Abstract
Introduction
A soluble (s) form of the endothelial protein C receptor (EPCR) circulates in plasma and inhibits activated protein C (APC) activities. The clinical impact of sEPCR and its involvement in the septic process is under investigation. This study determined the frequencies of EPCR haplotypes H1 and H3 to investigate possible associations with plasma admission levels of sEPCR in an intensive care unit (ICU) cohort of septic patients.
Methods
Three polymorphisms in the EPCR gene were genotyped in 239 Caucasian critically ill patients, and their plasma sEPCR levels were also measured at the time of admission to the ICU. Multivariate logistic regression analysis controlling for sepsis severity, age, acute physiology and chronic health evaluation (APACHE II) and sequential organ failure assessment (SOFA) scores, lactate level, sex, diagnostic category, length of ICU stay and hospital mortality was performed to determine the effect of EPCR haplotypes H1 and H3 on the levels of sEPCR.
Results
Individuals carrying at least one H3 allele had significantly higher levels of sEPCR than individuals with no H3 alleles (p < 0.001). No differences were found in the distribution of the H3 allele in the patient groups categorized using the pre-existing and current sepsis-3 definitions.
Conclusion
Using the preceding and current sepsis definitions, sEPCR levels and the H3 haplotype were not associated with sepsis severity and the risk of poor outcomes in septic patients; however, the EPCR H3 allele contributed to higher levels of sEPCR.
Keywords: Haplotypes, qSOFA, sEPCR, Sepsis
Introduction
Sepsis is defined as life-threatening organ dysfunction caused by a dysregulated host response to infections [1]. The protein C (PC) anticoagulant pathway controls thrombosis and inflammation [2], and activation of PC to activated PC (APC) is enhanced by the presence of the endothelial protein C receptor (EPCR) [3]. Apart from an anti-thrombotic role, APC exerts anti-inflammatory, anti-apoptotic and cytoprotective functions that are EPCR-dependent [2, 4–7]. The presence of a soluble form of EPCR (sEPCR) in human physiologic plasma, which is elevated in inflammatory conditions [8], strengthens the idea of EPCR shedding. sEPCR can be generated rapidly via metalloprotease tumor necrosis factor-α-converting enzyme/metallopeptidase domain 17 (TACE/ADAM17) shedding as a response to an inflammatory stimulus [9, 10] and slowly via alternative mRNA splicing [11]. sEPCR binds both PC and APC with similar affinity to membrane-bound EPCR [12, 13], blocking, however, the anti-thrombotic activity of the latter [12–14]. While the role of the membrane form of EPCR is clearly anti-thrombotic and anti-inflammatory, the physiologic significance of sEPCR in vivo remains to be clarified. Apart from sEPCR, functionally relevant mutations in the EPCR gene have been reported, resulting in either reduced expression or dysfunction of the receptor, which can induce thrombosis development. These include a rare 23-bp insertion in exon 3 (position 6367) [15] and single-nucleotide polymorphisms (SNPs) at coding and non-coding regions of the EPCR gene; based on the SNPs that are present, human EPCR genotypes can be divided into four haplotypes, H1, H2, H3 and the very rare H4. The H2 consists of all the common alleles [16, 17]. The functional importance of these mutations has been mainly studied in patients with venous and coronary thrombosis [18–24], and results from these studies have suggested that the H3 haplotype may be a candidate risk factor for deep vein thrombosis, possibly by increasing the levels of sEPCR and promoting its procoagulant effects because of APC sequestration [13, 16, 24]. On the other hand, the role of the EPCR gene haplotypes in sepsis has only been studied to some degree [25, 26], and even fewer have studied correlations between plasma sEPCR levels and EPCR gene polymorphisms in sepsis [26]. Therefore, the aim of the present study was to measure sEPCR levels in critically ill septic patients at admission to the intensive care unit (ICU) and to evaluate potential associations of sEPCR with patient genotype. We genotyped one haplotype-tagging single-nucleotide polymorphism (htSNP) belonging to H1 (C6333T) and two htSNPs belonging to H3 (A6936G and C1651G) (numbering according to GenBank accession no. AF106202) [15–17]. To investigate the role of the above EPCR mutations as predisposing factors to higher sEPCR levels, we performed multivariate logistic regression analysis controlling for potential confounding factors, such as disease severity, age, sex and diagnostic category.
Methods
Compliance with Ethics Guidelines
The study was approved by the Evangelismos Hospital Research Ethics Committee (76/4-3-2009). All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964, as revised in 2013. Informed consent was obtained from all patients or patients’ next-of-kin for being included in the study.
Study Population and Measurements
To determine the role of the EPCR haplotypes as possible contributors to higher levels of sEPCR in sepsis, we conducted a retrospective study on 239 patients with diagnosed sepsis admitted in the ICU of Evangelismos Hospital.
Patients were studied according to the preceding [27] and current sepsis-3 definitions [1]. Based on the former, patients were considered to have sepsis, severe sepsis or septic shock. We subsequently divided patients in two groups: (1) those who either had severe sepsis and/or septic shock at ICU admission or subsequently developed it during their ICU stay (severe sepsis/septic shock positive, N = 139) and (2) those who did not have severe sepsis and/or septic shock at ICU admission or develop it during their ICU stay, but had uncomplicated sepsis (severe sepsis/septic shock negative, N = 100). Based on sepsis-3 definitions and related scores and using patients’ files, our cohort was additionally grouped according to their quick SOFA (qSOFA) score prior to ICU admission.
Genetic Analysis
Genotyping had been previously performed in a larger cohort, and data generated were used in a different context [28]. Briefly, genotyping was performed by the 5′-nuclease/TaqMan assay using the polymerase chain reaction with fluorescent allele-specific oligonucleotide probes (Assay-by-Design/Assay-on-Demand, Life Technologies Corp., Carlsbad, CA, USA) and the Type-it Fast SNP probe PCR kit from Qiagen (QIAGEN GmbH, Hilden, Germany). Fluorescence endpoint reading for allelic discrimination was performed on a PTC-200 (MJ Research Inc., Waltham, MA, USA) using the Opticon Monitor 2 software. One htSNP belonging to H1 (C6333T—rs2069952) and two htSNPs belonging to H3 (C1651G—rs2069940 and A6936G—rs867186) were selected (Table 1). The presence of the minor allele determines the respective haplotype, while absence of all three minor alleles determines H2.
Table 1.
Nucleotide sequence surrounding the target SNPs
| SNP ID | Context sequence [VIC/FAM] | Position |
|---|---|---|
| rs2069952 | GCACCCTCTCTGCACAGTCCCCTGA[C/T]CCTGACTGTCTATCCACAGTTCCTC | Intron 2 |
| rs2069940 | GTTTCAAAAAGATTGCTTTAGGTAA[C/G]CAATCATCTTCTGAGATTTATACAG | 5′ UTR |
| rs867186 | GCTGGTCCTGGGCGTCCTGGTGGGC[A/G]GTTTCATCATTGCTGGTGTGGCTGT | Exon 4 |
The first nucleotide in the brackets corresponds to the VIC allele and the second to the FAM allele. Oligonucleotide probes were purchased from Applied Biosystems
SNP single-nucleotide polymorphism, 5′UTR 5′ untranslated region
sEPCR Antigen Measurement
Plasma sEPCR levels at ICU admission were measured by enzyme-linked immunosorbent assay (ELISA), according to the manufacturer’s instructions. The Human EPCR Immunoassay of R&D Systems was used (R&D Systems Inc., Minneapolis, MN, USA). The assay uses two different polyclonal antibodies against EPCR as the catching and tagging antibody. Samples were diluted 1/39 and assayed in triplicate.
Statistical Analysis
Data are presented as individual values and mean ± standard deviation (SD) for normally distributed variables and median with interquartile range (IQR) for variables with skewed distribution. Two-group comparisons were performed by the t test or the non-parametric Mann-Whitney test for skewed data. Associations between qualitative variables were examined by the Chi-square test or Fisher’s exact test when appropriate. Kruskal-Wallis ANOVA followed by Dunn’s multiple comparison post hoc test was used to examine differences between more than two groups. Multivariate logistic regression analysis controlling for APACHE II and SOFA scores, lactate level, length of ICU stay, age (continuous variables), sepsis severity, diagnostic category, hospital mortality, sex and haplotype (categorical variables) was performed to identify potential risk factors for elevated sEPCR levels as a binary outcome. Median sEPCR levels (192 ng/ml) were used as a cutoff point. Results are described by the odds ratio (OR) and 95% confidence interval (CI). All p values were two-sided; p < 0.05 was considered significant.
Results
Characteristics of the Study Population
Among the 239 critically ill septic patients studied, 166 were male (69.5%) and 73 were female (30.5%). Approximately half of the study sample consisted of surgical or trauma patients, while the other half had medical diagnoses. Severe sepsis and/or septic shock (SS/SS) occurred in 139 patients (58.16% of the total cohort). Of these, 104 patients had SS/SS at baseline, while 35 developed SS/SS during their ICU stay. Demographic characteristics of our critically ill septic cohort are given in Table 2.
Table 2.
Characteristics of the 239 critically ill septic patients enrolled in the study
| Parameters | All patients |
|---|---|
| Number of patients (N) | 239 |
| sEPCR (ng/ml) | 192 (136–245) |
| Diagnosis | |
| Medical | 88 (36.8%) |
| Surgical | 98 (41.0%) |
| Trauma | 53 (22.2%) |
| APACHE II score | 22 (16–27) |
| SOFA score | 8 (6–11) |
| Age (years) | 60 ± 18 |
| Sex | |
| Male | 166 (69.5%) |
| Female | 73 (30.5%) |
| ARDS | 46 (19.3%) |
| Mortality | 98 (41.0%) |
| Length of stay (days) | 10 (3–25) |
Data are expressed as number of patients (N) and percentages of total (%), mean ± SD or median (25–75% interquartile range). sEPCR, APACHE II and SOFA scores were measured at admission in the intensive care unit
APACHE acute physiology and chronic health evaluation, ARDS acute respiratory distress syndrome, sEPCR soluble endothelial protein C receptor, SOFA sequential organ failure assessment
EPCR Polymorphisms and sEPCR Levels
Allelic frequencies of the H1, H2 and H3 haplotypes are given in Table 3 for this critically ill study population. The frequencies of the H1, H2 and H3 haplotypes were similar in the SS/SS-positive and SS/SS-negative groups, as well as in the three qSOFA groups (Table 3). The distributions of the genotypes were in Hardy-Weinberg equilibrium (chi-square test, p > 0.05).
Table 3.
Allelic frequencies of the single-nucleotide polymorphisms
| Classification | SS/SS-positive (N = 139) [%] | SS/SS-negative (N = 100) [%] | qSOFA 1 (N = 7) [%] | qSOFA 2 (N = 197) [%] | qSOFA 3 (N = 35) [%] | All patients (N = 239) [%] | p value |
|---|---|---|---|---|---|---|---|
| Allele | |||||||
| H3 | 10.4 | 15.5 | 7.0 | 13.2 | 10.0 | 12.6 | ns |
| H1 | 34.2 | 34.5 | 50.0 | 32.5 | 34.3 | 33.3 | ns |
| H2 | 55.4 | 50.0 | 43.0 | 54.3 | 55.7 | 54.1 | ns |
Data are expressed as percentages of total (%). H1: 6333TC or CC; H3: 1651CG or GG and/or 6936AG or GG; H2: all common alleles
qSOFA quick sequential organ failure assessment, SS/SS severe sepsis and/or septic shock, ns non-significant (p > 0.05)
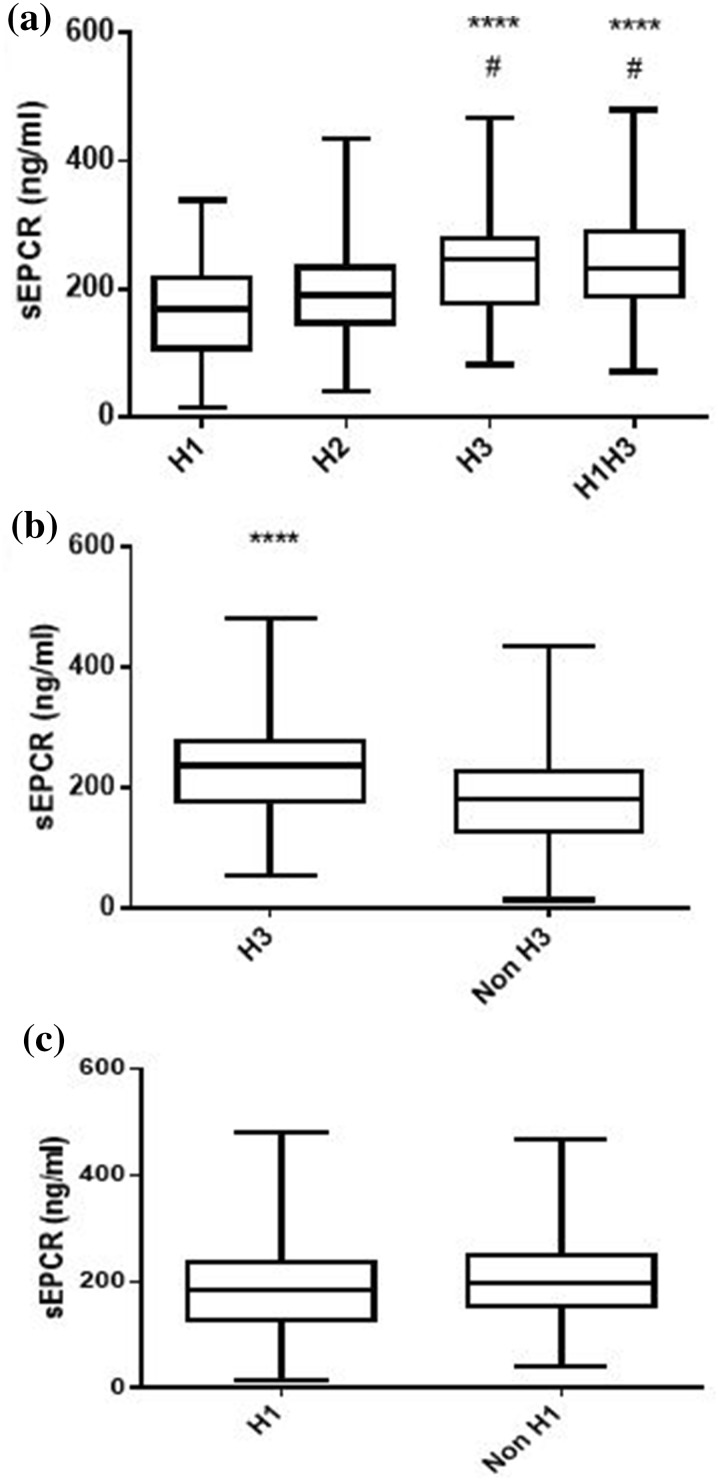
Soluble EPCR was quantified concurrently in all blood samples collected at ICU admission (N = 239). At the time of ICU admission, sEPCR levels were similar in the SS/SS-positive and -negative patients (Fig. 1a). When patients were categorized using the sepsis-3 classification, patients with a qSOFA score of 1, 2 or 3 had comparable sEPCR levels (Fig. 1b). However, when patients were grouped according to their genotypes, carriers of at least one H3 allele had significantly higher sEPCR levels (Fig. 2; p < 0.05).
Fig. 1.
sEPCR levels in patients classified according to the preceding and current sepsis-3 definitions. The 239 critically ill septic patients were classified according to a the preceding and b current sepsis-3 definitions. Soluble endothelial protein C receptor (sEPCR) was quantified in blood samples harvested at ICU admission (within the first 24 h). a Severe sepsis and/or septic shock (SS/SS) was present or subsequently developed in 139 patients (SS/SS-positive patients), while 100 suffered from uncomplicated sepsis and did not develop SS/SS (SS/SS-negative patients). b Patients were also assigned to groups according to their qSOFA score prior to ICU admission. qSOFA 1, N = 7 patients; qSOFA 2, N = 197 patients; qSOFA 3, N = 35 patients. Line in the box, median value; box edges, 25th to 75th centiles; whiskers, range of values. Two-group comparisons were performed by the Mann-Whitney test for skewed data, whereas Kruskal-Wallis ANOVA followed by Dunn’s multiple comparison post hoc test was used to examine differences between more than two groups
Fig. 2.
sEPCR levels and EPCR haplotypes. The 239 critically ill septic patients were grouped according to their genotype combination and presence of haplotype-tagging alleles. Soluble endothelial protein C receptor (sEPCR) was quantified in blood samples harvested at ICU admission (within the first 24 h). a Patients categorized according to their genotype combination. b Patients categorized according to carriage of at least one H3 allele. c Patients categorized according to carriage of at least one H1 allele. Line in the box, median value; box edges, 25th to 75th centiles; whiskers, range of values. a ****p < 0.0001 from the H1 group and #p < 0.05 from the H2 group with Kruskal-Wallis ANOVA followed by Dunn’s multiple comparison post hoc test; b ****p < 0.0001 with the Mann-Whitney test
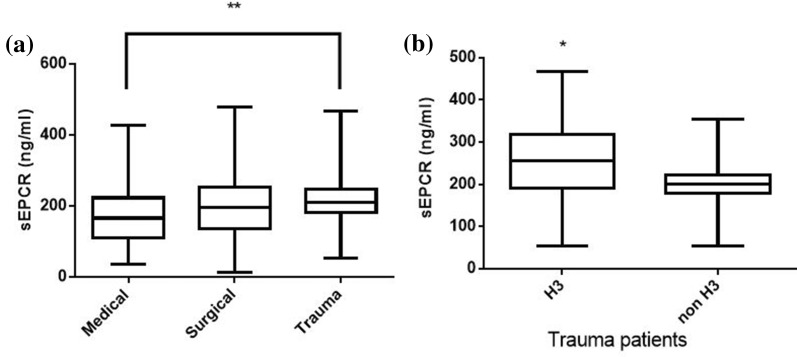
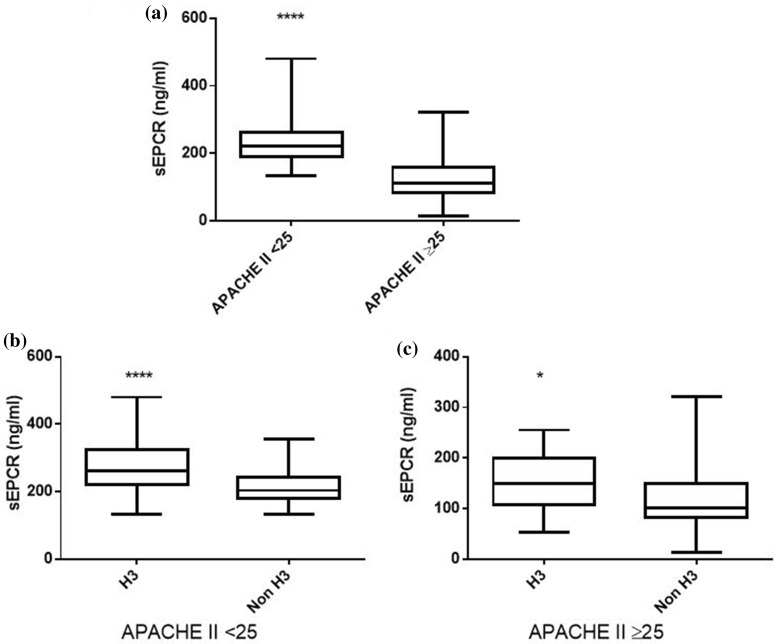
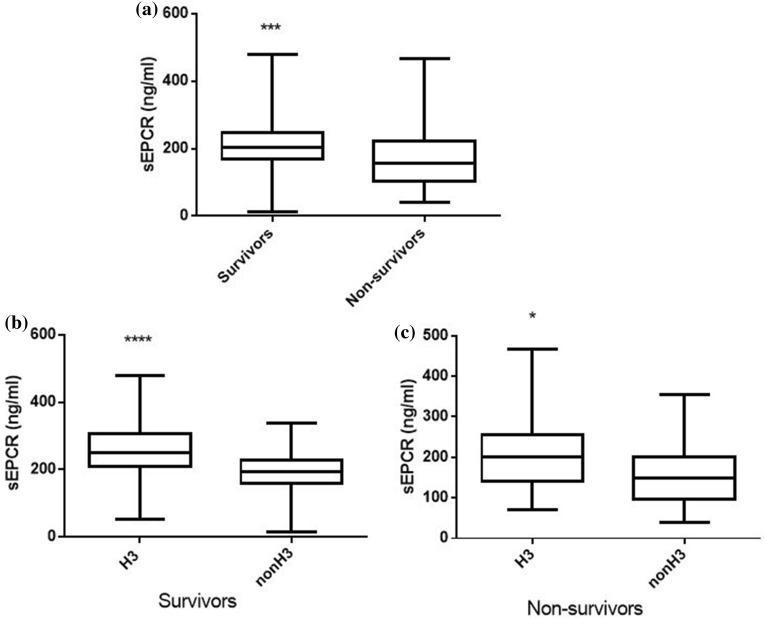
In our patient cohort, sEPCR levels were similar between male and female subjects; however, trauma patients exhibited higher sEPCR levels than patients with medical pathologies (Fig. 3a; p < 0.01). Nonetheless, in the trauma subgroup, H3 carriers had much higher sEPCR levels than non-H3 carriers (Fig. 3b; p < 0.05). Moreover, patients with very high APACHE II score levels (≥ 25) had lower sEPCR levels compared with patients with < 25 APACHE II scores (Fig. 4a; p < 0.0001). When the severely ill patients with an APACHE II score of 25 and over were assigned to groups based on their alleles, H3 carriers had higher sEPCR levels compared with non-H3 carriers (Fig. 4c; p < 0.05). Patients were additionally assigned to groups based on hospital mortality; sEPCR levels at ICU admission were higher in survivors compared with non-survivors (Fig. 5a; p < 0.001). When the survivors and non-survivors were divided according to their alleles, H3 carriers had higher sEPCR levels compared with non-H3 carriers in either case (Fig. 5b, c; p < 0.0001 for survivors and p < 0.05 for non-survivors).
Fig. 3.
sEPCR levels and diagnostic category. Soluble endothelial protein C receptor (sEPCR) was quantified in blood samples harvested at ICU admission (within the first 24 h). a Patients were categorized according to diagnostic category at admission. Line in the box, median value; box edges, 25th to 75th centiles; whiskers, range of values. **p < 0.01 with Kruskal-Wallis ANOVA followed by Dunn’s multiple comparison post hoc test. b Trauma patients (N = 53) were further divided according to the presence of at least one H3 allele. Line in the box, median value; box edges, 25th to 75th centiles; whiskers, range of values. *p < 0.05 with the Mann-Whitney test
Fig. 4.
sEPCR levels and APACHE II score. Soluble endothelial protein C receptor (sEPCR) was quantified in blood samples harvested at ICU admission (within the first 24 h). a Patients were categorized according to APACHE II scores at admission. A cut-off value of 25 was used. b Patients with APACHE II score < 25 (N = 137) and c patients with APACHE II score ≥ 25 (N = 92) were further divided according to the presence of at least one H3 allele. Line in the box, median value; box edges, 25th to 75th centiles; whiskers, range of values. *p < 0.05; ****p < 0.0001 with the Mann-Whitney test
Fig. 5.
sEPCR levels and hospital mortality. Soluble endothelial protein C receptor (sEPCR) was quantified in blood samples harvested at ICU admission (within the first 24 h). a Patients were categorized according to hospital mortality; ICU admission sEPCR levels were compared between survivors (N = 141) and non-survivors (N = 98). b Survivors and c non-survivors were further divided according to the presence of at least one H3 allele. Line in the box, median value; box edges, 25th to 75th centiles; whiskers, range of values. *p < 0.05, ***p < 0.001, ****p < 0.0001 with the Mann-Whitney test
Multivariate logistic regression analysis to identify potential risk factors for elevated sEPCR levels was performed using median sEPCR levels as a cut-off point (≥ 192 ng/ml) and controlling for possible confounding factors, showing that H3 haplotype, trauma and lower APACHE II scores are associated with increased levels of sEPCR (Table 4).
Table 4.
Effect on elevated sEPCR levels of the presence of alleles belonging only to the H1, or simultaneously to both the H1 and H3 haplotypes or all common alleles (H2) compared with the presence of only the H3 haplotype
| Variable | OR | 95% CI | p |
|---|---|---|---|
| Genotype combination | |||
| Only H3 | Reference | ||
| H2 | 0.266 | 0.094–0.754 | 0.013* |
| Only H1 | 0.222 | 0.082–0.600 | 0.003* |
| H1 and H3 | 0.717 | 0.177–2.912 | 0.642 |
| Alleles | |||
| H3 | Reference | ||
| H1 | 0.272 | 0.126–0.587 | 0.001* |
| Diagnosis | |||
| Trauma | Reference | ||
| Medical | 0.366 | 0.149–0.899 | 0.028* |
| Surgical | 0.276 | 0.115–0.665 | 0.004* |
| APACHE II score | 0.865 | 0.825–0.908 | < 0.0001* |
Multivariable logistic regression was performed to investigate associations between risk of high sEPCR levels (≥ 192 ng/ml) and genotypes, mutually adjusted for all other factors in the table. Multiple logistic regression analysis was performed with genotype combination used as key predictor variable (categorical: only H3, H2, only H1 and H1 + H3) and high (≥ 192 ng/ml) versus low (< 192 ng/ml) sEPCR plasma levels as a binary outcome, controlling for APACHE II and SOFA scores, lactate level, length of ICU stay, age (continuous variables), sepsis severity, diagnostic category, hospital mortality, sex and haplotype (categorical variables)
APACHE acute physiology and chronic health evaluation, CI confidence intervals, OR odds ratio, SOFA sequential organ failure assessment
Asterisk indicate p < 0.05 values are statistically significant
Discussion
To the best of our knowledge, this is the first study that investigated the association of the three most common haplotypes in the EPCR gene with the risk of elevated sEPCR levels in ICU septic patients. Our results demonstrated that critically ill septic patients who are carriers of at least one H3 allele have higher sEPCR levels at ICU admission, independent of sepsis severity or risk of poor outcomes, as predicted by their qSOFA score.
Since the question still remains whether sEPCR responds to changes in disease, and plasma sEPCR levels have been shown to be lower in patients with increased disease severity when controlling for rs867186-G variant (H3 haplotype) [29], we reasoned that EPCR mutations are solely responsible for increased sEPCR levels in critically ill septic patients rather than sepsis severity or the risk of poor outcomes. The H3 haplotype has been shown to be a candidate risk factor for deep vein thrombosis, possibly by increasing the levels of sEPCR and promoting the procoagulant effects due to APC sequestration [13, 16, 24]. In a previous investigation the frequency of H3 in critically ill septic patients was not associated with severe sepsis and/or septic shock development [28]. However, it has been shown that deep vein thrombosis patients and critically ill septic patients have a higher H3 haplotype frequency compared with healthy controls [16, 24, 28].
sEPCR increases in response to injury and selective regulatory mechanisms exist upon inflammation. The mechanisms controlling the expression of cell-bound and circulating soluble EPCR are still unclear and include chronic and pathological processes. There is evidence that the H3 haplotype-associated Gly219 involves increased EPCR shedding from the endothelium [30], and Guitton et al. [31] demonstrated the cumulative effect of gender and the H3 haplotype on sEPCR in primary cultures of vascular endothelial cells. In previous studies, sEPCR levels in septic patients were found to be significantly higher [8, 32], unchanged [33] or lower [34] compared with healthy volunteers. This inconsistency among studies could be partially explained by different frequencies of the EPCR H3 haplotype in the various study populations. Our laboratory has shown that sEPCR may act as an early indicator of sepsis prior to its clinical appearance [35], supporting the hypothesis that elevated sEPCR levels are implicated in the pathogenesis of the septic syndrome. The results of the present study indicate that severe sepsis, septic shock, higher qSOFA prior to ICU admission or high APACHE II scores are not associated with increased plasma sEPCR levels. Our results rather indicate that in our critically ill septic patient cohort H3 carriers have higher levels of sEPCR at ICU admission compared with non-H3 carriers. Our results additionally demonstrate that sEPCR levels at admission are not predictive of hospital mortality.
Our study has the following limitations: first, it was a single-center study with a modest sample size. Also, we did not obtain sEPCR measurements prior to sepsis appearance. We also only studied in-hospital mortality and not 30-day mortality. Finally, it was a pilot study, exploratory in nature and observational, not allowing conclusions about causative mechanisms. However, our patients were well characterized and all sEPCR measurements were made in septic patients. Our study was also conducted in a general ICU. We additionally genotyped a small non-septic patient cohort with systemic inflammatory response syndrome (SIRS) [27] and observed that their sEPCR levels are not dependent on haplotype (data not shown).
To our knowledge, this is the first study that demonstrated that in septic patients the EPCR H3 haplotype and sEPCR levels are not associated with sepsis severity or risk of worse outcomes; on the other hand, we were able to show that the presence of at least one H3 allele increases the levels of sEPCR in critically ill septic patients. The biological significance and function of the elevated sEPCR produced in H3 carriers are unknown.
Conclusion
In conclusion, we provide evidence that the EPCR H3 haplotype is strongly associated with sEPCR levels in septic patients. However, we observed no association of the H3 haplotype or greater sEPCR levels with risk of poor outcomes in septic patients.
Acknowledgements
Funding
This work was funded by the nonprofit institute “THORAX” Research Center for Intensive and Emergency Thoracic Medicine, Athens, Greece. No funding or sponsorship was received for the publication of the article.
Medical Writing, Editorial and Other Assistance
The authors would like to thank the nursing staff of the “Evangelismos” Hospital ICU for their assistance.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work as a whole and have given final approval to the version to be published.
Prior Presentation
This manuscript has been previously presented as an oral presentation at the Conference of the Hellenic Sepsis Study Group; Critically Ill Patients: From Translational Research to Clinical Management, 17–18 June 2017, Kalamata, Greece.
Disclosures
Alice G. Vassiliou, Anastasia Kotanidou, Zafeiria Mastora, Carlo Tascini, Gianluigi Cardinali and Stylianos E. Orfanos have nothing to disclose.
Compliance with Ethics Guidelines
The study was approved by the Evangelismos Hospital Research Ethics Committee (76/4-3-2009). All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964, as revised in 2013. Informed consent was obtained from all patients or patients’ next-of-kin for being included in the study.
Data Availability
The data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Footnotes
Enhanced content
To view enhanced content for this article go to 10.6084/m9.figshare.5873853.
References
- 1.Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orfanos SE, Maniatis NA, Kotanidou A. The effects of activated protein C on the septic syndrome. In: Vincent J-L, editor. Yearbook of intensive care and emergency medicine. Berlin: Springer; 2008. pp. 721–729. [Google Scholar]
- 3.Stearns-Kurosawa DJ, Kurosawa S, Mollica JS, Ferrell GL, Esmon CT. The endothelial cell protein C receptor augments protein C activation by the thrombin-thrombomodulin complex. Proc Natl Acad Sci USA. 1996;93(19):10212–10216. doi: 10.1073/pnas.93.19.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feistritzer C, Schuepbach RA, Mosnier LO, et al. Protective signaling by activated protein C is mechanistically linked to protein C activation on endothelial cells. J Biol Chem. 2006;281(29):20077–20084. doi: 10.1074/jbc.M600506200. [DOI] [PubMed] [Google Scholar]
- 5.Isermann B, Vinnikov IA, Madhusudhan T, et al. Activated protein C protects against diabetic nephropathy by inhibiting endothelial and podocyte apoptosis. Nat Med. 2007;13(11):1349–1358. doi: 10.1038/nm1667. [DOI] [PubMed] [Google Scholar]
- 6.Joyce DE, Gelbert L, Ciaccia A, DeHoff B, Grinnell BW. Gene expression profile of antithrombotic protein c defines new mechanisms modulating inflammation and apoptosis. J Biol Chem. 2001;276(14):11199–11203. doi: 10.1074/jbc.C100017200. [DOI] [PubMed] [Google Scholar]
- 7.Riewald M, Petrovan RJ, Donner A, Mueller BM, Ruf W. Activation of endothelial cell protease activated receptor 1 by the protein C pathway. Science. 2002;296(5574):1880–1882. doi: 10.1126/science.1071699. [DOI] [PubMed] [Google Scholar]
- 8.Kurosawa S, Stearns-Kurosawa DJ, Carson CW, D’Angelo A, Della Valle P, Esmon CT. Plasma levels of endothelial cell protein C receptor are elevated in patients with sepsis and systemic lupus erythematosus: lack of correlation with thrombomodulin suggests involvement of different pathological processes. Blood. 1998;91(2):725–727. [PubMed] [Google Scholar]
- 9.Qu D, Wang Y, Esmon NL, Esmon CT. Regulated endothelial protein C receptor shedding is mediated by tumor necrosis factor-alpha converting enzyme/ADAM17. J Thromb Haemost. 2007;5(2):395–402. doi: 10.1111/j.1538-7836.2007.02347.x. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Qu D, Esmon NL, Esmon CT. Metalloproteolytic release of endothelial cell protein C receptor. J Biol Chem. 2000;275(8):6038–6044. doi: 10.1074/jbc.275.8.6038. [DOI] [PubMed] [Google Scholar]
- 11.Saposnik B, Lesteven E, Lokajczyk A, Esmon CT, Aiach M, Gandrille S. Alternative mRNA is favored by the A3 haplotype of the EPCR gene PROCR and generates a novel soluble form of EPCR in plasma. Blood. 2008;111(7):3442–3451. doi: 10.1182/blood-2007-08-104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukudome K, Kurosawa S, Stearns-Kurosawa DJ, He X, Rezaie AR, Esmon CT. The endothelial cell protein C receptor. Cell surface expression and direct ligand binding by the soluble receptor. J Biol Chem. 1996;271(29):17491–17498. doi: 10.1074/jbc.271.29.17491. [DOI] [PubMed] [Google Scholar]
- 13.Liaw PC, Neuenschwander PF, Smirnov MD, Esmon CT. Mechanisms by which soluble endothelial cell protein C receptor modulates protein C and activated protein C function. J Biol Chem. 2000;275(8):5447–5452. doi: 10.1074/jbc.275.8.5447. [DOI] [PubMed] [Google Scholar]
- 14.Regan LM, Stearns-Kurosawa DJ, Kurosawa S, Mollica J, Fukudome K, Esmon CT. The endothelial cell protein C receptor. Inhibition of activated protein C anticoagulant function without modulation of reaction with proteinase inhibitors. J Biol Chem. 1996;271(29):17499–17503. doi: 10.1074/jbc.271.29.17499. [DOI] [PubMed] [Google Scholar]
- 15.Biguzzi E, Merati G, Liaw PC, et al. A 23 bp insertion in the endothelial protein C receptor (EPCR) gene impairs EPCR function. Thromb Haemost. 2001;86(4):945–948. [PubMed] [Google Scholar]
- 16.Saposnik B, Reny JL, Gaussem P, Emmerich J, Aiach M, Gandrille S. A haplotype of the EPCR gene is associated with increased plasma levels of sEPCR and is a candidate risk factor for thrombosis. Blood. 2004;103(4):1311–1318. doi: 10.1182/blood-2003-07-2520. [DOI] [PubMed] [Google Scholar]
- 17.Uitte de Willige S, Van Marion V, Rosendaal FR, Vos HL, de Visser MC, Bertina RM. Haplotypes of the EPCR gene, plasma sEPCR levels and the risk of deep venous thrombosis. J Thromb Haemost. 2004;2(8):1305–1310. doi: 10.1046/j.1538-7836.2004.00855.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen XD, Tian L, Li M, Jin W, Zhang HK, Zheng CF. Relationship between endothelial cell protein C receptor gene 6936A/G polymorphisms and deep venous thrombosis. Chin Med J (Engl). 2011;124(1):72–75. [PubMed] [Google Scholar]
- 19.Galligan L, Powell C, Livingstone W, Mynett-Johnston L, Smith OP. The G7763C endothelial protein C receptor (EPCR) gene mutation: prevalence and association with DVT in the Irish population. Thromb Haemost. 2002;88(1):163–165. [PubMed] [Google Scholar]
- 20.Medina P, Navarro S, Corral J, et al. Endothelial protein C receptor polymorphisms and risk of myocardial infarction. Haematologica. 2008;93(9):1358–1363. doi: 10.3324/haematol.13066. [DOI] [PubMed] [Google Scholar]
- 21.Medina P, Navarro S, Estelles A, Espana F. Polymorphisms in the endothelial protein C receptor gene and thrombophilia. Thromb Haemost. 2007;98(3):564–569. [PubMed] [Google Scholar]
- 22.Medina P, Navarro S, Estelles A, Vaya A, Bertina RM, Espana F. Influence of the 4600A/G and 4678G/C polymorphisms in the endothelial protein C receptor (EPCR) gene on the risk of venous thromboembolism in carriers of factor V Leiden. Thromb Haemost. 2005;94(2):389–394. doi: 10.1160/TH05-02-0089. [DOI] [PubMed] [Google Scholar]
- 23.Medina P, Navarro S, Estelles A, et al. Contribution of polymorphisms in the endothelial protein C receptor gene to soluble endothelial protein C receptor and circulating activated protein C levels, and thrombotic risk. Thromb Haemost. 2004;91(5):905–911. doi: 10.1160/TH03-10-0657. [DOI] [PubMed] [Google Scholar]
- 24.Navarro S, Medina P, Mira Y, et al. Haplotypes of the EPCR gene, prothrombin levels, and the risk of venous thrombosis in carriers of the prothrombin G20210A mutation. Haematologica. 2008;93(6):885–891. doi: 10.3324/haematol.12448. [DOI] [PubMed] [Google Scholar]
- 25.Vassiliou A, Orfanos S, Kotanidou A. Clinical assays in sepsis: prognosis, diagnosis, outcomes, and the genetic basis of sepsis. In: Kumar V, editor. Sepsis. Rijeka: Intech; 2017. pp. 93–129. [Google Scholar]
- 26.Liang Y, Huang X, Jiang Y, et al. Endothelial protein C receptor polymorphisms and risk of sepsis in a Chinese population. J Int Med Res. 2017;45(2):504–513. doi: 10.1177/0300060516686496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39(2):165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vassiliou AG, Maniatis NA, Kotanidou A, et al. Endothelial protein C receptor polymorphisms and risk of severe sepsis in critically ill patients. Intensive Care Med. 2013;39(10):1752–1759. doi: 10.1007/s00134-013-3018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shabani E, Opoka RO, Bangirana P, et al. The endothelial protein C receptor rs867186-GG genotype is associated with increased soluble EPCR and could mediate protection against severe malaria. Sci Rep. 2016;6:27084. doi: 10.1038/srep27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu D, Wang Y, Song Y, Esmon NL, Esmon CT. The Ser219→Gly dimorphism of the endothelial protein C receptor contributes to the higher soluble protein levels observed in individuals with the A3 haplotype. J Thromb Haemost. 2006;4(1):229–235. doi: 10.1111/j.1538-7836.2005.01676.x. [DOI] [PubMed] [Google Scholar]
- 31.Guitton C, Gerard N, Quillard T, Charreau B. Circulating endothelial cell protein C receptor: endothelial regulation and cumulative impact of gender and A3 haplotype. J Vasc Res. 2011;48(4):336–346. doi: 10.1159/000322582. [DOI] [PubMed] [Google Scholar]
- 32.Liaw PC, Esmon CT, Kahnamoui K, et al. Patients with severe sepsis vary markedly in their ability to generate activated protein C. Blood. 2004;104(13):3958–3964. doi: 10.1182/blood-2004-03-1203. [DOI] [PubMed] [Google Scholar]
- 33.Faust SN, Levin M, Harrison OB, et al. Dysfunction of endothelial protein C activation in severe meningococcal sepsis. N Engl J Med. 2001;345(6):408–416. doi: 10.1056/NEJM200108093450603. [DOI] [PubMed] [Google Scholar]
- 34.Borgel D, Bornstain C, Reitsma PH, et al. A comparative study of the protein C pathway in septic and nonseptic patients with organ failure. Am J Respir Crit Care Med. 2007;176(9):878–885. doi: 10.1164/rccm.200611-1692OC. [DOI] [PubMed] [Google Scholar]
- 35.Vassiliou AG, Kotanidou A, Mastora Z, et al. Elevated soluble endothelial protein C receptor levels at ICU admission are associated with sepsis development. Minerva Anestesiol. 2015;81(2):125–134. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.