Abstract
Functional polymorphisms in Linc-ROR may change its ability of regulation by regulating Linc-ROR expression. However, these functional polymorphisms in Linc-ROR and their associations with breast cancer (BC) susceptibility were scarcely reported. In this molecular epidemiological study, four SNPs (rs6420545, rs4801078, rs1942348 and rs9636089) were selected in Linc-ROR by bioinformatics method. Unconditional logistic regression model was performed to analyze the associations between four SNPs and BC susceptibility adjusted for reproductive factors. Quantitative real-time (qRT) PCR was used to evaluate relative expression of Linc-ROR in plasma. The interactions of gene reproductive factors were assessed by Multifactor Dimensionality Reduction (MDR) method. A novel finding showed TT (OR: 1.79; 95%CI: 1.20–2.68) genotype of rs4801078 in Linc-ROR had a significant association with the higher risk of BC and the expression of Linc-ROR mRNA was closely related with the alleles of rs4801078. In addition, we found the interaction of rs4801078, number of pregnancy and menopausal status might increase BC risk (OR: 2.78; 95%CI: 2.74–3.61). Our results suggest that interactions of SNPs in Linc-ROR and reproductive factors might contribute to BC risk, and alleles of rs4801078 might affect Linc-ROR expression level.
Introduction
Breast cancer is the most common diagnosed cancer and the leading cause of cancer death in Chinese women1. It alone is expected to account for fifteen percent of all new cancers in women with about 4292,000 newly diagnosed invasive cancer cases in 2015 in China2.
Noncoding RNAs (ncRNAs) which function by means other than directing the production of proteins were a distinguishing feature of metazoan genomes3. Numerous studies have underlined the regulatory role of microRNAs (miRNAs) in the development of cancers, and their variants are reported to be related to various cancer4–6. LncRNAs ranging from 200 nucleotides (nt) to over 10 kb are spliced, polyadenylated, and are roughly as diverse in a given cell type as protein-coding mRNAs7–10. In addition to the small regulatory RNAs, emerging studies indicate that lncRNAs play critical roles in various biological processes ranging from embryonic development to human diseases, including controlling cell cycle progression, apoptosis, invasion, and migration11. The aberrant expressions of several lncRNAs in various cancers indicate that lncRNAs may be play roles in tumor carcinogenesis12–14. Moreover, recent studies have shownthe important roles of lncRNAs and their genetic variants played in tumor carcinogenesis. Yan et al. found that TC genotype of rs10463297 in lncRNA SRA could increase BC risk compared with CC genotype15. Reactivation of the H19 gene has been observed in bladder tumors, and TC genotype of rs2839698 in H19 was found to decrease bladder cancer risk16. Yao et al. suggested that individuals with rs7958904 CC genotype in HOTAIR had asignificantlydecreasedriskofcolorectalcancer17.
Large intergenic noncoding RNA regulator of reprogramming (lincRNA-ROR, Linc-ROR), at 2.6 kb in length, was first identified as a regulator for reprogramming of differentiated cells to induced pluripotent stem cells (iPSCs)18. Linc-ROR also functions as a microRNA sponge to prevent the core transcription factors (TFs) Oct4, Nanog, and Sox2 from miRNA-mediated suppression in self-renewing human embryonic stem cells (ESCs)19 and studies indicated that Linc-ROR could act as a p53 repressor in response to DNAdamage20. All the evidence intrigued us to speculate that Linc-ROR might also have a role in cancer progression and we found several studies have focused the role of Linc-ROR in the development of cancers, including breast cancer21–24.
Reproductive factors are closely related to the development of breast cancer and the research on interaction between reproductive factors and susceptibility gene has been done25. However, up to now, research combining the SNPs in Linc-ROR and reproductive factors on BC risk has not been reported. In our current case-control study, we selected four tagger SNPs (rs6420545, rs4801078, rs1942348 and rs9636089) in Linc-ROR by bioinformatics method and explored a potential correlation between those potentially functional polymorphisms and risk of BC in Chinese women. Furthermore, analysis of qRT - PCR was applied to detect the relative expression level of Linc-ROR in plasma and the interactions of gene reproductive factors were analyzed using MDR method.
Results
Characteristics of patients and controls
The demographic and clinical characteristics of 968 subjects were displayed in Table 1. The mean age of patients and matched controls were 48.41 ± 10.21 and 49.23 ± 10.15 respectively (P = 0.21). The mean age at menarche of BC patients (14.48 ± 1.82) was significantly higher than mean age at menarche of controls (14.08 ± 1.78). More than 4 pregnancies for women increased the risk of BC (OR: 2.01, 95%CI: 1.32–3.06). However, there is no difference in the distributions of the other characteristics.
Table 1.
Thebaseline characteristics of total 484 BC patients and 484 cancer-free controls.
| Variables | Cases (%) | Controls (%) | P b | OR(95%CI) |
|---|---|---|---|---|
| n = 484 | n = 484 | |||
| Age (Mean ± SD) | 48.41 ± 10.21 | 49.23 ± 10.15 | 0.211a | |
| Age at menarche (Mean ± SD) | 14.48 ± 1.82 | 14.08 ± 1.78 | 0.001a | |
| Age at menopause | ||||
| ≤50 | 352 (0.73) | 337 (0.70) | 1 | |
| >50 | 132 (0.27) | 147 (0.30) | 0.287 | 0.86 (0.65–1.14) |
| Menopausal state | ||||
| Pre-menopausal | 291 (0.60) | 283 (0.59) | 1 | |
| Post-menopausal | 193 (0.40) | 201 (0.41) | 0.601 | 0.93 (0.72–1.21) |
| No. of abortions | ||||
| ≤1 | 336 (0.69) | 330 (0.68) | 1 | |
| 2~3 | 123 (0.25) | 138 (0.29) | 0.382 | 0.87 (0.65–1.18) |
| ≥4 | 25 (0.06) | 16 (0.03) | 0.189 | 1.59 (0.80–3.16) |
| No. of pregnancy | ||||
| ≤1 | 60 (0.12) | 87 (0.18) | 1 | |
| 2 | 116 (0.24) | 156 (0.33) | 0.727 | 1.08 (0.70–1.66) |
| 3 | 132 (0.27) | 114 (0.24) | 0.019 | 1.68 (1.09–2.60) |
| ≥4 | 176 (0.37) | 127 (0.26) | 0.001 | 2.01 (1.32–3.06) |
| Breast-feeding | ||||
| No | 27 (0.06) | 42 (0.09) | 1 | |
| Yes | 457 (0.94) | 442 (0.91) | 0.063 | 1.61 (0.98–2.65) |
| Family history | ||||
| No | 462 (0.95) | 463 (0.96) | 1 | |
| Yes | 22 (0.05) | 21 (0.04) | 0.876 | 1.05 (0.57–1.94) |
| ER | ||||
| Negative | 149 (0.31) | |||
| Positive | 293 (0.61) | |||
| PR | ||||
| Negative | 188 (0.39) | |||
| Positive | 253 (0.52) | |||
| HER-2 | ||||
| Negative | 120 (0.25) | |||
| Positive | 308 (0.64) | |||
aStudent’s t test.
bTwo-sided χ2 test, P < 0.05 was considered to be statistically significant.
The SNPs genotypes were associated with BC
Table 2 showed the distributions of genotypes in BC patients and control group for four selected SNPs. All of SNPs genotypes in control group meet Hardy-Weinberg equilibrium (P = 0.99 for rs1942348, 0.77 for rs4801078, 0.94 for rs6420545 and 0.99 for rs9636089). TT (OR: 1.79; 95%CI: 1.20–2.68) genotype of rs4801078in Linc-ROR increased BC risk incodominant model. Also, TT (OR: 1.84, 95%CI: 1.28–2.66) genotype of rs4801078 in Linc-ROR sho wed increased risk of BC in recessive model.
Table 2.
The association between four SNPs genotypes and risk of breast cancer.
| SNPs | Genetic model | Genotype | Case (%) | Control (%) | P a | Adjusted OR(95%CI)b | P b |
|---|---|---|---|---|---|---|---|
| n = 484 | n = 484 | ||||||
| rs1942348 | 0.99 | ||||||
| Codominant | TT | 184(0.38) | 183(0.38) | 1 | |||
| CT | 227(0.47) | 227(0.47) | 0.91 (0.67–1.23) | 0.529 | |||
| CC | 73(0.15) | 74(0.15) | 0.99 (0.65–1.51) | 0.959 | |||
| Dominant | TT | 1 | |||||
| CT + CC | 300(0.62) | 301(0.62) | 0.93 (0.70–1.23) | 0.602 | |||
| Recessive | TT + TC | 411(0.85) | 410(0.85) | 1 | |||
| CC | 1.04 (0.71–1.54) | 0.827 | |||||
| Over-dominant | TT + CC | 257(0.53) | 257(0.53) | 1 | |||
| TC | 0.91 (0.69–1.20) | 0.507 | |||||
| Allele | T | 595(0.61) | 593(0.61) | 1 | 0.981 | ||
| C | 373(0.39) | 375(0.39) | 1.01(0.83–1.20) | ||||
| rs4801078 | 0.77 | ||||||
| Codominant | CC | 162(0.33) | 176(0.36) | 1 | |||
| CT | 211(0.44) | 238(0.49) | 0.95 (0.70–1.30) | 0.740 | |||
| TT | 111(0.23) | 70(0.15) | 1.79(1.20–2.68) | 0.005 | |||
| Dominant | CC | 1 | |||||
| CT + TT | 322(0.67) | 308(0.64) | 1.14 (0. 85–1.52) | 0.386 | |||
| Recessive | CC + CT | 373(0.77) | 414(0.85) | 1 | |||
| TT | 1.85 (1.28–2.66) | 0.001 | |||||
| Over-dominant | CC + TT | 273(0.56) | 246(0.51) | 1 | |||
| CT | 0.78 (0.59–1.02) | 0.078 | |||||
| Allele | C | 535(0.55) | 590(0.61) | 1 | 0.022 | ||
| T | 433(0.45) | 378(0.39) | 1.24(1.03–1.48) | ||||
| rs6420545 | 0.94 | ||||||
| Codominant | CC | 126(0.26) | 120(0.25) | 1 | |||
| CT | 228(0.47) | 236(0.49) | 0.88(0.63–1.24) | 0.462 | |||
| TT | 130(0.27) | 128(0.26) | 1.05 (0.72–1.54) | 0.794 | |||
| Dominant | CC | 1 | |||||
| CT + TT | 358(0.74) | 364(0.75) | 0.94(0.68–1.29) | 0.695 | |||
| Recessive | CC + CT | 354(0.73) | 356 (0.74) | 1 | |||
| TT | 1.14 (0.83–1.57) | 0.408 | |||||
| Over-dominant | CC + TT | 256(0.53) | 248 (0.51) | 1 | |||
| CT | 0.86 (0.65–1.13) | 0.282 | |||||
| Allele | C | 480(0.50) | 476(0.49) | 1 | 0.518 | ||
| T | 488(0.50) | 492(0.51) | 0.94(0.79–1.23) | ||||
| rs9636089 | 0.99 | ||||||
| Codominant | TT | 187(0.39) | 191(0.39) | 1 | |||
| CT | 223(0.46) | 226(0.47) | 0.90(0.66–1.21) | 0.475 | |||
| CC | 74(0.15) | 67(0.14) | 1.03 (0.67–1.58) | 0.896 | |||
| Dominant | TT | 1 | |||||
| CT + CC | 297(0.61) | 293(0.61) | 0.93 (0.70–1.23) | 0.596 | |||
| Recessive | TT + TC | 410(0.85) | 417(0.86) | 1 | |||
| CC | 1.09(0.74–1.62) | 0.661 | |||||
| Over-dominant | TT + CC | 261(0.54) | 258(0.53) | 1 | |||
| TC | 0.89 (0.67–1.18) | 0.408 | |||||
| Allele | T | 597(0.62) | 608(0.63) | 1 | 0.680 | ||
| C | 371(0.38) | 360(0.37) | 1.04(0.86–1.25) |
aPvalue of Hardy-Weinberg equilibrium in controls;
bP value of logistic regression analysis with adjusted for age, age at menarche, menopausal status, number of pregnancy and abortion, breast-feeding status, and family history of BC in first-degree relatives.
Stratified analysis
We evaluated the relationship of SNPs genotypes and BC risk stratified by the reproductive factors (Table 3). The positive effect of rs6420545 CT + TT genotype was more significant in the subjects (P = 0.005) with age at menarche ≤13 (OR: 0.95, 95%CI: 0.92–0.99) and rs19 CT + TT genotype were more evident in the subjects (P = 0.045) with age >50 (OR: 0.60, 95%CI: 0.37–0.99). The protective role of CT + CC genotypes for rs9636089 were more obvious in the women (P = 0.021) with age > 50 (OR: 0.57, 95%CI: 0.35–0.92).In addition, the CT + TT genotypes of rs4801078revealed a significant higher risk of BC in the participant with age >50 (OR: 2.14, 95%CI: 1.33–3.45) and age of menarche >13 (OR: 1.52, 95%CI: 1.06–2.19).
Table 3.
Stratification analysis of the fiveSNPs and BC susceptibility.
| variables | case | control | rs6420545 | rs4801078 | rs1942348 | rs9636089 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) a | P a | OR (95%CI) a | P a | OR (95%CI) a | P a | OR (95%CI) a | P a | |||
| TT vs CT + CC | CC vs CT + TT | CC vs CT + TT | TT vs CT + CC | |||||||
| age | ||||||||||
| ≤50 | 302 | 285 | 0.70 (0.46–1.05) | 0.083 | 0.79(0.54–1.14) | 0.206 | 1.16(0.81–1.67) | 0.41 | 1.24 (0.87–1.78) | 0.238 |
| >50 | 182 | 199 | 1.50(0.90–2.52) | 0.125 | 2.14 (1.33–3.45) | 0.002 | 0.60(0.37–0.99) | 0.045 | 0.57(0.35–0.92) | 0.021 |
| Age at menarche | ||||||||||
| ≤13 | 152 | 201 | 0.95 (0.92–0.99) | 0.005 | 0.69(0.42–1.13) | 0.14 | 1.38(0.85–2.26) | 0.196 | 1.19(0.74–1.93) | 0.476 |
| >13 | 332 | 283 | 1.46 (0.98–2.16) | 0.061 | 1.52(1.06–2.19) | 0.025 | 0.77(0.54–1.11) | 0.162 | 0.83(0.58–1.19) | 0.31 |
| Age at menopause | ||||||||||
| ≤50 | 352 | 337 | 0.91(0.64–1.32) | 0.630 | 1.16 (0.83–1.62) | 0.4 | 1.01(0.72-0.1.42) | 0.946 | 1.03(0.74–1.44) | 0.85 |
| >50 | 132 | 147 | 1.13 (0.58–2.19) | 0.726 | 1.13(0.62–2.06) | 0.68 | 0.67(0.38–1.20) | 0.18 | 0.66 (0.37–1.16) | 0.147 |
| No. of pregnancy | ||||||||||
| ≤2 | 176 | 243 | 0.98 (0.58–1.66) | 0.95 | 1.32 (0.82–2.12) | 0.155 | 0.96(0.61–1.52) | 0.866 | 0.86 (0.55–1.35) | 0.512 |
| >2 | 308 | 241 | 0.95(0.63–1.43) | 0.807 | 1.08 (0.74–1.59) | 0.679 | 0.89(0.61–1.31) | 0.577 | 0.96(0.66–1.41) | 0.848 |
| No. of abortion | ||||||||||
| ≤2 | 417 | 438 | 0.95(0.68–1.33) | 0.776 | 1.16 (0.86–1.57) | 0.339 | 0.91(0.68–1.24) | 0.559 | 0.93(0.69–1.25) | 0.622 |
| >2 | 67 | 46 | 1.10(0.44–2.73) | 0.843 | 1.13(0.46–2.77) | 0.784 | 0.86(0.37–2.02) | 0.729 | 0.84(0.36–1.98) | 0.691 |
| Breast-feeding | ||||||||||
| no | 27 | 42 | 46.67 (2.46–884.21) | 0.010 | 18.72(1.17–300.7) | 0.039 | 0.56(0.16–1.96) | 0.366 | 0.60(0.17–2.11) | 0.422 |
| yes | 457 | 442 | 0.89 (0.64–1.23) | 0.473 | 1.11 (0.82–1.49) | 0.513 | 0.95(0.71–1.28) | 0.735 | 0.94(0.70–1.27) | 0.696 |
| Family history | ||||||||||
| no | 462 | 463 | 1.01(0.73–1.40) | 0.947 | 1.21(0.90–1.64) | 0.212 | 0.92(0.69–1.24) | 0.599 | 0.90 (0.67–1.21) | 0.473 |
| yes | 22 | 21 | 0.29(0.05–1.55) | 0.147 | 0.52(0.10–2.80) | 0.448 | 0.65(0.14–2.92) | 0.573 | 1.33 (0.31–5.77) | 0.701 |
aPvalue of logistic regression analysis with adjusted for age, age at menarche, menopausal status, number of pregnancy and abortion, breast-feeding status, family history of BC in first-degree relatives(the stratified factor in each stratum excluded).
The association of receptor status and the four SNPs genotypes
The association between SNPs in Linc-ROR and the receptors status in cases were displayed in Table 4. After adjusted for reproductive factors, only a boundary significant association between rs6420545 CT (OR: 0.66; 95%CI: 0.99–2.76) genotype with PR status of patients was detected.
Table 4.
The Associations between four SNPs and ER, PR and HER-2 Status of Breast Cancer Patients.
| genotypes | ER | P a | OR (95%CI)a | PR | P a | OR (95%CI)a | Her-2 | P a | OR (95%CI)a | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative (n = 149) | Positive (n = 293) | Negative (n = 188) | Positive (n = 253) | Negative (n = 120) | Positive (n = 308) | |||||||
| rs6420545 | ||||||||||||
| CC | 40 | 78 | 1 | 55 | 63 | 1 | 36 | 77 | 1 | |||
| CT | 64 | 137 | 0.389 | 1.26 (0.75–2.13) | 75 | 125 | 0.051 | 0.66 (0.99–2.76) | 53 | 144 | 0.402 | 1.27(0.73–2.22) |
| TT | 45 | 78 | 0.826 | 0.94 (0.53–1.65) | 58 | 65 | 0.957 | 0.99(0.57–1.70) | 31 | 87 | 0.313 | 1.38(0.74–2.58) |
| rs4801078 | ||||||||||||
| CC | 54 | 110 | 1 | 71 | 82 | 1 | 45 | 102 | 1 | |||
| CT | 61 | 123 | 0.770 | 1.08 (0.66–1.77) | 75 | 109 | 0.145 | 1.43 (0.88–2.04) | 46 | 134 | 0.478 | 1.22 (0.71–2.09) |
| TT | 34 | 70 | 0.774 | 1.09(0.62–1.92) | 42 | 62 | 0.370 | 1.28(0.74–2.21) | 29 | 72 | 0.713 | 1.12(0.61–2.07) |
| rs1942348 | ||||||||||||
| TT | 59 | 110 | 1 | 75 | 94 | 1 | 48 | 117 | 1 | |||
| CT | 64 | 140 | 0.361 | 1.25 (0.78–2.01) | 78 | 125 | 0.183 | 1.37 (0.86–2.16) | 55 | 144 | 0.789 | 0.93 (0.56–1.57) |
| CC | 26 | 43 | 0.761 | 0.91 (0.49–1.69) | 35 | 34 | 0.344 | 0.75 (0.41–1.36) | 17 | 47 | 0.723 | 0.88(0.45–1.75) |
| rs9636089 | ||||||||||||
| TT | 61 | 113 | 1 | 76 | 98 | 1 | 48 | 118 | 1 | |||
| CT | 63 | 137 | 0.301 | 1.28(0.80–2.05) | 78 | 121 | 0.276 | 1.28(0.82–2.02) | 55 | 142 | 0.936 | 0.98(0.59–1.63) |
| CC | 25 | 43 | 0.932 | 1.03(0.55–1.92) | 34 | 34 | 0.475 | 0.80(0.44–1.47) | 17 | 48 | 0.935 | 1.03(0.51–2.06) |
aP value of logistic regression analysis with adjusted for age, age at menarche, menopausal status, number of pregnancy and abortion, breast-feeding status, family history of BC in first-degree relatives.
Haplotype analysis
The Haplotype analysis for polymorphisms in Linc-ROR was showed in Table 5.Four haplotypes were showed in the table (a total of 16 haplotypes) and all those frequencies < 0.03 in cases or controls has been dropped in this analysis. Trs6420545Trs4801078Trs1942348Trs9636089 was the most frequent haplotype in the cases (38.5%) and controls (35.3%). Compared with the controls, the frequency of haplotype Trs6420545Crs4801078Trs1942348Trs9636089 was lower in cases (OR: 0.72, 95%CI: 0.54–0.97).
Table 5.
Haplotype analysis of four SNPs in Linc-ROR.
| Haplotypea | Cases (%) | Controls (%) | χ 2 | P | OR (95%CI) |
|---|---|---|---|---|---|
| CCCC | 318.36(0.33) | 328.80(0.34) | 0.028 | 0.867 | 0.98(0.81–1.19) |
| CCTT | 96.25(0.10) | 103.46(0.11) | 0.143 | 0.705 | 0.95(0.70–1.27) |
| TCTT | 87.43(0.09) | 119.52(0.12) | 4.87 | 0.030 | 0.72(0.54–0.97) |
| TTTT | 372.34(0.39) | 341.89(0.35) | 3.44 | 0.060 | 1.20(0.99–1.45) |
aSNPs sequence: rs6420545, rs4801078, rs1942348 and rs9636089.
The others haplotypes were ignored in analysis for the frequencies < 0.03.
The analysis of Gene reproductive factors interactions
The interactions of gene and reproductive factors among four SNPs (rs6420545, rs4801078, rs1942348 and rs9636089) and reproductive factors were revealed in Table 6. Four models in MDR were showed and we found the interaction of rs4801078, number of pregnancy and menopausal status (Trs4801078, number of pregnancy ≥2 and post-menopausal) might increase risk for breast cancer by 2.78 times (OR: 2.78; 95%CI: 2.74–3.61).
Table 6.
Interaction results between the SNPs and reproductive factors by MDR.
| Model | TBAa | CVCb | χ 2 | P | OR(95%CI) |
|---|---|---|---|---|---|
| number of pregnancy | 0.52 | 10/10 | 20.03 | <0.01 | 1.81(1.39–2.35) |
| number of pregnancy, menopausal status | 0.61 | 10/10 | 51.83 | <0.01 | 2.57(1.98–3.33) |
| rs4801078, number of pregnancy, menopausal status | 0.62 | 5/10 | 60.56 | <0.01 | 2.78(2.74–3.61) |
| rs1942348, age at menarche, number of pregnancy | 0.65 | 3/10 | 90.91 | <0.01 | 3.73(2.82–4.92) |
aTesting balance accuracy.
bCross-validation consistency.
The results of Benjamini-Hochberg (BH) correction for false discovery rate (FDR)
We applied the BH correction to control FDR (Table 7). The q-value indicated TT genotype and T allele of rs4801078in Linc-ROR could still increase BC risk. Besides, the relationship between CT + TT genotypes of rs4801078and BC was still noteworthy in the participant with age >50.
Table 7.
Results of Benjamini-Hochberg (BH) correction.
| Genotype | Stratified factors | Adjusted P | q-value | |
|---|---|---|---|---|
| rs4801078 | CC/TT | all subjects | 0.005 | 0.015 |
| CC + CT/TT | all subjects | 0.001 | 0.006 | |
| C/T | all subjects | 0.022 | 0.044 | |
| rs6420545 | TT/CT + CC | age at menarche ≤ 13 | 0.005 | 0.070 |
| TT/CT + CC | no Breast-feeding | 0.010 | 0.070 | |
| rs4801078 | CC/CT + TT | age >50 | 0.002 | 0.028 |
| CC/CT + TT | age at menarche >13 | 0.025 | 0.175 | |
| CC/CT + TT | no Breast-feeding | 0.039 | 0.182 | |
| rs1942348 | CC/CT + TT | age >50 | 0.045 | 0.630 |
| rs9636089 | TT/CT + CC | age >50 | 0.021 | 0.294 |
The relative expression of Linc-ROR in plasma
We investigated the association between rs4801078 genotypes and Linc-ROR mRNA expression level by the real-time PCR amplification reactions in 150 subjects. There were 38 subjects with CC genotype, 64 with CT genotype and 48 with TT genotype (Fig. 1). The relative expression of Linc-ROR mRNA in CT + TT group (1.94 ± 0.27) was significantly higher than the CC group (1.21 ± 0.19) and that indicated the SNPs in Linc-ROR might play a role in the expression level of Linc-ROR mRNA.
Figure 1.
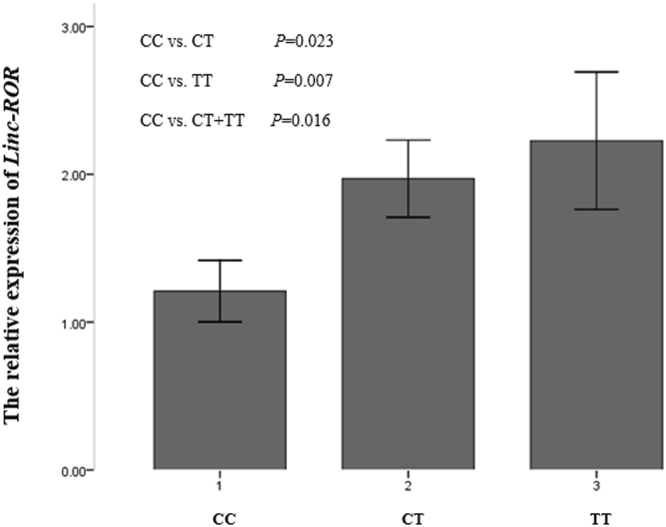
The relative expression of linc-ROR in plasma Note: linc-ROR expression was assessed by qRT-PCR in plasma. Data was evaluated statistically usingthe one-way ANOVA with the Tukey method and represent the mean ± SD from the experiments in triplicate.
Discussion
This case-control study was the first to demonstrate the association between potential regulatory variants in Linc-ROR and BC risk, and we found TT (OR: 1.79; 95%CI: 1.195–2.68) genotype of rs4801078 in Linc-ROR had a significant association with the higher risk of BC in Chinese population. Furthermore, analysis of qRT-PCR showed the relative expression of Linc-ROR mRNA in rs4801078CT + TT group (1.94 ± 0.27) was higher than the CC group (1.21 ± 0.19). In addition, we found the frequency of haplotype Trs6420545Crs4801078Trs1942348Trs9636089was lower in cases than in controls (OR: 0.72, 95%CI: 0.54–0.97) and the interactions of rs4801078, number of pregnancy and menopausal status might increase BC risk (OR: 2.78; 95%CI: 2.74–3.61).
LncRNAs have received widespread attention and are observed to play pivotal roles in tumorigenesis and progression of human cancers. It has already been revealed that some lncRNAs, such as HOTAIR, H19 and MALAT1, are potential biomarkers in cancer diagnosis and prognosis. Among them, Linc-ROR, first discovered in 2010, are also found to have strong association with tumorigenesis, metastasis and poor therapeutic response of malignant tumors26–28. Recent studies found that Linc-ROR was upregulated in pancreatic cancer tissues and decreased Linc-ROR expression could inhibit pancreatic cancer cell proliferation, invasion, and tumourigenicity22,29,30. In another study, the researchers found Linc-ROR confers gemcitabine resistance to pancreatic cancer cells at least partly via inducing autophagy31. Zhou et al. found that Linc-ROR had an important role during endometrial carcinogenesis by acting as a miR-145 “sponge” to inhibit mediation of the differentiation of endometrial tumorspheres32. A recent study suggested the function of Linc-ROR exerted in LAD cells depended on the sponging of miR-145 and it led to the chemotherapy resistance and EMT phenotypes of docetaxel-resistant LAD cells33. The qRT-PCR showed a significant up-regulation of Linc-ROR and its variants 2 (P = 0.025) and 4 (P = 0.0002) in esophageal squamous cell carcinoma34. Li et al. found that Linc-ROR was significantly upregulated in nasopharyngeal carcinoma tissues and the enrichment of Linc-ROR played acritical functional role in chemoresistance by suppressing P53 signal pathway35. A recent study also provided several new mechanistic insights into acquired chemoresistance in HCC and they found Linc-ROR acting as mediators are involved in modulation of cellular responses to chemotherapy36. However, the role of Linc-ROR in glioma is the opposite of other tumors. Feng et al. suggested that Linc-ROR might act as a novel tumor suppressor gene in glioma by inhibiting the proliferation of cancer cell, self-renewal of GSCs and the KLF4 expression37.
In addition to the mentioned malignant tumor, the role of Linc-ROR in breast cancer has also been reported. In 2014, Hou et al. found that Linc-ROR could function as an important regulator of epithelial-to-mesenchymal transition and promote breast cancer progression and metastasis through regulation of miRNAs21. In 2016, the study of Chen et al. investigated the role of Linc-ROR in the chemotherapy resistance of human BC cells and its mechanism23. The effect of the Linc-ROR on epithelial-to-mesenchymal transition was verified to contribute to the chemotherapy resistance and invasion of breast cancer cells23. In the same year, One study concluded that Linc-ROR suppressed gemcitabine-induced autophagy and apoptosis in breast cancer cells by silencing miR-34a expression38. Recently, Zhang et al. found down-regulated Linc-ROR could enhance the sensibility of breast cancer cells to tamoxifen by increasing miR-205 expression and suppressing the expressions of ZEB1 and ZEB239. Zhao et al. suggested the expression levels of Linc-ROR were significantly higher in breast cancer and combination of the Linc-ROR with the conventional biomarkers might produce better diagnostic ability40. Together, these results indicate that Linc-ROR might have a crucial impact on the development of BC and it is necessary to investigate the association between regulatory variants in Linc-ROR and BC.
In recent years, a large amount of SNPs in lncRNAs have been found to be related to carcinogenesis41. For example, genetic variants in MALAT1A were suggested to be associated with breast cancer42 and colorectal cancer risk27. A combined analysis of genome-wide association study (GWAS) and meta-analysis identified a novel and significant association between rs16941835 in lncRNARP11-58A18.1 and CRC susceptibility43. Not only that, but those SNPs in lncRNAs were implied to have function in regulating the expression level of lncRNAs in the process of cancer. A genome-wide association study indicated that variant SNPs in a long noncoding RNA MIR2052HG showed a dose-dependent increase in MIR2052HG expression as well as increased binding of ERα to the EREs by performing estradiol treatment44. Another research showed the rs2147578 in lnc-LAMC2–1:1 were significantly associated with increased CRC risk by influencing the binding of lnc-LAMC2–1:1 and miR-128–3p45. In addition, the latest findings were reported that the SNP rs2027701 of Linc-ROR in the lncRNA-p53 regulatory network had significant associations with the risk of neutropaenia42. These results inspire us to study the role of Linc-ROR tagger SNPs in the expression of Linc-ROR mRNA and the the process of breast cancer.
However, to date, no study about the tagger SNPs of Linc-ROR has ever been reported in BC. In our case-control study, we found TT (OR: 1.79; 95%CI: 1.20–2.68) genotype of rs4801078 in Linc-ROR had a significant association with the higher BC risk in codominant model (OR: 1.79; 95%CI: 1.20–2.68) and recessive model (OR: 1.80, 95%CI: 1.28–2.66). Moreover, the expression of Linc-ROR mRNA was closely related with the alleles of rs4801078. The results of qRT-PCR revealed the relative expression of Linc-ROR mRNA in rs4801078 CT + TT group (1.94 ± 0.27) was significantly higher than the CC group (1.21 ± 0.19). No meaningful association between the other three SNPs (rs1942348, rs6420545 and rs9636089) and the risk of BC was observed in the major models, however, women with the mutant alleles of rs1942348, rs6420545 and rs9636089 had a lower BC risk in the subgrounps with age at menarche ≤ 13 (OR: 0.95, 95%CI: 0.92–0.99 for rs6420545) and age >50 (OR: 0.60, 95%CI: 0.37–0.99 for rs1942348; OR: 0.57, 95%CI: 0.35–0.92 for rs9636089). In addition, the Haplotype analysis showed haplotype Trs6420545Crs4801078Trs1942348Trs9636089 could decrease BC risk (OR: 0.72, 95%CI: 0.54–0.97). Our results suggest that the regulatory polymorphisms in Linc-ROR might influence breast cancer risk and large sample studies carrying in other races are needed to verify our discovery.
BC is a complex disease and epidemiological studies suggest that the occurrence of BC is usually due to the gene-environmental factors interaction46. The reproductive factors have been proved to be related with BC, including menarche age, menopausal state, number of pregnancy, number of abortion, breast-feeding history for born baby47,48. In addition, a large number of SNPs in susceptibility genes have been found to be related to BC risk. There have been few reports of the interactions between SNPs in susceptibility genes and breast cancer influence factors46,49. However, the analysis of such gene-environment interaction is rarely reported in Chinese population and no study about the interaction between variants in Linc-ROR and the reproductive factors was found. In the current study, we found the interaction of rs4801078, number of pregnancy and menopausal status might increase BC risk by 2.78 times. We all know that reproductive factors usually represent a difference in hormone levels for women. Recent research suggested that Linc-ROR functions as an onco-lncRNA to promote estrogen-independent growth of ER+ breast cancer by the MAPK/ERK signaling pathway24. Moreover, another research showed the Linc-ROR expression levels in plasma were correlated with estrogen receptor (P < 0.05) and progesterone receptor (P < 0.05)40. Considering that the result of our study “functional polymorphisms in Linc-ROR might be associated with expression of Linc-ROR”, we assume that functional polymorphisms in Linc-ROR might have interaction with reproductive factors and the MDR analysis of our study proved the idea. Our results suggested that the gene-reproductive factors interaction might contribute to BC risk and further studies about the gene-reproductive factors interaction were warranted.
The strengths and limitations of the article are as follows: the new and pathological diagnosed BC patients avoids the prevalence-incidence bias; the selection bias is effectively controlled by random selection of the control group in the community; age and gender-matched controls and strict inclusion criteria reduce confounding bias; data was collected through the questionnaire by uniformly trained investigators and information was double entered in the database; moreover, randomly selected ten percent DNA samples were directly sequenced and each sample was repeated three times independently in qRT-PCR tests for the repeatability; nevertheless, in some subgroups, the statistical power is reduced due to the relatively small sample size; in addition, since early menarche was an established risk factor for breast cancer, the higher mean age at menarche in cases showed that there was still selection bias in our study.
To sum up, we found rs4801078 TT genotype in Linc-ROR had a significant association with the higher risk of BC and analysis of qRT -PCR showed the expression of Linc-ROR mRNA was closely related with the alleles of rs4801078. Rs4801078 might be related to BC by interacting with the number of pregnancy and menopausal status. Our results suggested that tagger SNPs in Linc-ROR and the interaction of tagger SNP-reproductive factors might contribute to BC risk, and the alleles of rs4801078 have positive correlation with Linc-ROR mRNA expression. Prospective studies with large sample and furthermechanism research are warranted to explore the role of the variants in Linc-ROR.
Methods
Subjects
A total of 968 participants were enrolled in the genetic epidemiology study between 2013 and 2015 from a community-based study in Henan province attended by 20000 individuals. Due to the low prevalence of breast cancer in the program, most of BC patients came from the First Affiliated and the Third Affiliated Hospital of Zhengzhou University. The inclusion criteria included the newly pathological diagnosed BC patients in Henan province without any other malignant tumor. 484 frequency matched cancer-free controls were selected from the program randomly. The inclusion criteria of the control group was healthy people without chronic diseases history and age-appropriate frequency matched (±2 years). All the participants were unrelated.
The demographic data and some potential BC risk factors were collected from a structured questionnaire by face to face interviews. The information of receptor status (estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER-2) status) came from pathological report. Some potential BC risk factors in the current study included age, age of menarche, menopause age, menopausal state (premenopausal, postmenopausal), gravidity, number of abortions, history of breast-feeding (yes, no) and BC family history in first-degree relatives (yes, no). Written informed consents were obtained from all participants. The study was approved by the ethical review committee of Zhengzhou University Committee for Medical and Health Research Ethics and all experiments were performed in accordance with relevant guidelines and regulations.
DNA extraction
We collected 5 ml venous blood from each subject in ethylene diamine tetra acetic acid (EDTA), part of the blood cells were used to extract genomic DNA with the DNA Extraction Kit (TIANGEN BIOTECH, Beijing). Separated plasma and DNA samples were stored at −80 °C.
SNP selection and genotyping
All tagger SNPs of Linc-ROR were selected by Haploview software basing on Chinese Han population data of HapMap Project (HapMapRel 28, NCBI B36) as well as 1000 Genome Project with minor allele frequency (MAF) > 0.1 in Chinese Han population (Table 8).
Table 8.
PCR information of the four SNPs.
| SNP ID | Allele | MAF | Genotyping assay | Tm(°C) | Primers(5′-3′) |
|---|---|---|---|---|---|
| rs1942348 | T/C | 0.44 | CRS-RFLP | 59.6 | Sense: TTTCCCTCTTGGCTAATGCTGCTGA |
| Antisense:TTACATAACTGTGGCAGAATGAAGG | |||||
| rs6420545 | C/T | 0.45 | PCR-RFLP | 56.1 | Sense: CTCCAGCCTAGATGACAGA |
| Antisense: CACAGCAGCACTATTCCTAT | |||||
| rs4801078 | C/T | 0.41 | CRS-RFLP | 56.1 | Sense: ATTTCAAGCTCAGATCACTATAGAG |
| Antisense: TCTAAGGGACAGAATAAATAATCGT | |||||
| rs9636089 | T/C | 0.41 | PCR-RFLP | 59.6 | Sense: GCACAGTTCACAGATGGA |
| Antisense: CAGGAGATTGGCTTGGTT |
rs6420545 and rs9636089were genotyped by polymerase chain reaction-restriction fragment-length polymorphism (PCR-RFLP) while the genotyping of rs1942348 and rs4801078were conducted by created restriction site PCR (CRS-RFLP) assays in our research. Primer 5.0 software was used to design the primer pairs of the four SNPs for PCR amplification and the NCBI BLAST was performed to evaluate the specificity of primers. Gradient PCR reactions was performed to standardize the DNA amplification conditions and optimize the annealing temperature for the primers set (Table 1). Rs6420545, rs9636089 rs1942348 and rs4801078 were digested by restriction enzymes of NsiI, MwoI, BlpI and BsmAI (Thermo Scientific) respectively which were selected by WATCUT website (http://watcut.uwaterloo.ca/watcut/watcut/template.php).In order to control the quality, we randomly selected 10% of the DNA samples to be sequenced (BGI Sequencing, Beijing).
The relative expression of the Linc-ROR in plasma
One hundred and fifty individuals from the controls were selected and qRT-PCR test was used to detect the relative expression of Linc-ROR in plasma by the Eco Real-Time PCR System (Illumina, USA). The conditions for the reaction of real-time PCR are as follows: 1) initial denaturation: 95 °C for 30 s; 2) 40 cycles of denaturation: 95 °C for 10 s; 3) anneal: 60 °C for 30 s. Our assay provided GAPDH as a reference gene and the 2−ΔΔCt method was performed to calculate the relative expression of Linc-ROR. The real-time PCR amplification of each sample was repeated three times.
Analysis
The sample size (n = 459) of the study was calculated by the software power analysis and sample size (PASS)with the minimum allele frequency (0.25) and the study power (0.9). We assess the representativeness of the control population using a goodness-of-fit χ2-test (Hardy–Weinberg equilibrium).Categorical variables and continuous variables were calculated by Chi-squared and Student’s t test respectively to assess distribution departure in two groups. Unconditional logistic regression analysis was applied to estimate the relationship between SNPs and BC (or receptor status) with adjusted for the potential BC risk factors. Stratified analysis for the potential risk factors mentioned was made in different subgroups to estimate the relationship between SNPs and BC risk. MDR method was conducted to calculate the gene-reproductive factors interactions and online SHEsis (http://analysis.bio-x.cn/myAnalysis.php) was used to analyze the difference of haplotype frequencies in both patients and controls. Relative expressions of the gene Linc-ROR were presented as mean ± standard deviation ( ± SD) and the one-way ANOVA was applied to assess the difference. The Benjamini-Hochberg (BH) correction was used to control false discovery rate (FDR).The SPSS 21.0 statistical software package (Analysis software, Shanghai, co., LTD, 6761805c6989326cbf14) was used for all statistics analyses, and two-side P value less than 0.05 was regarded as significant.
Data availability
The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant (U1604168); Major Science and Technology programs in Henan Province under Grant (161100311400); and Medical Science and Technology Key Projects of Henan Province and Zhengzhou under Grant (201602295 and 20150374). We also acknowledge the Key Laboratory of cancer epidemiology in Henan for providing research platform.
Author Contributions
Conception and design: ChenglinLuo, Chunhua Song. Development of methodology: ChenglinLuo, Jingjing Cao, Chunhua Song, RuiPeng. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Jingjing Cao, ChenglinLuo, Chunhua Song,RuiPeng, QiaoyunGuo, Kaijuan Wang, Peng Wang, Hua Ye. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): ChenglinLuo, Jingjing Cao, Chunhua Song, QiaoyunGuo. Writing, review and/or revision of the manuscript: Jingjing Cao, ChenglinLuo, Chunhua Song. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Chunhua Song, Kaijuan Wang, Peng Wang, Hua Ye. Study supervision: Chunhua Song, Kaijuan Wang, Peng Wang, Hua Ye.
Competing Interests
The authors declare no competing interests.
Footnotes
Chenglin Luo and Jingjing Cao contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fan L, et al. Breast cancer in China. The Lancet Oncology. 2014;15:e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.John L, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Non-Coding RNAs. cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, J. et al. MiRNA-binding site functional polymorphisms in DNA repair genes RAD51, RAD52, and XRCC2 and breast cancer risk in Chinese population. Tumor Biol, 10.1007/s13277-016-5459-2 (2016). [DOI] [PubMed]
- 5.Cao J, et al. rs15869 at miRNA binding site in BRCA2 is associated with breast cancer susceptibility. Med Oncol. 2016;33:135. doi: 10.1007/s12032-016-0849-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, et al. Associations of Il-1 Family-Related Polymorphisms With Gastric Cancer Risk and the Role of Mir-197 In Il-1f5 Expression. Medicine. 2015;94:e1982. doi: 10.1097/MD.0000000000001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 8.Group), T. F. C. a. R. G. E. R. G. a. G. S. G. G. N. P. C. The Transcriptional Landscape of the Mammalian Genome. science309, 1559–1563, 10.1126/science.1112014 (2005). [DOI] [PubMed]
- 9.Kapranov P, et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doerks, T., Copley, R. R., Schultz, J., Ponting, C. P. & Bork, P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res12, 47–56, https://doi.org/10.1101/ (2002). [DOI] [PMC free article] [PubMed]
- 11.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai MC, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 13.Braconi C, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun H, et al. H19 lncRNA mediates 17beta-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol Rep. 2015;33:3045–3052. doi: 10.3892/or.2015.3899. [DOI] [PubMed] [Google Scholar]
- 15.Yan R, Wang K, Peng R, Wang S. Genetic variants in lncRNA SRA and risk of breast cancer. oncotarget. 2016;7:22486–22496. doi: 10.18632/oncotarget.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaegh GW, et al. Polymorphisms in the H19 gene and the risk of bladder cancer. Eur Urol. 2008;54:1118–1126. doi: 10.1016/j.eururo.2008.01.060. [DOI] [PubMed] [Google Scholar]
- 17.Xue Y, et al. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30:303–310. doi: 10.1093/mutage/geu076. [DOI] [PubMed] [Google Scholar]
- 18.Loewer S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nature genetics. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang A, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou P, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan HX, et al. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016;374:261–271. doi: 10.1016/j.canlet.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Chen YM, Liu Y, Wei HY, Lv KZ, Fu P. Linc-ROR induces epithelial-mesenchymal transition and contributes to drug resistance and invasion of breast cancer cells. Tumour Biol. 2016;37:10861–10870. doi: 10.1007/s13277-016-4909-1. [DOI] [PubMed] [Google Scholar]
- 24.Peng WX, Huang JG, Yang L, Gong AH, Mo YY. Linc-ROR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Molecular cancer. 2017;16:161. doi: 10.1186/s12943-017-0727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi AD, et al. Additive interactions between susceptibility single-nucleotide polymorphisms identified in genome-wide association studies and breast cancer risk factors in the Breast and Prostate Cancer Cohort Consortium. Am J Epidemiol. 2014;180:1018–1027. doi: 10.1093/aje/kwu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arunkumar G, et al. Expression profiling of long non-coding RNA identifies Linc-ROR as a prognostic biomarker in oral cancer. Tumour Biol. 2017;39:1010428317698366. doi: 10.1177/1010428317698366. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, et al. Associations between novel genetic variants in the promoter region of MALAT1 and risk of colorectal cancer. Oncotarget. 2017;8:92604–92614. doi: 10.18632/oncotarget.21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lou Y, et al. Linc-ROR induces epithelial-to-mesenchymal transition in ovarian cancer by increasing Wnt/beta-catenin signaling. Oncotarget. 2017;8:69983–69994. doi: 10.18632/oncotarget.19545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Gao PW, Hua Y, Xi H, Meng Z, Te Liu ROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancer. oncotarget. 2015;7:1608–1618. doi: 10.18632/oncotarget.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Z, et al. Endogenous miRNA Sponge LincRNA-ROR promotes proliferation, invasion and stem cell-like phenotype of pancreatic cancer cells. Cell Death Discov. 2017;3:17004. doi: 10.1038/cddiscovery.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Zhao Z, Zhou Z, Liu R. Linc-ROR confers gemcitabine resistance to pancreatic cancer cells via inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis. Cancer Chemother Pharmacol. 2016;78:1199–1207. doi: 10.1007/s00280-016-3178-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X, et al. Linc-RNA-RoR acts as a “sponge” against mediation of the differentiation of endometrial cancer stem cells by microRNA-145. Gynecol Oncol. 2014;133:333–339. doi: 10.1016/j.ygyno.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 33.Pan Y, et al. Long noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget. 2017;8:33144–33158. doi: 10.18632/oncotarget.16562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reza Sahebi MM, et al. Seyed Javad Mowla Linc-ROR and its spliced variants 2 and 4 are significantly up-regulated in esophageal squamous cell carcinoma. Iranian Journal of Basic Medical Sciences. 2015;19:1131–1135. [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, et al. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. 2016;107:1215–1222. doi: 10.1111/cas.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng S, et al. Expression and Functional Role of Reprogramming-Related Long NoncodingRNA (lincRNA-ROR) in Glioma. Journal of Molecular Neuroscience. 2015;56:623–630. doi: 10.1007/s12031-014-0488-z. [DOI] [PubMed] [Google Scholar]
- 38.Yao-Min Chen YL, Hai-Yan Wei K-ZL, Pei-Fen F. Large intergenic non-coding RNA-ROR reverses Gemcitabine-induced autophagy and apoptosis in breast cancer cells. oncotarget. 2016;7:59604–59617. doi: 10.18632/oncotarget.10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang HY, et al. Effects of long noncoding RNA-ROR on tamoxifen resistance of breast cancer cells by regulating microRNA-205. Cancer Chemother Pharmacol. 2017;79:327–337. doi: 10.1007/s00280-016-3208-2. [DOI] [PubMed] [Google Scholar]
- 40.Zhao T, Wu L, Li X, Dai H, Zhang Z. Large intergenic non-coding RNA-ROR as a potential biomarker for the diagnosis and dynamic monitoring of breast cancer. Cancer Biomarkers. 2017;20:165–173. doi: 10.3233/CBM-170064. [DOI] [PubMed] [Google Scholar]
- 41.Gao, P. & Wei, G. H. Genomic Insight into the Role of lncRNA in Cancer Susceptibility. International journal of molecular sciences18, 10.3390/ijms18061239 (2017). [DOI] [PMC free article] [PubMed]
- 42.Peng, R. et al. Association analyses of genetic variants in long non-coding RNA MALAT1 with breast cancer susceptibility and mRNA expression of MALAT1 in Chinese Han population. Gene, 10.1016/j.gene.2017.11.013 (2017). [DOI] [PubMed]
- 43.Al-Tassan NA, et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Scientific reports. 2015;5:10442. doi: 10.1038/srep10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ingle JN, et al. Genetic Polymorphisms in the Long Noncoding RNA MIR2052HG Offer a Pharmacogenomic Basis for the Response of Breast Cancer Patients to Aromatase Inhibitor Therapy. Cancer research. 2016;76:7012–7023. doi: 10.1158/0008-5472.CAN-16-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong J, et al. A functional polymorphism in lnc-LAMC2-1:1 confers risk of colorectal cancer by affecting miRNA binding. Carcinogenesis. 2016;37:443–451. doi: 10.1093/carcin/bgw024. [DOI] [PubMed] [Google Scholar]
- 46.Milne RL, et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study. Breast cancer research: BCR. 2010;12:R110. doi: 10.1186/bcr2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becher H, Schmidt S, Chang-Claude J. Reproductive factors and familial predisposition for breast cancer by age 50 years. A case-control-family study for assessing main effects and possible gene-environment interaction. International Journal of Epidemiology. 2003;32:38–48. doi: 10.1093/ije/dyg003. [DOI] [PubMed] [Google Scholar]
- 48.Andrieu N, et al. Variation in the interaction between familial and reproductive factors on the risk of breast cancer according to age, menopausal status, and degree of familiality. International Journal of Epidemiology. 2000;29:214–223. doi: 10.1093/ije/29.2.214. [DOI] [PubMed] [Google Scholar]
- 49.Campa D, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. Journal of the National Cancer Institute. 2011;103:1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.


