Abstract
N-methyl-D-aspartate receptors (NMDARs), a subtype of glutamate-gated ion channels, play a central role in epileptogenesis. Recent studies have identified an increasing number of GRIN2A (a gene encoding the NMDAR GluN2A subunit) mutations in patients with epilepsy. Phenotypes of GRIN2A mutations include epilepsy-aphasia disorders and other epileptic encephalopathies, which pose challenges in clinical treatment. Here we identified a heterozygous GRIN2A mutation (c.1341T>A, p.N447K) from a boy with Rolandic epilepsy by whole-exome sequencing. The patient became seizure-free with a combination of valproate and lamotrigine. Functional investigation was carried out using recombinant NMDARs containing a GluN2A-N447K mutant that is located in the ligand-binding domain of the GluN2A subunit. Whole-cell current recordings in HEK 293T cells revealed that the N447K mutation increased the NMDAR current density by ~1.2-fold, enhanced the glutamate potency by 2-fold, and reduced the sensitivity to Mg2+ inhibition. These results indicated that N447K is a gain-of-function mutation. Interestingly, alternative substitutions by alanine and glutamic acid at the same residue (N447A and N447E) did not change NMDAR function, suggesting a residual dependence of this mutation in altering NMDAR function. Taken together, this study identified human GluN2A N447K as a novel mutation associated with epilepsy and validated its functional consequences in vitro. Identification of this mutation is also helpful for advancing our understanding of the role of NMDARs in epilepsy and provides new insights for precision therapeutics in epilepsy.
Keywords: Epilepsy, NMDA receptors, GRIN2A, Mutation
Introduction
N-methyl-D-aspartate receptors (NMDARs) are one of the major ionotropic glutamate receptors that play a critical role in excitatory synaptic transmission, synaptic plasticity, and brain development [1]. Dysfunction of NMDARs is associated with a spectrum of neurological and psychiatric disorders [2, 3]. Functional NMDARs are heterotetramers usually composed of two glycine/D-serine-binding obligatory GluN1 subunits and two glutamate-binding regulatory GluN2 subunits (GluN2A-2D), which are encoded by GRIN1 and GRIN2A-2D, respectively. Each NMDAR subunit has four discrete modules: an extracellular amino-terminal domain, a ligand-binding domain (LBD), a transmembrane domain, and a C-terminal domain [1]. For the past few years, advances in genomic technologies have led to a rapid increase in the discovery of novel epilepsy-associated genes. Among them, a large proportion comprises ion channels and neurotransmitter receptors [4–6]. Interestingly, recent studies have demonstrated an association between epilepsies and mutations in GRIN1 [7–9], GRIN2A [10–17], GRIN2B [18, 19], and GRIN2D [20].
GRIN2A, which encodes the GluN2A subunit, is one of the most relevant epilepsy genes. More than 70 GRIN2A gene mutations have been identified in patients with epilepsy, commonly focal epilepsy with speech disorder (epilepsy-aphasia syndromes) [12, 15, 16]. Recently, the phenotypic spectrum of the GRIN2A mutation has been extended to more severe epilepsies such as early-onset epileptic encephalopathy [10, 11]. Functional studies on GRIN2A mutations have indicated a diversity of functional alterations [10, 12, 18]. Several mutations lead to gain-of-function of NMDARs [10–12, 14], while others have revealed a loss-of-function effect or lack functional deficits [13, 17]. The mechanisms underlying the diverse functional alterations are mostly unknown. Distinct correlations between phenotypes and functional changes have not been defined, either.
We recently identified a GRIN2A missense mutation (c.1341T>A, p.N447K, hereafter referred to as GluN2A-N447K) from a pediatric patient with Rolandic epilepsy (focal epilepsy with centro-temporal spikes) by whole-exome sequencing. Whole-cell current recordings were performed to determine the functional effect of this mutation. To further understand the mechanism underlying the functional changes, the impact of alternative substitutions at residue N447 was examined.
Materials and Methods
Whole-Exome Sequencing
The patient with the GRIN2A mutation c.1341T>A, p.N447K was treated at the Epilepsy Center of the Second Affiliated Hospital of Guangzhou Medical University in 2009. This patient was clinically diagnosed as having Rolandic epilepsy of unknown etiology. In order to determine the etiology of the epilepsy, whole-exome sequencing was performed.
A blood sample was obtained after the patient and his guardian had given written informed consent. Genomic DNA was extracted from peripheral blood using a QuickGene DNA whole blood kit L (Fujifilm, Tokyo, Japan). To systematically screen for the disease-associated variants in this patient, exome sequencing was conducted on the Illumina HiSeq 2500/4000 platform by BGI-Shenzhen (Shenzhen, China). The exome library was constructed with 3 μg of gDNA, which was randomly sheared by sonication and hybridized to the Nimblegen SeqCap EZ Library for enrichment in targeting exonic DNA, according to the manufacturer’s instructions. Paired-end reads with a length of 90 bp were generated by massively parallel sequencing with >125× average depth and >98% coverage of the target region, which fulfilled the quality test. Variants having potential clinical significance were confirmed by Sanger sequencing.
Molecular Modeling of NMDAR Domains
The LBD-located residue N447, at which the mutation had occurred, was modeled based on the crystal structure of the GluN1/GluN2A LBD (PDB ID 5I57) [21] using Chimera software (University of California, San Francisco). The schematic architecture of the GluN1/GluN2A NMDAR was drawn using Adobe Illustrator CS6.
cDNA Construction, Cell Culture, and Transfection
N447K, N447A, and N447E exchange mutants of rat GluN2A cDNA (GluN2A-N447K, GluN2A-N447A, and GluN2A-N447E) were generated by site-directed mutagenesis [22] from the plasmid pcDNA1.1+-GluN2A. The GFP-tagged version of N447K (GFP-GluN2A-N447K) was made from the plasmid GFP-GluN2A [23]. These vectors were all confirmed by DNA sequencing. Human embryonic kidney (HEK) 293 and 293T cells were grown in Dulbecco’s Modified Eagle’s Medium (Gibco, Grand Island, NY), supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco) in a 5% CO2 incubator at 37 °C. The cDNA constructs were transfected into the cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. To protect the cells from NMDAR-mediated toxicity, 200 μmol/L D,L-2-amino-5-phosphonovaleric acid (Sigma) and 1 mmol/L kynurenic acid (Sigma) were added to the culture medium.
Whole-Cell Recordings
HEK 293T cells were co-transfected with the cDNA constructs EGFP, GluN1-1a, and wild-type GluN2A (hereafter referred to as GluN2A-WT) or mutant GluN2A, at a ratio of 1:30:40. Twenty-four hours after transfection, the EGFP-positive cells were patch-clamped at room temperature (22 °C–25 °C) using an Axopatch 200B amplifier (Molecular Devices, Union City, CA). We set whole-cell parameters (capacitance and series resistance) and adjusted the series resistance compensation to ~80% during the whole-cell experiments. The recording chamber was continuously perfused with an extracellular solution composed of (in mmol/L) 135 NaCl, 5 KCl, 2 CaCl2, 10 HEPES at pH 7.4 with NaOH and the osmolarity was adjusted to 300 mOsm with sucrose. Electrodes were made on a vertical pipette puller (Narishige PC-10, Tokyo, Japan) and those with a resistance of 3 MΩ–5 MΩ were used. The internal solution filling in the electrodes contained (in mmol/L) 140 CsCl, 2 MgATP, 10 EGTA, 10 HEPES at pH 7.3 with CsOH, and the osmolarity was ~300–310 mOsm [24]. Drugs were applied to the lifted cells using an RSC-160 system (Biologic Science Instruments, Claix, France). The drug solutions for glutamate dose-response experiments were made from glutamate stock solution (100 mmol/L) each time and were diluted in the extracellular solution. The percentage current was normalized to the current value obtained at 300 μmol/L glutamate and 20 μmol/L glycine. The Mg2+ current-voltage relationships of the co-agonist-induced current were recorded at 15 mV intervals ranging from −90 mV to +30 mV and normalized to the value obtained at +30 mV. The drug solutions for the Mg2+ (1 μmol/L–300 μmol/L) and Zn2+ (3 nmol/L–1000 nmol/L) concentration-response experiments were prepared fresh daily from a MgCl2 stock solution (1 mol/L) and a ZnSO4 stock solution (0.1 mol/L)). The percentage current was normalized to the current value obtained at 100 μmol/L glutamate and 20 μmol/L glycine [11]. HEK 293T cells were clamped at −60 mV, and 100 μmol/L glutamate and 20 μmol/L glycine were used in all of the whole-cell recordings unless otherwise stated. The agonist concentration-response data and the inhibitory concentration-response data were fitted with equations 1 and 2, respectively, as follows.
| 1 |
| 2 |
where nH is the Hill slope, EC50 is the agonist concentration that elicited a half-maximal response, and IC50 is the concentration that produced a half-maximal inhibition [25]. All reagents were from Sigma. Off-line data analysis, curve fitting, and figure preparation were performed with Clampfit 9 (Axon Instruments), GraphPad Prism 5.0, and Adobe Illustrator CS6.
Surface Immunofluorescence Staining and Quantitative Analysis
The protocols used for surface immunofluorescence staining and quantitative analysis were as described previously [23]. GluN1-1a/GFP-Glu2A or GluN1-1a/GFP-Glu2A-N447K cDNAs were co-transfected into HEK 293 cells. Twenty-four hours after transfection, cells were rinsed once with PBS, and then incubated with rabbit anti-GFP antibody (Chemicon) for 7 min, rinsed 3 times in the extracellular solution, and then incubated with goat anti-rabbit Alexa-546-conjugated secondary antibody (Invitrogen) for another 7 min. After rinsing 3 times again in the extracellular solution, the cells were immediately fixed in 4% paraformaldehyde for 10 min. All procedures were performed at room temperature. Images were acquired with a fluorescence microscope (BX51, Olympus). Intensity measurements were performed using MetaMorph image analysis software (Universal Imaging, West Chester, PA). The average intensity of the surface fluorescence was determined for the regions outlined around the transfected HEK 293 cells. The surface intensities of wild-type and mutant NMDARs were both normalized to the average surface intensity of the wild-type NMDARs. Means were obtained from three different cultures.
Statistical Analysis
All data values are expressed as mean ± SEM derived from at least three separate transfections. GraphPad Prism software was used for statistical analysis. Whole-cell NMDAR current density, EC50 values of glutamate-concentration response curves, IC50 values of Mg2+ or Zn2+ concentration–response curves, and surface expression levels of wild-type and N447K mutant receptors were compared by the unpaired t-test. Current density and EC50 values in wild-type and different mutant receptors (N447K, N447A, and N447E) were compared by one-way ANOVA with the Bonferroni post hoc multiple comparison test. A P value of <0.05 was considered to be statistically significant.
Results
Identification of the Mutation
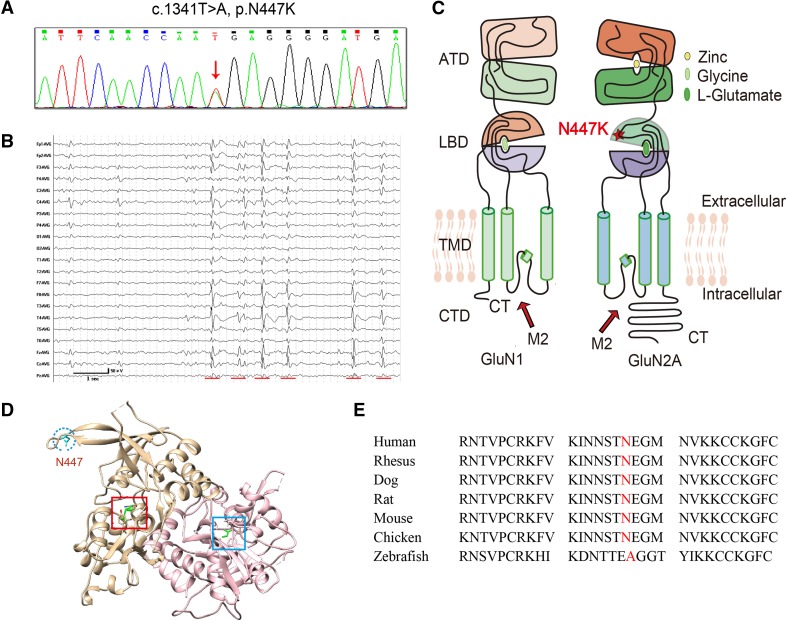
A heterozygous GRIN2A mutation, c.1341T>A (N447K) (Fig. 1A), was identified in a patient with Rolandic epilepsy. The patient was a 17-year-old boy with normal delivery and development. He presented his first seizure during sleep at nine years of age. It was described as a convulsive seizure, most likely a generalized tonic-clonic seizure, and it lasted for about one minute. Later, he presented two partial seizures during sleep, starting with opening of the eyes, followed by head raising. Electroencephalogram monitoring showed remarkable interictal high-voltage spikes and spike-and-slow waves in the bilateral central-temporal regions, predominantly on the right (Fig. 1B). The discharges were aggravated by sleep, with spike-and-slow wave indexes ~50%–60%, which did not achieve the criterion for continuous spike-and-wave during sleep (≥80%). A diagnosis of Rolandic epilepsy was made. The patient was given valproate at 25 mg/kg per day initially. After being seizure-free for three months, the seizures recurred. Lamotrigine (2.5 mg/kg per day) was added, and the patient has been seizure free from that time.
Fig. 1.
Identification of a GRIN2A missense mutation in a patient with Rolandic epilepsy. A GRIN2A sequencing schematic of the mutation c.1341T>A, p.N447K. B Interictal electroencephalogram obtained at nine years old showing frequent high-voltage spikes and spike-and-slow waves in the bilateral central-temporal regions, predominantly on the right. Red lines indicate abnormalities. C Topological model of a GluN1 and a GluN2A subunit. Residue N447 in GluN2A lies within the LBD. D Model of the glutamate- and glycine-bound LBDs of GluN1/GluN2A NMDARs (PDB ID: 5I57). The position of Asn447 is colored cyan and is indicated by the blue circle. The ligand glutamate in the GluN2 subunit and glycine in the GluN1 subunit are both colored green and are indicated by the red square and blue square, respectively. E Protein sequence alignment showing that residue N447 is highly conserved across higher vertebrates. An alternative substitution (N447A) was found in lower vertebrates.
His mother complained of his mild hyperactivity since the onset of seizures, but no speech and language problems were presented. The family history of epilepsy and febrile seizures was negative. The physical examination and intelligence test were normal. Brain magnetic resonance imaging was negative.
Residue N447 is located in the S1 segment of the extracellular LBD of the GluN2A subunit and in a loose loop at the edge of the LBD, which is somewhat distant from the glutamate-binding core (Fig. 1C, D). The mutation c.1341T>A (N447K) was predicted not to damage the function of the protein by PolyPhen-2 and MutationTaster. However, this mutation occurs in the population at a frequency of <0.5% (0.004143% in the general population and 0.0583% in East Asians) in the ExAC database (http://exac.broadinstitute.org/variant/16-9934949-A-T). Further physiological studies were performed to determine its impact on channel function. Residue N447 is highly conserved across higher vertebrates (Fig. 1E). Alternative substitutions with an uncharged or negatively-charged amino-acid were found in lower vertebrates (anamniotes), such as a substitution by alanine (N447A) in zebrafish, on which further functional studies were also performed.
GluN2A-N447K Increases NMDAR Current Density
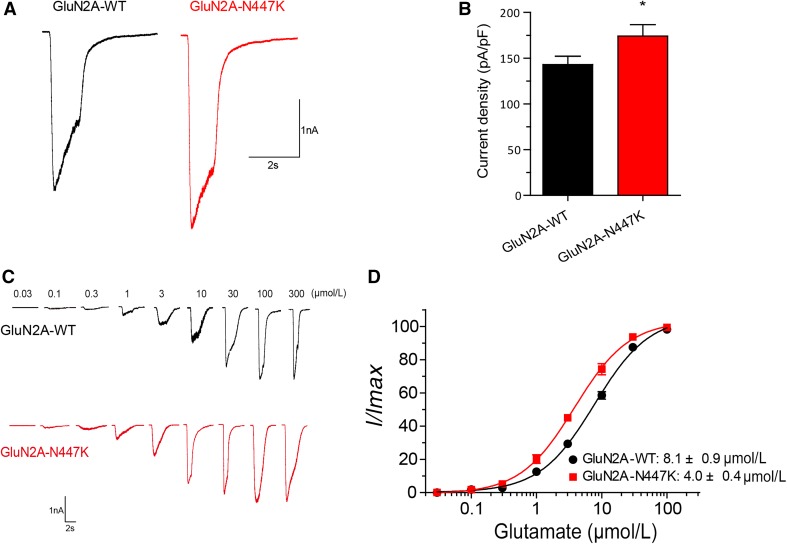
With co-expression of GluN1 and GluN2A subunits in HEK 293T cells, we found that the average current density of GluN1/GluN2A-N447K NMDARs recorded at a holding potential of −60 mV was 22% higher than that of the GluN1/GluN2A-WT NMDARs (174.1 ± 12.5 pA/pF, n = 22 versus 143.0 ± 9.1 pA/pF, n = 25 for wild-type; P = 0.0470), which was evoked by 100 μmol/L glutamate and 20 μmol/L glycine (Fig. 2A, B; Table 1), suggesting that the N447K mutation results in a mild gain-of-function of NMDARs.
Fig. 2.
GluN2A-N447K increases NMDAR whole-cell current density and glutamate potency. A Representative current traces of GluN1/GluN2A-WT and GluN1/GluN2A-N447K NMDARs evoked by 100 μmol/L glutamate and 20 μmol/L glycine at −60 mV (current scale bar, 1 nA; time scale bar, 2 s). B Quantitative analysis of whole-cell current density induced by co-agonists. The peak current density of the mutant was increased by ~1.2-fold. C Representative current traces of wild-type and mutant NMDARs at different glutamate concentrations measured after heterologous expression in HEK 293T cells recorded by voltage-clamping. D Glutamate concentration-response curves of GluN1/GluN2A-WT (black) and GluN1/GluN2A-N447K (red) NMDARs. Note that the N447K mutation enhances glutamate potency 2-fold.
Table 1.
Summary of pharmacological data for NMDARs.
| GluN2A-WT | GluN2A-N447K | |
|---|---|---|
| Current density, pA/pF, (n)a | 143.0 ± 9.1 (25) | 174.1 ± 12.5 (22)* |
| Glutamate, EC50, μmol/L (n)a | 8.1 ± 0.9 (6) | 4.0 ± 0.4 (6)** |
| Mg2+, I-V (n)b | 61.5 ± 4.0% (9) | 72.1 ± 4.6% (11) |
| Mg2+, IC50, μmol/L (n)a | 28.6 ± 4.3 (6) | 48.9 ± 5.2 (6)* |
| Zn2+, IC50, nmol/L (n)a | 79.0 ± 6.5 (7) | 110.1 ± 15.8 (6) |
The data are expressed as mean ± SEM.
a At the holding potential of −60 mV.
b Percentage current remaining in the presence of 300 μmol/L MgCl2 at −30 mV.
* P < 0.05 compared with GluN2A-WT, unpaired t-test.
** P < 0.01 compared with GluN2A-WT, unpaired t-test.
GluN2A-N447K Enhances Glutamate Potency
To test whether the mutant changes glutamate sensitivity, we performed glutamate concentration-response assessments for GluN1/GluN2A-WT and GluN1/GluN2A-N447K NMDARs. They showed that GluN2A-N447K-containing NMADRs enhanced glutamate potency, with a significantly lower half-maximally effective concentration (EC50) than that of GluN1/GluN2A-WT NMDARs (4.0 ± 0.4 μmol/L, n = 6 versus 8.1 ± 0.9 μmol/L, n = 6 for wild-type; P = 0.0016) (Fig. 2C, D; Table 1). This indicated that the GluN2A-N447K mutant can be activated at a lower glutamate concentration.
GluN2A-N447K Decreases Voltage-Dependent Mg2+ Block
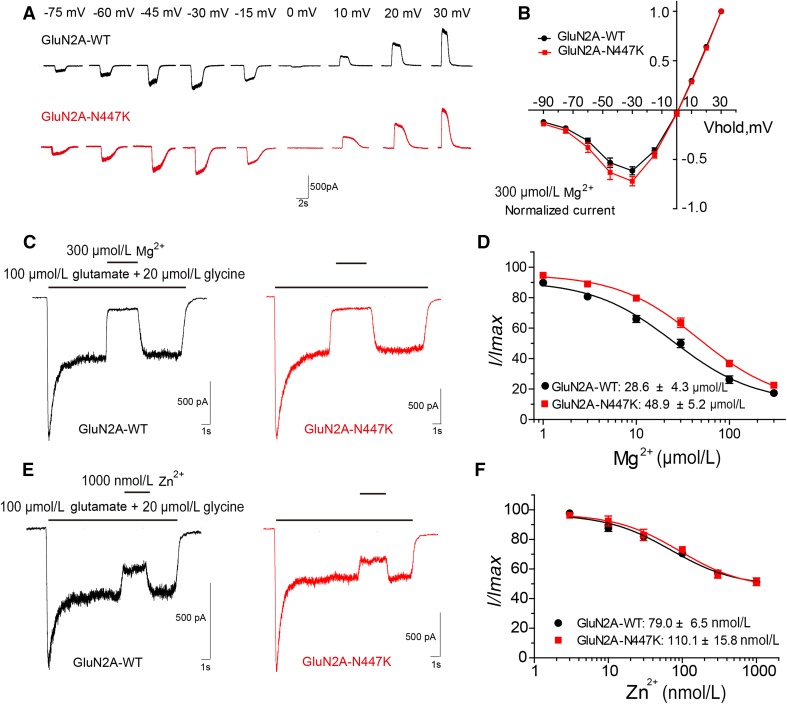
Voltage-dependent Mg2+ blockade is a distinct functional property of NMDARs [1, 26, 27]. We evaluated the Mg2+ sensitivity of NMDARs by generating current-voltage curves and concentration-response curves. In the presence of 300 μmol/L Mg2+, a slightly larger current flow was recorded in the mutant NMDARs at −30 mV (72.1 ± 4.6%, n = 11 versus 61.5 ± 4.0%, n = 9 for wild-type; P = 0.1097), but it was not statistically significant (Fig. 3A, B; Table 1). The Mg2+ concentration-response curve for the mutant shifted to the right, with a significantly higher IC50 (48.9 ± 5.2 μmol/L, n = 6 versus 28.6 ± 4.3 μmol/L, n = 6 for wild-type; P = 0.0138), when the membrane potential was held at −60 mV (Fig. 3C, D; Table 1). These data indicated that GluN2A-N447K decreases Mg2+ inhibition of NMDARs.
Fig. 3.
Effects of GluN2A-N447K on Mg2+ and Zn2+ sensitivity of NMDARs. A Representative evoked current traces of wild-type and mutant NMDARs at different holding potentials (current scale bar, 500 pA; time scale bar, 2 s). B Mg2+ current-voltage (I-V) relationship of GluN1/GluN2A-WT (black) and GluN1/GluN2A-N447K (red) NMDARs. C Representative current traces of wild-type and mutant NMDARs induced by co-agonists in the presence of 300 μmol/L Mg2+. Other doses of Mg2+ were applied in the same way (current scale bar, 500 pA; time scale bar, 1 s). D Mg2+ concentration-response curves reveal a decreased Mg2+ block for the GluN1/GluN2A-N447K NMDARs. E Representative evoked current traces of wild-type and mutant NMDARs inhibited by 1000 nmol/L Zn2+. Other doses of Zn2+ were applied in the same way (current scale bar, 500 pA; time scale bar, 1 s). F Zn2+ concentration-response curves show that Zn2+ inhibition of mutant NMDARs is undistinguishable from that of the WT.
GluN2A-N447K Does Not Affect Negative Allosteric Modulation
The function of GluN2A-containing NMDARs can be allosterically modulated by endogenous Zn2+ [28, 29]. The GluN2A-D731N mutation located within the LBD domain changes Zn2+ inhibition [17]. We therefore tested whether the N447K mutation affects Zn2+ inhibition of NMDARs. Concentration-response analysis showed that there was no detectable difference in percentage NMDAR current between the mutant and wild-type NMDAR at any concentration of Zn2+. The IC50 values were comparable (110.1 ± 15.8 nmol/L, n = 6 versus 79.0 ± 6.5 nmol/L, n = 7 for wild-type; P = 0.0802) (Fig. 3E, F; Table 1).
GluN2A-N447K Does Not Alter Surface Expression
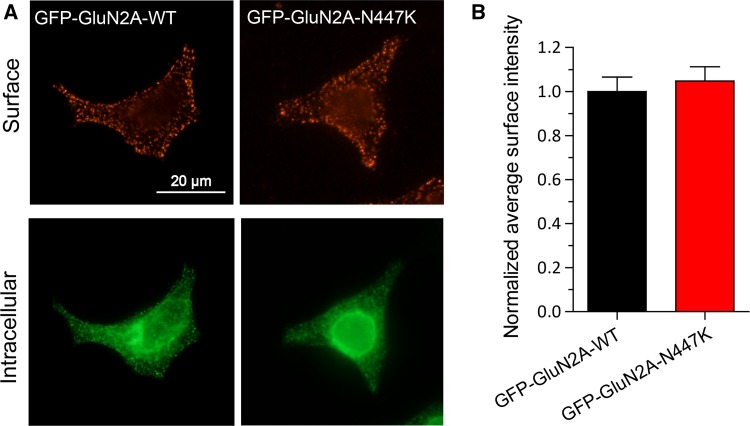
We quantified the surface expression level of GluN2A-N447K-containing NMDARs by immunofluorescence staining. Transfection of GluN1/GluN2A and GluN1/GluN2A-N447K into HEK 293 cells both led to abundant distribution in the membrane and cytoplasm. The normalized average intensity of GluN1/GluN2A-N447K NMDARs on the cell surface showed no significant alteration when compared to wild-type NMDARs (1.05 ± 0.06, n = 48 versus 1.00 ± 0.07, n = 68 for wild-type; P = 0.3225) (Fig. 4A, B). This result further suggested that the increased whole-cell NMDAR current density is not due to changes in surface expression, and enhanced glutamate sensitivity may be the main reason.
Fig. 4.
Surface expression of wild-type and mutant NMDARs. A Representative images of surface (red, upper row) and intracellular (green, lower row) expression of GluN1/GluN2A-WT or GluN1/GluN2A-N447K NMDARs in HEK 293 cells (scale bar, 20 μm). B Summary data showing that the mutant and wild-type NMDARs have similar surface expression levels.
Impact of Alternative Substitutions at N447 on NMDAR Function
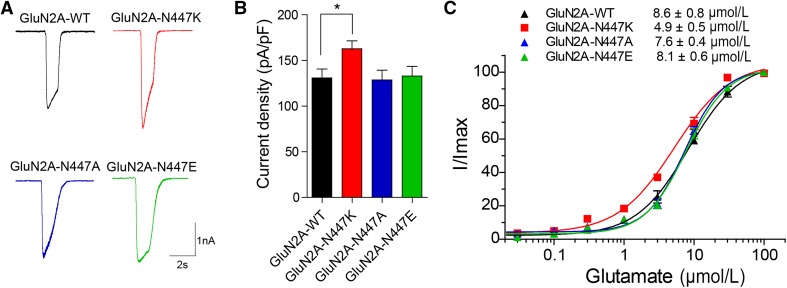
Whether a mutation has a functional impact is useful in evaluating its pathogenicity. Diverse functional changes have been detected for epilepsy-associated missense mutations, and the locations of the mutations are one of the determinants [10–17]. Alternative substitution is another potential consideration. Sequence alignment showed that amino-acids at the homologous site of this residue in different NMDAR subunits are all uncharged or negatively-charged. To determine whether alternative substitutions at the N447 residue have different functional consequences, the effects of N447A and N447E mutations, which are uncharged and negatively-charged, respectively, were tested. The current density of GluN1/GluN2A-N447K NMDARs was ~20% higher than that of GluN1/GluN2A-WT NMDARs (162.8 ± 8.6 pA/pF, n = 20 versus 130.9 ± 9.7 pA/pF, n = 20 for wild-type; P <0.05), however, the current density of GluN1/GluN2A-N447A and GluN1/GluN2A-N447E NMDARs was similar to that of GluN1/GluN2A-WT NMDARs (N447A: 128.7 ± 10.6 pA/pF, n = 14; P >0.05; N447E: 133.0 ± 10.5 pA/pF, n = 14; P >0.05) (Fig. 5A, B; Table 2). The concentration-response curves showed that only GluN2A-N447K increased the glutamate potency (EC50 = 4.9 ± 0.5 μmol/L, n = 5 versus 8.6 ± 0.8 μmol/L, n = 5 for wild-type; P <0.001), and neither GluN2A-N447A nor GluN2A-N447E changed the glutamate-evoked NMDAR current (N447A: EC50 = 7.6 ± 0.4 μmol/L, n = 5; P >0.05; N447E: EC50 = 8.1 ± 0.6 μmol/L, n = 5; P >0.05) (Fig. 5C; Table 2). This result suggested that the properties of the residue at site 447 are important for NMDAR function.
Fig. 5.
Impact of alternative substitutions at N447 on NMDAR function. A Representative current traces of GluN1/GluN2A-WT, GluN1/GluN2A-N447K, GluN1/GluN2A-N447A, and GluN1/GluN2A-N447E NMDARs evoked by 100 μmol/L glutamate and 20 μmol/L glycine at −60 mV (current scale bar, 1 nA; time scale bar, 2 s). B Quantitative analysis of whole-cell current density induced by co-agonists as in (A). C The glutamate concentration-response curves of GluN1/GluN2A-WT (black), GluN1/GluN2A-N447K (red), GluN1/GluN2A-N447A (blue), and GluN1/GluN2A-N447E (green) NMDARs were plotted by measuring the peak current evoked by graded doses of glutamate and 20 μmol/L glycine. The recordings were performed by whole-cell patch clamp at a holding potential of −60 mV after heterologous expression of the mutants and GluN1 in HEK 293T cells.
Table 2.
Current density and glutamate EC50 values for NMDARs with alternative substitutions at GluN2A-N447.
| GluN2A-WT | GluN2A-N447K | GluN2A-N447A | GluN2A-N447E | |
|---|---|---|---|---|
| Current density, pA/pF, (n)a | 130.9 ± 9.7 (20) | 162.8 ± 8.6 (20)* | 128.7 ± 10.6 (14) | 133.0 ± 10.5 (14) |
| Glutamate EC50, μmol/L (n)a | 8.6 ± 0.8 (5) | 4.9 ± 0.5 (5)*** | 7.6 ± 0.4 (5) | 8.1 ± 0.6 (5) |
The data are expressed as mean ± SEM.
a At the holding potential of −60 mV.
* P < 0.05 compared with GluN2A-WT, one-way ANOVA.
*** P < 0.001 compared with GluN2A-WT, one-way ANOVA.
Discussion
Epilepsy is a disorder with diverse clinical manifestations and etiology. About 70%–80% of cases are believed to be due to one or more genetic factors [30]. Thus, the identification and functional analysis of the causative genes of epilepsy will lead to a better understanding of its pathogenic mechanisms, which would contribute to precision treatment in the future [11, 31, 32]. Recent studies have identified an increasing number of GRIN2A mutations in patients with epilepsy [12, 15]. However, the majority of the mutations have been found in families with incomplete penetrance or intra-familial variability, leading to suspicions concerning the pathogenicity of the mutations [12, 15, 16]. Functional studies are used to determine the impairments caused by gene mutations and thus they are helpful in evaluating the pathogenicity of mutations in the GRIN2A gene. In the present study, the GluN2A mutation N447K revealed a mild gain-of-function effect due to increased NMDAR current density and enhanced glutamate potency as well as decreased Mg2+ inhibition, supporting its pathogenic potential in epilepsy. In contrast, alternative substitutions at the residue N447 by alanine and glutamic acid (N447A and N447E) did not impact NMDAR function, providing a possible explanation for the diverse functional effects of GRIN2A variants.
Previous studies have demonstrated a gain-of-function effect in several GRIN2A mutations, including P552R [33, 34], N615K [10], F652V [15], L812M [11], and M817V [14, 35]. For the most part, these mutations present more severe phenotypes and apparent functional changes. The patient with N447K in this study presented relatively mild Rolandic epilepsy without aphasia. Functional studies revealed a relatively milder gain-of-function effect featuring a ~1.2-fold increase in NMDAR current density and a 2-fold enhancement of glutamate potency, coincident with the mild clinical phenotype. Topologically, residue N447 is located in a loose loop at the edge of the LBD, which is relatively distant from the glutamate-binding core. This might explain the less severe functional impairment and subsequently milder clinical phenotype.
Two recent studies have identified GRIN2A mutations with loss-of-function effects or without apparent physiological deficits [13, 17], indicating a complex relationship between GRIN2A mutation and epilepsy. Considering the possibility of other pathogenic mechanisms like a dominant negative effect, the pathogenicity of mutants without functional deficits could not be excluded. Further studies are required to determine the correlations between functional alterations and phenotypes of GRIN2A mutations and the underlying mechanisms. In the current study, alternative substitutions at residue N447 by alanine and glutamic acid (N447A and N447E) did not show functional deficits, suggesting that the chemical properties of the substituted amino-acid are also one of the determinants for functional changes. Sequence alignment showed that the amino-acids at the homologous site of this residue in different NMDAR subunits are all uncharged or negatively-charged. It is likely that substitution of a positively-charged lysine (K) may increase the attraction to glutamate through conformational changes and thus enhance the glutamate potency. In contrast, alanine is uncharged and glutamic acid is negatively-charged, in accordance with the polarity of the residue in wild-type NMDAR subunits. Therefore, a change in the polarity of the amino-acid in missense mutations may help to predict their functional alterations.
In conclusion, we identified a GRIN2A mutation N447K in a patient with Rolandic epilepsy without language or intelligence disability. Biophysiological studies of the mutant revealed a relatively mild functional change, which may explain the mild clinical phenotype. Further functional studies on alternative substitutions at N447 provided insight into the mechanism underlying the diverse functional effects of GRIN2A variants. Our study leads to a better understanding of the role of NMDARs in epilepsy and will be helpful in the genetic diagnosis of epilepsy as well as in developing precision therapies for patients who carry GRIN2A mutations.
Acknowledgements
This work was supported by the grants of the National Natural Science Foundation of China (81671162, 81521062, 81561168, and 81571273), the National Basic Research Development Program of China (2014CB910300 and 2013CB530904), and Key Research Project of the Ministry of Science and Technology of China (2016YFC0904400).
Footnotes
Xing-Xing Xu and Xiao-rong Liu have contributed equally to this work.
Contributor Information
Jian-Hong Luo, Email: luojianhong@zju.edu.cn.
Wei-Ping Liao, Email: wpliao@tom.com.
References
- 1.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, et al. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: Impact on receptor properties, synaptic plasticity and disease. Nature Reviews Neuroscience. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 3.Burnashev N, Szepetowski P. NMDA receptor subunit mutations in neurodevelopmental disorders. Curr Opin Pharmacol. 2015;20:73–82. doi: 10.1016/j.coph.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Myers CT, Mefford HC. Advancing epilepsy genetics in the genomic era. Genome Med. 2015;7:91. doi: 10.1186/s13073-015-0214-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan H, Low CM, Moody OA, Jenkins A, Traynelis SF. Ionotropic GABA and Glutamate Receptor Mutations and Human Neurologic Diseases. Mol Pharmacol. 2015;88:203–217. doi: 10.1124/mol.115.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei F, Yan LM, Su T, He N, Lin ZJ, Wang J, et al. Ion channel genes and epilepsy: functional alteration, pathogenic potential, and mechanism of epilepsy. Neurosci Bull. 2017;33:455–477. doi: 10.1007/s12264-017-0134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemke JR, Geider K, Helbig KL, Heyne HO, Schutz H, Hentschel J, et al. Delineating the GRIN1 phenotypic spectrum: A distinct genetic NMDA receptor encephalopathy. Neurology. 2016;86:2171–2178. doi: 10.1212/WNL.0000000000002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohba C, Shiina M, Tohyama J, Haginoya K, Lerman-Sagie T, Okamoto N, et al. GRIN1 mutations cause encephalopathy with infantile-onset epilepsy, and hyperkinetic and stereotyped movement disorders. Epilepsia. 2015;56:841–848. doi: 10.1111/epi.12987. [DOI] [PubMed] [Google Scholar]
- 10.Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 11.Yuan H, Hansen KB, Zhang J, Pierson TM, Markello TC, Fajardo KV, et al. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat Commun. 2014;5:3251. doi: 10.1038/ncomms4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemke JR, Lal D, Reinthaler EM, Steiner I, Nothnagel M, Alber M, et al. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat Genet. 2013;45:1067–1072. doi: 10.1038/ng.2728. [DOI] [PubMed] [Google Scholar]
- 13.Addis L, Virdee JK, Vidler LR, Collier DA, Pal DK, Ursu D. Epilepsy-associated GRIN2A mutations reduce NMDA receptor trafficking and agonist potency–molecular profiling and functional rescue. Sci Rep. 2017;7:66. doi: 10.1038/s41598-017-00115-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, Tankovic A, Burger PB, Kusumoto H, Traynelis SF, Yuan H. Functional evaluation of a de novo GRIN2A mutation identified in a patient with profound global developmental delay and refractory epilepsy. Mol Pharmacol. 2017;91:317–330. doi: 10.1124/mol.116.106781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lesca G, Rudolf G, Bruneau N, Lozovaya N, Labalme A, Boutry-Kryza N, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet. 2013;45:1061–1066. doi: 10.1038/ng.2726. [DOI] [PubMed] [Google Scholar]
- 16.Carvill GL, Regan BM, Yendle SC, O’Roak BJ, Lozovaya N, Bruneau N, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet. 2013;45:1073–1076. doi: 10.1038/ng.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao K, Tankovic A, Zhang Y, Kusumoto H, Zhang J, Chen W, et al. A de novo loss-of-function GRIN2A mutation associated with childhood focal epilepsy and acquired epileptic aphasia. PLoS One. 2017;12:e0170818. doi: 10.1371/journal.pone.0170818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemke JR, Hendrickx R, Geider K, Laube B, Schwake M, Harvey RJ, et al. GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann Neurol. 2014;75:147–154. doi: 10.1002/ana.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swanger SA, Chen W, Wells G, Burger PB, Tankovic A, Bhattacharya S, et al. Mechanistic insight into NMDA receptor dysregulation by rare variants in the GluN2A and GluN2B agonist binding domains. Am J Hum Genet. 2016;99:1261–1280. doi: 10.1016/j.ajhg.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, Yuan H, Ortiz-Gonzalez XR, Marsh ED, Tian L, McCormick EM, et al. GRIN2D recurrent de novo dominant mutation causes a severe epileptic encephalopathy treatable with NMDA receptor channel blockers. Am J Hum Genet. 2016;99:802–816. doi: 10.1016/j.ajhg.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi F, Mou TC, Dorsett KN, Volkmann RA, Menniti FS, Sprang SR, et al. Structural basis for negative allosteric modulation of GluN2A-containing NMDA receptors. Neuron. 2016;91:1316–1329. doi: 10.1016/j.neuron.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W, Jiang LH. Site-directed mutagenesis to study the structure-function relationships of ion channels. Methods Mol Biol. 2013;998:257–266. doi: 10.1007/978-1-62703-351-0_20. [DOI] [PubMed] [Google Scholar]
- 23.Luo JH, Fu ZY, Losi G, Kim BG, Prybylowski K, Vissel B, et al. Functional expression of distinct NMDA channel subunits tagged with green fluorescent protein in hippocampal neurons in culture. Neuropharmacology. 2002;42:306–318. doi: 10.1016/S0028-3908(01)00188-5. [DOI] [PubMed] [Google Scholar]
- 24.Chen Q, He S, Hu XL, Yu J, Zhou Y, Zheng J, et al. Differential roles of NR2A- and NR2B-containing NMDA receptors in activity-dependent brain-derived neurotrophic factor gene regulation and limbic epileptogenesis. J Neurosci. 2007;27:542–552. doi: 10.1523/JNEUROSCI.3607-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL. Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J Neurosci. 1998;18:6163–6175. doi: 10.1523/JNEUROSCI.18-16-06163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309:261–263. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- 27.Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307:462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- 28.Paoletti P, Ascher P, Neyton J. High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J Neurosci. 1997;17:5711–5725. doi: 10.1523/JNEUROSCI.17-15-05711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erreger K, Traynelis SF. Zinc inhibition of rat NR1/NR2A N-methyl-D-aspartate receptors. J Physiol. 2008;586:763–778. doi: 10.1113/jphysiol.2007.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hildebrand MS, Dahl HH, Damiano JA, Smith RJ, Scheffer IE, Berkovic SF. Recent advances in the molecular genetics of epilepsy. J Med Genet. 2013;50:271–279. doi: 10.1136/jmedgenet-2012-101448. [DOI] [PubMed] [Google Scholar]
- 31.Pierson TM, Yuan H, Marsh ED, Fuentes-Fajardo K, Adams DR, Markello T, et al. GRIN2A mutation and early-onset epileptic encephalopathy: Personalized therapy with memantine. Ann Clin Transl Neurol. 2014;1:190–198. doi: 10.1002/acn3.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kearney JA. Precision medicine: NMDA receptor-targeted therapy for GRIN2D encephalopathy. Epilepsy Curr. 2017;17:112–114. doi: 10.5698/1535-7511.17.2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 34.Ogden KK, Chen W, Swanger SA, McDaniel MJ, Fan LZ, Hu C, et al. Molecular mechanism of disease-associated mutations in the Pre-M1 helix of NMDA receptors and potential rescue pharmacology. PLoS Genet. 2017;13:e1006536. doi: 10.1371/journal.pgen.1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkateswaran S, Myers KA, Smith AC, Beaulieu CL, Schwartzentruber JA, Consortium FC, et al. Whole-exome sequencing in an individual with severe global developmental delay and intractable epilepsy identifies a novel, de novo GRIN2A mutation. Epilepsia 2014, 55: e75–79. [DOI] [PubMed]