Abstract
Gastric cancer is one of the most fatal cancers in the world. Many efforts in recent years have attempted to find effective proteins in gastric cancer. By using a comprehensive list of proteins involved in gastric cancer, scientists were able to retrieve interaction information. The study of protein-protein interaction networks through systems biology based analysis provides appropriate strategies to discover candidate proteins and key biological pathways.
In this study, we investigated dominant functional themes and centrality parameters including betweenness as well as the degree of each topological clusters and expressionally active sub-networks in the resulted network. The results of functional analysis on gene sets showed that neurotrophin signaling pathway, cell cycle and nucleotide excision possess the strongest enrichment signals. According to the computed centrality parameters, HNF4A, TAF1 and TP53 manifested as the most significant nodes in the interaction network of the engaged proteins in gastric cancer. This study also demonstrates pathways and proteins that are applicable as diagnostic markers and therapeutic targets for future attempts to overcome gastric cancer.
Keywords: Gastric cancer, Systems biology, Neurotrophin signaling pathway, HNF4A, TAF1, TP53
Highlights
-
•
A systematic study of protein-protein interaction networks through comprehensive extracted list of proteins involved in gastric cancer.
-
•
Dominant functional theme and pathways of each topological clusters and expressionally active subnetworks were reported.
-
•
The most effective proteins in gastric cancer formation were proposed according to the computed centrality parameters.
-
•
HNF4A, TAF1and TP53 were mentioned as the key proteins in gastric cancer.
1. Introduction
Gastric cancer is the third cause of death by cancer and the fifth most common cancer worldwide [1]. Like most other types of cancer, in addition to genetic factors non-genetic factors such as smoking, alcohol consumption, poor diet, physical inactivity, viral infections and stress increase the risk of being affected by this type of cancer [2], [3]. Furthermore, the role of H. pylori infection in gastric cancer has been proven [4], [5].
Numerous studies investigate the causes and genetic factors involved in gastric cancer, where effective proteins have been identified in the cancer's pathogenesis, and most often the expression level of receptor tyrosine-protein kinase erbB-2 (ERBB2) which increases its levels in gastric cancer [6], [7]. Likewise, cellular tumor antigen p53 involved predominantly in cell division regulation and apoptosis induction, mutates in most cancers [8], [9], [10]. As such, Gastrokine (GKN1) reducing the expression of gastrin-CCKBR signaling pathway is capable of preventing gastric cancer [11], [12], [13], [14]. This protein can also prevent the invasion of cancer cells into other tissues through inactivation of NF-kappaB pathway [15].
In addition to the aforementioned proteins, other biological molecules involved in cancer were detected, including miR-145 which prevents the tumor formation through a vitamin D3-dependent pathway, and its expression level decreases in gastric cancer cells [16].
As intracellular operator units, proteins interact with other molecules for their function in the cell. Disease or health condition of an organism can be determined by such interactions [17]. Deployment of interactions between proteins and their related networks remains a determinative method in biological cell studies. Investigating and constructing such networks improves our knowledge of physiological mechanisms in disease and health conditions [18], [19].
In high throughput based methods applied to identify the potential treatment or diagnosis targets, only genes or proteins with significant expressional changes are applicable; a single criteria cannot be an indicator, because proteins such as various types of kinases have not shown significant expressional changes while their participation in a variety of cancers is certified [20]. On the other hand, it has been proven that elevated levels of varied proteins is not due to cancer, rather a result of increased physiological requirements [21].
Defining a threshold for output data in these methods would result in excluding unchanged level proteins from the study and considering less effective proteins. In this study, we have tried to pass through these problems using a new perspective based on recent systems biology methods. We hypothesized that the involved proteins in cancer are concentrated in limited numbers of cellular signaling pathways which lead to subsequent cellular changes.
To date, the only curing method for gastric cancer therapy is surgery, where chemotherapy has very limited effect, if any, lowering the quality of life. An urgent need for alternative curing strategies leads us to study the protein-protein interaction networks through systems biology-based approaches as an appropriate methodology to discover candidate proteins and key biological pathways in this mortal disease, which claims over 700,000 deaths each year [22]. Identifying these proteins and pathways according to the proposed systems biology enables us to introduce potential therapeutic targets as well as key diagnostic markers for gastric cancer.
2. Materials and methods
2.1. Collecting proteins involved in gastric cancer
PubMed database (www.ncbi.nlm.nih.gov/pubmed) search was performed using “gastric cancer” keyword, limited to the article title and excluding case reports. Validated introduced proteins were extracted from published articles since 2014.
2.2. Interaction network construction
Interaction information of introduced proteins in literature was gathered from well-known protein-protein interaction databases including Reactome [23], [24], KEGG [25], BIND [26], CCSB [27], DIP [28], GRID [29], HPRD [30], IntAct [31], MINT [32] and MDC [33] using MiMI algorithm in Cytoscape 2.8.3 platform [34]. Information from different sources was retrieved and merged in MiMI repository based on an intelligent integration strategy [35].
2.3. Identification of sub-networks
In order to identify topologically highly dense areas in the network, clusters were determined using Cytoscape plugins including AllegroMCODE and clusterMaker which is based on Community Clustering (GLay) and Markov Clustering (MCL) algorithms. The clustering algorithm of AllegroMCODE plugin is Molecular COmplex DEtection (MCODE), based on node weighting according to the local neighborhood density. This algorithm performs in three steps including vertex weighting, molecular complex prediction and post-processing to filter or add proteins to the predicted complex [36]. MCODE parameters included Degree cutoff: 2, Node score cutoff: 0.5, K-score: 5 and Max. Depth: 100.
GLay by dynamically linking highly optimized C functions JAVA program, provides assorted collection of versatile community structure algorithms and graph layout functions for network clustering and structured visualization [37].
The Markov Cluster (MCL) defines a sequence of matrices by alternation of two operators on a generating matrix. It is basically all that is needed for the clustering of graphs, but it is useful to distinguish between the algorithm and the algebraic process employed by the algorithm. This algorithm performs subnet recognition based on network simulation [38].
2.4. Annotating network with microarray data
Arrayexpress database (www.ebi.ac.uk/arrayexpress) was searched using multiple criteria such as Affymetrix HGU133 plus platform, RNA assay, and simultaneous access to both cancer and non-cancer samples. Expression data of the series GSE19826 were applied for annotating the network, performing quality control and pre-processing using ArrayAnalysis modules (http://www.arrayanalysis.org) [39]. Normalizing statistical analysis, multiple-testing corrections on p-value and generating annotations using GEO2R software (www.ncbi.nlm.nih.gov/geo/geo2r). Were performed using imported expression data into the network, jActiveModules 3.1 detected active expression sub-networks [40], [41].
2.5. Gene set analysis
The Database for Annotation, Visualization and Integrated Discovery, DAVID (https://david.ncifcrf.gov) is one of the most efficient online tools to organize and annotate heterogeneous data from high-throughput techniques such as microarray. This program includes 68 gene enrichment tools representing the gene sets in four different modules including Annotation tools, GOchart, KEGGchart and Domainchart [42], [43], [44]. Gene Ontology (GO) terms and pathways of detected sub-networks and clusters were retrieved using DAVID functional annotation tool.
2.6. Computing centrality parameters
In order to identify hubs, centrality parameters including Betweenness and Degree for each node were calculated with CentiScaPe 2.1 in the network and sub-networks were detected by jActiveModules, AllegroMCODE, GLay and MCL. The Degree index shows the number of directly connected edges to each node. The Role of a node in the linking with the rest of the network nodes is evaluated by the Betweenness index [45].
3. Results
3.1. Collecting the proteins involved in gastric cancer
We retrieved a total of 3500 articles based on the defined criteria from PubMed, which over 600 recently published articles. Sixty articles referred to 72 different proteins involved in the pathogenesis of gastric cancer (Supplementary Table 1).
3.2. Drawing interactive network and determining sub-networks
MiMI algorithm has the ability to search multiple databases. In addition to displaying interactions (edges) between seeds (protein input) and first neighbors, it can show the identified edges between first neighbors of different seeds. The primary interactive network with 1673 nodes (proteins) and 21,548 edges (interactions) was created using 72 seed proteins obtained from our bibliographic data with Cytoscape software and its MiMI plugin. We used AllegroMCODE (Table 1), GLay and MCL plugins (see Supplementary Table 2) to identify high-density areas of our constructed network, which can be sets of protein acting as a complex within the cell, and this calculation resulted in 4 subnetworks.
Table 1.
Sub-networks created by AllegroMCODE.
| Cluster name | Score | Nodes | Edges |
|---|---|---|---|
| 1 | 27.323 | 254 | 6940 |
| 2 | 7.571 | 182 | 1378 |
| 3 | 5.48 | 98 | 537 |
| 4 | 4.456 | 296 | 1319 |
3.3. Loading microarray data in the network
With regard to the designated filters, we obtained only two datasets from ArrayExpress database, including E-GEOD-19826 and E-TABM-424. E-TABM-424 datasets were excluded because of the low number of samples (a tumor sample and a normal sample). After qualitative analysis of E-GEOD-19826 datasets with ArrayAnalysis, we found 7 slides lacking the necessary criteria for our statistical analyses (GSM495053 due to the paint stains on the slide, and GSM495051, GSM495057, GSM495063, GSM495071, GSM495072, and GSM495073 due to the contamination with other tissue cells), and hence they were excluded from our review set, and the rest of the slides were analyzed using GEO2R software embedded in GEO (normalization, statistical tests, p-value correction, and adding commentary). After adding expression data to our network nodes, calculation and classification using jActiveModules plugin, and considering the amount of adj. P.Val, the active expression sub-networks were identified.
3.4. Study of functional relationship and analysis of gene sets
DAVID functional annotation tool was utilized in order to identify related biological pathways and functions within each of the sub-networks.
Evaluation of the main network using KEGG pathways showed that the neurotrophin signaling pathway, mitotic cell cycle, and ERBB pathways were mostly involved in gastric cancer (Table 2). The enrichment on subnetworks showed that the pathways of nucleotide excision repair, cell cycle, pathways in cancer, neurotrophin signaling pathway, and focal adhesion manifested themselves as the highly involved pathways in the development of gastric cancer.
Table 2.
Results of enrichment of nodes.
| Main network | |||
|---|---|---|---|
| KEGG Code | KEGG Term | Count | P Value |
| hsa04110 | Cell cycle | 102 | 2.95E-52 |
| hsa05200 | Pathways in cancer | 179 | 1.73E-50 |
| hsa04722 | Neurotrophin signaling pathway | 86 | 1.77E-34 |
| hsa05212 | Pancreatic cancer | 62 | 4.82E-34 |
| hsa05220 | Chronic myeloid leukemia | 62 | 4.94E-32 |
| hsa04510 | Focal adhesion | 109 | 8.68E-30 |
| hsa05215 | Prostate cancer | 63 | 5.82E-26 |
| hsa04012 | ErbB signaling pathway | 60 | 8.46E-24 |
| hsa04914 | Progesterone-mediated oocyte maturation | 58 | 2.89E-22 |
| hsa04662 | B cell receptor signaling pathway | 53 | 8.65E-22 |
| AllegroMCODE Cluster 1 | |||
| hsa03420 | Nucleotide excision repair | 27 | 5.41E-30 |
| hsa04110 | Cell cycle | 36 | 2.33E-26 |
| hsa03030 | DNA replication | 23 | 6.34E-26 |
| hsa03010 | Ribosome | 23 | 1.17E-15 |
| hsa03430 | Mismatch repair | 11 | 2.33E-10 |
| hsa03022 | Basal transcription factors | 10 | 3.87E-07 |
| hsa04810 | Regulation of actin cytoskeleton | 19 | 3.54E-05 |
| hsa03410 | Base excision repair | 7 | 4.12E-04 |
| hsa04662 | B cell receptor signaling pathway | 9 | 0.001264 |
| hsa00240 | Pyrimidine metabolism | 10 | 0.001487 |
| jActiveModules Subnetwork 1 | |||
| hsa05200 | Pathways in cancer | 70 | 1.47E-16 |
| hsa04110 | Cell cycle | 39 | 1.04E-14 |
| hsa04722 | Neurotrophin signaling pathway | 35 | 7.62E-12 |
| hsa04660 | T cell receptor signaling pathway | 28 | 1.10E-08 |
| hsa05220 | Chronic myeloid leukemia | 23 | 1.11E-08 |
| hsa03420 | Nucleotide excision repair | 17 | 3.99E-08 |
| hsa04662 | B cell receptor signaling pathway | 22 | 5.97E-08 |
| hsa05215 | Prostate cancer | 24 | 7.18E-08 |
| hsa05219 | Bladder cancer | 16 | 1.42E-07 |
3.5. Network evaluation indices
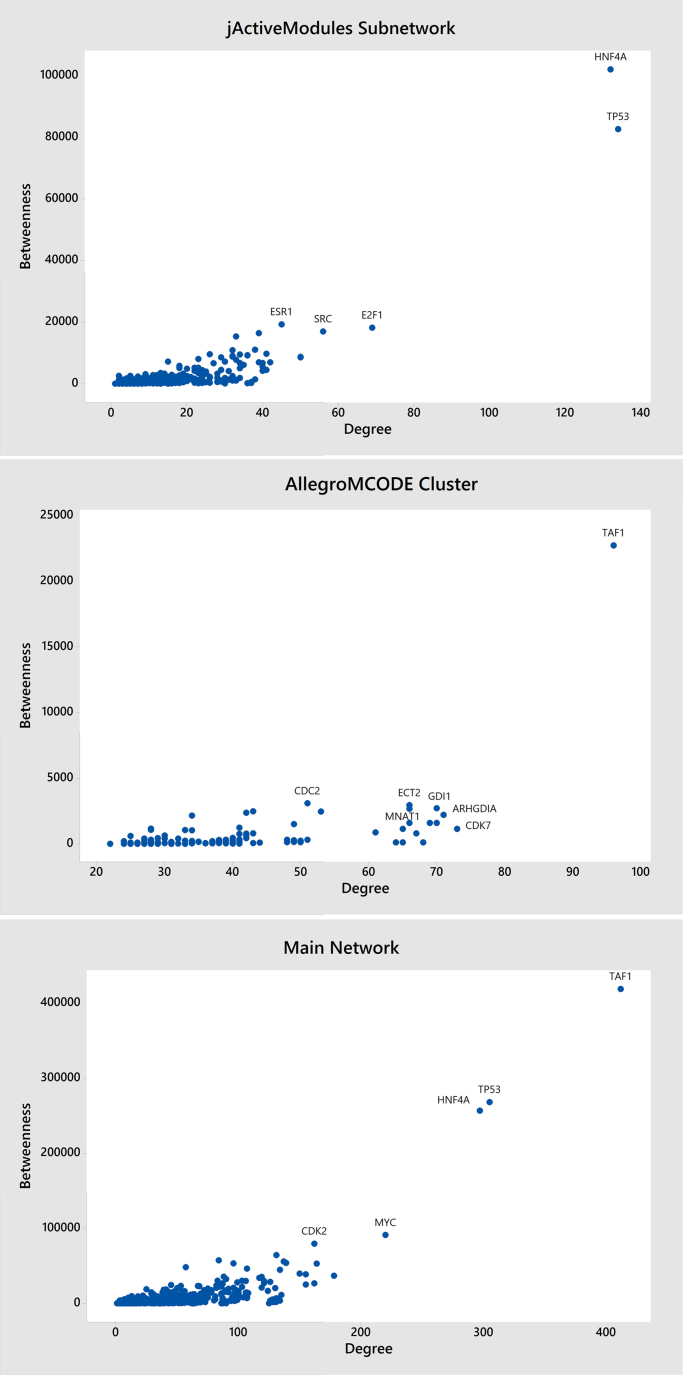
Evaluation of the central criteria of network including Betweenness and Degree using Centiscape indicated that the highest values of these indices in the core network belonged to TAF1, TP53, HNF4A, MYC and CDK2. These calculations were repeated through the classification by AllegroMCODE, GLay and MCL algorithms as well, before adding the expression data and by jActiveModules algorithm after adding the expression data; we found that TAF1 is in the highest scored cluster obtained from those three algorithms and HNF4A and TP53 in the most expressed active subnetwork obtained from jActiveModules, GLay (cluster1,2) and MCL (cluster 7, 1) which had the highest Degree and Betweenness values (Fig. 1).
Fig. 1.
Scatterplot of Betweenness vs Degree in main network, jActiveModules sub-network and AllegroMCODE cluster.
4. Discussion
According to our study which was conducted on the network and its subnetworks, proteins TAF1, HNF4A and TP53 had the highest values of central indices.
TBP-associated factor1 (TAF1) is the largest subunit of TAFIID, composes of TBP and 13 TAFs, plays a crucial role in cell growth and regulation and its kinase activity may have a pivotal role in tumor suppressor [46]. TAF1 also plays a role in determining the concentration of ATP within the cell, in disabling TP53 at high concentrations of ATP, and in inducing apoptosis through p27. It seems that in cancer, increased concentrations of ATP by TP53 becomes inactive by TAF1 which decreases the expression of p27 and provides a useful condition for the cancer cells to escape from apoptosis by anti-cancer drugs. It has also been illustrated that TAF1 phosphorylates P53 at Thr55, leads to a dissociation of P53 from p27 and hence deactivates transcription in the DNA damage response or apoptosis [46], [47].
Although studies showed that TAF1 is not a potential target for RNAi or chemical inhibition due to its leading role in reducing apoptosis and increasing the cell survival, it has not been mentioned that it is a distinct target for gastric cancer treatment. Since our study has been performed using protein interaction databases with experimental background this capability should be experimentally validated with more caution [46].
As a specific protein of gastrointestinal cells, Hepatocyte nuclear factor 4 alpha (HNF4A) is a transcription factor which plays a role in final differentiation of embryonic endoderm tissue along with other proteins such as HNF1β, albumin, and surfactant protein C [48]. A survey conducted by Bolotin et al. showed that this protein was associated with processes such as immune response, stress response, apoptosis, metabolism regulation, and cancer related pathways through targeting more than 240 proteins [49].
HNF4A is expressed in gastric carcinoma but is never expressed in breast carcinoma hence it remains a specific biomarker for gastric cancer [50], can be introduced as an excellent marker for differentiating breast cancer from gastric cancer. Walesky et al. showed that the elimination of HNF4A gene increases the susceptibility of these cells to cancer [51], while Jung et al. demonstrated that decreased expression of this protein by metformin reduces the rate of tumor growth [52]. Inhibition of HNF4A with RNA and pharmacological inhibitors demonstrated antineoplastic activity via down regulation of cyclins, cell cycle arrest and apoptosis.
[53].
Assessment of expression data showed that the expression of this gene decreased in gastric cancer cells. Wang et al. demonstrated that berberin extracted from Coptis chinensis can increase the expression of HNF4A [54], [55]. Walesky et al. showed that increased expression of HNF4A can reduce the growth of cancer cells [56]. According to the mentioned studies, the combined use of metformin and plants with the active ingredient of beberin are effective in reducing the growth rate of tumor cells. Assessment of mouse hepatocytes treated with berberin and metformin respectively, showed that berberin promotes HNF4A and glucokinase [57].
HNF4A is an important protein among the 240 interplay in cell protein-protein interaction network with a definite role in gastric cell differentiation and plays an essential role in cell growth and division [49]. So based on mentioned points, this protein could be a potential target for inhibition as stated by Chang et al. HNF4A RNAi and/or its pharmacological inhibition could lead to cell cycle arrest and tumor growth inhibition [53]. McDonald et al. also showed that HNF4A protein could have a relationship (Synthetic lethality) with other proteins in distinct cancers such as gastric [58].
Recently, synthetic lethality studies have shown, gastric cancer cells with ATM deficiencies along with pharmacological inhibitors affect ATR and could induce death in this cells [59]
TP53, inhibiting the cell growth remains the main protein in apoptosis, mutated in more than half of different types of cancer [60]. For this reason, TP53 is one of the potential therapeutic targets in various cancers, targeted through different means. Wang et al. pointed out different methods such as reactivation of mutant proteins as well as the inhibition of the wild type protein through a combination of drugs or targeting cells carrying the mutated protein [61].
Gene set enrichment analysis on the network showed that pathways such as Cell cycle, Neurotrophin signaling pathway, Nucleotide Excision Repair (NER), and Focal adhesion can contribute to the development of gastric cancer. This is partly due to the fact that the rate of cell growth and division increases in tumor formation, causing the activation of cell cycle pathway in this cancer.
Neurotrophin signaling pathway consists of four receptor proteins with conserved structures and growth factor function [62], [63]. Du et al. demonstrated the increased expression of these receptors and their ligands which were secreted from tumor cells [64]. Okugawa et al. showed the increased expression of BNDF/TrkB axis in gastric cancer cells and inhibition of this receptor by specific drug resulting in tumor growth inhibition in mouse models [65]. Enrichment analysis of 72 basic proteins revealed that 6 proteins were involved in this pathway and after ranking them based on the p-value, it demonstrated that the pathway was located at the 21th rank. After the network expansion process, the study showed that this pathway included 86 proteins, promoted to the 3rd rank. This represents the accumulation of proteins involved in the interaction network in this signaling pathway which can be a sign of its importance in the incidence or spread of gastric cancer.
Liu et al. showed that polymorphisms in the nucleotide excision repair, responsible for repair of mutations, was significantly associated with gastric cancer [66].
A number of our introduced candidates including CDK7, CCNH, and PCNA are involved in both NER pathway and cell cycle. These results were consistent with the study of Stoimenov et al. who showed that PCNA has a key role in DNA replication events determining tumor progression and cancer development [67]. Our results were also in line with the study of Czyzewska et al. who showed that this protein correlated with the varied degrees of malignancy of gastric cancer [68]. In a study by Wang et al., researchers found that in gastric cancer CDK7 is overexpressed, attributing this problem to an increased division of cancerous cells [69]. However, no study yet examines the relationship of CCNH and gastric cancer, whereas our results give a relational clue about this protein was potentially important as a biomarker in gastric cancer. Liu et al. showed that in breast cancer, CtBP2 affected by CCNH/CDK7 complexes and as a result this protein is more stable against proteasome degradation, correlated with more invasive potential of the cells. It is not far-fetched that the same mechanism is responsible in gastric cancer, resulting in the necessity to study further this protein based on the suggested systems biology approach [70].
On the other hand, the evaluation of HNF4A gene in GENE database in NCBI showed that this gene consists of a miRNA (miR-3646) where its expression is altered in cancers such as colon, lung, bladder, and breast, according to Meiri et al. [71]. This study also showed that it plays a key role in drug resistance, cell division, and tissue invasion, but no study has been performed on this miRNA and gastric cancer. It is likely that the miRNA causes contradictory results regarding the effect of HNF4A in gastric cancer.
We have shown a comprehensive collection of pathways and key proteins which remains unique to this study, whereas no other study has yet investigated such a colllection, except where they have been sporadically presented. On the other hand the involvement of some of these proteins have been approved in other cancers. But the exact role of these proteins in gastric cancer has not been examined, which shows the merit of further investigation in a systemic and comprehensive manner. The results of the present study shows the pathways and proteins which are important in the occurrence of gastric cancer and can be introduced as therapeutic targets and significant biomarkers in this disease. In fact, our methodology offers a systematic insight into the involvement of particular proteins in gastric cancer which can be applied in order to identify key proteins and pathways in other diseases.
Acknowledgments
The authors would like to give their gratitude to the bioinformatics lab group of the National Institute of Genetic Engineering and Biotechnology for providing us the facility and friendly environment to perform this study, and Dr. Pardis Minuchehr for scientifically reading and editting the text.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.01.001.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.01.001.
Appendix A. Transparency document
Supplementary material
Appendix B. Supplementary material
Supplementary material
Supplementary material
References
- 1.Cancer I.Af.Ro. WHO; Geneva: 2014. World Cancer Report 2014. [Google Scholar]
- 2.Compare D., Rocco A., Nardone G. Risk factors in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2010;14:302–308. [PubMed] [Google Scholar]
- 3.Guggenheim D.E., Shah M.A. Gastric cancer epidemiology and risk factors. J. Surg. Oncol. 2013;107:230–236. doi: 10.1002/jso.23262. [DOI] [PubMed] [Google Scholar]
- 4.Wroblewski L.E., Peek R.M., Wilson K.T. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin. Microbiol. Rev. 2010;23:713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song X., Xin N., Wang W., Zhao C. Wnt/β-catenin, an oncogenic pathway targeted by H. pylori in gastric carcinogenesis. Oncotarget. 2015;6:35579–35588. doi: 10.18632/oncotarget.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito T., Kondo C., Shitara K., Ito Y., Saito N., Ikehara Y., Yatabe Y., Yamamichi K., Tanaka H., Nakanishi H. Comparison of intratumoral heterogeneity of HER2 expression between primary tumor and multiple organ metastases in gastric cancer: clinicopathological study of three autopsy cases and one resected case. Pathol. Int. 2015 doi: 10.1111/pin.12290. [DOI] [PubMed] [Google Scholar]
- 7.Rüschoff J., Hanna W., Bilous M., Hofmann M., Osamura R.Y., Penault-Llorca F., van de Vijver M., Viale G. HER2 testing in gastric cancer: a practical approach. Modern Pathol. 2012;25:637–650. doi: 10.1038/modpathol.2011.198. [DOI] [PubMed] [Google Scholar]
- 8.Starzynska T., Bromley M., Ghosh A., Stern P.L. Prognostic significance of p53 overexpression in gastric and colorectal carcinoma. Br. J. Cancer. 1992;66:558. doi: 10.1038/bjc.1992.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenoglio-Preiser C., Wang J., Stemmermann G., Noffsinger A. TP53 and gastric carcinoma: a review. Hum. Mutat. 2003;21:258–270. doi: 10.1002/humu.10180. [DOI] [PubMed] [Google Scholar]
- 10.Azarhoush R., Keshtkar A.A., Amiriani T., Kazemi-Nejad V. Relationship between p53 expression and gastric cancers in cardia and antrum. Arch. Iran Med. 2008;11:502–506. [PubMed] [Google Scholar]
- 11.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 12.Kim O., Yoon J.H., Choi W.S., Ashktorab H., Smoot D.T., Nam S.W., Lee J.Y., Park W.S. Gastrokine 1 inhibits gastrin-induced cell proliferation. Gastric Cancer. 2015:1–11. doi: 10.1007/s10120-015-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao W., Chen J., Peng T.-L., Yin X.-F., Chen L.-Z., Chen M.-H. Role of trefoil factor 1 in gastric cancer and relationship between trefoil factor 1 and gastrokine 1. Oncol. Rep. 2012;28:1257–1262. doi: 10.3892/or.2012.1939. [DOI] [PubMed] [Google Scholar]
- 14.Choi B.J., Yoon J.H., Choi W.S., Kim O., Nam S.W., Lee J.Y., Park W.S. GKN1 and mir-185 are associated with CpG island methylator phenotype in gastric cancers. Mole. Cell. Toxicol. 2013;9:227–233. [Google Scholar]
- 15.Xing R., Li W.-M., Cui J.-T., Xia N., Lu Y.-Y. GKN1 inhibits cell invasion in gastric cancer by inactivating the NF-kappaB pathway. Discov. Med. 2015;19:65–71. [PubMed] [Google Scholar]
- 16.Chang S., Gao L., Yang Y., Tong D., Guo B., Liu L., Li Z., Song T., Huang C. miR-145 mediates the antiproliferative and gene regulatory effects of vitamin D3 by directly targeting E2F3 in gastric cancer cells. Oncotarget. 2015;6:7675–7685. doi: 10.18632/oncotarget.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez M.W., Kann M.G. Chapter 4: protein interactions and disease. PLoS Comput. Biol. 2012;8:002819. doi: 10.1371/journal.pcbi.1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kann M.G. Protein interactions and disease: computational approaches to uncover the etiology of diseases. Brief. Bioinform. 2007;8:333–346. doi: 10.1093/bib/bbm031. [DOI] [PubMed] [Google Scholar]
- 19.Safari-Alighiarloo N., Taghizadeh M., Rezaei-Tavirani M., Goliaei B., Peyvandi A.A. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol. Hepatol. Bed Bench. 2014;7:17. [PMC free article] [PubMed] [Google Scholar]
- 20.Lin W.-c., Kao H.-W., Robinson D., Kung H.-J., Wu C.-W., Chen H.-C. Tyrosine kinases and gastric cancer. Oncogene. 2000;19:5680–5689. doi: 10.1038/sj.onc.1203924. [DOI] [PubMed] [Google Scholar]
- 21.Gottlieb E., Tomlinson I.P. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat. Rev. Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 22.Corso S., Giordano S. How can gastric cancer molecular profiling guide future therapies? Trends Mole. Med. 2016;22:534–544. doi: 10.1016/j.molmed.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Croft D., Mundo A.F., Haw R., Milacic M., Weiser J., Wu G., Caudy M., Garapati P., Gillespie M., Kamdar M.R. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. The Reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bader G.D., Betel D., Hogue C.W. BIND: the biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rual J.-F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 28.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitkreutz B.-J., Stark C., Tyers M. The GRID: the general repository for interaction datasets. Genome Biol. 2003;4:R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasad T.K., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A. Human protein reference database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermjakob H., Montecchi‐Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Licata L., Briganti L., Peluso D., Perfetto L., Iannuccelli M., Galeota E., Sacco F., Palma A., Nardozza A.P., Santonico E. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 34.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J., Ade A.S., Tarcea V.G., Weymouth T.E., Mirel B.R., Jagadish H. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics. 2009;25:137–138. doi: 10.1093/bioinformatics/btn501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su G., Kuchinsky A., Morris J.H., States D.J., Meng F. GLay: community structure analysis of biological networks. Bioinformatics. 2010;26:3135–3137. doi: 10.1093/bioinformatics/btq596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.S.M. Van Dongen, Graph Clustering by Flow Simulation, 2001.
- 39.Eijssen L.M., Jaillard M., Adriaens M.E., Gaj S., de Groot P.J., Müller M., Evelo C.T. User-friendly solutions for microarray quality control and pre-processing on ArrayAnalysis. org. Nucleic Acids Res. 2013;41:W71–W76. doi: 10.1093/nar/gkt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ideker T., Ozier O., Schwikowski B., Siegel A.F. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18:S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 41.Saito R., Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Lotia S., Pico A.R., Bader G.D., Ideker T. A travel guide to Cytoscape plugins. Nat. Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:1. [PubMed] [Google Scholar]
- 45.Scardoni G., Petterlini M., Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura J., Nguyen S.T., Liu H., Taira N., Miki Y., Yoshida K. A functional genome-wide RNAi screen identifies TAF1 as a regulator for apoptosis in response to genotoxic stress. Nucleic Acids Res. 2008;36:5250–5259. doi: 10.1093/nar/gkn506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y., Lin J.C., Piluso L.G., Dhahbi J.M., Bobadilla S., Spindler S.R., Liu X. Phosphorylation of p53 by TAF1 inactivates p53-dependent transcription in the DNA damage response. Molecular Cell. 2014;53:63–74. doi: 10.1016/j.molcel.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Grapin-Botton, Endoderm Specification, 2008. [PubMed]
- 49.Bolotin E., Liao H., Ta T.C., Yang C., Hwang‐Verslues W., Evans J.R., Jiang T., Sladek F.M. Integrated approach for the identification of human hepatocyte nuclear factor 4α target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koyama T., Sekine S., Taniguchi H., Tsuda H., Ikegami M., Hano H., Kushima R. Hepatocyte nuclear factor 4A expression discriminates gastric involvement by metastatic breast carcinomas from primary gastric adenocarcinomas. Hum. Pathol. 2011;42:1777–1784. doi: 10.1016/j.humpath.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Walesky C., Edwards G., Borude P., Gunewardena S., O'Neil M., Yoo B., Apte U. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine‐induced hepatocellular carcinoma in rodents. Hepatology. 2013;57:2480–2490. doi: 10.1002/hep.26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung H.R., Chang H.R., Seo H.-H., Lemos R., Park H.S., Liang H., Powis G., Kim Y.H. Metformin increases AMPK {alpha} activity by inhibition of AMPK {alpha} and cell cycle proliferation in Asian gastric cancer. Cancer Res. 2013;73:5534. [Google Scholar]
- 53.Chang H.R., Nam S., Kook M.-C., Kim K.-T., Liu X., Yao H., Jung H.R., Lemos R., Seo H.H., Park H.S. HNF4α is a therapeutic target that links AMPK to WNT signalling in early-stage gastric cancer. Gut. 2014 doi: 10.1136/gutjnl-2014-307918. (gutjnl-2014-307918) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung J., Choi J.S., Jeong C.-S. Inhibitory activities of palmatine from coptis chinensis against helicobactor pylori and gastric damage. Toxicol. Res. 2014;30:45. doi: 10.5487/TR.2014.30.1.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z.-Q., Lu F.-E., Leng S.-H., Fang X.-S., Chen G., Wang Z.-S., Dong L.-P., Yan Z.-Q. Facilitating effects of berberine on rat pancreatic islets through modulating hepatic nuclear factor 4 alpha expression and glucokinase activity. World J. Gastroenterol. 2008;14:6004–6011. doi: 10.3748/wjg.14.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walesky C., Apte U. Role of hepatocyte nuclear factor 4α (HNF4α) in cell proliferation and cancer. Gene Expression. 2015;16:101–108. doi: 10.3727/105221615X14181438356292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan Z., Leng S., Lu F., Lu X., Dong H., Gao Z. [Effects of berberine on expression of hepatocyte nuclear factor 4alpha and glucokinase activity in mouse primary hepatocytes] Zhongguo Zhong yao za zhi= Zhongguo zhongyao zazhi= China J. Chin. Materia Med. 2008;33:2105–2109. [PubMed] [Google Scholar]
- 58.Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M.C., Zimmerman L.J., Shaddox K.F., Kim S. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H., Liu T., Zhang Z., Payne S.H., Zhang B., McDermott J.E., Zhou J.-Y., Petyuk V.A., Chen L., Ray D. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 2016;166:755–765. doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogelstein B., Sur S., Prives C. p53: the most frequently altered gene in human cancers. Nat. Educ. 2010;3:6. [Google Scholar]
- 61.Wang Z., Sun Y. Targeting p53 for novel anticancer therapy. Trans. Oncol. 2010;3:1–12. doi: 10.1593/tlo.09250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chopin V., Lagadec C., Toillon R.-A., Le Bourhis X. Neurotrophin signaling in cancer stem cells. Cell. Mole. Life Sci. 2016;73:1859–1870. doi: 10.1007/s00018-016-2156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kostrzewa R.M. Springer; 2014. Handbook of Neurotoxicity. [Google Scholar]
- 64.Du J.-J., Dou K.-F., Peng S.-Y., Qian B.-Z., Xiao H.-S., Liu F., Wang W.-Z., Guan W.-X., Gao Z.-Q., Liu Y.-B. Expression of NGF family and their receptors in gastric carcinoma: a cDNA microarray study. World J. Gastroenterol.: WJG. 2003;9:1431–1434. doi: 10.3748/wjg.v9.i7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okugawa Y., Tanaka K., Inoue Y., Kawamura M., Kawamoto A., Hiro J., Saigusa S., Toiyama Y., Ohi M., Uchida K. Brain-derived neurotrophic factor/tropomyosin-related kinase B pathway in gastric cancer. Br. J. Cancer. 2013;108:121–130. doi: 10.1038/bjc.2012.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J., Sun L., Xu Q., Tu H., He C., Xing C., Yuan Y. Association of nucleotide excision repair pathway gene polymorphisms with gastric cancer and atrophic gastritis risks. Oncotarget. 2016 doi: 10.18632/oncotarget.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stoimenov I., Helleday T. PCNA on the crossroad of cancer. Biochem. Soc. Trans. 2009;37:605–613. doi: 10.1042/BST0370605. [DOI] [PubMed] [Google Scholar]
- 68.Czyzewska J., Guzińska-Ustymowicz K., Lebelt A., Zalewski B., Kemona A. Evaluation of proliferating markers Ki-67, PCNA in gastric cancers. Roczniki Akademii Medycznej w Bialymstoku. 1995;49(2003):64–66. [PubMed] [Google Scholar]
- 69.Wang Q., Li M., Zhang X., Huang H., Huang J., Ke J., Ding H., Xiao J., Shan X., Liu Q. Upregulation of CDK7 in gastric cancer cell promotes tumor cell proliferation and predicts poor prognosis. Exp. Mole. Pathol. 2016;100:514–521. doi: 10.1016/j.yexmp.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y., Liu F., Mao F., Hang Q., Huang X., He S., Wang Y., Cheng C., Wang H., Xu G. Interaction with cyclin H/cyclin-dependent kinase 7 (CCNH/CDK7) stabilizes C-terminal binding protein 2 (CtBP2) and promotes cancer cell migration. J. Biol. Chem. 2013;288:9028–9034. doi: 10.1074/jbc.M112.432005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meiri E., Levy A., Benjamin H., Ben-David M., Cohen L., Dov A., Dromi N., Elyakim E., Yerushalmi N., Zion O. Discovery of microRNAs and other small RNAs in solid tumors. Nucleic Acids Res. 2010:gkq376. doi: 10.1093/nar/gkq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material