Abstract
Introduction
Data processing is one of the biggest problems in metabolomics, given the high number of samples analyzed and the need of multiple software packages for each step of the processing workflow.
Objectives
Merge in the same platform the steps required for metabolomics data processing.
Methods
KniMet is a workflow for the processing of mass spectrometry-metabolomics data based on the KNIME Analytics platform.
Results
The approach includes key steps to follow in metabolomics data processing: feature filtering, missing value imputation, normalization, batch correction and annotation.
Conclusion
KniMet provides the user with a local, modular and customizable workflow for the processing of both GC–MS and LC–MS open profiling data.
Keywords: Metabolomics, Data processing, GC–MS, LC–MS
Introduction
Among the several analytical techniques employed within metabolomics, gas and liquid chromatography coupled with mass spectrometry (GC– and LC–MS) are the most commonly used in metabolomics studies as they allow the identification of a large number of diverse molecular species. However, the plethora of samples analyzed during high-throughput screenings, the number of processing steps, and the required computational competences and resources often represent a bottleneck that renders these analyses slow and potentially inaccurate. Hence, utilization of standardized procedures is fundamental for reliable and reproducible results (Meier et al. 2017; Rocca-Serra et al. 2016; Sandve et al. 2013). Several protocols have been proposed or are currently being developed (Beisken et al. 2014; Di Guida et al. 2016; Dunn et al. 2011a; Giacomoni et al. 2015; Guitton et al. 2017; Rocca-Serra 2017; Southam et al. 2017; Weber et al. 2017). However, they are not free from pitfalls, the main ones being related to a high level of computational expertise needed for their local installation, utilization and implementation. The alternative provided by web-based services can be affected by inadequate stability, security and performance in handling a large number of samples, or sensitive data.
For these reasons, the KNIME Analytics Platform (Berthold et al. 2007) was used to build a vendor-independent processing workflow. KniMet (Liggi 2017) joins several steps required to process GC– and LC–MS metabolomics data, outputting a data matrix normalized, annotated and filtered from inconsistently detected features in a semi-automated, documented and reproducible analysis.
KniMet features
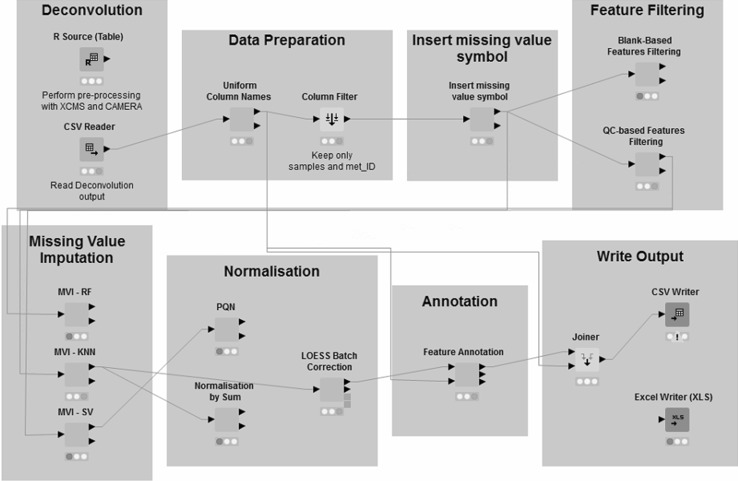
The steps performed by KniMet comprise data deconvolution, feature filtering, missing value imputation, normalization and features annotation. For each one of these steps there are several options, as shown in Fig. 1 and described below, allowing users to utilize the most appropriate tool for the specific case study at hand.
Fig. 1.
The KniMet pipeline comprises different steps for the post-processing of metabolomics data each one enclosed in a square in this representation. Most of these steps can be performed with multiple tools, allowing the user to combine them in the most appropriate way for the specific dataset studied
Data deconvolution
GC– and LC–MS data in mzXML or CDF format (previously converted with, for instance, Proteowizard [14]) can be deconvoluted internally with the R (R Core Team 2014) library XCMS (Smith et al. 2006), or by integrating into KniMet the OpenMS nodes (Pfeuffer et al. 2017). Alternatively, this step can be performed externally with either the locally installed R instance, XCMS online [17] or a vendor software. In this case, the obtained data matrix can then be imported in the pipeline and subjected to further analysis. For instance, a dataset obtained using the Agilent 6560 Ion Mobility Q-TOF LC–MS was deconvoluted with MassProfiler from the MassHunter Workstation Software suite (Agilent Technologies, Santa Clara, USA), fed into KniMet and then subsequently processed using downstream tools.
Feature filtering
Periodic injections of pooled samples, also known as quality controls (QCs) are used to account and correct for analytical variation, based on the assumption that QCs should contain all the signals present in the samples. Hence, if the instrument performance is stable, these signals should be consistently detected across the run, while only unstable metabolites or contaminants would be detected inconsistently (Dunn et al. 2011a). According to these principles, all features whose signal is missing in more than a given percentage of QCs (defined by the user, default 50%) and whose Relative Standard Deviation across the QCs is higher than a threshold (set by the user, default 20%) are deleted. An alternative method not based on pooled samples was implemented to account for experimental setups in which QCs are missing and/or the user would rather perform feature filtering based on other samples, such as blanks. In this case, only features whose average intensity in the samples is higher than their average intensity in blanks multiplied by a user-defined factor are retained. Moreover, features are filtered if they are missing in more than a user-defined percentage of samples.
Missing values handling
Missing values in the data matrix can occur for several reasons, such as (i) missingness of a feature in one (class of) sample(s) and not in another, (ii) concentration of a metabolite in a sample lower than the analytical limit of detection (iii), or inaccurate pre-processing with lack of deconvolution of a feature. An appropriate evaluation of the reasons behind the presence of missing values in the data matrix, and their consecutive imputation, is fundamental to avoid biased statistical results (Di Guida et al. 2016; Gromski et al. 2014). In this application, missing values imputation can be performed with either Random Forest (RF) or K-Nearest Neighbour (KNN) algorithms, implemented as R scripts using the libraries missForest (Stekhoven and Buhlmann 2012) and impute (Hastie et al. 2016) respectively, or Small Value replacement (SV), i.e. half of the minimum value found for a given feature in given sample.
Normalization
Among the several normalisation methods available, Probabilistic Quotient Normalization (PQN) (Dieterle et al. 2006) and Sum Normalisation have been implemented in KniMet as they are the most commonly used in MS-based metabolomics data (Di Guida et al. 2016). PQN consists of: (i) calculation of a reference spectrum (or vector) as the median of each signal in the entire set of samples or, if available, in the QCs; (ii) division of each signal found in the samples by the value for the same signal in the reference spectrum to obtain a list of quotients; (iii) division of the original data matrix for the median of these quotients. On the other hand, in Sum normalization each feature in a given sample is divided by the sum of all features in that sample and multiplied by 100.
Peak drift is an issue in metabolomics data obtained from LC–MS instruments, as a number of factors which vary with time can affect the results. In the case of batch-effects being present, batch-correction normalization can be performed to merge samples measured in different analytical blocks. Among the several methods available, the robust locally estimated scatterplot smoothing (LOESS) signal correction (RLSC) method based either on QCs or all samples (Dunn et al. 2011b; Thévenot et al. 2015) were implemented utilizing the R scripts developed by the Workflow4metabolomics team (Giacomoni et al. 2015).
Metabolite annotation
Metabolite annotation based on accurate mass match with the Human Metabolome Database (Wishart et al. 2017) and the LIPID MAPS database (Fahy et al. 2007; Sud et al. 2006) was implemented by integrating the AccurateMassSearch functionality of OpenMS.
Conclusions
KniMet is a KNIME-based pipeline for the analysis of metabolomics MS data. This platform is easy to install and run locally, providing the user with full control of the analysis. Indeed, the modular structure of the platform allows the pipeline to be modified based on the nature of the data to be processed, and hence be applied to datasets derived from different analytical and/or experimental setups. The resulting tables containing all the analyzed samples and the detected metabolic features can be exported and are ready for further statistical analysis. A recent and published example of its application is the processing of both GC– and LC–MS data of fecal samples from patients affected by Inflammatory Bowel Diseases compared with a population of healthy subjects, with the aim to identify new biomarkers for the disease (Santoru et al. 2017).
Moreover, KniMet is fast and does not require particularly high computational power: the post-processing of the R data package faahKO (Saghatelian et al. 2004) as described in the user guide, takes less than 10 s and a peak of 1331.65 MB of memory consumption on a PC with Intel® Core™ i7.
In conclusion, with the KniMet application we provide the user with a highly flexible, fully customizable and user-friendly platform which includes the key processing steps of metabolomics data.
Availability and implementation
KniMet is freely available under the 3-Clause BSD License at https://github.com/sonial/KniMet along with usage instructions and example data.
Acknowledgements
We thank Evelina Charidemou for providing some of the example data.
Funding
This study was funded by Agilent Technologies, Regione Autonoma della Sardegna (L.R.7/2007, Grant Number F71J12001180002), and the Medical Research Council UK (Grant Number MR/P011705/1).
Compliance with ethical standards
Conflict of interest
Authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Contributor Information
Sonia Liggi, Phone: +44 1223 333600, Email: sl584@cam.ac.uk.
Luigi Atzori, Phone: +39 070 6754125, Email: latzori@unica.it.
Julian L. Griffin, Phone: +44 1223 764922, Email: jlg40@cam.ac.uk
References
- Beisken S, Earll M, Portwood D, Seymour M, Steinbeck C. MassCascade: Visual programming for LC–MS data processing in metabolomics. Molecular Informatics. 2014;33(4):307–310. doi: 10.1002/minf.201400016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold MR, Cebron N, Dill F, Gabriel TR. Studies in classification, data analysis, and knowledge organization (GfKL 2007) New York: Springer; 2007. KNIME: The Konstanz information miner; pp. 319–326. [Google Scholar]
- Di Guida R, Engel J, Allwood JW, Weber RJM, Jones MR, Sommer U, et al. Non-targeted UHPLC-MS metabolomic data processing methods: A comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics. 2016;12(5):1–14. doi: 10.1007/s11306-016-1030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in1H NMR metabonomics. Analytical Chemistry. 2006;78(13):4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols. 2011;6(7):1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Goodacre R, Neyses L, Mamas M. Integration of metabolomics in heart disease and diabetes research: Current achievements and future outlook. Bioanalysis. 2011;3(19):2205–2222. doi: 10.4155/bio.11.223. [DOI] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Research. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomoni F, Le Corguillé G, Monsoor M, Landi M, Pericard P, Pétéra M, et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics. 2015;31(9):1493–1495. doi: 10.1093/bioinformatics/btu813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromski P, Xu Y, Kotze H, Correa E, Ellis D, Armitage E, et al. Influence of missing values substitutes on multivariate analysis of metabolomics data. Metabolites. 2014;4(2):433–452. doi: 10.3390/metabo4020433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton Y, Tremblay-Franco M, Le Corguillé G, Martin JF, Pétéra M, Roger-Mele P, et al. Create, run, share, publish, and reference your LC-MS, FIA-MS, GC-MS, and NMR data analysis workflows with the Workflow4Metabolomics 3.0 Galaxy online infrastructure for metabolomics. International Journal of Biochemistry and Cell Biology. 2017 doi: 10.1016/j.biocel.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Hastie, T., Tibshirani, R., Narasimhan, B., & Chu, G. (2016). Impute: Impute—imputation for microarray data. 10.18129/B9.bioc.impute.
- Liggi S. First release of KniMet (version v1.0.0) Zenodo. 2017 [Google Scholar]
- Meier R, Ruttkies C, Treutler H, Neumann S. Bioinformatics can boost metabolomics research. Journal of Biotechnology. 2017 doi: 10.1016/j.jbiotec.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Pfeuffer J, Sachsenberg T, Alka O, Walzer M, Fillbrunn A, Nilse L, et al. OpenMS: A platform for reproducible analysis of mass spectrometry data. Journal of Biotechnology. 2017;261:142–148. doi: 10.1016/j.jbiotec.2017.05.016. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- Rocca-Serra, P. (2017). PhenoMeNal: Virtual e-Infrastructure supporting clinical research and metabolism studies. Retrieved 20 September 2017, from http://www.oerc.ox.ac.uk/news/PhenoMeNal-datahack.
- Rocca-Serra P, Salek RM, Arita M, Correa E, Dayalan S, Gonzalez-Beltran A, et al. Data standards can boost metabolomics research, and if there is a will, there is a way. Metabolomics. 2016;12(1):1–13. doi: 10.1007/s11306-015-0879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43(45):14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- Sandve GK, Nekrutenko A, Taylor J, Hovig E. Ten simple rules for reproducible computational research. PLoS Computational Biology. 2013;9(10):1–4. doi: 10.1371/journal.pcbi.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S, et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Scientific Reports. 2017;7(1):9523. doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Southam AD, Weber RJM, Engel J, Jones MR, Viant MR. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics. Nature Protocols. 2017;12(2):255–273. doi: 10.1038/nprot.2016.156. [DOI] [PubMed] [Google Scholar]
- Stekhoven DJ, Buhlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28(1):112–118. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Brown A, Dennis E, Glass C, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Research. 2006;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenot EA, Roux A, Xu Y, Ezan E, Junot C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. Journal of Proteome Research. 2015;14(8):3322–3335. doi: 10.1021/acs.jproteome.5b00354. [DOI] [PubMed] [Google Scholar]
- Weber RJM, Lawson TN, Salek RM, Ebbels TMD, Glen RC, Goodacre R, et al. Computational tools and workflows in metabolomics: An international survey highlights the opportunity for harmonisation through Galaxy. Metabolomics. 2017;13(2):1–5. doi: 10.1007/s11306-016-1147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Research. 2017;2017:1–10. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]