Abstract
The ability of Listeria monocytogenes to invade non-phagocytic cells is important for development of a systemic listeriosis infection. The authors previously reported that a L. monocytogenes ΔsigB strain is defective in invasion into human intestinal epithelial cells, in part, due to decreased expression of a major invasion gene, inlA. To characterize additional invasion mechanisms under the control of σB, mutants were generated carrying combinations of in-frame deletions in inlA, inlB and sigB. Quantitative assessment of bacterial invasion into the human enterocyte Caco-2 and hepatocyte HepG-2 cell lines demonstrated that σB contributes to both InlA and InlB-mediated invasion of L. monocytogenes. Previous identification of the σB-dependent P2prfA promoter upstream of the major virulence gene regulator, positive regulatory factor A (PrfA), suggested that the contributions of σB to expression of various virulence genes, including inlA, could be at least partially mediated through PrfA. To test this hypothesis, relative invasion capabilities of ΔsigB and ΔprfA strains were compared. Exponential-phase cells of the ΔsigB and ΔprfA strains were similarly defective at invasion; however, stationary-phase ΔsigB cells were significantly less invasive than stationary-phase ΔprfA cells, suggesting that the contributions of σB to invasion extend beyond those mediated through PrfA in stationary-phase L. monocytogenes. TaqMan quantitative reverse-transcriptase PCRs further demonstrated that expression of inlA and inlB was greatly increased in a σB-dependent manner in stationary-phase L. monocytogenes. Together, results from this study provide strong biological evidence of a critical role for σB in L. monocytogenes invasion into non-phagocytic cells, primarily mediated through control of inlA and inlB expression.
INTRODUCTION
The Gram-positive facultative intracellular food-borne pathogen Listeria monocytogenes is associated with serious invasive infections in humans and animals (Farber & Peterkin, 1991). Its ability to invade and multiply in a wide range of mammalian cells (Vazquez-Boland et al., 2001) is essential for development of systemic listeriosis. For example, the ability of L. monocytogenes to invade non-phagocytic cells plays an important role in this organism’s traversal of the intestinal barrier (Lecuit et al., 2001; MacDonald & Carter, 1980; Racz et al., 1972), and its ability to multiply in hepatocytes is essential for causing a systemic infection (Conlan & North, 1991; Gaillard et al., 1996). Several bacterial factors that mediate internalization events have been identified. Critical among these are two cell-wall-anchored proteins, internalin A (InlA) and internalin B (InlB). InlA mediates L. monocytogenes entry into the Caco-2 human colon adenocarcinoma cell line (Gaillard et al., 1991), while InlB mediates entry into hepatocytes and several endothelial and epithelial cell lines of various human and animal origins, including HepG-2 (human hepatocyte), TIB73 (mouse hepatocyte), HUVEC (human endothelial) and Vero (African green monkey epithelial) cells (Dramsi et al., 1995; Ireton et al., 1996; Parida et al., 1998). Expression of inlA and inlB is regulated by both positive regulatory factor A (PrfA)-dependent and -independent mechanisms (Dramsi et al., 1993; Lingnau et al., 1995; Sokolovic et al., 1993). Other bacterial factors, including ActA, p60, FbpA, Iap, and the ClpC ATPase, also have been reported to contribute to L. monocytogenes invasion (Alvarez-Dominguez et al., 1997; Dramsi et al., 2004; Kuhn & Goebel, 1989; Nair et al., 2000a; Wuenscher et al., 1993).
Recently, the alternative sigma factor, σB, which was initially identified as responsible for general stress responses in Gram-positive bacteria (Hecker & Volker, 2001), has also been associated with invasion capabilities in L. monocytogenes. Specifically, a σB-dependent promoter has been identified upstream of inlA (P4inlA) (Kazmierczak et al., 2003). Loss of σB resulted in reduced inlA expression and InlA levels in stationary-phase cells (Kim et al., 2004). However, the presence of a putative σB-dependent promoter upstream of inlB (P2inlB) (Kazmierczak et al., 2003) suggests that contributions of σB to L. monocytogenes invasion may not be solely limited to modulation of inlA expression.
To further study the role of σB in L. monocytogenes invasion, we analysed invasion capabilities of various mutant strains bearing combinations of in-frame deletions in inlA, inlB and sigB in the human enterocyte Caco-2 and hepatocyte HepG-2 cell lines. Previous identification of the σB-dependent P2prfA promoter (Nadon et al., 2002) suggested that the contributions of σB to L. monocytogenes virulence gene expression might be at least partially mediated through PrfA. To quantify the relative functional contributions of σB and PrfA, invasion capabilities of ΔsigB and ΔprfA strains were compared. We also measured σB-mediated contributions to expression of multiple genes reported to contribute to L. monocytogenes invasion and virulence using TaqMan quantitative reverse transcriptase polymerase chain reactions (qRT-PCR). Specifically, relative expression of inlA, inlB, prfA, iap, act A and clpC was measured in both the wild-type and ΔsigB backgrounds. Here, we present evidence that σB is a major contributor to L. monocytogenes invasion, primarily through modulation of expression of inlA and inlB.
METHODS
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 1. L. monocytogenes cells were grown overnight at 37 °C prior to use in the invasion assays to optimize PrfA-mediated gene expression (Johansson et al., 2002). Specifically, stationary-phase bacteria were prepared by growth in brain heart infusion broth (BHI) overnight at 37 °C with constant shaking (250 r.p.m.). Exponential-phase bacteria were prepared by passaging overnight cultures 1 : 100 into BHI and then growing the resulting culture to OD600 0·8 under the same conditions. For invasion assays, bacteria were harvested by centrifugation, washed and diluted in PBS.
Table 1.
L. monocytogenes strains
| Strain | Genotype | Reference; source |
|---|---|---|
| 10403S | Wild-type | Camilli et al. (1993); provided by D. Portnoy* |
| FSL A1-254 | ΔsigB | Wiedmann et al. (1998) |
| DP-L4137 | ΔprfA | Cheng & Portnoy (2003); provided by J. Miller* |
| DP-L4405 | ΔinlA | Bakardjiev et al. (2004); provided by J. Miller |
| FSL B2-042 | ΔinlAΔsigB | This study |
| HEL-137 | ΔinlB | Kim et al. (2004) |
| FSL K4-008 | ΔinlBΔsigB | This study |
| FSL K4-009 | ΔinlAB | This study |
| FSL K4-010 | ΔinlABΔsigB | This study |
Dr Daniel Portnoy, Department of Molecular & Cell Biology, University of California, Berkeley, CA, USA; Dr Jeffrey Miller, Department of Microbiology, Immunology, & Molecular Genetics, UCLA, Los Angeles, CA, USA.
Construction of L. monocytogenes mutant strains
An internal in-frame deletion in the inlAB operon, which inactivated both genes, was generated by SOE (site-directed mutagenesis by overlap extension) PCR (Ho et al., 1989). Primers used were 5′-AAC TGC AGC TTT GGG AGT GAC ATG C-3′ (inlAB-SOEA), 5′-TGC CCT TAA ATT AGC TGC TCT CAC TAT ATA CAC TCC-3′ (inlAB-SOEB), 5′-GGA GTG TAT ATA GTG AGA GCA GCT AAT TTA AGG GCA-3′ (inlAB-SOEC) and 5′-CCG GAT CCA GTG AAA TTA TTG CTG GT-3′ (inlAB-SOED) (Dramsi et al., 1995). Primers inlAB-SOEB and inlAB-SOEC are complementary, and primers inlAB-SOEA and inlAB-SOED contain PstI and BamHI sites, respectively. Briefly, two fragments were amplified by PCR from 10403S chromosomal DNA using either primer pair inlAB-SOEA and inlAB-SOEB, or primer pair inlAB-SOEC and inlAB-SOED. The products were gel-purified and combined in a second PCR with primers inlAB-SOEA and inlAB-SOED. The resulting product was digested with PstI and BamHI and ligated between the PstI and BamHI sites of the shuttle vector pKSV7 (Camilli et al., 1993) to yield plasmid pHK2. The recombinant sequence in pHK2 was used to replace the wild-type inlAB sequence in the chromosome of the L. monocytogenes 10403S strain by allelic exchange, as previously described (Camilli et al., 1993), to create strain FSL K4-009. ΔinlAΔsigB (strain FSL B2-042), ΔinlBΔsigB (strain FSL K4-008), and ΔinlABΔsigB (strain FSL K4-010) mutants were generated from strains DP-L4405 (Bakardjiev et al., 2004), HEL-137 (Kim et al., 2004) and FSL K4-009 (this study), respectively, by replacing the chromosomal allele of sigB with the ΔsigB allele of pTJA-57, as previously described (Wiedmann et al., 1998).
Cell culture and invasion assay
The human colorectal epithelial cell line Caco-2 (ATCC HTB-37) and human hepatic epithelial cell line HepG-2 (ATCC HB-8065) were cultivated at 37 °C in a cell culture incubator at 80–95 % relative humidity under 5 % CO2. Caco-2 cells were cultured in EMEM (Eagle’s Minimum Essential Medium with Earle’s Salts) supplemented with 20 % fetal bovine serum (FBS), 1 % non-essential amino acids, 1 % sodium pyruvate, and antibiotics (penicillin G 100 units ml−1; streptomycin 100 μg ml−1). HepG-2 cells were cultured in DMEM (Dulbecco’s Modified Eagle’s Medium) supplemented with 10 % FBS, 1 % non-essential amino acids, 1 % sodium pyruvate, and antibiotics (penicillin G 100 units ml−1, streptomycin 100 μg ml−1). Two days prior to infection, 1·5×105 Caco-2 and 7·5×105 HepG-2 cells in media without antibiotics were seeded into each of six (35 mm diameter) tissue culture plate wells that contained three 12 mm glass cover-slips. Host cells were grown to confluence for 2 days. Thirty minutes before infection, the medium in each well was replaced with pre-warmed fresh medium without antibiotics. For infection, approximately 108 c.f.u. of exponential- or stationary-phase bacteria were inoculated onto the host cell monolayer in each well. Host cells were washed with PBS at 30 min post-infection and prewarmed fresh medium containing 50 μg gentamicin sulfate ml−1 was added. The number of internalized bacteria per coverslip was determined at 1 h post-infection by lysing infected cells in distilled water and plating appropriate serial dilutions of lysates onto LB (Luria–Bertani) agar plates.
Total RNA preparation and TaqMan qRT-PCR
Total RNA was purified from exponential- and stationary-phase bacterial cells using the RNAprotect/RNeasy Midi kit (Qiagen) and treated with RNase-free DNase as described by Sue et al. (2004). qRT-PCR was performed as described previously (Sue et al., 2004) using the ABI Prism 7000 Sequence Detection System (Applied Biosystems). TaqMan primers and probes were designed using Primer Express software (Applied Biosystems) according to the manufacturer’s guidelines. The primers and probes for rpoB and inlA were reported previously (Sue et al., 2004); those created for this study are listed in Table 2. All primers were tested in PCRs with 10403S genomic DNA as template and the amplification products were evaluated by gel electrophoresis. For each RNA sample, the control transcript (rpoB or gap mRNA) and target gene transcripts (prfA, clpC, inlA, inlB, actA or iap mRNAs) were transcribed in the same 96-well plate, and the resulting cDNAs were quantified by real-time PCR. Specifically, RT-PCR reactions were performed using the TaqMan One-Step RT-PCR Master Mix Reagents kit according to the manufacturer’s instructions (Applied Biosystems) using 25 ng total RNA with the following reaction conditions: 1 cycle at 48 °C for 30 min, 1 cycle at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Transcript levels for each gene (i.e. cDNA copy numbers) were determined as the difference between the experimental reactions and the corresponding reverse-transcriptase-negative controls, which were used to quantify the amount of contaminating L. monocytogenes DNA in each reaction. Standard curves for each gene were generated by using serial dilutions of 10403S genomic DNA template that had been prepared as described by Flamm et al. (1984). Absolute cDNA copy numbers, which were calculated based on genomic DNA standard curves to reflect mRNA levels for each gene present in each RNA sample, were used for subsequent analyses.
Table 2.
TaqMan primer and probe sequences
| Gene | Forward primer (5′→3′) | Taqman probes (5′→3′)* | Reverse primer (5′→3′) |
|---|---|---|---|
| gap | AAAGCTGGCGCTAAAAAAGTTG | FAM-ATCTCCGCTCCAGCAACTGGCGATAT† | TTCATGGTTTACATTGTAAACGATTG |
| prfA | CAATGGGATCCACAAGAATATT-GTAT | FAM-TGTAAATTCATGATGGTCCCGTTCTC-GCT† | AATAAAGCCAGACATTATAACGAAAGC |
| clpC | CGGCGAAAGCTCTCTATGAACT | FAM-TCCCTCTACCTCTTGCTGCACTTTTT-CAGA† | GGTGTATATTGGATCGTCGTCACA |
| inlB | GCAAATTTTTCCAGATGATGCT-TT | FAM-CAGAAACAATCAAAGACAAT†-MGB‡ | TGTCACTGCATCTGTCACACTTTT |
| actA | TGCGTGCGATGATGGTAGTT | FAM-CCAACTGCATTACGATTAACCCCGA-CATAA† | TTCGCTATCTGTCGCTGCAA |
| iap | AGCTGGGATTGCGGTAACAG | FAM-TGCTGCTCCAACAATCGCATCCG† | CAAAGAGTATCACCAGCTTCGACTAC |
FAM represents the reporter dye, 6-carboxyfluorescein.
Represents the non-fluorescent quenching dye, QSY7.
MGB is a minor groove binder.
Statistical analyses
For qRT-PCR data, expression levels of targeted genes were normalized using expression levels for housekeeping genes that had been processed in parallel with the targeted genes. rpoB, which encodes the β subunit of RNA polymerase (Milohanic et al., 2003), and gap, which encodes glyceraldehyde-3-phosphate dehydrogenase, were chosen as two independent housekeeping genes for data normalization (M. Kazmierczak & M. Wiedmann, unpublished data). Target gene expression level was normalized to a housekeeping gene expression level in the same sample by dividing (target gene cDNA copy number) by (rpoB cDNA copy number) or (gap cDNA copy number). Normalized target gene expression levels were then scale-transformed using their natural logarithms (ln) to stabilize the variance to approximate normality and expressed as ln[(target gene cDNA copy number)/(rpoB cDNA copy number)] or ln[(target gene cDNA copy number)/(gap cDNA copy number)]. Relative gene expression was evaluated by analysis of variance for strain, growth phase and interaction effects. Individual comparisons were done by the Bonferroni multiple comparison test. One-sample t tests were used to compare bacterial invasion abilities between the wild-type 10403S and each mutant strain. For all analyses, statistical significance was declared at P<0·05. All statistical analyses were done with Statistix 7 (Analytical Software).
RESULTS AND DISCUSSION
Relative contributions of InlA, InlB, σB and PrfA to L. monocytogenes invasion of Caco-2 and HepG-2 cells
L. monocytogenes invasion of human non-professional phagocytic cells is predominantly mediated by two surface proteins, InlA and InlB (Dramsi et al., 1995; Drevets et al., 1995; Gaillard et al., 1987, 1991, 1996; Lingnau et al., 1995; Mengaud et al., 1996). Recognition of the invasion-defective phenotype of the ΔsigB strain in non-phagocytic cells (Kim et al., 2004), of P4inlA as a σB-dependent promoter, and of P2inlB as a putative σB-dependent promoter (Kazmierczak et al., 2003) identified σB as an important factor contributing to regulation of inlA and inlB. The inlA and inlB genes are transcribed both individually and in an operon (Lingnau et al., 1995). To date, six promoters have been identified in the inlAB locus, which include a confirmed PrfA-regulated promoter (P3inlA) and a confirmed σB-dependent promoter (P4inlA) upstream of inlA, and a putative σB-dependent promoter (P2inlB) upstream of inlB (Kazmierczak et al., 2003; Lingnau et al., 1995). The PrfA-regulated promoter, P3inlA, is the only promoter reported to generate a bicistronic transcript (Lingnau et al., 1995); therefore, we reasoned that any σB-mediated effects on inlB expression would probably occur through P2inlB. To quantify contributions of σB to L. monocytogenes invasion, we analysed invasion capabilities of various mutant strains bearing combinations of in-frame deletions in inlA, inlB and sigB (Table 1) using Caco-2 and HepG-2 cells (Table 3). We rationalized that additional effects of a ΔsigB mutation in a ΔinlAB background could be interpreted as the contribution of σB beyond that which is mediated by InlA or InlB.
Table 3. Bacterial invasion.
Results are reported as percentages relative to wild-type strain invasion, which was arbitrarily set to 100 %. Means and standard deviations from three independent experiments are shown. The 100 % values correspond to absolute invasiveness values of 1·27×10−4±0·30×10−4 for Caco-2 exponential-phase; 4·37×10−4±1·27×10−4 for Caco-2 stationary-phase; 8·76×10−5±1·42×10−5 for HepG-2 exponential-phase; 6·89×10−4±2·12×10−4 for HepG-2 stationary-phase.
| Strain | Caco-2
|
HepG-2
|
||
|---|---|---|---|---|
| Exponential-phase | Stationary-phase | Exponential-phase | Stationary-phase | |
| Wild-type | 100 | 100 | 100 | 100 |
| ΔsigB | 29·2±14·6* | 26·8±6·2** | 17·9±10·5** | 1·7±0·7*** |
| ΔprfA | 30·7±3·2*** | 64·8±31·1 | 13·3±7·2** | 57·8±14·1* |
| ΔinlA | 5·8±1·3*** | 2·4±1·6*** | 15·6±15·3* | 2·3±0·5*** |
| ΔinlAΔsigB | 1·8±0·9***(*) | 1·1±0·5*** | 13·7±20·0* | 1·8±1·4*** |
| ΔinlB | 48·6±14·7* | 52·4±2·9** | 26·2±6·4** | 20·1±5·2** |
| ΔinlBΔsigB | 18·5±9·1**(*) | 23·4±12·9**(*) | 9·9±8·9** | 1·4±0·5***(**) |
| ΔinlAB | 2·6±0·3*** | 2·3±2·3*** | 10·8±10·8** | 0·8±0·4*** |
| ΔinlABΔsigB | 2·0±0·7*** | 1·3±1·3*** | 6·3±7·9** | 0·8±1·0*** |
P values for comparison by one-sample t test of invasion capabilities between a strain bearing a single mutation (e.g. ΔsigB) and the wild-type strain are indicated by asterisks: *, <0·05; **, <0·01; ***, <0·001. In parentheses are P values for comparison by one-sample t test of invasion capabilities between ΔinlAΔsigB and ΔinlA, ΔinlBΔsigB and ΔinlB, ΔinlABΔsigB and ΔinlAB (*, <0·05; **, <0·01).
Invasion of the ΔinlA strain was significantly reduced in both Caco-2 and HepG-2 cells (Table 3), confirming the importance of InlA to L. monocytogenes invasion into both cell lines. The invasion defect of the ΔinlA strain was more pronounced with stationary-phase than with exponential-phase bacteria (Table 3): invasion capability of the ΔinlA strain was reduced by 17- and 42-fold in Caco-2 cells, and by 6- and 43-fold in HepG-2 cells, with exponential- and stationary-phase bacteria, respectively. These findings are in agreement with previous reports of the importance of InlA in L. monocytogenes invasion of these host cell lines (Dramsi et al., 1995; Gaillard et al., 1991; Lecuit et al., 1999), and further demonstrate that the contributions of InlA to invasion are more critical in stationary-phase cells than in exponential-phase cells.
The ΔinlB strain was more defective in invasion of HepG-2 cells than of Caco-2 cells, as previously reported (Dramsi et al., 1995) (Table 3); however, the ΔinlB invasion defect was less severe than that of ΔinlA in both cell lines (Table 3). Further, in contrast to the ΔinlA strain, the relative invasion defect associated with the ΔinlB strain was similar regardless of growth phase (Table 3). Specifically, invasion of the ΔinlB strain was reduced 2- and 2-fold in Caco-2 cells and 4- and 5-fold in HepG-2 cells with exponential- and stationary-phase bacteria, respectively. In the absence of both inlA and inlB, L. monocytogenes invasion was reduced 38- and 43-fold in Caco-2 cells, and 9- and 125-fold in HepG-2 cells with exponential- and stationary-phase bacteria, respectively (Table 3). While loss of both InlA and InlB greatly reduced L. monocytogenes invasion of Caco-2 cells independently of growth phase, the effects of their loss on HepG-2 invasion were more pronounced with stationary-phase bacteria.
InlB is required for L. monocytogenes entry into hepatocytes (Dramsi et al., 2004). As previously reported (Dramsi et al., 1995), we also found that the L. monocytogenes 10403S ΔinlB strain was more defective in invasion of HepG-2 cells than that of Caco-2 cells. However, our ΔinlB strain was less defective at invasion than the ΔinlA strain in both HepG-2 and Caco-2 cells (Table 3). This observation contrasts with the results of Dramsi et al. (1995), who showed a threefold reduced invasion capacity for a L. monocytogenes EGD ΔinlB strain relative to that of an EGD ΔinlA strain in HepG-2 cells. The most likely explanations for this discrepancy are: (i) as InlA-mediated invasion is affected by bacterial growth phase (Table 3), differences in bacterial growth and harvest conditions between the experiments are likely to affect relative strain invasion capacity; and (ii) the relative roles of the internalin proteins in mediating host cell entry may differ between L. monocytogenes EGD and 10403S.
Invasion by the ΔsigB strain was significantly reduced compared with that of the wild-type strain (Table 3). Loss of σB resulted in a greater bacterial invasion defect with HepG-2 cells than with Caco-2 cells. In Caco-2 cells, the ability of the ΔsigB strain to invade was reduced 3- and 4-fold with exponential- and stationary-phase bacteria, respectively. In HepG-2 cells, invasion of the ΔsigB strain was decreased 6- and 59-fold with exponential- and stationary-phase bacteria, respectively, which is essentially equivalent to the invasion defect resulting from the ΔinlA mutation in this host cell line.
Loss of σB in the ΔinlA background resulted in a further reduction of exponential-phase bacterial invasion in Caco-2 cells (Table 3), while loss of σB in the ΔinlB background resulted in a further reduction in L. monocytogenes invasion in both host cell lines, regardless of bacterial growth phase (Table 3). Loss of σB in the ΔinlAB background did not contribute to a further reduction in L. monocytogenes invasion (Table 3). Taken together, these results suggest that σB contributes to invasion of L. monocytogenes into Caco-2 and HepG-2 cells predominantly by directly affecting InlA-and InlB-mediated invasion pathways rather than through indirect mechanisms, such as those that might be mediated by PrfA. Further, the invasion defects resulting from additional loss of σB are essentially equivalent to those resulting from loss of InlA in the ΔinlB background for L. monocytogenes invasion into HepG-2 cells.
Invasion capabilities of the ΔprfA strain were reduced relative to those of the wild-type strain in both Caco-2 and HepG-2 cells (3- and 2-fold decrease in Caco-2 cells, and 8-and 2-fold decrease in HepG-2 cells with exponential- and stationary-phase bacteria, respectively; Table 3). The ability of the ΔprfA strain to invade these host cells was similar to that of the ΔsigB strain for exponential-phase bacteria, but greater than that of the ΔsigB strain for stationary-phase bacteria (Table 3). These results suggest that the contribution of PrfA to L. monocytogenes invasion differs with growth phase, with a greater relative contribution in exponential-phase than in stationary-phase bacteria.
σB modulates expression of inlA and inlB
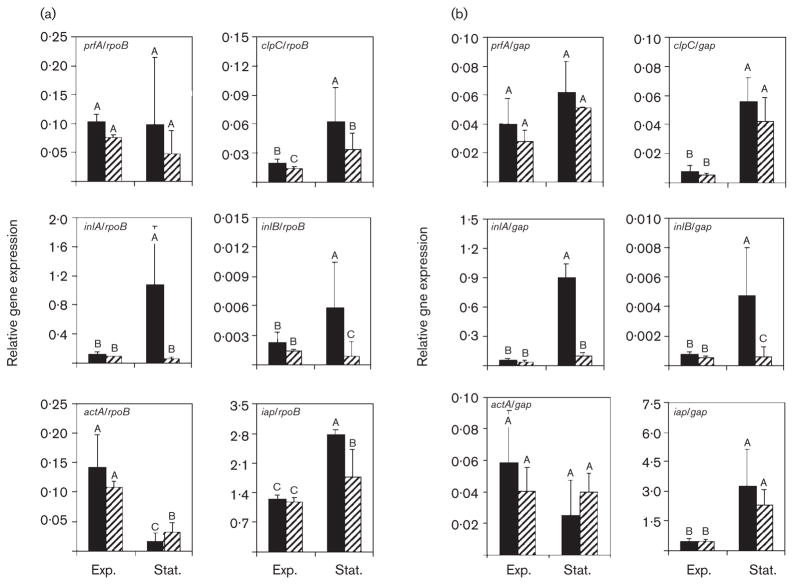
To determine the effect of σB on the expression of genes responsible for L. monocytogenes entry into non-phagocytic cells, we analysed relative expression of six selected genes (prfA, clpC, inlA, inlB, actA and iap) in the wild-type and ΔsigB strains using TaqMan qRT-PCR. To provide two independent assessments of relative gene expression patterns, mRNA collected from two different housekeeping genes, rpoB and gap, was used to normalize target gene expression data. Although expression patterns generated by normalizing target gene transcripts with those of each housekeeping gene were similar, they were not identical (Fig. 1). To provide the most conservative interpretation of the data, transcript levels representing a target gene under a given condition were only deemed different from those of the gene under a different condition (e.g. exponential- vs stationary-phase) or in a different background (wild-type vs ΔsigB) if levels were statistically significantly different by both normalizing analyses.
Fig. 1.
Relative expression of six virulence genes in exponential-phase (Exp.) and stationary-phase (Stat.) cells of wild-type and ΔsigB backgrounds. Relative gene expression, reported as [target gene mRNA level/rpoB mRNA level] (a) or [target gene mRNA level/gap mRNA level] (b) on the y-axis for prfA, clpC, inlA, inlB, actA and iap in the wild-type (black bars) and ΔsigB (striped bars) strains. Error bars represent standard deviations from three independent experiments. Differing upper-case letters within the same graph (i.e. A, B, C) indicate statistically significant differences in relative gene expression (P<0·05) by ANOVA on ln-transformed data (ln[mRNA level of gene of interest/mRNA level of housekeeping gene, rpoB (a) or gap (b)]) for strain, growth phase and interaction effect in a given gene of interest.
σB-dependent inlA expression has been reported previously (Kim et al., 2004); however, several lines of evidence suggest that σB-mediated effects on inlA and inlB expression may be direct or indirect, and that several factors affect inlAB expression. For example, σB could indirectly contribute to inlAB locus transcription through its control of prfA expression, as transcription initiated from P2prfA is σB-dependent (Nadon et al., 2002). Further, both prfA and clpC have been shown to modulate transcription of the inlAB locus (Dramsi et al., 1993; Lingnau et al., 1995; Nair et al., 2000a; Sokolovic et al., 1993).
qRT-PCR analyses showed that in exponential-phase L. monocytogenes, levels of inlA expression were similarly low in the wild-type and the ΔsigB strains (Fig. 1). In stationary phase, however, inlA expression was significantly up-regulated in the wild-type strain (9–19-fold) (P<0·05), but remained at a level similar to that in exponential phase in the ΔsigB strain (Fig. 1). These results show that σB plays a critical role for stationary-phase up-regulation of inlA. As with inlA (Fig. 1), exponential-phase inlB expression was similarly low in the wild-type and the ΔsigB strains (Fig. 1), and stationary-phase inlB expression was significantly up-regulated in the wild-type (3–6-fold) (P<0·05), but not in the ΔsigB strain (Fig. 1). These findings demonstrate that σB contributes to inlB expression as well as to inlA expression in stationary-phase bacteria.
Relative expression of prfA was evaluated for the wild-type and ΔsigB strains, as the P2prfA promoter has been shown to be σB-dependent (Nadon et al., 2002) and as regulation of inlA and inlB also is influenced by a PrfA-dependent mechanism (Dramsi et al., 1993; Lingnau et al., 1995). Although prfA expression appeared higher in the wild-type strain than in the ΔsigB strain, both in exponential and in stationary phase (Fig. 1), the differences were not statistically significant. Further, prfA expression also was not statistically different in exponential- and stationary-phase cells. These results suggest that the σB-regulated P2prfA promoter does not play a predominant role in prfA expression under the conditions examined in this study, and that increased transcriptional activation of prfA is not required for increased expression of inlA and inlB in stationary-phase L. monocytogenes cells. These data provide additional support for the conclusion that σB contributes to L. monocytogenes invasion primarily by directly affecting inlA and inlB expression, rather than through indirect effects mediated by PrfA.
σB does not make major contributions to clpC, actA or iap expression under the conditions examined in this study
To quantify contributions of σB to expression of multiple L. monocytogenes invasion genes, iap, actA and clpC transcripts were measured using qRT-PCR in both wild-type and ΔsigB backgrounds. iap encodes a major surface protein, p60, which is indirectly involved in invasion (Wuenscher et al., 1993) and actA also participates in L. monocytogenes invasion (Alvarez-Dominguez et al., 1997). clpC reportedly contributes to L. monocytogenes virulence (Rouquette et al., 1996, 1998) and influences expression of inlA, inlB and actA (Nair et al., 2000a). Regulation of clpC appears to be very complex, involving several regulators, including CtsR, PrfA and σB (Nair et al., 2000b; Ripio et al., 1998).
Expression of clpC and iap was significantly affected by bacterial growth phase (Fig. 1). Specifically, transcripts for both genes were present at significantly higher levels in stationary-phase bacteria than in exponential-phase bacteria for both the wild-type and ΔsigB strains (P<0·05). Although clpC and iap transcripts appeared to be present at higher levels in the wild-type strain than in the ΔsigB strain (Fig. 1a), the differences were not statistically significant at the 95 % confidence level when expression data were normalized by gap (Fig. 1b), suggesting that σB is not a predominant contributor to clpC or iap expression under the conditions examined in this study. In contrast, while actA also appeared to be affected by bacterial growth phase, actA transcripts were present at higher levels in exponential-phase than in stationary-phase bacteria for both strains when data were normalized by rpoB (Fig. 1). Transcript levels were not lower in the ΔsigB strain, and did not differ significantly between the wild-type and ΔsigB strains in data normalized by gap, suggesting that σB is not a positive regulator of actA expression in L. monocytogenes. These results suggest that any contributions of Iap, ActA and ClpC to L. monocytogenes invasion are predominantly independent of σB, providing further support for the hypothesis that σB-mediated invasion effects occur primarily through its regulation of expression of inlA and inlB.
Conclusions
The ability of L. monocytogenes to invade non-phagocytic cells allows the organism to breach host barriers, and hence is critical for systemic listeriosis. Our results demonstrate that σB significantly contributes to L. monocytogenes invasion of human enterocytes and hepatocytes, predominantly through InlA- and InlB-mediated pathways, as shown by both invasion and TaqMan qRT-PCR assay results. Specifically, we have shown that while stationary-phase expression of inlA and inlB is significantly enhanced (9–18-fold for inlA expression; 3–6-fold for inlB expression) in the wild-type strain relative to that in exponential phase (Fig. 1), stationary-phase expression of inlA and inlB does not increase in the ΔsigB strain. Further, loss of σB did not significantly reduce expression of Iap, ActA or ClpC, each of which have been associated with L. monocytogenes invasion. Our data support a model in which invasion defects associated with loss of σB result from loss of σB-mediated transcription of the inlAB locus, with relatively minor, if any, indirect effects resulting from σB-dependent expression of prfA. In support of this hypothesis, we have demonstrated dramatically reduced expression of both inlA and inlB in stationary-phase ΔsigB cells despite essentially wild-type expression levels for prfA (Fig. 1). However, our results do not rule out the possibility that the relative role of PrfA in invasion reflects growth-phase-dependent changes in PrfA activity to a greater extent than it reflects changes in prfA transcriptional activation, as PrfA is known to exist in both active and inactive forms (Renzoni et al., 1997). While additional studies will be necessary to fully attribute the relative contributions of PrfA- and σB-mediated mechanisms to invasion, the results presented in this study clearly highlight critical contributions of σB to L. monocytogenes invasion into non-phagocytic cells.
Acknowledgments
We thank Martin Wiedmann for helpful discussions. This work was supported in part by the National Institutes of Health Award No. RO1-AI052151-01A1 (to K. J. B.).
Abbreviation
- qRT-PCR
quantitative reverse-transcriptase polymerase chain reaction
References
- Alvarez-Dominguez C, Vazquez-Boland JA, Carrasco-Marin E, Lopez-Mato P, Leyva-Cobian F. Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect Immun. 1997;65:78–88. doi: 10.1128/iai.65.1.78-88.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakardjiev AI, Stacy BA, Fisher SJ, Portnoy DA. Listeriosis in the pregnant guinea pig: a model of vertical transmission. Infect Immun. 2004;72:489–497. doi: 10.1128/IAI.72.1.489-497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli A, Tilney LG, Portnoy DA. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol. 1993;8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LW, Portnoy DA. Drosophila S2 cells: an alternative infection model for Listeria monocytogenes. Cell Microbiol. 2003;5:875–885. doi: 10.1046/j.1462-5822.2003.00327.x. [DOI] [PubMed] [Google Scholar]
- Conlan JW, North RJ. Neutrophil-mediated dissolution of infected host cells as a defense strategy against a facultative intracellular bacterium. J Exp Med. 1991;174:741–744. doi: 10.1084/jem.174.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramsi S, Kocks C, Forestier C, Cossart P. Internalin-mediated invasion of epithelial cells by Listeria monocytogenes is regulated by the bacterial growth state, temperature and the pleiotropic activator prfA. Mol Microbiol. 1993;9:931–941. doi: 10.1111/j.1365-2958.1993.tb01223.x. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Biswas I, Maguin E, Braun L, Mastroeni P, Cossart P. Entry of Listeria monocytogenes into hepatocytes requires expression of InlB, a surface protein of the internalin multigene family. Mol Microbiol. 1995;16:251–261. doi: 10.1111/j.1365-2958.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Bourdichon F, Cabanes D, Lecuit M, Fsihi H, Cossart P. FbpA, a novel multifunctional Listeria mono-cytogenes virulence factor. Mol Microbiol. 2004;53:639–649. doi: 10.1111/j.1365-2958.2004.04138.x. [DOI] [PubMed] [Google Scholar]
- Drevets DA, Sawyer RT, Potter TA, Campbell PA. Listeria monocytogenes infects human endothelial cells by two distinct mechanisms. Infect Immun. 1995;63:4268–4276. doi: 10.1128/iai.63.11.4268-4276.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm RK, Hinrichs DJ, Thomashow MF. Introduction of pAM beta 1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect Immun. 1984;44:157–161. doi: 10.1128/iai.44.1.157-161.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard JL, Berche P, Mounier J, Richard S, Sansonetti P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun. 1987;55:2822–2829. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- Gaillard JL, Jaubert F, Berche P. The inlAB locus mediates the entry of Listeria monocytogenes into hepatocytes in vivo. J Exp Med. 1996;183:359–369. doi: 10.1084/jem.183.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Volker U. General stress response of Bacillus subtilis and other bacteria. Adv Microb Physiol. 2001;44:35–91. doi: 10.1016/s0065-2911(01)44011-2. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Ireton K, Payrastre B, Chap H, Ogawa W, Sakaue H, Kasuga M, Cossart P. A role for phosphoinositide 3-kinase in bacterial invasion. Science. 1996;274:780–782. doi: 10.1126/science.274.5288.780. [DOI] [PubMed] [Google Scholar]
- Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. Listeria monocytogenes σB regulates stress response and virulence functions. J Bacteriol. 2003;185:5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Boor KJ, Marquis H. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect Immun. 2004;72:7374–7378. doi: 10.1128/IAI.72.12.7374-7378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M, Goebel W. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect Immun. 1989;57:55–61. doi: 10.1128/iai.57.1.55-61.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit M, Dramsi S, Gottardi C, Fedor-Chaiken M, Gumbiner B, Cossart P. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 1999;18:3956–3963. doi: 10.1093/emboj/18.14.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, Babinet C, Cossart P. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science. 2001;292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- Lingnau A, Domann E, Hudel M, Bock M, Nichterlein T, Wehland J, Chakraborty T. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect Immun. 1995;63:3896–3903. doi: 10.1128/iai.63.10.3896-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald TT, Carter PB. Cell-mediated immunity to intestinal infection. Infect Immun. 1980;28:516–523. doi: 10.1128/iai.28.2.516-523.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J, Ohayon H, Gounon P, Mege RM, Cossart P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- Milohanic E, Glaser P, Coppee JY, Frangeul L, Vega Y, Vazquez-Boland JA, Kunst F, Cossart P, Buchrieser C. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol. 2003;47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- Nadon CA, Bowen BM, Wiedmann M, Boor KJ. σB contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect Immun. 2002;70:3948–3952. doi: 10.1128/IAI.70.7.3948-3952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Milohanic E, Berche P. ClpC ATPase is required for cell adhesion and invasion of Listeria monocytogenes. Infect Immun. 2000a;68:7061–7068. doi: 10.1128/iai.68.12.7061-7068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Derre I, Msadek T, Gaillot O, Berche P. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol Microbiol. 2000b;35:800–811. doi: 10.1046/j.1365-2958.2000.01752.x. [DOI] [PubMed] [Google Scholar]
- Parida SK, Domann E, Rohde M, Muller S, Darji A, Hain T, Wehland J, Chakraborty T. Internalin B is essential for adhesion and mediates the invasion of Listeria monocytogenes into human endothelial cells. Mol Microbiol. 1998;28:81–93. doi: 10.1046/j.1365-2958.1998.00776.x. [DOI] [PubMed] [Google Scholar]
- Racz P, Tenner K, Mero E. Experimental Listeria enteritis. I. An electron microscopic study of the epithelial phase in experimental listeria infection. Lab Invest. 1972;26:694–700. [PubMed] [Google Scholar]
- Renzoni A, Klarsfeld A, Dramsi S, Cossart P. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect Immun. 1997;65:1515–1518. doi: 10.1128/iai.65.4.1515-1518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripio MT, Vazquez-Boland JA, Vega Y, Nair S, Berche P. Evidence for expressional crosstalk between the central virulence regulator PrfA and the stress response mediator ClpC in Listeria monocytogenes. FEMS Microbiol Lett. 1998;158:45–50. doi: 10.1111/j.1574-6968.1998.tb12798.x. [DOI] [PubMed] [Google Scholar]
- Rouquette C, Ripio MT, Pellegrini E, Bolla JM, Tascon RI, Vazquez-Boland JA, Berche P. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol Microbiol. 1996;21:977–987. doi: 10.1046/j.1365-2958.1996.641432.x. [DOI] [PubMed] [Google Scholar]
- Rouquette C, de Chastellier C, Nair S, Berche P. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol Microbiol. 1998;27:1235–1245. doi: 10.1046/j.1365-2958.1998.00775.x. [DOI] [PubMed] [Google Scholar]
- Sokolovic Z, Riedel J, Wuenscher M, Goebel W. Surface-associated, PrfA-regulated proteins of Listeria monocytogenes synthesized under stress conditions. Mol Microbiol. 1993;8:219–227. doi: 10.1111/j.1365-2958.1993.tb01566.x. [DOI] [PubMed] [Google Scholar]
- Sue D, Fink D, Wiedmann M, Boor KJ. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology. 2004;150:3843–3855. doi: 10.1099/mic.0.27257-0. [DOI] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol. 1998;180:3650–3656. doi: 10.1128/jb.180.14.3650-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuenscher MD, Kohler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol. 1993;175:3491–3501. doi: 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]