Abstract
Introduction
FENDRR is a long non-coding RNA (lncRNA) that mediates the modification of the epigenetic landscape of target promoters by binding to polycomb repressive complex 2. However, the role of FENDRR in breast cancer remains unknown.
Materials and methods
We detected the expression of FENDRR in 52 breast cancer patients’ tissues and five breast cancer cell lines. The association between FENDRR expression and clinicopathological features and prognosis of breast cancer patients was also analyzed. Moreover, cell proliferation assays, flow cytometry analysis, wound-healing assays, and transwell migration assays were performed to detect the biological effects of FENDRR in the breast cancer cells. A xenograft model was used to explore the role of FENDRR expression on tumor growth.
Results
We found that FENDRR expression was lower in breast cancer cell lines and cancerous tissues than in the adjacent normal tissues. Low expression of FENDRR was associated with a shorter overall survival and a shorter progression-free survival in breast cancer patients (p<0.001, p<0.001, respectively). We found that FENDRR inhibits breast cancer cell proliferation and migration and promotes cell apoptosis, while FENDRR knockdown promotes breast cancer cell proliferation and migration and suppresses cell apoptosis. Finally, we also detected that FENDRR overexpression could inhibit tumor growth in a xenograft model.
Conclusion
Our data suggested that FENDRR inhibits breast cancer cell proliferation, promotes cell apoptosis, and is associated with good prognosis in breast cancer. Thus, FENDRR plays an important role in the growth and progression of breast cancer.
Keywords: breast cancer, FENDRR, lncRNA, prognosis, proliferation
Introduction
The vast majority of the human genome is made up of non-coding RNA (ncRNA), apart from about 2% protein-coding genes.1 Long non-coding RNAs (lncRNAs) are a class of ncRNAs, which have a length of more than 200 nucleotides. So far, a large number of lncRNAs have been recognized and found to play functional regulatory roles in the expression of nearby genes, the activation and localization of proteins, and the organization of small RNAs.2 Accumulated findings implicate that the dysregulated expression of lncRNAs plays an important role in the cancer development process, including proliferation, apoptosis, and metastasis.3,4 In addition, recent reports have reported that lncRNAs exhibit distinct gene-expression patterns and play functional roles in various types of carcinomas, including colon cancer, prostate cancer, leukemias, hepatocellular cancer, and breast cancer.5–9 Therefore, functional lncRNAs could be used for clinical cancer diagnosis, and serve as potential therapeutic targets.10
FENDRR located at chr3q13.31 has a length of 3,099 nucleotides and consists of four exons. It was reported that FENDRR is a lateral mesoderm-specific lncRNA and is essential for heart and body wall development in a mouse model.11 FENDRR was confirmed to bind to both Trithorax group/MLL protein complexes (TrxG/MLL) and polycomb repressive complex 2 (PRC2), which play significant roles in the regulation of chromatin structure and gene activity.12,13 Grote et al reported that embryos lacking FENDRR displayed increased expression of several transcription factors regulating differentiation, accompanied by a reduction in PRC2 occupancy along with decreased H3K27 trimethylation and/or an increase in H3K4 trimethylation at their promoters.11 Xu et al reported that low expression of FENDRR occurs in gastric cancer and is associated with poor prognosis.14 FENDRR overexpression could suppress migration and invasion in gastric cancer cells by downregulating fibronectin1 (FN1) and matrix metalloproteinase 2/9 (MMP2/MMP9) expression.14 However, the role of FENDRR in breast cancer remains yet to be found.
In the present study, we found that FENDRR expression was lower in breast cancer cell lines and cancerous tissues than in the adjacent normal tissues. Low expression of FENDRR was associated with a shorter overall survival (OS) and a shorter progression-free survival (PFS) in breast cancer patients. Ectopic expression of FENDRR in breast cancer cells significantly inhibited cell proliferation and migration and promoted cell apoptosis, while FENDRR knockdown promoted cell proliferation and migration and suppressed cell apoptosis. Moreover, we also detected that FENDRR over-expression could inhibit tumor growth in a xenograft model. These results suggested that FENDRR plays an important role in the growth and progression of breast cancer and could be used as a new therapeutic target for breast cancer patients.
Materials and methods
Clinical samples
Breast cancer tissues were collected from The First Affiliated Hospital of Nanjing Medical University (Nanjing, People’s Republic of China) between July 2011 and March 2012. None of the patients received chemotherapy or radiotherapy prior to surgery. All patients whose tissue samples were used in this research provided written informed consent. Experiments were approved by the Ethics Committee of Jinling Hospital and were conducted in compliance with the Helsinki Declaration. Histological parameters were determined in accordance with the criteria of the World Health Organization. Pathologic staging was performed in accordance with the current International Union against Cancer tumor-lymph node metastasis classification.
Immunohistochemistry
Immunohistochemistry (IHC) of 52 breast tumor tissue samples was performed as previously described.15,16
Cell lines and mice
Cell lines MCF-7, BT-474, SK-BR-3, HCC1937, and MDA-MB-231 were purchased in 2015–2016 from the Chinese Academy of Science Committee Type Culture Collection Cell Bank (Shanghai, People’s Republic of China). Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 or DMEM medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37°C in a humidified atmosphere containing 5% CO2. BALB/c athymic nude mice (female, 4–6 weeks) were purchased from the Department of Comparative Medicine, Jinling Hospital (Nanjing, People’s Republic of China), and maintained in a pathogen-free facility. Animal studies were performed in accordance with institutional guidelines.
Plasmids and transient transfection
Plasmids pcDNA-FENDRR and shRNA-FENDRR were all chemically synthesized by Realgene (Nanjing, People’s Republic of China). Cells (1×106 cells/well) were plated in six-well plates 24 h prior to transfection. Plasmids were then transfected into cells using TurboFect Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. Following incubation at 37°C for 24 h, cells were collected. Transient transfection was performed as previously described.15–18
Cell survival (MTT) assay
A total of 1×104 cells per well were seeded into 96-well plates and transfected with required plasmids for 48 h. Following addition of 20 µL of 0.5 mg/mL MTT solution (Sigma-Aldrich, St Louis, MO, USA) to each well, the medium was replaced with 200 µL dimethyl sulfoxide after 4 h and vortexed for 10 min. Absorbance was measured at 490 nm with a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA) to determine the relative numbers of viable cells. Assays were performed independently three times.
Cell apoptosis analysis
Cells were treated with plasmids for 48 h, then harvested by trypsinization (no EDTA), and washed with phosphate-buffered saline. Analysis of the cell apoptosis was performed as previously described.15–18 Each sample was tested in triplicate, and untreated cells were used as controls.
Quantitative real-time PCR
Total cellular RNA was extracted from different cell types using TRIzol (Thermo Fisher Scientific) and reversely transcribed according to the manufacturer’s instruction using the Step One System (Applied Biosystems, Waltham, MA, USA). Relative gene expression was determined by the ΔΔCt method based on glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels, and results were expressed as fold change over different conditions. Primer sequences (forward and reverse, respectively) were as follows: FENDRR, 5′-TAAAATTGCAGATCCTCCG-3′ and 5′-AACGTTCGCATTTAGC-3′; GAPDH, 5′-GGGAGCCAAAAGGGTCAT-3′ and 5′-GAGTCCTTCCACGATACCAA-3′. Quantitative real-time PCR was performed as previously described.15–18
Colony formation assay
Cells were seeded in six-well plates at a density of 500 cells per well. After 14 days, the colonies were fixed with 70% ethanol and stained with 0.1% crystal violet. Then, the number of colonies larger than 1 mm was manually counted. These experiments were repeated at least three times.
Wound-healing assay
Cell migration was measured using a wound-healing assay. In brief, cells were seeded in 12-well plates and cultured to confluence. Wounds of 1.0 mm width were created with a plastic scriber, and cells were washed and incubated in a serum-free medium. After 48 h of wounding, cultures were fixed and observed under a microscope. A minimum of five randomly chosen areas was measured and the distance of cell migration to the wound area was determined.
Transwell migration assays
Cell migration was assessed with modified Boyden chamber (Becton Dickinson, Franklin Lakes, NJ, USA) assays. Briefly, approximately 1.0×105 cells were plated into the upper chamber of a polycarbonate Transwell filter chamber (Corning Inc., Corning, NY, USA). After 24 h, cells that did not migrate were removed from the top side of the inserts with a cotton swab. Migrated cells were stained with crystal violet and fixed in 4% paraformaldehyde followed by counting under a microscope. Cells were counted in five random fields per insert. Three independent experiments were carried out.
Xenograft transplantation
Approximately 5.0×106 MDA-MB-231 cells transfected with pcDNA-FENDRR vector or control vector were subcutaneously transplanted into the right side of the posterior flank of nude mice (n=4 mice/group). After 4 weeks, mice were euthanized and tumors were harvested.
Statistical analysis
SPSS Statistics 21.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis. Data were analyzed using one-way analysis of variance (ANOVA) or Student’s t-test. Data are presented as means ± SD of three independent experiments. The log-rank test was used to assess statistical significance of Kaplan–Meier plots. The chi-square test was used for IHC data. A p-value of <0.05 was considered statistically significant.
Results
FENDRR had low expression in breast cancer tissues and cell lines, and was associated with good prognosis in breast cancer patients
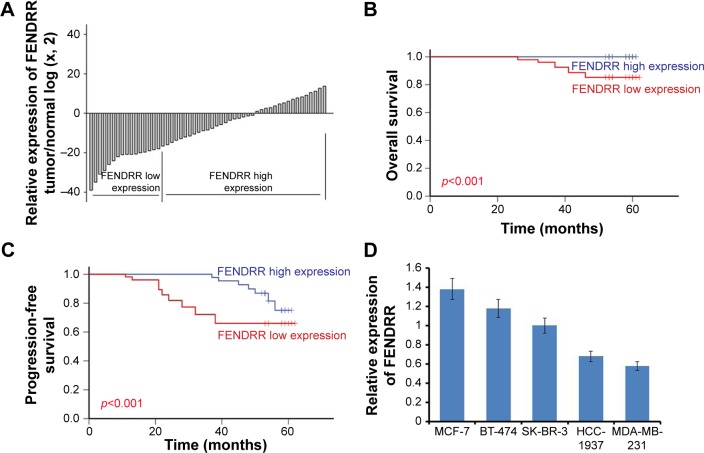
To investigate the role of FENDRR in breast cancer, we detected the expression levels of FENDRR in 52 breast cancer samples using quantitative polymerase chain reaction assays. As shown in Figure 1A, FENDRR expression was significantly lower in tumor tissues than in the adjacent normal tissues (p<0.05). The patients were divided into a low FENDRR expression group (16) and a high FENDRR expression group (36) according to the mean value of relative FENDRR expression (15-fold, adjacent normal/tumor). Relationship between expression of FENDRR and clinicopathological features of the 52 breast cancer patients is shown in Table 1. We found that the expression of FENDRR was significantly associated with the lymph nodes status, and progesterone receptor (PR), and HER-2 expression (p=0.006, 0.016, 0.008, respectively). The low expression levels of FENDRR were associated with more lymph nodes metastasis, PR negative expression, and HER-2 positive expression. However, there was no significant correlation between FEN-DRR expression and other clinicopathological features such as age, tumor size, histological grade, and estrogen receptor (ER) expression status (p>0.005, Table 1). We also analyzed the association between FENDRR expression levels and the prognosis of the 52 breast cancer patients. As shown in Figure 1B and C, FENDRR expression and the clinicopathological characteristics on PFS and OS were evaluated. The results showed that patients with a low FENDRR expression had a shorter OS and a shorter PFS (p<0.001, Figure 1B; p<0.001, Figure 1C, respectively). Moreover, univariate and multivariate analyses of clinical variables considered as potential predictors of survival are shown in Tables 2 and 3. Analysis in both univariate and multivariate Cox proportional hazards model showed that FENDRR expression was strongly associated with OS (p=0.001, p=0.047, respectively; Table 2). Furthermore, the expression of FENDRR was an independent prognostic indicator of PFS (hazard ratio [HR] =0.578; 95% confidence interval [CI], 0.454–0.735; p=0.001) and OS (HR =0.001; 95% CI, 0.001–0.675; p=0.047) in patients with breast cancer (Tables 2 and 3). Next, we detected the expression of FENDRR in five breast cancer cell lines; SK-BR-3, MCF-7, HCC-1937, BT-474, and MDA-MB-231. We found significantly lower expression of FENDRR in triple-negative breast cancer (TNBC) cell lines HCC-1937 and MDA-MB-231, while the expression levels of FENDRR in the non-TNBC cell lines SK-BR-3, BT-474, and MCF-7 were high (Figure 1D). Taken together, these results clearly showed that FENDRR had low expression in breast cancer tissues and cell lines, and was associated with good prognosis in breast cancer patients.
Figure 1.
FENDRR had low expression in breast cancer tissues and cell lines, and was associated with good prognosis in breast cancer patients.
Notes: (A) FENDRR expression is examined by quantitative real-time PCR in 52 paired human breast cancer tissues and adjacent noncancerous tissues. Data are represented as log 2-fold change (tumor/normal). The patients were divided into a FENDRR low expression group (16) and a FENDRR high expression group (36) according to the mean value of relative FENDRR expression. (B) Overall survival in high and low FENDRR-expressing breast cancer tissues. (C) Progression-free survival in high and low FENDRR-expressing breast cancer tissues. (D) Real-time PCR analysis of FENDRR expression in five breast cancer cell lines. The error bars represent the standard deviation.
Abbreviation: PCR, polymerase chain reaction.
Table 1.
Relationship between expression of FENDRR and clinicopathological features of breast cancer patients
| Variables | Total (n=52)
|
FENDRR low (n=16)
|
FENDRR high (n=36)
|
p-value | |||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| Age | 0.704 | ||||||
| <45 years | 43 | 82.7 | 14 | 87.5 | 29 | 80.6 | |
| ≥45 years | 9 | 17.3 | 2 | 12.5 | 7 | 19.4 | |
| Tumor size, cm | 0.071 | ||||||
| <2 | 16 | 30.8 | 2 | 12.5 | 14 | 38.9 | |
| 2–5 | 33 | 63.5 | 12 | 75.0 | 21 | 58.3 | |
| ≥5 | 3 | 5.8 | 2 | 12.5 | 1 | 2.8 | |
| Histological grade | 0.064 | ||||||
| I–II | 21 | 40.4 | 3 | 18.8 | 18 | 50.0 | |
| III | 31 | 59.6 | 13 | 81.3 | 18 | 50.0 | |
| Lymph nodes | 0.006* | ||||||
| 0 | 27 | 51.9 | 4 | 25.0 | 23 | 63.9 | |
| 1–3 | 11 | 21.2 | 3 | 18.8 | 8 | 22.2 | |
| ≥4 | 14 | 26.9 | 9 | 56.3 | 5 | 13.9 | |
| ER | 0.179 | ||||||
| Positive | 33 | 63.5 | 8 | 50.0 | 25 | 69.4 | |
| Negative | 19 | 36.5 | 8 | 50.0 | 11 | 30.6 | |
| PR | 0.016* | ||||||
| Positive | 27 | 51.9 | 4 | 25.0 | 23 | 63.9 | |
| Negative | 25 | 48.1 | 12 | 75.0 | 13 | 36.1 | |
| HER-2 | 0.008* | ||||||
| Positive | 14 | 26.9 | 9 | 56.3 | 7 | 19.4 | |
| Negative | 38 | 73.1 | 7 | 43.8 | 29 | 80.6 | |
Note:
p<0.05 shown in bold.
Abbreviations: HR, hazard ratio; CI, confidence interval; ER, estrogen receptor; PR, progesterone receptor.
Table 2.
Univariate and multivariate Cox regression analysis of overall survival in breast cancer patients
| Variables | Univariate analysis
|
Multivariate analysis
|
||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Age | 1.020 (1.005–1.035) | 0.009* | 9.438 (3.081–28.91) | 0.001* |
| Histological grade (I + II vs III) | 0.021 (0.004–0.115) | 0.001* | 0.001 (0.000–1.8E4) | 0.185 |
| Tumor size (<2 cm vs 2–5 cm vs ≥5 cm) | 1.493 (1.388–1.606) | 0.001* | 0.008 (0.000–0.363) | 0.013* |
| Lymph nodes (0 vs 1–3 vs ≥4) | 2.387 (1.863–3.059) | 0.001* | 131.2 (4.625–3,720.4) | 0.004* |
| ER (negative vs positive) | 2.045 (1.256–3.331) | 0.004* | 1.2E9 (4.79–2.8E17) | 0.034* |
| PR (negative vs positive) | 0.014 (0.003–0.074) | 0.001* | 0.001 (0.000–2.367) | 0.053 |
| HER-2 (negative vs positive) | 14.13 (7.854–25.42) | 0.001* | 2.2E9 (225–2.2E16) | 0.009* |
| FENDRR (negative vs positive) | 0.003 (0.001–0.028) | 0.001* | 0.001 (0.001–0.675) | 0.047* |
Note:
p<0.05 shown in bold.
Abbreviations: HR, hazard ratio; CI, confidence interval; ER, estrogen receptor; PR, progesterone receptor.
Table 3.
Univariate and multivariate Cox regression analysis of progress free survival in breast cancer patients
| Variables | Univariate analysis
|
Multivariate analysis
|
||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Age | 0.983 (0.976–0.989) | 0.001* | 1.003 (0.995–1.012) | 0.411 |
| Histological grade (I + II vs III) | 0.503 (0.416–0.608) | 0.001* | 0.569 (0.452–0.716) | 0.001* |
| Tumor size (<2 cm vs 2–5 cm vs ≥5 cm) | 1.744 (1.650–1.843) | 0.001* | 1.638 (1.527–1.757) | 0.001* |
| Lymph nodes (0 vs 1–3 vs ≥4) | 1.871 (1.697–2.063) | 0.001* | 1.526 (1.332–1.749) | 0.001* |
| ER (negative vs positive) | 0.426 (0.360–0.506) | 0.001* | 1.001 (0.756–1.326) | 0.993 |
| PR (negative vs positive) | 0.366 (0.305–0.439) | 0.001* | 0.536 (0.396–0.727) | 0.001* |
| HER-2 (negative vs positive) | 3.186 (2.687–3.776) | 0.001* | 2.481 (2.027–3.037) | 0.001* |
| FENDRR (negative vs positive) | 0.486 (0.408–0.578) | 0.001* | 0.578 (0.454–0.735) | 0.001* |
Note:
p<0.05 shown in bold.
Abbreviations: HR, hazard ratio; CI, confidence interval; ER, estrogen receptor; PR, progesterone receptor.
FENDRR inhibits breast cancer cell proliferation and promotes cell apoptosis
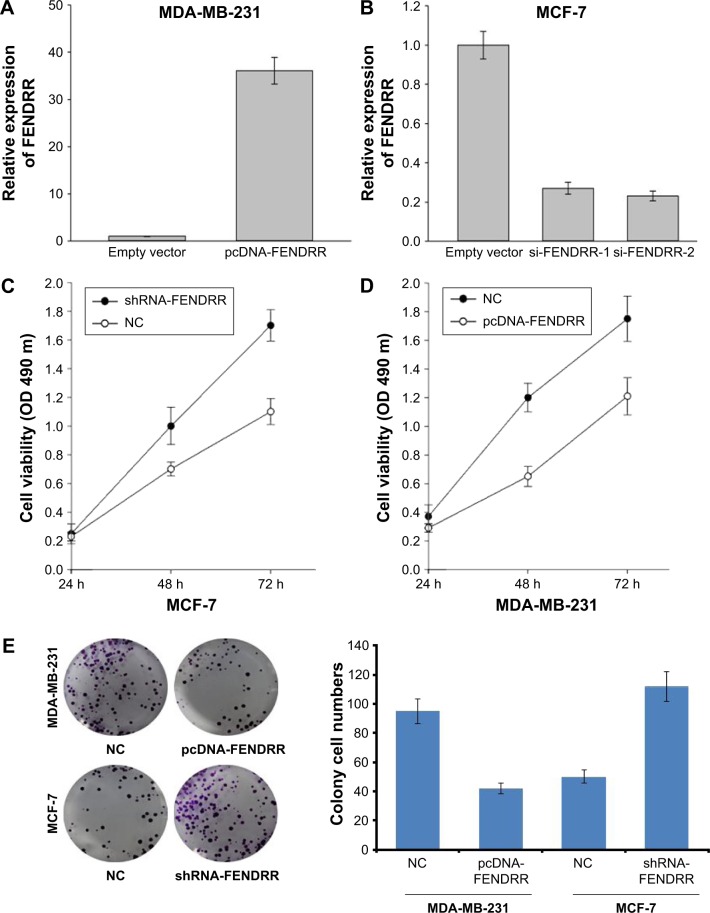
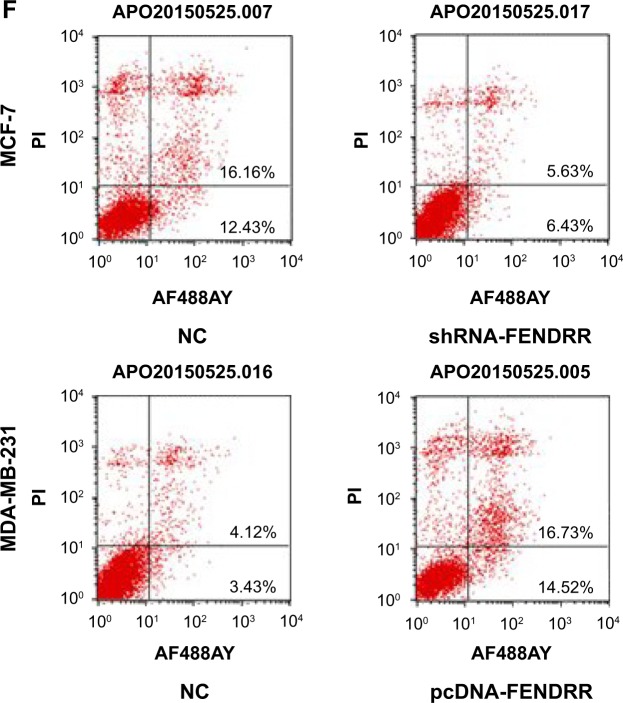
As shown in Figure 1D, the expression of FENDRR was high in the MCF-7 cell line and was low in MDA-MB-231 cell line. Thus, we transfected MDA-MB-231 cells with pcDNA-FEN-DRR vector to induce the expression of FENDRR, and MCF-7 cells with shRNA-FENDRR vector to inhibit FENDRR expression (Figure 2A and B). Next, using MTT assay and cell clone formation assay, we detected cell proliferation in the MDA-MB-231 and MCF-7 cell lines after being transfected with FENDRR or shRNA-FENDRR vector. As shown in Figure 2C and D, FENDRR overexpression inhibited cell viability, while suppression of FENDRR expression promoted cell viability in the breast cancer cell lines. We also detected that FENDRR overexpression inhibited cell clone formation, and inhibition of FENDRR expression promoted cell clone formation (Figure 2E). Next, flow cytometry analysis was used to detect the effect of FENDRR on cell apoptosis in the breast cancer cell lines. As shown in Figure 2F, FENDRR promoted MDA-MB-231 cells apoptosis and knockdown of FENDRR suppressed MCF-7 cell apoptosis. Taken together, our observation indicated that FENDRR inhibits breast cancer cell proliferation and promotes cell apoptosis.
Figure 2.
FENDRR inhibits breast cancer cell proliferation and promotes cell apoptosis.
Notes: (A) RT-PCR analysis of FENDRR expression in the MDA-MB-231 cells transfected with a control or pcDNA-FENDRR vector. (B) RT-PCR analysis of FENDRR expression in the MCF-7 cells transfected with a control, si-FENDRR-1, or si-FENDRR-2 vector. (C) Cell viability of MCF-7 cells transfected with a control or shRNA-FENDRR vector. (D) Cell viability of MDA-MB-231 cells transfected with a control or pcDNA-FENDRR vector. (E) Colony formation assay of MCF-7 or MDA-MB-231 cells transfected with a normal control, pcDNA-FENDRR, or shRNA-FENDRR. (F) Flow cytometry assay of MCF-7 or MDA-MB-231 cells transfected with a normal control, pcDNA-FENDRR, or shRNA-FENDRR. The data represent the mean ± standard deviation.
Abbreviations: PI, propidium iodide; NC, normal control; RT-PCR, real-time polymerase chain reaction.
FENDRR inhibits breast cancer cell migration
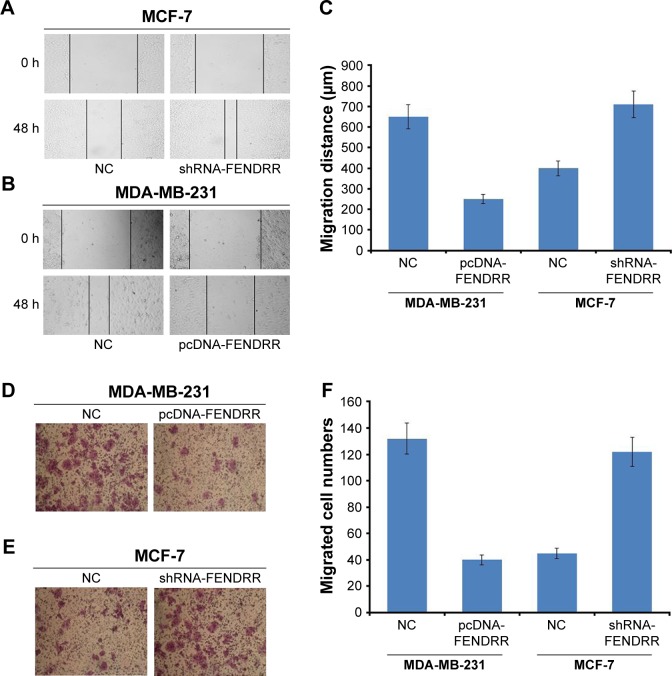
To investigate the effect of FENDRR on the motility of breast cancer cells, we detected cell migration in the MDA-MB-231 and MCF-7 cell lines after being transfected with FENDRR or shRNA-FENDRR vector using the wound-healing assay. As shown in Figure 3A–C, FENDRR overexpression inhibited cell migration, while suppression of FENDRR expression promoted cell migration in the breast cancer cell lines. Transwell migration assay was also used to detect the effect of FENDRR on cell motility in the breast cancer cell lines. As shown in Figure 3D–F, FENDRR suppressed MDA-MB-231 cells migration and knockdown of FENDRR promoted MCF-7 cell migration. Taken together, our observation indicated that FENDRR inhibits breast cancer cell migration.
Figure 3.
FENDRR inhibits breast cancer cell migration.
Notes: (A) Wound-healing assay of MCF-7 cells transfected with a control or shRNA-FENDRR vector. (B) Wound-healing assay of MDA-MB-231 cells transfected with a control or pcDNA-FENDRR vector. (C) Quantified analysis of wound healing of MCF-7 and MDA-MB-231 cells. The data represent the mean ± SD. (D) Migration assay of MDA-MB-231 cells transfected with a control or pcDNA-FENDRR vector. (E) Migration assay of MCF-7 cells transfected with a control or shRNA-FENDRR vector. (F) Quantified analysis of cell migration of MCF-7 and MDA-MB-231 cells. The data represent the mean ± standard deviation.
Abbreviation: NC, normal control.
FENDRR inhibits tumor growth in a xenograft model
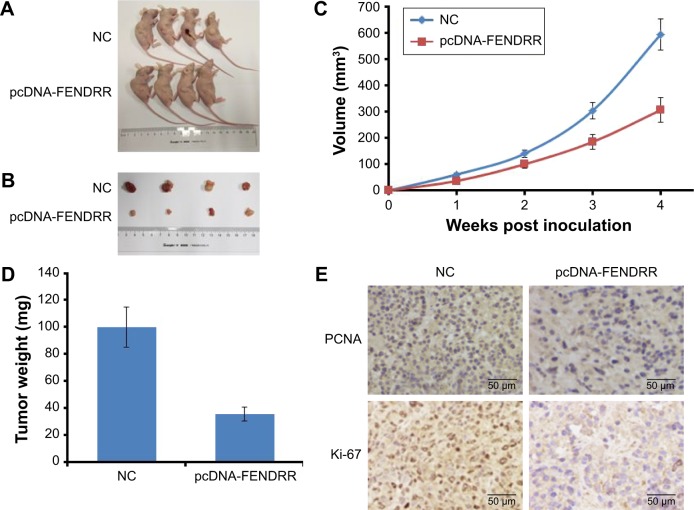
In order to explore the effect of FENDRR in the breast cancer cells in vivo, we used BALB/c mice to establish a breast cancer subcutaneous transplantation tumor model with MDA-MB-231 cells transfected with pcDNA-FENDRR vector or control vector. After 4 weeks of transplantation, FENDRR dramatically inhibited the growth of MDA-MB-231 tumor in vivo, in comparison with the control treatment (Figure 4A and B). Moreover, we also found that FENDRR transfection could result in a significantly reduced tumor size and weight (Figure 4C and D). Tumor sections from each treatment were analyzed by IHC of proliferating cell nuclear antigen (PCNA) and proliferation index Ki-67. As shown in Figure 4E, the expression levels of PCNA and Ki-67 were low in the FENDRR transfection group, in comparison with the control group (Figure 4E). Collectively, FENDRR inhibits tumor growth in a xenograft model.
Figure 4.
FENDRR inhibits tumor growth in a xenograft model.
Notes: (A, B) Representative image of xenograft tumors after being infected with MDA-MB-231 cells transfected with a control or pcDNA-FENDRR vector. (C) Tumor growth curve. Tumor growth was measured every week after injecting tumor cells. The error bars represent SD. (D) Tumor weight when the tumors were harvested. The data represent the mean ± standard deviation. (E) Representative immunohistochemical staining of proliferating cell nuclear antigen and Ki-67 in the MDA-MB-231 xenografts.
Abbreviations: NC, normal control; PCNA, proliferating cell nuclear antigen.
Discussion
In the present study, we investigated the role of FENDRR in breast cancer. We detected the expression of FENDRR in 52 breast cancer patients’ tissues and five breast cancer cell lines. We found significantly lower expression of FENDRR in TNBC cell lines HCC-1937 and MDA-MB-231, while the expression levels of FENDRR in the non-TNBC cell lines SK-BR-3, BT-474, and MCF-7 were high. We also detected that FENDRR levels were lower in breast cancer cell lines and cancerous tissues than in the adjacent normal tissues. The expression of FENDRR was significantly associated with the lymph nodes status and PR and HER-2 expression. Low expression of FENDRR was associated with more lymph nodes metastasis, PR negative expression, and HER-2 positive expression. However, there was no significant correlation between FENDRR expression and other clinicopathological features such as age, tumor size, histological grade, and ER expression status. Moreover, low expression of FENDRR was significantly associated with a shorter OS and a shorter PFS in breast cancer patients. Univariate and multivariate analyses of clinical variables also showed that FENDRR expression was strongly associated with OS and PFS in our study cohort suggesting that FENDRR expression is an independent prognostic indicator of PFS and OS in patients with breast cancer.
We also detected the biological effects of FENDRR in the breast cancer cells using cell proliferation assays, flow cytometry analysis, wound-healing assays, and transwell migration assays. We found that ectopic expression of FENDRR inhibited breast cancer cell proliferation and migration and promoted cell apoptosis, whereas knockdown of endogenous FENDRR expression significantly enhanced breast cancer cells proliferation and migration and suppressed cell apoptosis. We also observed that FENDRR overexpression could inhibit tumor growth in a xenograft model. These results suggested that lncRNA FENDRR inhibits breast cancer cell proliferation, promotes cell apoptosis, and is associated with good prognosis in breast cancer. Thus, FENDRR plays an important role in the growth and progression of breast cancer.
FENDRR is a lateral mesoderm-specific lncRNA and is essential for proper development of tissues derived from lateral mesoderm, specifically the heart and the body wall.11,19 It was reported to act by modifying the chromatin signatures of genes involved in the formation and differentiation of the lateral mesoderm lineage through binding to both the PRC2 and TrxG/MLL complexes.11 Embryos lacking FENDRR displayed increased expression of several transcription factors regulating lateral plate or cardiac mesoderm differentiation, accompanied by a drastic reduction in PRC2 occupancy along with decreased H3K27 trimethylation and/or an increase in H3K4 trimethylation at their promoters.11 Recently, it was also reported that FENDRR contributes to invasion and metastasis in gastric cancer by downregulating FN1 and MMP2/MMP9 expression.14 Sauvageau et al reported that FENDRR was expressed at high levels in adult lungs, and lower levels are detectable in colon, liver, spleen, and brain.20 In the present study, we found that FENDRR has low expression in breast cancer cell lines and cancerous tissues, and could inhibit cell proliferation, promote cell apoptosis, and is associated with good prognosis in breast cancer. These results highlight the role of FENDRR on the regulation of growth and progression of solid cancer. However, the underlying mechanism of FENDRR in cell proliferation should be investigated in further study.
In conclusion, our results suggested that FENDRR has low expression in breast cancer cell lines and cancerous tissues, and inhibits cell proliferation, promotes cell apoptosis, and is associated with good prognosis in breast cancer. Thus, FENDRR plays an important role in the growth and progression of breast cancer. To the best of our knowledge, this is the first report revealing the role of lncRNA FENDRR in breast cancer patients and cell lines. Our results would certainly be helpful for studying FENDRR expression and its effect on the growth and progression of breast cancer and support its use as a marker in personalized therapy for breast cancer. However, large studies on underlying mechanisms involving FENDRR expression, associated with the prognosis of breast cancer patients, are needed to validate our findings.
Acknowledgments
This project was supported by National Natural Science Foundation of China (No 81470357, 81773102).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. 2010;19(R2):R162–R168. doi: 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muers M. RNA: genome-wide views of long non-coding RNAs. Nat Rev Genet. 2011;12(11):742. doi: 10.1038/nrg3088. [DOI] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long non-coding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Pibouin L, Villaudy J, Ferbus D, et al. Cloning of the mRNA of overexpression in colon carcinoma-1: a sequence overexpressed in a subset of colon carcinomas. Cancer Genet Cytogenet. 2002;133(1):55–60. doi: 10.1016/s0165-4608(01)00634-3. [DOI] [PubMed] [Google Scholar]
- 6.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25(3):135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 8.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26(6):851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 9.Douglass S, Ali S, Meeson AP, Browell D, Kirby JA. The role of FOXP3 in the development and metastatic spread of breast cancer. Cancer Metastasis Rev. 2012;31(3–4):843–854. doi: 10.1007/s10555-012-9395-3. [DOI] [PubMed] [Google Scholar]
- 10.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 11.Grote P, Wittler L, Hendrix D, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu TP, Huang MD, Xia R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. doi: 10.1186/s13045-014-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim T, Lee H, Han S, Oh D, Im S, Bang Y. The comparison of the benefits obtained from platinum-containing chemotherapy between triple-negative and non-triple-negative metastatic breast cancer. [Accessed January 08, 2018]. Available from: http://ascopubs.org/doi/abs/10.1200/jco.2010.28.15_suppl.1071.
- 16.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bussard KM, Mutkus L, Stumpf K, Gomez-Manzano C, Marini FC. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016;18(1):84. doi: 10.1186/s13058-016-0740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorusso G, Rüegg C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem Cell Biol. 2008;130(6):1091–1103. doi: 10.1007/s00418-008-0530-8. [DOI] [PubMed] [Google Scholar]
- 19.Grote P, Herrmann BG. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013;10(10):1579–1585. doi: 10.4161/rna.26165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauvageau M, Goff LA, Lodato S, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]