Abstract
We demonstrate the use of the nematode Caenorhabditis elegans as a facile and inexpensive model host for several Gram-positive human bacterial pathogens. Enterococcus faecalis, Streptococcus pneumoniae, and Staphylococcus aureus, but not Bacillus subtilis, Enterococcus faecium, or Streptococcus pyogenes, kill adult C. elegans. Focusing our studies on the enterococcal species, we found that both E. faecalis and E. faecium kill C. elegans eggs and hatchlings, although only E. faecalis kills the adults. In the case of adults, a low inoculum of E. faecalis grows to a high titer in the C. elegans intestine, resulting in a persistent infection that cannot be eradicated by prolonged feeding on E. faecium. Interestingly, a high titer of E. faecium also accumulates in the nematode gut, but does not affect the longevity of the worms. Two E. faecalis virulence-related factors that play an important role in mammalian models of infection, fsr, a putative quorum-sensing system, and cytolysin, are also important for nematode killing. We exploit the apparent parallels between Gram-positive infection in simple and more complex organisms by using the nematode to identify an E. faecalis virulence factor, ScrB, which is relevant to mammalian pathogenesis.
Gram-positive pathogens in the genera Streptococcus, Staphylococcus, and Enterococcus are leading causes of human infections, causing diseases such as pneumonia, meningitis, bacteremia, endocarditis, and necrotizing fasciitis. The genus Enterococcus, the main focus of this article, is particularly problematic because of multidrug resistance, including resistance to vancomycin, often the antibiotic of last resort. Nevertheless, only a limited number of enterococcal virulence-related factors have been described, including cytolysin (Cyl), a factor called aggregation substance (AS), a zinc metalloprotease (gelatinase), and fsr, a putative quorum-sensing system thought to be involved in gelatinase and/or serine protease regulation (1–5). As the development of antibiotic resistance continues to erode one of the greatest advances in modern health care, it is crucial to identify bacterial targets that can form the basis of novel anti-infective therapies.
One reason that relatively little is known about enterococcal virulence factors is that the mammalian models used to study enterococcal infections are cumbersome and expensive. Using a mammalian host to screen enterococcal mutant libraries for avirulent mutants, for example, would be prohibitively time consuming and expensive because of the large number of animals involved. Therefore, we have sought to develop alternative nonvertebrate hosts for Enterococcus and other Gram-positive human pathogens.
Previously, our laboratory and others have shown not only that the Gram-negative human pathogens Pseudomonas aeruginosa and Salmonella enterica kill the nematode Caenorhabditis elegans, but also that P. aeruginosa and S. enterica virulence factors required for mammalian pathogenesis also are required for efficient killing of C. elegans (6–10). In this article, we demonstrate the suitability of using C. elegans as a model host for Gram-positive infection. We show that clinical isolates of Enterococcus faecalis, Streptococcus pneumoniae, and Staphylococcus aureus kill adult C. elegans, and that several features of the E. faecalis–C. elegans model that we have studied suggest that some aspects of human Gram-positive pathogenesis can be modeled successfully in a simple nonvertebrate host.
Materials and Methods
Assay of C. elegans Killing by Bacteria.
Bacterial strains were grown on brain heart infusion (BHI) agar medium (Difco) in 35-mm tissue-culture plates (Falcon). Appropriate antibiotics were added to the medium to selectively prevent growth of Escherichia coli. Tetracycline (12.5 μg/ml) was used for E. faecalis strains E001, E002, E006, and E009 and for Enterococcus faecium strains E003 and E007. Gentamicin (25–50 μg/ml) was used for E. faecalis strains V583, OG1RF, and OG1X, and 5 μg/ml polymyxin B (American Research Products, Belmont, MA) was used for the experiment shown in Fig. 1A with S. aureus A003, E. faecium E007, E. faecalis E002, S. pneumoniae SP003, Streptococcus pyogenes GAS002, and Bacillus subtilis PY79. Lawns of bacteria were grown as follows: 2 ml of BHI was inoculated with a single colony of the appropriate strain and grown at 37°C for 4–5 hr, and 10 μl of the culture was spread on each plate. The plates were incubated at 37°C overnight. Streptococcus plates were grown in a 5% CO2/95% air atmosphere.
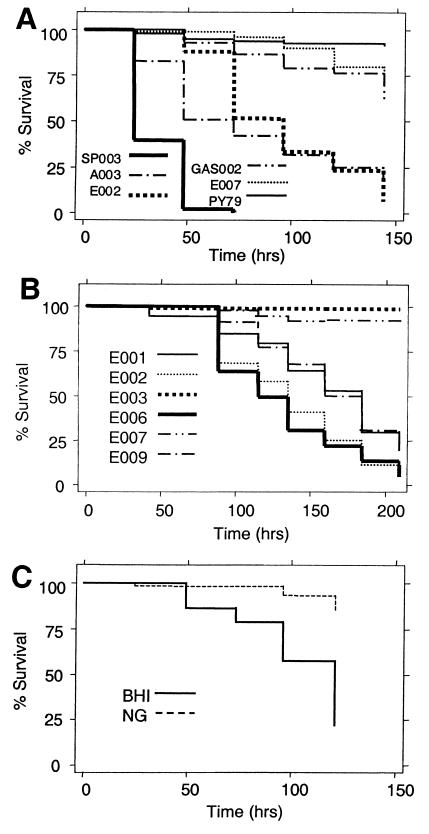
Figure 1.
C. elegans killing by Gram-positive pathogens. (A) Survival of C. elegans fed on clinical isolates of S. pneumoniae (SP003), S. aureus (A003), E. faecalis (E002), S. pyogenes (GAS002), E. faecium (E007), and nonpathogenic B. subtilis (PY79). (B) Survival of C. elegans fed on lawns of E. faecalis (E001, E002, E006, and E009) and E. faecium (E003 and E007) clinical isolates. (C) Survival of C. elegans fed on lawns of Escherichia coli strain OP50 grown on BHI medium or nematode growth (NG) medium.
C. elegans strain N2 was maintained and propagated on Escherichia coli strain OP50 with standard techniques (11). Between 20 and 30 C. elegans L4 or young adult hermaphrodites were transferred from a lawn of Escherichia coli OP50 to a lawn of the bacterium to be tested, incubated at 25°C, and examined at ≈24-hr intervals with a dissecting microscope for viability. Worms were considered dead when they did not respond to touch with a platinum wire pick. Each experimental condition was tested in duplicate or triplicate. Nematode survival was plotted with the Kaplan–Meier method using STATA 6 statistical software (Stata, College Station, TX). The same software was used for testing equality of survival (log-rank and Wilcoxon tests). P values of <0.05 were considered statistically significant.
Measuring the Number of Bacteria Within the C. elegans Digestive Tract.
Five C. elegans were picked at the times indicated in Fig. 2, and the surface bacteria were removed by washing the worms twice in 4-μl drops of M9 medium on a BHI agar plate containing 25 μg/ml gentamicin. The nematodes were placed in a 1.5-ml Eppendorf tube containing 20 μl of M9 medium with 1% Triton X-100 and were mechanically disrupted by using a pestle. The volume was adjusted to 50 μl with M9 medium containing 1% Triton X-100 which was diluted and plated on BHI agar containing 15 μg/ml vancomycin, 50 μg/ml ampicillin, or 12.5 μg/ml tetracycline to select for strains V583, E007, or E003, respectively. Each data point represents the mean cfu from triplicate samples, and the error bars represent the standard deviation.
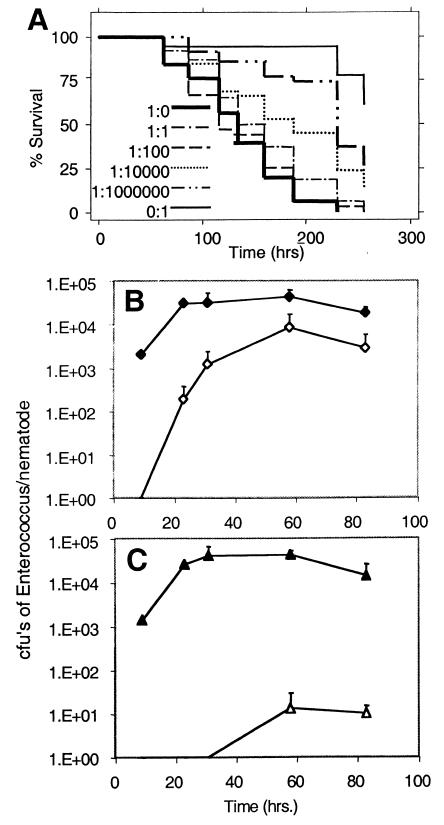
Figure 2.
E. faecalis proliferates in the nematode gut. (A) Various ratios of E. faecalis V583 to E. faecium E007 were plated to form bacterial lawns. The survival of C. elegans fed on these lawns was measured. (B) C. elegans were placed on a lawn of E. faecalis V583 and E. faecium E007 in the ratio of 1:1,000. After 12 hr, the C. elegans were washed and moved to a plate of E007 (time 0). At the times indicated, nematodes were washed and ground, and dilutions of the resulting suspension were plated on selective media. The number of colony-forming units (cfu) of V583 (⋄) and E007 (♦) per worm was calculated. (C) The same protocol as described for B was followed except that two E. faecium strains with different drug sensitivities were used. C. elegans were initially plated on a lawn of E003 (▵) and E007 (▴) in a ratio of 1:1,000. Note that the error bars correspond to the standard deviations calculated from three experiments and are present on only the upper side of the curves because of the logarithmic scale.
Microscopic Pictures of C. elegans.
Nematodes were exposed for 3 days to bacteria on BHI plates and then placed on a pad of 2% agarose in a 5-μl drop of 30 mM NaN3 in M9 medium. The worms were examined at 100× magnification with an Axioplan2 microscope (Zeiss) with Nomarski optics.
Screening for E. faecalis Tn917 Mutants Attenuated in Nematode Killing.
A library consisting of ≈1,000 OG1RF Tn917 insertion mutants was constructed as described (12). The mutants were assayed for their ability to kill C. elegans as described above, except that only eight worms were placed on each lawn, and the number of live worms was determined only on days 2 and 6. Mutants that caused less than average killing were tested twice more with the standard assay to confirm the attenuated phenotype.
Mouse Peritonitis Model.
For the experiment shown in Fig. 4C, inoculum preparation, colony counts, and use of animals were carried out as described (4), except that each inoculum was diluted a final 1:10 in 35% sterile rat fecal extract (SRFE; ref. 13). In the experiment shown in Fig. 4D, the cells were resuspended in saline to an optical density of 1.7 to 2.0 at 600 nm and diluted 1:10 in 40% SRFE. ICR-CD1 mice (Charles River Breeding Laboratories) were used. Survival curves were plotted and analyzed as described above for nematode killing. Guidelines of the Animal Welfare and the Animal Care Committees of the University of Texas Health Science Center and the Institutional Animal Care and Use Committee of Massachusetts General Hospital, respectively, were followed throughout the course of the animal experiments.
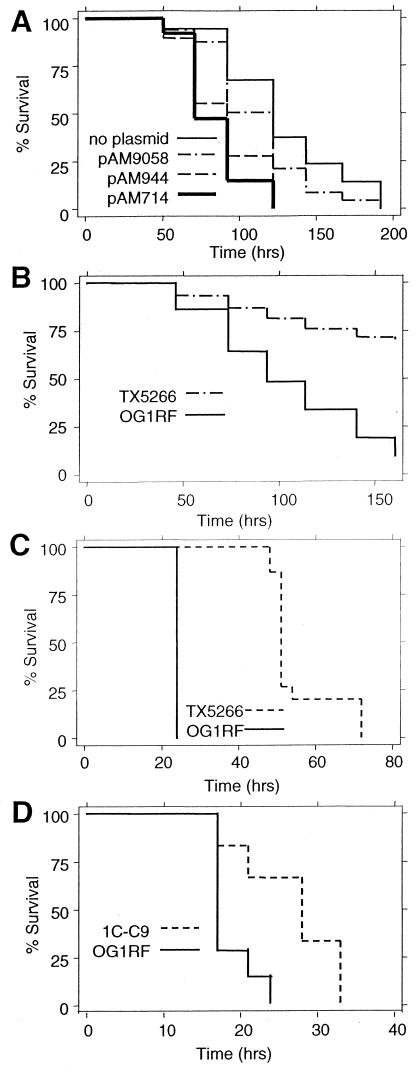
Figure 4.
Mammalian virulence factors enhance E. faecalis killing of C. elegans. (A) Survival of C. elegans placed on lawns of E. faecalis OG1X containing no plasmids (Cyl−, AS−), OG1X + pAM9058 (Cyl− AS+), OG1X + pAM944 (Cyl+ AS−), and OG1X + pAM714 (Cyl+ AS+). (B) Survival of nematodes feeding on lawns of OG1RF or TX5266 (fsrBΔ). (C) Survival of mice infected peritoneally with 1.3 × 108 cfu of OG1RF or 2.1 × 108 cfu of TX5266 (fsrBΔ). (D) Survival of mice infected peritoneally with 2.20 × 108 cfu of OG1RF or 2.86 × 108 cfu of 1C-C9.
Results
E. faecalis, Staphylococcus aureus, and Streptococcus pneumoniae Kill Adult C. elegans.
We tested whether Gram-positive clinical isolates obtained from the Massachusetts General Hospital Clinical Laboratory could kill C. elegans as described in Materials and Methods. All strains of E. faecalis, S. aureus, and S. pneumoniae tested killed C. elegans. Representative data are shown in Fig. 1 A and B. The most virulent S. pneumoniae strains killed quickly, with an LT50 (time for half of the worms to die) of ≈1 day. The most virulent S. aureus and E. faecalis strains had LT50s of ≈2 days and 4 days, respectively (Fig. 1 A and B). In contrast, B. subtilis [strain PY79 (14)] and several clinical isolates of Streptococcus pyogenes and E. faecium did not cause significant killing (Fig. 1 A and B). Interestingly, Escherichia coli strain OP50, the food source used by most laboratories studying C. elegans (11), caused significant killing when grown on BHI (LT50 = 6 days; Fig. 1C). Perhaps the rich BHI medium induces the expression of Escherichia coli factors that are lethal for C. elegans but are not expressed on the commonly used NG medium.
We studied the E. faecalis–C. elegans model in detail to investigate further the feasibility of using C. elegans as a model host for Gram-positive pathogens. We chose E. faecalis, in part, because we observed that both E. faecalis and E. faecium killed C. elegans' eggs and hatchlings in a manner that seems to be mechanistically distinct from the killing of the adult nematodes (data not shown). It was technically convenient that C. elegans failed to produce a brood on enterococcal lawns, as it was easier to keep track of the original adult nematodes. We used the previously studied E. faecalis isolates V583 (15) (recently sequenced and available at www.tigr.org) and OG1RF (16) for most of the following experiments because they killed C. elegans at approximately the same rate as the most virulent new isolates that were tested.
A Small Initial Inoculum of E. faecalis Can Proliferate in the Nematode Gut.
To determine whether a relatively small number of E. faecalis cells could kill C. elegans, we took advantage of the fact that E. faecium did not kill C. elegans under the conditions tested. L4 C. elegans were placed on mixed lawns of E. faecalis and E. faecium in ratios of 1:1, 1:102, 1:104, and 1:106. As shown in Fig. 2A, even at a ratio of 1:106 E. faecalis to E. faecium cells, a significant amount of killing of C. elegans was observed compared with feeding on 100% E. faecium. Indeed, there was no diminution in killing until the E. faecalis to E. faecium ratio was less than 1:102. In general, as the percentage of E. faecalis decreased, we observed an increasingly longer lag time before killing eventually commenced. We hypothesized that the lag in killing corresponded to the time required for a small initial inoculum of E. faecalis that the nematodes had ingested to proliferate in the C. elegans intestine. To test this hypothesis, C. elegans were fed on a mixed lawn of E. faecalis and E. faecium in the ratio of 1:103. After 12 hr, the worms were transferred to a lawn consisting only of E. faecium. At various times, the nematodes were washed to remove bacteria from their surfaces and then disrupted to recover bacteria from inside their digestive tracts. By plating on appropriate selective media, E. faecalis and E. faecium cfu could be distinguished readily. As shown in Fig. 2B, the number of E. faecalis cfu within the nematode gut increased over the course of a couple of days from undetectable levels to about 104 bacteria per worm. This latter result indicates that E. faecalis not only proliferates in the C. elegans intestine but also can cause a persistent infection.
Surprisingly, as shown in Fig. 2B, we found that the worms also had a stable population of about 105 E. faecium cells in their intestines during the course of the experiment, although E. faecium does not cause death. In contrast, when C. elegans feeds on Escherichia coli, essentially no live bacteria can be recovered from the intestine (6, 10). Thus, in the experiment shown in Fig. 2B, whereas the number of E. faecium cells per worm remained approximately constant, the ratio of E. faecalis to E. faecium increased from less than 1:103 to 1:10. In other similar experiments, the ratio reached 1:1. The increase of E. faecalis within the gut is not simply a reflection of E. faecalis overgrowing the bacterial lawn on which the C. elegans were feeding. Random sampling of the lawns showed that the ratio of E. faecalis to E. faecium never exceeded 1:103 (data not shown).
To investigate the accumulation of E. faecium in the C. elegans intestine further, we carried out a transfer experiment with the E. faecium strains E003 and E007, which can be distinguished by their antibiotic resistance phenotypes, plated at an initial ratio of 1:103 E003 to E007 cells. When the worms on these plates were transferred to a lawn of only E007 cells, the ratio of E003 to E007 in the digestive tract never exceeded the initial ratio of 1:103 (Fig. 2C). We also experimented with moving nematodes from a lawn of pure E003 to a lawn of pure E007. After 24 hr, most of the E003 cells had been cleared from the nematode's gut, suggesting that the high titer of E. faecium observed in the gut is not primarily a consequence of a persistent colonization (data not shown).
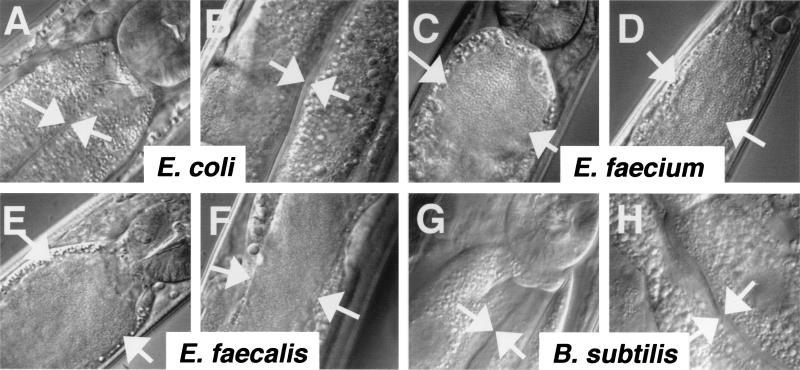
To confirm the observation of high titers of bacteria in the nematodes' intestines in the above experiments, we examined microscopically worms fed on Escherichia coli, B. subtilis, E. faecalis, and E. faecium lawns. As observed (6, 10), worms fed on Escherichia coli had no visible intact bacteria present in their intestinal lumen, and the lumen was slender in appearance (Fig. 3 A and B). In marked contrast, the lumens of worms fed on E. faecalis (Fig. 3 E and F) and E. faecium (Fig. 3 C and D) were full of intact bacteria, and the intestines were grossly distended. It is unlikely that the nematodes fail to grind E. faecalis and E. faecium efficiently because of their thick Gram-positive cell walls. Worms feeding on B. subtilis (strain PY79) contained no visible bacteria and had slender intestinal lumens (Fig. 3 G and H).
Figure 3.
Intact bacteria and gut distension visualized in C. elegans feeding on E. faecalis and E. faecium but not Escherichia coli or B. subtilis. Micrographs were taken of worms fed on lawns of bacteria on BHI plates for 3 days. Arrows point to the borders of the intestinal lumen. (A, E, C, and G) Beginning of intestinal tracts of worms fed on Escherichia coli, E. faecalis, E. faecium, and B. subtilis, respectively. In the upper right-hand corner of each image, a round structure is visible that corresponds to the grinder organ, which functions to disrupt ingested bacteria. (B, F, D, and H) Middle of the intestinal tracts.
Cyl Causes Faster Killing of C. elegans.
E. faecalis has many naturally occurring conjugative plasmids, some of which encode a Cyl that lyses both eukaryotic and prokaryotic cells (17) and functions as a virulence factor in mouse models of endophthalmitis (5, 18) and peritonitis (2, 4). These plasmids also produce a cell surface protein called AS that promotes plasmid transfer (17) and also is thought to play a role in pathogenesis by mediating E. faecalis binding to host tissues (19). There is a significant increase in mortality in a rabbit model of endocarditis when both AS and Cyl are expressed (1).
To determine whether Cyl or AS plays a role in C. elegans killing, we tested isogenic strains of E. faecalis OG1X (20) carrying mutant derivatives of pAD1 that encode both AS and Cyl. Plasmid pAM944 has a Tn917 disruption of the AS gene and is therefore Cyl+ AS−, whereas Tn917 disrupts the Cyl operon in pAM9058 (Cyl− AS+). pAM714 has a nondisruptive Tn917 insertion (Cyl+ AS+) (1) which was used as a control. As shown in Fig. 4A, OG1X strains containing plasmid pAM714 (Cyl+ AS+) or pAM944 (Cyl+ AS−) killed C. elegans faster than OG1X (P < 0.001). There was no significant difference between OG1X containing no plasmids and OG1X containing pAM9058 (Cyl− AS+). These data suggest that Cyl is a virulence factor that affects C. elegans killing independently of the presence or absence of AS.
A Deletion in fsrB Causes Attenuation in the Mouse and the Nematode.
Previous work has shown that a disruption of E. faecalis gelE, which encodes gelatinase, causes attenuation of E. faecalis strain OG1RF in a mouse peritonitis model (4). Three genes located directly upstream of gelE—fsrA, fsrB, and fsrC—seem to be involved in gelE regulation. FsrA and FsrB are homologous to two component response regulators and sensor kinases (3), respectively, and a nonpolar fsrB deletion blocks the production of gelatinase (21). Based on the homology of FsrA, FsrB, and FsrC to the S. aureus quorum-sensing system encoded by agrA, agrB, and agrC, fsrB may encode a processor of a putative E. faecalis signal peptide (3).
Fig. 4B shows that C. elegans died much more slowly when placed on lawns of the fsrBΔ gelatinase regulatory mutant (strain TX5266) than when placed on lawns of the isogenic parental strain OG1RF (P < 0.001). E. faecalis OG1RF mutants containing disruptions in fsrA, fsrB, and gelE also caused prolonged survival of C. elegans, which was nearly identical to that observed with fsrB (data not shown). As shown in Fig. 4C, the fsrBΔ mutant is also less virulent in the mouse i.p. injection model (P < 0.0009).
An Avirulent E. faecalis Mutant Identified by Using C. elegans Is Also Attenuated in the Mouse.
In an attempt to identify previously unknown enterococcal virulence factors, we screened an E. faecalis transposon Tn917 library for mutants that were attenuated in killing nematodes. About 20 such mutants were identified, one of which, designated 1C-C9, was studied in further detail. When 1C-C9 was tested in the mouse i.p. infection model described above, the mice exhibited prolonged survival relative to the mice injected with parental strain OG1RF (P < 0.0104; Fig. 4D). DNA blot hybridization with a Tn917-specific probe indicated that mutant 1C-C9 contains a single transposon insertion (data not shown). By using primers specific to the transposon in combination with arbitrary primers, a DNA fragment flanking the transposon insertion was amplified (22) and sequenced, thereby identifying the gene in which Tn917 had been inserted. The translated sequence was highly homologous to sucrose-6-phosphate hydrolases (ScrB) of other bacterial species (48% identity and 64% similarity with ScrB of Streptococcus sobrinus).
Discussion
Several Gram-negative human pathogens, including P. aeruginosa and S. enterica, have been shown to kill C. elegans when presented to the nematodes as a source of food (6–8, 10). In this article, we demonstrate that Gram-positive human pathogens including E. faecalis, S. aureus, and S. pneumoniae also kill C. elegans, and that E. faecium and S. pyogenes do not exhibit significant killing under the conditions tested. We do not understand why some human pathogens kill C. elegans whereas others do not. Initially, we thought that human pathogens able to infect and kill C. elegans would tend to be unspecialized, opportunistic species such as P. aeruginosa and E. faecalis. However, in this article, we show that S. pneumoniae, with an ecological niche thought to be limited to the human nasal-pharynx, also kills C. elegans. As illustrated in Fig. 1C, one factor that has a critical effect on C. elegans killing is the medium used to grow the bacterial lawn. Escherichia coli does not kill C. elegans when grown on NG medium, but it is an efficient killer when grown on BHI medium. Thus, species such as E. faecium and S. pyogenes, which did not exhibit killing on BHI medium, may kill C. elegans if grown on the appropriate medium. The uses and limitations of C. elegans as a model host need to be clarified and defined with further research; we predict that the list of pathogens able to use C. elegans as a model host will expand as different condition are tested.
In the case of P. aeruginosa, two mechanistically distinct types of C. elegans killing have been identified in our laboratory. “Slow-killing” requires live bacteria and occurs when C. elegans are fed P. aeruginosa PA14 grown on low-osmolarity minimal medium (10). In contrast, “fast-killing” occurs on high-osmolarity medium, does not require live bacteria, and has been shown to be mediated by low molecular weight toxins (8). In the case of E. faecalis, secreted toxins are probably not the primary cause of killing adults, because antibiotic-treated E. faecalis did not kill (data not shown). With the exception of the P. aeruginosa toxin-mediated fast killing of worms, the mechanisms by which live P. aeruginosa, S. enterica, and E. faecalis kill worms is not clear. In all three cases, the worms do not appear to become rapidly paralyzed, as is the case in another P. aeruginosa–C. elegans model system involving P. aeruginosa strain PA01 (7).
When C. elegans are feeding on Escherichia coli, essentially no live bacteria can be recovered from and no intact green fluorescent protein-labeled bacteria can be detected in the digestive system of the nematodes. In contrast, E. faecalis not only accumulates in the intestinal lumen, reaching steady-state titers as high as 105 cfu per worm (6, 10), but as shown in Fig. 2B, proliferates in the gut, resulting in a long-lasting infection that persists for the entire lifespan of the worms. Similar results were obtained previously with S. enterica (6). Overgrowth of E. faecalis and S. enterica in the intestine of C. elegans to the animal's apparent detriment has many characteristics of a true infection. In contrast, although P. aeruginosa also accumulates in the C. elegans gut, it is not able to establish a persistent infection (10). Interestingly, as shown in Fig. 2B and Figs. 3 C and D, E. faecium also accumulates to high titers in the intestinal lumen, although it does not kill adult worms and does not form a persistent infection (Fig. 2C).
P. aeruginosa and S. enterica containing mutations in well known virulence factors such as lasR and phoPQ, respectively, are less virulent in C. elegans, validating the use of C. elegans to identify and study virulence factors relevant for mammalian pathogenesis (6, 10) In this article, we show that two known enterococcal virulence factors, Cyl and FsrB, which have been shown to enhance pathogenesis in various mammalian model systems, also enhance the rate at which C. elegans are killed by E. faecalis. Cyl disrupts membranes of both eukaryotic and prokaryotic cells (23) and is thought to increase tissue damage in mammalian models of endocarditis (1). fsrB putatively encodes a peptide processor necessary for quorum sensing (3). Polar mutations in fsrB previously have been shown to increase the survival time for i.p.-infected mice (3), and in this article we show that a nonpolar deletion in fsrB increases survival time in both the mouse and the nematode. In the C. elegans model, because noncytolytic E. faecalis and a fsrB deletion mutant are significantly attenuated but remain lethal (Fig. 4), it seems that Cyl and FsrB contribute to, but are not sufficient for, C. elegans killing.
In the case of P. aeruginosa strain PA14, we used TnphoA mutagenesis to identify killing-related factors by simply screening for PA14 mutants that exhibited a decreased rate of killing. This procedure resulted in the discovery of a variety of virulence factors that also are relevant for mouse pathogenesis (8, 9, 24). As shown in Fig. 4D, our preliminary experiments show that C. elegans also can be used to identify E. faecalis mutants that are attenuated in a mammalian model. Specifically, we have found that a mutant with an insertion in the scrB gene is less virulent in both the mouse (Fig. 4D) and the nematode (data not shown) models. Although it is not clear whether there is an appreciable amount of sucrose in the mouse peritoneal cavity, sucrose utilization has been shown to play a role in the formation of dental caries and in endocarditis in the related bacterium Streptococcus mutans. Sucrose catabolites are used to synthesize insoluble glucans, exopolymers that may promote adherence to the tooth surface and to the heart valve (25, 26). An alternative explanation for these results is that the Tn917 insertion in scrB exerts a polar effect on a downstream virulence factor.
In contrast to Cyl, FsrB, and ScrB, E. faecalis AS seems to be a virulence factor only in mammalian infections. Because we have very little information concerning the mechanism by which E. faecalis (as well as P. aeruginosa and S. enterica) kill C. elegans, we do not understand why some virulence factors important for mammalian infection play a role in C. elegans killing, whereas others do not. Another factor that limits our mechanistic understanding of nematode–bacterial interactions is the lack of information concerning the C. elegans innate immune response. Although C. elegans seem to have homologs of some Toll signaling pathway components important to innate immunity in organisms as diverse as humans and Drosophila melanogaster, experiments to date have failed to demonstrate a significant role for the Toll pathway in a C. elegans defense response (27). We can hypothesize that shared virulence factors exert their effect by acting on conserved targets shared among diverse organisms. Conversely, virulence factors that have an effect in one species, but not in another, may be targeting factors specific to that species. Therefore, the use of C. elegans as a model host for mammalian pathogenesis likely will be limited to studying virulence factors involved in targeting evolutionarily conserved cellular mechanisms.
In conclusion, we have shown that the clinically important Gram-positive pathogens E. faecalis, S. aureus, and S. pneumoniae kill adult C. elegans. Killing of adult C. elegans by E. faecalis has characteristics of an infectious process including colonization and proliferation of a small inoculum of bacteria. We show that two previously characterized enterococcal factors necessary for virulence in mammalian models, Cyl and FsrB, a protein that seems to be involved in quorum sensing, increase the rate of C. elegans killing. Finally, we provide evidence that C. elegans may be a useful model host for identifying novel Gram-positive virulence factors by showing that at least one E. faecalis mutation found in an initial screen for attenuation in C. elegans killing seems to prolong survival in a mouse model.
Acknowledgments
We thank D. B. Clewell, G. M. Dunny, M. S. Gilmore, W. Haas, and L. E. Hancock for generous gifts of strains and advice and M. S. Gilmore and A. Grossman for helpful comments about the manuscript. This work was supported by a grant from Aventis to Massachusetts General Hospital, by National Institutes of Health Grants AI42399 and AI47923 from the Division of Microbiology and Infectious Diseases and the National Institute of Allergy and Infectious Diseases (to B.E.M.), by a postdoctoral fellowship from the Irvington Institute for Immunological Research (to D.A.G.), and by postdoctoral fellowships from the Howard Hughes Medical Institute (to C.D.S. and E.M.).
Abbreviations
- cfu
colony-forming units
- Cyl
cytolysin
- AS
aggregation substance
- ScrB
sucrose-6-phosphate hydrolases
References
- 1.Chow J W, Thal L A, Perri M B, Vazquez J A, Donabedian S M, Clewell D B, Zervos M J. Antimicrob Agents Chemother. 1993;37:2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ike Y, Hashimoto H, Clewell D B. Infect Immun. 1984;45:528–530. doi: 10.1128/iai.45.2.528-530.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin X, Singh K V, Weinstock G M, Murray B E. Infect Immun. 2000;68:2579–2586. doi: 10.1128/iai.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh K V, Qin X, Weinstock G M, Murray B E. J Infect Dis. 1998;178:1416–1420. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 5.Stevens S X, Jensen H G, Jett B D, Gilmore M S. Invest Ophthalmol Visual Sci. 1992;33:1650–1656. [PubMed] [Google Scholar]
- 6.Aballay A, Yorgey P, Ausubel F M. Curr Biol. 2000;10:1539–1542. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- 7.Darby C, Cosma C L, Thomas J H, Manoil C. Proc Natl Acad Sci USA. 1999;96:15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan-Miklos S, Tan M W, Rahme L G, Ausubel F M. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 9.Tan M W, Ausubel F M. Curr Opin Microbiol. 2000;3:29–34. doi: 10.1016/s1369-5274(99)00047-8. [DOI] [PubMed] [Google Scholar]
- 10.Tan M W, Mahajan-Miklos S, Ausubel F M. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sulston J, Hodgkin J. The Nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 12.Gutierrez J A, Crowley P J, Brown D P, Hillman J D, Youngman P, Bleiweis A S. J Bacteriol. 1996;178:4166–4175. doi: 10.1128/jb.178.14.4166-4175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chenoweth C E, Robinson K A, Schaberg D R. Antimicrob Agents Chemother. 1990;34:1800–1802. doi: 10.1128/aac.34.9.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youngman P, Perkins J B, Losick R. Mol Gen Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- 15.Sahm D F, Kissinger J, Gilmore M S, Murray P R, Mulder R, Solliday J, Clarke B. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunny G M, Brown B L, Clewell D B. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clewell D B. Cell. 1993;73:9–12. doi: 10.1016/0092-8674(93)90153-h. [DOI] [PubMed] [Google Scholar]
- 18.Jett B D, Jensen H G, Nordquist R E, Gilmore M S. Infect Immun. 1992;60:2445–2452. doi: 10.1128/iai.60.6.2445-2452.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreft B, Marre R, Schramm U, Wirth R. Infect Immun. 1992;60:25–30. doi: 10.1128/iai.60.1.25-30.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y A, Sulavik M C, He P, Makinen K K, Makinen P L, Fiedler S, Wirth R, Clewell D B. Infect Immun. 1991;59:415–420. doi: 10.1128/iai.59.1.415-420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin X, Singh K V, Weinstock G M, Murray B E. J Bacteriol. 2001;183:3372–3382. doi: 10.1128/JB.183.11.3372-3382.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Toole G A, Pratt L A, Watnick P I, Newman D K, Weaver V B, Kolter R. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 23.Jett B D, Huycke M M, Gilmore M S. Clin Microbiol Rev. 1994;7:462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan M W, Rahme L G, Sternberg J A, Tompkins R G, Ausubel F M. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loesche W J. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munro C L, Macrina F L. Mol Microbiol. 1993;8:133–142. doi: 10.1111/j.1365-2958.1993.tb01210.x. [DOI] [PubMed] [Google Scholar]
- 27.Pujol N, Link E M, Liu L X, Kurz C L, Alloing G, Tan M, Ray K P, Solari R, Johnson C D, Ewbank J J. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]