactor (Fig. 4A). In fact, cryoelectron microscopy has demonstrated that particles assembled from this protein are structurally very similar to authentic immature virions (6, 12). However, these particles do not exhibit trypsin resistance in CA unless they are assembled in the presence of IPs (Fig. 4B). One hypothesis that is consistent with these observations is that residues 16–99 interfere with proper assembly unless they are bound by IPs. The results also suggest that trypsin resistance is not necessarily a reliable indicator of proper assembly. There may be a binding site(s) for IPs between residues 16 and 99, leading to correct particle assembly, and an additional site(s) elsewhere in Gag, leading to trypsin resistance in either wild-type or Δ16–99 Gag VLPs.
It is interesting that cell lysates or IPs are not required for the in vitro assembly of correctly sized particles in the case of other retroviruses (Rous sarcoma virus, Mason–Pfizer monkey virus, or MoMuLV; refs. 2 and 3, and S.C. and A.R., unpublished data). We do not yet know whether other lentiviruses share this requirement with HIV-1.
Although we have no direct evidence that IPs modulate HIV-1 particle assembly in vivo, the similarity between the particles assembled in their presence in vitro and authentic immature HIV-1 particles strongly suggests that such cofactors are involved in normal HIV-1 assembly. Estimates of the level of IP5 in mammalian cells vary widely, but are generally considerably greater than the concentration (2 μM) that we found was sufficient for correct assembly of VLPs from 1 mg/ml Gag protein (13–15). However, it is important to note that our assays are performed in solution, whereas HIV-1 particle assembly normally occurs at the plasma membrane. Thus, it is intriguing that dibutyryl PIP3 is active in the in vitro assembly assays: this compound is an analog of PIP3, which is found in the plasma membrane and which is a key element in many signal-transduction pathways in the cell (16). The activity of the PIP analog raises the possibility that PIPs in the plasma membrane, rather than soluble IPs, act as cofactors in HIV-1 assembly in vivo.
Remarkably, the presence of a single IP5 molecule is sufficient to correct the assembly of ≈10 HIV-1 Gag molecules (Table 1). It is possible that IP5 interacts transiently or “catalytically” with Gag molecules before assembly, inducing the correct conformation in all of them; alternatively, binding of IP5 to a single Gag molecule may alter it in such a way that it and its immediate neighbors assemble correctly. It would be important to know whether the cofactor is incorporated into the assembled particle, but this is technically very difficult to answer. A molar ratio of 1 IP5 per 10 Gag molecules corresponds to a mass ratio of 1:1000; detection of IP5 at this level would require extremely rigorous purification of VLPs. It is particularly difficult in the case of authentic HIV-1 virions because they contain plasma membrane that, in turn, contains PIPs. Another study (17) measured the level of phosphatidylinositols in HIV-1 particles, but did not distinguish between different phosphatidylinositol species. It seems likely that the number of phosphate groups on the inositol ring is a crucial determinant of the activity of a PIP in virus assembly, as it is with IPs (Table 1). If this is so, then the analysis did not distinguish between active and inactive PIP derivatives, and cannot tell us the level of active PIP molecules in the particles.
Inositol derivatives have previously been shown to help organize large macromolecular complexes within cells (16). For example, PI(4,5)P2 recruits adaptor proteins to the plasma membrane before nucleation of clathrin lattices (18, 19); indeed, these adaptor proteins can regulate the size of the resulting clathrin cages (20). Our findings suggest that particle assembly by the HIV-1 Gag protein may involve a somewhat analogous interaction with inositol derivatives.
Our data suggest that HIV-1 differs from some other retroviruses in that it requires IPs for assembly of correctly sized particles. However, it is possible that IPs play a distinct role in the assembly of many retroviruses. All retroviral Gag proteins (except those of spumaretroviruses) are targeted to the plasma membrane of the virus-producing cell. It is known that the membrane affinity of these proteins results from a combination of N-terminal myristoylation and clusters of basic residues in the MA domain (21). However, we have no insight at present into how they are specifically directed to the plasma membrane, rather than other cellular membranes. One possibility is that PIPs in the plasma membrane are responsible for this specific targeting, just as they are with a wide variety of cellular proteins (16, 18, 19, 22). It will thus be of interest to determine whether Gag proteins from retroviruses other than HIV-1 bind inositol phosphate derivatives. It is striking that Δ16–99 Gag protein, which does not interact with IPs or PIPs in the same way as its wild-type counterpart (Fig. 4), is not targeted to the plasma membrane in vivo: cells expressing this protein form particles by budding into the endoplasmic reticulum (5).
We should also note the possibility that the presence in the cell of HIV-1 Gag protein, which binds IP6 (unpublished data) and, presumably, related molecules, disrupts IP- and/or PIP-based cell signaling pathways. It is conceivable that this interaction contributes to the pathogenicity of HIV-1 infection.
Acknowledgments
We thank L. Cromwell for rabbit reticulocytes, D. Harvin for help with factor purification, Hans-Georg Krausslich for the Δ16–99 construct, D. Ott and J. Mirro for immature HIV-1 and MoMuLV virions, respectively, R. Sowder and L. Henderson for N-terminal sequencing of the trypsin-resistant fragment, G. Chmurny and J. Hrabie for help with chemical analysis, J. Levin, L. Henderson, and S. Sukumar for reviews of the manuscript, and T. Balla, L. Greene, and S. Shears for helpful comments.
Abbreviations
- VLP
virus-like particle
- MA
matrix
- CA
capsid
- PR
protease
- IP
inositol phosphate in general
- IPn
inositol with n phosphates
- PIP
phosphatidylinositol phosphate
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- MoMuLV
Moloney murine leukemia virus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Swanstrom R, Wills J W. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 263–334. [Google Scholar]
- 2.Campbell S, Vogt V M. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klikova M, Rhee S S, Hunter E, Ruml T. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell S, Rein A. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Facke M, Janetzko A, Shoeman R L, Krausslich H G. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Krausslich H G. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson R J, Hunt T. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- 8.Muriaux D, Mirro J, Harvin D, Rein A. Proc Natl Acad Sci USA. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrlich L S, Agresta B E, Gelfand C A, Jentoft J, Carter C A. Virology. 1994;204:515–525. doi: 10.1006/viro.1994.1565. [DOI] [PubMed] [Google Scholar]
- 10.Loyet K M, Kowalchyk J A, Chaudhary A, Chen J, Prestwich G D, Martin T F. J Biol Chem. 1998;273:8337–8343. doi: 10.1074/jbc.273.14.8337. [DOI] [PubMed] [Google Scholar]
- 11.Turner B G, Summers M F. J Mol Biol. 1999;285:1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- 12.Wilk T, Gross I, Gowen B E, Rutten T, de Haas F, Welker R, Krausslich H G, Boulanger P, Fuller S D. J Virol. 2001;75:759–771. doi: 10.1128/JVI.75.2.759-771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pittet D, Schlegel W, Lew D P, Monod A, Mayr G W. J Biol Chem. 1989;264:18489–18493. [PubMed] [Google Scholar]
- 14.Oliver K G, Putney J W, Jr, Obie J F, Shears S B. J Biol Chem. 1992;267:21528–21534. [PubMed] [Google Scholar]
- 15.Guse A H, Greiner E, Emmrich F, Brand K. J Biol Chem. 1993;268:7129–7133. [PubMed] [Google Scholar]
- 16.Cockroft S. Biology of Phosphoinositides. New York: Oxford Univ. Press; 2000. [Google Scholar]
- 17.Aloia R C, Tian H, Jensen F C. Proc Natl Acad Sci USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- 19.Ford M G, Pearse B M, Higgins M K, Vallis Y, Owen D J, Gibson A, Hopkins C R, Evans P R, McMahon H T. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 20.Ye W, Lafer E M. J Neurosci Res. 1995;41:15–26. doi: 10.1002/jnr.490410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W, Parent L J, Wills J W, Resh M D. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santagata S, Boggon T J, Baird C L, Gomez C A, Zhao J, Shan W S, Myszka D G, Shapiro L. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]