Abstract
To understand the physiological responses of the brown macroalga Macrocystis integrifolia during the marine tidal cycle, two RNA libraries were prepared from algal frond samples collected in the intertidal zone (0 m depth) and subtidal zone (10 m depth). Samples collected from intertidal zone during low tide was considered as abiotic stressed (MI0), while samples collected from subtidal zone was considered as control (MI10). Both RNA libraries were sequenced on Illumina NextSeq 500 which generated approx. 46.9 million and 47.7 million raw paired-end reads for MI0 and MI10, respectively. Among the representative transcripts (RTs), a total of 16,398 RTs (39.20%) from MI0 and 21,646 RTs (39.24%) from MI10 were successfully annotated. A total of 535 unigenes (271 upregulated and 264 downregulated) showed significantly altered expression between MI0 and MI10. In abiotic-stressed condition (MI0), the relative expression levels of genes associated with antioxidant defenses (vanadium-dependent bromoperoxidase, glutathione S-transferase, lipoxygenase, serine/threonine-protein kinase, aspartate Aminotransferase, HSPs), water transport (aquaporin), photosynthesis (light-harvesting complex) protein were significantly upregulated, while in control condition (MI10) most of the genes predominantly involved in energy metabolism (NADH-ubiquinone oxidoreductase/NADH dehydrogenase, NAD(P)H-Nitrate reductase, long-chain acyl-CoA synthetase, udp-n-acetylglucosamine pyrophosphorylase) were overexpressed.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1204-4) contains supplementary material, which is available to authorized users.
Keywords: Macrocystis integrifolia (Phaeophyceae), Abiotic stress, Transcriptome analysis, De novo assembly, Differential gene expression
Introduction
Brown algae (Kingdom: Chromista; class: Phaeophyceae) are probably the most abundant photosynthetic inhabitants of the intertidal zone. Like other organisms of the intertidal zone, brown alga is also subjected to recurring, harsh changes in the environment associated with life in the interface of terrestrial and marine habitats. These changes include, but are not limited to, desiccation, osmotic pressure, temperature, nutrients, light, and tidal flow (Kupper et al. 2008). However, little is known about the physiological processes in the brown algae which allow them to adapt to such abiotic stress conditions. In macroalgae, the intensity of the effects generated by the abiotic stresses is related to the vertical distribution range of these species in the intertidal zone (López-Cristoffanini 2013). Several species of brown macroalga are economically very important. The economic interest of brown macroalga is related mainly due to their use in the industrial production of polysaccharides (McHugh 2003; Bixler and Porse 2011; Synytsya et al. 2015). For example, alginate, a brown algal cell wall polysaccharide, is used both for pharmaceutical purposes (Tonnesen and Karlsen 2002; Thanh-Sang Vo et al. 2012) and in the food industry (Jensen 1993; Hafting et al. 2012; Fleurence 2016; Chapman 2015). Economically important Macrocystis integrifolia is one of four morphotypes of kelp (large brown algae) in the genus Macrocystis which lives in intertidal zone along the Pacific coast of North America, Peru and Chile (Alveal 1995) and is moderately tolerant to abiotic stresses. However, to date, the cellular and molecular responses, as well as abiotic stress tolerance factors of brown algae are not well-studied. Hence, the main objective of this study is to analyze the effect of abiotic stresses in brown algae M. integrifolia under intertidal condition using global transcriptome data.
Data description
Sample collection and maintenance
Samples from six individual plants of M. Integrifolia, three of each condition were collected at two sampling points in the rocky intertidal zone of Punta San Juanito, Ica, Peru (Table 1). Samples collected from intertidal zone (0 m) during low tide after 2 h of air exposure as well as from subtidal zone (10 m). Small pieces of tissues (approx. 2 cm2) were cut from fresh fronds and were promptly cleaned with ethanol for removing epiphytes and immediately frozen in liquid nitrogen until further analysis. In this study, the samples from 0 and 10 m were marked as MI0 and MI10, respectively. MI10 represented the control samples, while MI0 represented the natural abiotic-stressed samples.
Table 1.
MixS information of Macrocystis integrifolia
| Item | Definition |
|---|---|
| General feature of classification | |
| Classification | Eukaryota; Heterokonta; Phaeophyceae; Laminariales; Laminariaceae; Macrocystis integrifolia |
| Investigation type | Eukaryote transcriptome |
| Project name | Bioproject PRJNA322132 |
| Environment | |
| Geographic location | Zona Rocosa de Punta San Juanito, Ica, Perú |
| Latitude, longitude | 15°14′43.2″S, 75°15′31.9″W |
| Collection date | 2015-04 |
| Isolation environment | Ocean |
| Depth | 0 and 10 m of water depth |
| Biome | ENVO:01000320 (marine environment) |
| Collector (s) | Erika Salavarría and Sujay Paul |
| Sequencing | |
| Sequencing | Illumina NextSeq 500; Paired-end (2 × 151 bp Max) |
| Assembly | |
| Method | De novo assembly |
| Program | trinityrnaseq_r20140717 |
| Finishing strategy | High-quality transcriptome assembly |
| Data accessibility | |
| Sequence information | Genbank accession SRA426960 |
RNA extraction and preparation of cDNA library for RNAseq
Total RNA was extracted from the samples MI0 and MI10 using RNAeasy mini kit (Quiagen) and treated with RNase free DNase (Quiagen) to remove the residual genomic DNA. RNA quantity and quality were assessed with NanoDropTM 1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and 2100 Bioanalyzer (AGILENT). Library preparation was performed by Illumina TruSeq RNA library protocol. Briefly, 1 g of Total RNA was subjected to Poly A purification of mRNA. Purified mRNA was fragmented for 2 min at elevated temperature (94 °C) in the presence of divalent cations and reverse transcribed with Superscript III Reverse transcriptase by priming with Random Hexamers. Second strand cDNA was synthesized in the presence of DNA Polymerase I and RnaseH. The cDNA was cleaned up using HighPrep PCR (MAGBIO, Cat# AC-60050) and Illumina adapters were ligated to the cDNA molecules after end repair and addition of A base. SPRI cleanup was performed after ligation. Each of the two libraries was amplified using 8 cycles of PCR for the enrichment of adapter-ligated fragments. The prepared library was quantified using Qubit and validated for quality by running an aliquot on High Sensitivity Bioanalyzer Chip (Agilent). Based on the population observed in the profile, the libraries were sequenced on Illumina NextSeq 500 at Genotypic Technology Private Limited, Bangalore, India.
Data filtering, de novo transcriptome assembly, and functional annotation
The Illumina NextSeq 500 paired-end raw reads were first quality checked using FastQC (Andrews, 2010) and then processed by the in-house script for adapters and low-quality bases trimming towards 3′- end. De novo assembly of Illumina NextSeq 500 processed data was performed using trinityrnaseq (Grabherr et al. 2011) with default K-mer = 25. Transcripts having length ≥ 300 bp have been considered, followed by clustering of these transcripts with 95% identity using CD-HIT (Li and Godzik 2006) which resulted into COG’s. The assembled transcripts were then annotated using NCBI BLAST 2.2.31 (Altschul et al. 1990) with the protein sequences of brown algae Ectocarpus siliculosus available at UniProt Protein Database. The same Uniprot protein database was used for Gene Ontology (GO) annotation and transcripts were assigned GO subcategories under biological process (BP), molecular function (MF) and cellular component (CC). Differential gene expression (DGE) between the samples was performed by employing a negative binomial distribution model with DeSeq v1.8.1 tool (Anders and Huber 2010). Briefly, transcripts of both MI0 and MI10 samples were combined to the size ≥ 300 bp and were clustered together using CD-HIT at 95% identity. Consequently, master control transcriptome data (unigenes) were generated. The RPKM measurement (readings per kilobase of transcribed per million mapped readings) was then performed using “Bowtie2 tool” to generate the read count profile. Transcripts having log2 (fold change) value ≥ 1 and ≤ − 1 were considered to be upregulated and downregulated, respectively. Moreover, transcripts showing significant variation in fold change expression (corrected P value ≤ 0.05) in these sets were also identified. The overall transcriptome assembly and annotation procedure, as well as DGE analysis performed in this study, have been graphically represented in Supplementary Figure S1 and S2.
A total of approx. 46.9 million and 47.7 million raw paired-end reads were generated from Illumina NextSeq 500 platform for MI0 and MI10 samples, respectively. The raw paired-end sequences data in FASTQ format was deposited in the National Centre for Biotechnology Information’s (NCBI) Short Read Archive (SRA) database under the accession number SRA426960. Raw reads were subjected to quality control using FastQC and the results showed that the average per sequence Phred quality score was above 30 in both the reads indicating high-quality sequencing run. Post quality filtering for low-quality regions, adaptors and sequencing tags, a total read count of approx. 42.4 million for MI0 and 42.3 million for MI10 were withdrawn for further processing. De novo assembly of Illumina NextSeq 500 processed data was performed using trinity at a hash length of 25, which generated 43,952 and 57,516 transcripts for MI0 and MI10, respectively (Table 2). A total of 11,756 and 12,524 transcripts with more than 1 kb in length as well as 25,982 and 30,709 transcripts with more than 500 bp in length were generated in MI0 and MI10 (Table 2). Higher N50 values (1159 and 986 for MI0 and MI10, respectively) also indicate the high quality of sequence assembly.
Table 2.
De novo assembly summary of sample MI0 and MI10 using Trinity tool
| Samples | MI0 | MI10 |
|---|---|---|
| Tool used | Trinity | |
| Hash/k-mer length | 25 | |
| Transcripts/isotigs(contigs) generated | 43,952 | 57,516 |
| Maximum transcript/isotig length | 10,538 bp | 10,627 |
| Minimum transcript/isotig length | 301 bp | 301 bp |
| Average transcript/isotig length | 869.4 bp | 784.3 bp |
| Median transcript/isotig length | 2329 bp | 441.5 bp |
| Total transcript length | 38,210,788 bp | 45,109,937 bp |
| Transcripts ≥ 500 bp | 25,982 | 30,709 |
| Transcripts > 1 kb | 11,756 | 12,524 |
| Transcripts > 10 kb | 2 | 1 |
| N50 value | 1159 | 986 |
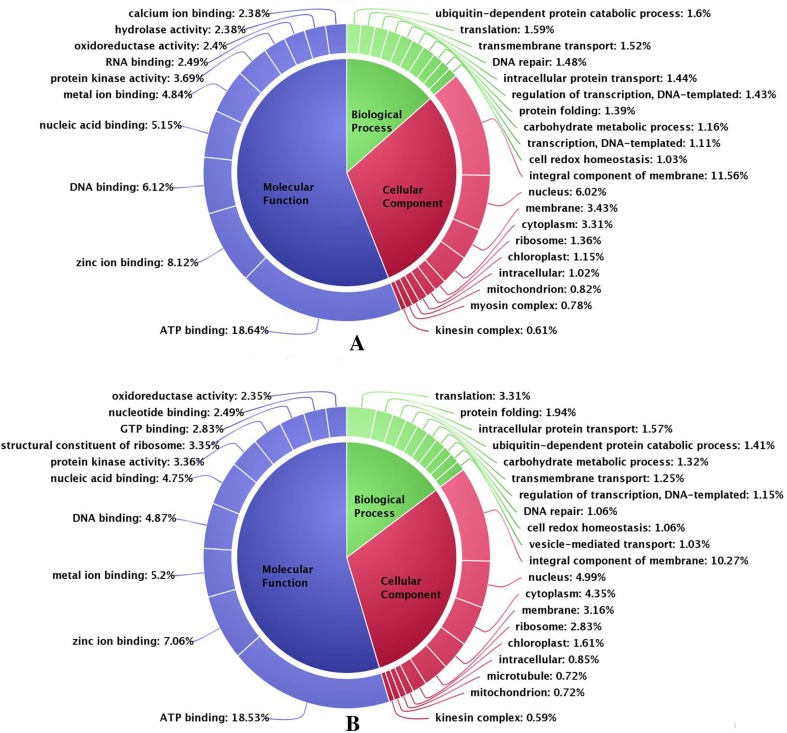
It is always challenging to predict accurate annotations for the transcripts from the non-model organism. In this study, representative transcripts (RTs) were annotated using NCBI BLAST 2.2.31 with the protein sequences of brown alga Ectocarpus siliculosus. Transcripts greater than 30% identities were considered to be suitable to assign annotation based on high degree of sequence identity. GO annotation revealed that the annotated RTs represent various genes which are involved in various metabolic pathways (Fig. 1). In the molecular function (MF) category, the most frequently occurring terms were ATP binding (18.64%) and zinc ion binding (8.12%) followed by DNA binding (6.12%). Ubiquitin-dependent protein catabolic process (1.60%) and translation (1.59%) were the most frequently occurring terms in biological processes (BP) category followed by transmembrane transport (1.52%).The most frequently occurring terms in cellular component (CC) category was integral component of the membrane (11.56%) and nucleus (6.02%) followed by membrane (3.31%) (Fig. 1a). In control condition (MI10), the most frequently occurring terms under MF category were found also ATP binding (18.53%) followed by zinc ion binding (7.06%) and metal ion binding (5.20%). Translation (3.31%), followed by protein folding (1.94%) and intracellular protein transport (1.57%) were found to be the most represented terms under BP category. The most frequently occurring terms in CC category were also integral component of membrane (10.27%) and nucleus (4.99%) followed by cytoplasm (4.35%) (Fig. 1b).
Fig. 1.
Gene ontology (GO) analysis. a MI0, b MI10
Differential gene expression (DGE) analysis
In the transcriptomic analysis, the differentially expressed genes between MI0 and MI10 were estimated by clusters or hierarchical groupings considering q value < 0.05, FDR ≤ 0.05 and minimum twofold change (log 2 ≥ + 1 or ≤ − 1). A total of 9519 unigenes expressed differentially between MI0 and MI10 among which 271 upregulated and 264 downregulated genes are significantly altered. To find out the possible molecular mechanisms involved in the stress responses, we manually classified and examined several genes that exhibited major significant changes in the stress conditions (MI0) compared to the MI10 control (Table 3). Many of these genes altered were related to the synthesis of chaperone proteins (molecular chaperones HSP70/HSC70, HSP70 superfamily), electron transport (Rieske (2Fe-2S), essential components in physiological processes including enzymes for carbon fixation (fructose-bisphosphate aldolase, glycine dehydrogenase), alginate and cellulose synthesis (mannuronan C-5-epimerase, UDP glucose 6-dehydrogenase), amino acid metabolism (shikimate kinase), transmembrane transport systems (aquaporins), genes involved in nitrogen fixation (glutamine synthetase), processes related to photosynthesis (light-harvesting complex protein), classical pathways to stress (vanadium-dependent bromoperoxidase), aprataxin synthesis (FHA-HIT protein), fatty acid metabolism (lipoxygenase), oxidative stress (alkaline phosphatase, HSPs, glutathione S-transferase), phytohormone biosynthesis (S-adenosylmethionine synthase, gibberellin 2-beta-dioxygenase), ascorbate biosynthesis (L-galactono-1,4-lactone dehydrogenase) and other processes (Table 3).
Table 3.
List of some significantly altered genes in MI0 as compared to MI10 in M. integrifolia during abiotic stress response
| Gene/protein name | Regulation | Fold change | p value | Function |
|---|---|---|---|---|
| Vanadium-dependent bromoperoxidase | UP in MI0 | 6.8914 | 0.0222098808 | Antioxidant defenses |
| Urease accessory protein UreF | UP in MI0 | 22.4184 | 0.0317399721 | Urea cycle |
| Glutathione S-transferase (EC 2.5.1.18) | UP in MI0 | 9.5782 | 0.0096688523 | Glutathione synthesis and antioxidant defenses |
| Light harvesting complex protein | UP in MI0 | 16.4412 | 0.0017643654 | Photosynthesis |
| Rieske (2Fe-2S) region | UP in MI0 | 39.0188 | 0.0003381751 | Photosystem 2, electron transport, light sensing and carotenoid synthesis |
| Aquaporin | UP in MI0 | 30.4891 | 0.0091832762 | Transportation system (activation of water and ion transporters) |
| Molecular chaperones HSP70/HSC70, HSP70 superfamily | UP in MI0 | 12.3671 | 0.0040478809 | Response to heat and oxidative stress (desiccation). Synthesis of chaperones |
| Branched-chain alpha-keto acid dehydrogenase E1 beta subunit | UP in MI0 | 6.2733 | 0.0350976028 | Branched-chain amino acids |
| Serine/threonine-protein kinase CTR1 | UP in MI0 | 11.8631 | 0.0415284001 | Branched-chain amino acids, antioxidant defenses |
| Aspartate aminotransferase (EC 2.6.1.1) | UP in MI0 | 11.1982 | 0.0075979684 | Branched-chain amino acids, antioxidant defenses |
| Similar to ubiquitin specific protease 34 | UP in MI0 | 18.3831 | 0.0151442253 | Recycling proteins |
| Similar to ATP synthase mitochondrial F1 complex assembly factor 2 | UP in MI0 | 6.5190 | 0.0458993508 | Energy metabolism |
| Monophenol monooxygenase | UP in MI0 | 27.0267 | 0.000428844 | Betalaina biosynthesis |
| Alkaline phosphatase | UP in MI0 | 19.2381 | 0.0022184014 | Pathway: NAD/NADH phosphorylation and dephosphorylation( responses to oxidative stress) |
| Oxidoreductase (arsenate reductase. glutaredoxin) | UP in MI0 | 10.3163 | 0.0069709376 | Arsenate detoxification |
| DNA helicase (EC 3.6.4.12) | UP in MI0 | 2.6621 | 0.0090710198 | DNA helicases, utilize the energy from ATP hydrolysis |
| 5-Methyltetrahydropteroyltriglutamate–homocysteine S-methyltransferase | UP in MI0 | 12.4255 | 0.0092518168 | Methionine metabolism, seleno compound metabolism, l-metionine biosynthesis |
| DEAD box helicase (EC 3.6.4.13) | UP in MI0 | 9.9354 | 0.0363002914 | A nucleoside triphosphate + H2O = a nucleoside diphosphate + phosphate catalytic mechanism |
| Lactoylglutathione lyase, putative | UP in MI0 | 9.8005 | 0.0141131854 | Pyruvate metabolism, methylglyoxal degradation I |
| Ubiquitin-conjugating enzyme, putative | UP in MI0 | 5.5672 | 0.04170213 | Protein ubiquitylation pathway |
| Ankyrin 3.6.1.15 (NTPase) | UP in MI0 | 7.3078 | 0.02370033 | Oxidative phosphorylation |
| Gamma-glutamyl transpeptidase (EC 2.3.2.2) | UP in MI0 | 8.8739 | 0.03232579 | Glutatione metabolism |
| Monogalactosyldiacylglycerol synthase, family GT28 (EC 2.4.1.46) | UP in MI0 | 6.9442 | 0.03697278 | Galactolipid biosynthesis I |
| Serine/threonine dehydratase (EC 4.3.1.-) l-serine ammonia-lyase | UP in MI0 | 6.4735 | 0.02875467 | Serine metabolism, threonine metabolism |
| Patatin(EC. 3.1.1.4) | UP in MI0 | 8.9673 | 0.04265161 | Phospholipase metabolism |
| Acetoacetate–CoA ligase (EC 6.2.1.16) | UP in MI0 | 6.7772 | 0.04515125 | Valine and leucine degradation |
| Carbohydrate kinase 2.7.1.14 | UP in MI0 | 5.6828 | 0.04656553 | Aerobic respiration I–III |
| Dihydroorotate dehydrogenase (quinone), mitochondrial (DHOdehase) (EC 1.3.5.2) | UP in MI0 | 5.3654 | 0.04693179 | Pyrimidine metabolism |
| Lipoxygenase | UP in MI0 | 18.02 | 0.001319908 | Fatty acid metabolism and antioxidant defenses |
| ABC transporter-like | DOWN in MI0 | 0.1472 | 0.03404662 | Cell signaling |
| NADH-ubiquinone oxidoreductase chain 1/NADH dehydrogenase (Ubiquinone) (EC 1.6.5.3) | DOWN in MI0 | 0.0026 | 1.40E−07 | Aerobic respiration I–III |
| Shikimate kinase | DOWN in MI0 | 0.1832 | 0.04200643 | Aromatic amino acids |
| Udp-n-acetylglucosamine pyrophosphorylase | DOWN in MI0 | 0.0557 | 0.00209323 | Energy metabolism |
| Galactose-4-epimerase, UDP | DOWN in MI0 | 0.932 | 0.00831223 | UDP-d-galactose biosynthesis |
| Long-chain acyl-CoA synthetase | DOWN in MI0 | 0.1441 | 0.02197075 | Energy metabolism |
| Casein kinase II alpha subunit | DOWN in MI0 | 0.0813 | 0.02875841 | Branched-chain amino acids |
| Magnesium chelatase subunit H, putative chloroplast (EC 6.6.1.1) | DOWN in MI0 | 0.0006 | 4.834E−09 | Chlorophyll metabolism |
| Mannose-6-phosphate isomerase | DOWN in MI0 | 0.0463 | 0.0170957 | l-ascorbate biosynthesis I (l-galactose pathway) Smirnoff wheeler pathway |
| Transposase | ONLY in MI0 | 0.01483208 | Replicative transposition mechanism | |
| FHA-HIT protein | ONLY in MI0 | 0.00128322 | Aprataxin synthesis | |
| Gibberellin 2-beta-dioxygenase | ONLY in MI0 | 0.004783 | Diterpenoid biosynthesis; gibberellin inactivation I | |
| Isocitrate lyase | ONLY in MI0 | 1.63E−06 | Gluconeogenesis | |
| Polymorphic Outer membrane protein G/I family | ONLY in MI0 | 5.38E−06 | Components of the membrane | |
| Polymorphic outer membrane protein | ONLY in MI0 | 1.2758E−05 | Components of the membrane | |
| Light harvesting complex protein | ONLY in MI0 | 0.02008396 | Photosynthesis | |
| NAD(P)H-nitrate reductase (EC 1.7.1.1) | ONLY in MI10 | 8.64E−07 | Nitrogen assimilation | |
| Glycine dehydrogenase (decarboxylating) | ONLY in MI10 | 2.328782E−10 | Photorespiration and carbon availability | |
| Phosphoenolpyruvate carboxykinase (ATP) | ONLY in MI10 | 0.00097953 | Branched-chain amino acids | |
| Pyruvate dehydrogenase (EC 1.2.4.1) | ONLY in MI10 | 0.00083066 | Branched-chain amino acids | |
| Pyruvate kinase (EC 2.7.1.40) | ONLY in MI10 | 0.00013577 | Pyruvate metabolism. Branched chain amino acids | |
| Serine hydroxymethyltransferase | ONLY in MI10 | 0.03295779 | Photorespiration and carbon availability | |
| S-adenosylmethionine synthase | ONLY in MI10 | 0.00431818 | Ethylene synthesis | |
| Glyceraldehyde-3-phosphate dehydrogenase | ONLY in MI10 | 1.1215E−05 | Pathway: glycolysis IV (plant cytosol) | |
| Triosephosphate isomerase/glyceraldehyde-3-phosphate dehydrogenase | ONLY in MI10 | 8.47E−11 | Gluconeogenesis. Photosynthesis | |
| Aldehyde dehydrogenase | ONLY in MI10 | 2.9078E−05 | Glycolysis/gluconeogenesis | |
| Fructose-bisphosphate aldolase | ONLY in MI10 | 5.89E−10 | Glycolysis/gluconeogenesis | |
| l-Galactono-1,4-lactone dehydrogenase | ONLY in MI10 | 0.02630423 | l-Ascorbate biosynthesis I (l-galactose pathway) Smirnoff-wheeler pathway | |
| Formaldehyde dehydrogenase (EC 1.1.1.284) | ONLY in MI10 | 0.00160783 | Methane metabolism, formaldehyde oxidation | |
| Ribulose bisphosphate carboxylase large chain (RuBisCO large subunit) (EC 4.1.1.39) | ONLY in MI10 | 0.00013577 | Involved in the first step of the calvin cycle for fixing carbon in photosynthetic systems, photorespiration |
This is the first global transcriptome analysis of brown algae Macrocystis integrifolia (Phaeophyceae) under marine intertidal conditions. This study could be an important resource for subsequent genomic studies in brown algae to identify functional genes involved in different metabolic processes related to stress tolerance.
Data accessibility
The raw paired-end sequences data in FASTQ format was deposited in the National Centre for Biotechnology Information’s (NCBI) Short Read Archive (SRA) database under the accession number SRA426960.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Grant no 352-PNICP-PIBA-2014 from Programa Nacional de Innovación para la Competitividad y Productividad (INNOVATE PERU), Ministry of Production of Peru. Erika Salavarría was funded by scholarship program “Academia 2010”. SENESCYT. Ecuador. We acknowledge Peruvian Seaweeds S.R.L. and Gunter Villena for logistic facilitated for the sampling. We acknowledge Genotypic Technology Pvt. Ltd Bengaluru, India for additional analysis. We want to thank Dr. Marcel Gutiérrez-Correa (RIP) for his extraordinary support and useful advice during the development of this work.
Compliance with ethical standards
Conflict of interest
The authors declare that they don’t have any conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1204-4) contains supplementary material, which is available to authorized users.
References
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alveal K. Manejo de algas marinas. In: Alveal K, Ferrario M, Oliveira E, Sar E, editors. Manual de métodos ficológicos. Concepción: Ediciones Universidad de Concepción; 1995. pp. 825–863. [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010) FastQC: A quality control tool for high throughput sequence data. http://www.Bioinformatics.Babraham.Ac.Uk/Projects/Fastqc/
- Bixler HJ, Porse H. A decade of change in the seaweed hydrocolloids industry. J Appl Phycol. 2011;23:321–335. doi: 10.1007/s10811-010-9529-3. [DOI] [Google Scholar]
- Chapman AS, Stévant P, Larssen WE. Food or fad? challenges and opportunities for including seaweeds in a Nordic diet. Bot Mar. 2015;58:423–433. doi: 10.1515/bot-2015-0044. [DOI] [Google Scholar]
- Fleurence J. Seaweeds as food: seaweed in health and disease prevention. Cambridge: Academic Press; 2016. pp. 149–167. [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafting JT, Critchley AT, Cornish ML, et al. On-land cultivation of functional seaweed products for human usage. J Appl Phycol. 2012;24:385–392. doi: 10.1007/s10811-011-9720-1. [DOI] [Google Scholar]
- Jensen A. Present and future needs for algae and algal products. Hydrobiologia. 1993;260–261:15–23. doi: 10.1007/BF00048998. [DOI] [Google Scholar]
- Kupper FC, Carpenter LJ, McFiggans GB, et al. Iodide accumulation provides kelp with an inorganic antioxidant impacting atmospheric chemistry. Proc Natl Acad Sci. 2008;105:6954–6958. doi: 10.1073/pnas.0709959105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- López-Cristoffanini C, Tellier F, Otaíza R, et al. Tolerance to air exposure: A feature driving the latitudinal distribution of two sibling kelp species. Bot Mar. 2013;56:431–440. doi: 10.1515/bot-2013-0042. [DOI] [Google Scholar]
- McHugh DJ (2003) A Guide to the Seaweed Industry. FAO Fisheries Technical Paper. ISBN 92-5-104958-0
- Synytsya A, Copiková J, Kim WJ, Park YI. Springer handbook of marine biotechnology. Berlin: Springer; 2015. Cell wall polysaccharides of marine algae; pp. 543–590. [Google Scholar]
- Tønnesen HH, Karlsen J. Alginate in drug delivery systems. Drug Dev Ind Pharm. 2002;28:621–630. doi: 10.1081/DDC-120003853. [DOI] [PubMed] [Google Scholar]
- Vo TS, Ngo DH, Kim SK. Marine algae as a potential pharmaceutical source for anti-allergic therapeutics. Process Biochem. 2012;47:386–394. doi: 10.1016/j.procbio.2011.12.014. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw paired-end sequences data in FASTQ format was deposited in the National Centre for Biotechnology Information’s (NCBI) Short Read Archive (SRA) database under the accession number SRA426960.