Abstract
The accumulation of sequenced Francisella strains has made it increasingly apparent that the 16S rRNA gene alone is not enough to stratify the Francisella genus into precise and clinically useful classifications. Continued whole-genome sequencing of isolates will provide a larger base of knowledge for targeted approaches with broad applicability. Additionally, examination of genomic information on a case-by-case basis will help resolve outstanding questions regarding strain stratification. We report the complete genome sequence of a clinical isolate, designated here as F. novicida-like strain TCH2015, acquired from the lymph node of a 6-year-old male. Two features were atypical for F. novicida: exhibition of functional oxidase activity and additional gene content, including proposed virulence determinants. These differences, which could potentially impact virulence and clinical diagnosis, emphasize the need for more comprehensive methods to profile Francisella isolates. This study highlights the value of whole-genome sequencing, which will lead to a more robust database of environmental and clinical genomes and inform strategies to improve detection and classification of Francisella strains.
Keywords: Whole Genome Sequencing, Clinical Microbiology, Comparative Genomics
1.1. INTRODUCTION
The number of recognized species within the genus Francisella has increased from two in 2005 to nine: F. guangzhouensis (also Allofrancisella guangzhouensis), F. halioticida, F. hispaniensis, F. noatunensis, F. novicida, F. persica, F. philomiragia, F. piscicida, and F. tularensis (Sjöstedt, 2005; Bacterio.net; DSMZ.de). Not included in this list are four recently proposed species: F. opportunistica, F. salina, F. uliginis, and F. frigiditurris (Challacombe et al., 2016). In 2010, F. novicida was formally reclassified as a subspecies within F. tularensis based on 99.8% sequence identity between F. novicida and F. tularensis 16S rDNA, along with DNA-DNA hybridization studies (Hollis et al., 1989; Forsman et al., 1994; Huber et al., 2010; Busse et al., 2010). Objections were raised which appealed to differences in disease manifestation, along with distinct evolutionary, metabolic, phenotypic, and regulatory attributes of F. novicida (Larsson et al., 2009; Johannson et al., 2010; Kingry and Petersen, 2014). Both arguments have merit, leading some to name F. novicida as its own species in the literature, and others to name it as F. tularensis subspecies (subsp.) novicida. It should be noted that NCBI only allows genome assemblies to be deposited under the latter; however, here we will use the designation F. novicida as it allows for a clear separation of virulent Francisella species from less virulent, opportunistic ones.
The Gram-negative, intracellular bacterium F. tularensis is the etiological agent of the zoonotic disease tularemia and is considered a Tier 1 select agent due to a low infectious dose, high fatality rate via the inhalational route, and lack of a licensed vaccine. Prior to 2010, F. tularensis included F. tularensis subsp. tularensis (type A), F. tularensis subsp. holarctica (type B), and F. tularensis subsp. mediasiatica. As type A and type B are virulent in humans, clinical samples that test positive for F. tularensis necessitate further testing by reference laboratories to rule out type A and type B prior to continued handling. In contrast, F. novicida is generally regarded as avirulent except in the context of immune-compromised individuals where it can cause opportunistic infections. Twelve human infections with F. novicida have been reported, with fatalities resulting only in cases of predisposing medical conditions (Brett et al., 2012, 2014). As of 2014, all F. novicida strains are excluded from select agent regulations, giving rise to its common use as a model organism to study Francisella (selectagents.gov). In addition to nomenclature irregularities in research literature, the high sequence identity (>97%) within Francisella species makes rapid and accurate classification of isolates difficult (Gunnell et al., 2012). Progress has been made to develop more sensitive clinical assays for detection of Francisella directly from blood or specific antibodies from serum (Banada et al., 2017; Nakajima et al., 2016). While these tools are immediately useful, they are designed more broadly to distinguish Francisella from other infectious agents. Other assays rely on previously characterized reference strains, and in some cases well-annotated genomes, that may not correctly call new isolates. Genomics can provide insights into differences in Francisella pathogenesis, which may ultimately lead to new DNA-based assays.
The number of available clinical and environmental Francisella genomes have increased due to widespread access to sequencing technologies and decreased costs. Recent genomic analyses have even gone so far as to specify loci that distinguish between virulent and avirulent isolates (Challacombe et al., 2016). Here we characterize a novel F. novicida-like strain, TCH2015, isolated from a human lymph node. To our knowledge, this is the first Francisella strain to be identified from Guatemala, and is the only non-F. tularensis genome reported to possess homologs of the F. tularensis type A FTT0794–FTT0796 locus involved in capsule biosynthesis. Finally, we address the strain-to-strain variability observed in F. novicida isolates for oxidase activity, a common screening test for clinical laboratories. Further examination and comparison of these genomes will improve our ability to anticipate the disease-causing potential of emerging isolates and may provide targets for therapeutic intervention against tularemia. Development and maintenance of a robust database containing clinical isolates is crucial for epidemiological monitoring and updating relevant clinical assays.
1.2. MATERIALS AND METHODS
1.2.1. Clinical isolate and reference strain source information
Clinical isolate TCH2015 was acquired from the lymph node of an afebrile 6-year-old male who resides in Guatemala and presented to Texas Children’s Cancer Center with a 5-week history of left cervical lymphadenopathy that was firm and non-tender (refer to supplemental materials for case report details). An excisional biopsy was performed and lymph node tissue was processed as described in section 1.2.2. Comparisons of the isolate were performed against F. novicida strain U112T which was originally isolated from Ogden Bay in Utah in 1950 (Larson et al., 1955; refer to www.straininfo.net or the product sheet for ATCC® 15482™). Sequencing and assembly of the U112T genome (DNA provided by USAMRIID) was previously performed by Los Alamos National Laboratory (Johnson et al., 2015; CP009633).
1.2.2. Clinical laboratory workup
Lymph node tissue was ground and streaked on chocolate agar, sheep’s blood agar, and MacConkey agar (Remel) and incubated at 37 °C in 5% CO2 and room air. At 72 hours, small white colonies were observed on the chocolate agar only, and Gram-stain revealed Gram-negative coccobacilli. Upon subculture, the isolate grew on sheep’s blood agar and chocolate agar but not MacConkey agar. The isolate tested indole and catalase negative (or weakly catalase positive depending on laboratory methodology) but weakly oxidase positive. Automated and rapid identification methods yielded no definitive identification: Vitek MS (MALDI-TOF, bioMérieux) returned a 50% call for Moraxella osloensis; Vitek 2 NH panel (bioMérieux), an 86% call for Oligella urethralis; and Remel NH RapID, no identification. Subsequent sequencing of 16S rRNA gene hypervariable regions V1, V3, and V6 was performed against the Ribosomal Database Project (RDP, rdp.cme.msu.edu) and SmartGene’s bacterial module (www.smartgene.com/mod_bacteria.html). V1 resulted in 38 bases matching over 20 organisms with 100% identity. V3 yielded 51 bases of high quality sequence with 100% identity to F. tularensisT and four other F. tularensis subspecies via RDP. SmartGene data yielded no organisms with 100% identity across the 51 bases, but F. tularensis had the highest overall identity of 98.04%. V6 resulted in 34 bases with 100% identity to five different Francisella species (including F. tularensis), and to Wolbachia persica. Thus, the only organism yielding 100% identity (or 98% via SmartGene) across both V3 and V6 regions was F. tularensis, at which point the City of Houston regional lab was notified and no further workup was performed.
The Houston Health Department identified a Gram-negative coccobacillus that tested negative for urease and oxidase (tetramethyl) and positive for F. tularensis by PCR (proprietary CDC formulation). Several characteristics were inconsistent with F. tularensis. Specifically, results were negative for cysteine requirement, slide agglutination, and direct fluorescent antibody (DFA). The isolate was therefore sent to the Centers for Disease Control and Prevention (CDC) for further testing due to discrepant results between PCR and culture, where multi-gene sequencing results were consistent with F. novicida.
1.2.3. Extraction, sequencing, and assembly of isolate genome
The isolate was grown in modified Mueller-Hinton cation-adjusted (MHII) broth (Becton Dickinson) supplemented with sterile 0.1% glucose, sterile 0.025% ferric pyrophosphate, and 2% reconstituted IsoVitaleX (Becton Dickinson) at 37 °C. Genomic DNA was extracted by phenol-chloroform as previously described (Atkins et al., 2015) and sequenced on the Pacific Biosciences RS II platform according to the manufacturer’s protocols (pacb.com). Briefly, adapters were ligated to size-selected (>10 kb) DNA ends and the generated libraries were sequenced using the C4 sequencing kit with p6 enzyme. Pacific Biosciences SMRT Cell read data was assembled using the RS_HGAP_Assembly.2 protocol available in the SMRT Analysis 2.3.0.140936 package (pacb.com). NCBI and RAST were used for genome annotation (ncbi.nlm.nih.gov; Aziz et al., 2008).
1.2.4. Comparative genomic analyses
Nucleotide alignments were generated using Mauve 2.3.1 (Darling et al., 2004, 2010) from FASTA files. Data files generated by Mauve were extracted to create a Circos plot of the alignment (Krzywinski et al., 2009). An un-rooted de novo phylogenetic tree was generated in PhyloPhlAn from a user-specified set of genomes (Segata et al., 2013). Briefly, peptide sequence files from each genome were downloaded from NCBI and compared against a reference dataset of 400 universal proteins. Of those, 275–295 marker proteins were identified in the user-defined set of genomes and used to build the tree. RAxML (Stamatakis, 2014) was used for tree optimization. Two-way average nucleotide identity was determined using the ANI calculator with default settings (http://enve-omics.ce.gatech.edu/ani/index). For function-based comparisons, genomes were uploaded and annotated in RAST and visualized in SeedViewer (rast.nmpdr.org; Overbeek et al., 2005).
1.3. RESULTS
1.3.1. Relationship of TCH2015 relative to other published Francisella genomes
Assembly of the isolate, designated TCH2015, resulted in a complete circular genome of 2,000,087 base pairs with 1,879 CDS, 32.4% G+C content, and mean coverage of 260.51 (Figure S1). BLAST® analysis of the isolate’s genome against NCBI’s non-redundant database revealed F. novicida as the top hit, with 98% sequence identity to strain U112T across 90% of the isolate’s genome, as well as to strains Fx1 and PA10-7858. Sequence-based comparisons of TCH2015 to F. novicida strain U112T (CP009633) revealed 1,671 bidirectional best hits corresponding to shared protein-coding genes. The 16S rRNA gene of TCH2015 is 99.94% identical to that of F. novicida U112T (1538/1539 bp). The same is true of F. novicida strains AZ06-7470, AL97-2214, and F6168. The next closest match is to F. novicida strain PA10-7858 (99.87%, CP016635). In agreement with the notion that 16S comparisons alone do not adequately resolve species of this genus, 16S rRNA gene alignment to F. tularensis type A and type B strains still yield greater than 99% sequence identity. Although TCH2015 deviates from typical F. novicida strains for reasons discussed below, it nonetheless grouped with published F. novicida genomes in a phylogenic tree generated de novo from PhyloPhlAn and shares the highest average nucleotide identity (ANI) with these genomes (Figure 1 and Table S2; Goris et al., 2007). Two-way ANI percentages compared to TCH2015 ranged from 98.31–98.74 for F. novicida strains, 98.02–98.23 for all other F. tularensis subspecies, and 90.73–90.83 for F. hispaniensis. A single copy of the Francisella Pathogenicity Island (FPI) is present in TCH2015. This is the case for all F. novicida strains described, whereas a duplication event of the FPI has occurred in virulent F. tularensis genomes (Nano and Schmerk, 2007). Also found in TCH2015 is a 144-bp insert within the pdpD gene (encoding the Pathogenicity Determinant Protein D) that is found in F. novicida strains but not in F. tularensis type A, while type B lacks most of pdpD (Brett et al., 2014; Nano et al., 2004). The clinical presentation was consistent with F. novicida infections, consisting of lymphadenopathy without fever or other symptoms (Kingry and Petersen, 2014). Taken together, these data indicate that TCH2015 is most similar to F. novicida.
Figure 1. Phylogenetic tree of isolate TCH2015 in relation to 21 Francisella strains.
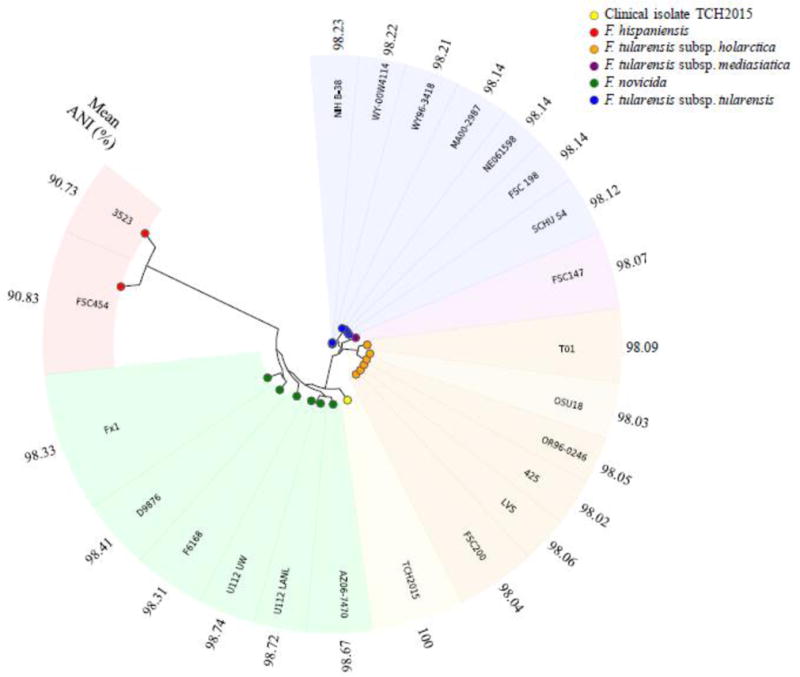
Clinical isolate TCH2015 is shown in yellow. Genomes are labeled by strain name and grouped by species/subspecies as shown in the legend. Two separate U112T genomes are shown corresponding to University of Washington (UW) and Los Alamos National Laboratories (LANL) sequencing projects.
1.3.2. Additional gene content is present in TCH2015 which includes F. tularensis exopolysaccharide genes
High-level assessment of genome topology revealed similar genome preservation between TCH2015 and F. novicida U112T, as demonstrated by consistent spatial distribution of genomic content with few syntenic block rearrangements. However, TCH2015 possesses almost 90 kb of additional gene content distributed throughout the genome (Figure S2). Upon manual curation, this extra gene content contains 106 open reading frames (excluding mobile elements or transposases). Predicted functions of the proteins encoded by these additional genes include an ABC-type multidrug transport system, sugar-modifying enzymes, thiamine biosynthesis, and 53 hypothetical proteins, while 14 do not yield hits to any Francisella proteins (Table 1; see Table S1 for full list and protein BLAST® hits, and Tables S3 and S4). Of clinical importance, several of these genes were recently described as “features that may be associated with tularemia virulence or other defining functions,” including the SpeADE and AguAB operons, and a three-ORF cluster encoding a phosphoserine phosphatase, methyltransferase, and cholinephosphotransferase (Table 2; Challacombe et al., 2016). This last cluster is orthologous to three genes in F. tularensis type A that are part of a larger 12-gene cluster (Figure 2; Larsson et al., 2005). First identified from whole-genome sequencing (WGS), portions of this polysaccharide locus have since been shown to play a role in the formation of a capsule-like complex (CLC) in F. tularensis. Capsule is important for host-adaptation, immune complement evasion and bacterial uptake (Bandara et al., 2011; Clay et al., 2008; Zarrella, et al., 2011). Remarkably, this is the first report of the FTT0794–FTT0796 locus in a non-F. tularensis strain. The TCH2015 locus shares 99% identity with this cluster across 99%, 100%, and 100% of the FTT0794, FTT0795, and FTT0796 genes, respectively. As this cluster is hypothesized to be partially responsible for glycan structure differences observed between Francisella species, further examination of TCH2015 is warranted (Thomas et al., 2011). We are aware that Challacombe et al. recently asserted that atypical F. novicida strains termed “novicida-like” do not constitute a separate species, and thus should be classified as F. novicida to remove ambiguity (Hollis et al., 1989; Clarridge et al., 1996; Whipp et al., 2003). However, due to the unprecedented finding of the FTT0794–FTT0796 locus, we have designated TCH2015 as an F. novicida-like isolate pending further examination. Finally, with regards to restriction-modification (R-M) loci, TCH2015 closely resembles F. novicida strains, which have only one or two of four R-M systems intact (apart from strain U112T which has four, and strain D9876 which has none).
Table 1.
Genes Present in TCH2015 but Lacking in F. novicida U112 (LANL)
| Gen e # |
Prote in Lengt h (aa) |
Annotated Function | Gen e # |
Prote in Lengt h (aa) |
Annotated Function |
Gen e # |
Prote in Lengt h (aa) |
Annotated Function |
|---|---|---|---|---|---|---|---|---|
| 100 | 145 | Putative Nudix hydrolase YfcD (EC 3.6.-.-) | 1013 | 47 | Di-/tripeptide transporter | 1424 | 373 | ABC-type multidrug transport system, permease component |
| 169 | 340 | ATP-binding region, ATPase-like | 1130 | 326 | Inositol oxygenase (EC 1.13.99.1) | 1438 | 213 | 4′-phosphopantethe inyl transferase (EC 2.7.8.-) |
| 176 | 293 | 2-hydroxy-3-oxopropionate reductase (EC 1.1.1.60) | 1132 | 473 | Uronate isomerase (EC 5.3.1.12) | 1464 | 42 | Cell division protein FtsI [Peptidoglycan synthetase] (EC 2.4.1.129) |
| 447 | 109 | ATP-binding protein p271 | 1133 | 183 | 4-Hydroxy-2-oxoglutarate aldolase (EC 4.1.3.16) @ 2-dehydro-3-deoxyphosphogluconate aldolase (EC 4.1.2.14) | 1553 | 287 | N-carbamoylputres cine amidase (3.5.1.53) / Aliphatic amidase AmiE (EC 3.5.1.4) |
| 450 | 199 | ThiJ/PfpI | 1135 | 397 | Mannonate dehydratase (EC 4.2.1.8) | 1554 | 329 | Agmatine deiminase (EC 3.5.3.12) |
| 549 | 263 | UDP-glucose 4-epimerase (EC 5.1.3.2) | 1136 | 491 | D-mannonate oxidoreductase (EC 1.1.1.57) | 1555 | 550 | Biosynthetic arginine decarboxylase (EC 4.1.1.19) |
| 550 | 279 | Glycosyl transferase, group 2 family protein | 1231 | 869 | FIG045374: Type II restriction enzyme, methylase subunit YeeA | 1556 | 290 | Spermidine synthase (EC 2.5.1.16) |
| 556 | 375 | Glycosyl transferase, family 2 | 1232 | 1253 | ATPase involved in DNA replication | 1557 | 163 | S-adenosylmethionine decarboxylase proenzyme (EC 4.1.1.50), prokaryotic class 1B |
| 557 | 350 | Capsular polysaccharide biosynthesis protein | 1233 | 46 | fic family protein | 1558 | 72 | Beta-galactosidase (EC 3.2.1.23) |
| 560 | 194 | dTDP-4-dehydrorhamnose 3,5-epimerase (EC 5.1.3.13) | 1236 | 642 | FIG006126: DNA helicase, restriction/modification system component YeeB | 1576 | 291 | Homoserine O-succinyltransferase (EC 2.3.1.46) |
| 729 | 423 | O-antigen ligase | 1237 | 401 | YeeC-like protein | 1577 | 437 | O-acetylhomoserine sulfhydrylase (EC 2.5.1.49) |
| 769 | 429 | Phosphoserine phosphatase (EC 3.1.3.3) | 1239 | 324 | Type I restriction-modification system, specificity subunit S (EC 3.1.21.3) | 1579 | 122 | Secreted effector protein |
| 770 | 210 | Methyltransferase | 1240 | 169 | Antirestriction protein | 1594 | 535 | Choline dehydrogenase (EC 1.1.99.1) |
| 771 | 228 | Lipopolysaccharide cholinephosphotransf erase LicD1 (EC 2.7.8.-) | 1242 | 1095 | Reticulocyte binding-like protein 2b | 1595 | 409 | Gluconate dehydratase (EC 4.2.1.39) |
| 810 | 594 | Hydroxymethylpyrimidine phosphate synthase ThiC (EC 4.1.99.17) | 1243 | 241 | putative mobilization protein mobC | 1596 | 413 | Transporter, MFS superfamily |
| 811 | 351 | Glycine oxidase ThiO (EC 1.4.3.19) | 1245 | 417 | DNA primase (EC 2.7.7.-) | 1870 | 845 | Phosphoenolpyru vate carboxylase (EC 4.1.1.31) |
| 813 | 260 | Thiazole biosynthesis protein ThiG | 1250 | 419 | HipA protein | |||
| 814 | 486 | Phosphomethylpyrimidine kinase/Thiamin-phosphate pyrophosphorylase | 1339 | 370 | Alanine dehydrogenase (EC 1.4.1.1) |
Table 2.
Frequency of features among different Francisella species/subspecies
| Number of Francisella genomes with feature present/total genomes | ||||
|---|---|---|---|---|
|
| ||||
| Locus/Feature | F. tularensis | F. novicida* | F. philomiragia | Other |
| OppABCDF | 4/7 | 9/10 | 4/4 | 0/10 |
| SpeADE, AguAB | 7/7 | 6/10 | 0/4 | 0/10 |
| FTT0794 – 0796 | 7/7 | 1/10 | 0/4 | 0/10 |
| Oxidase Activity | No | 4/10 | Yes | Varies consistently by species |
Includes F. novicida-like clinical isolate TCH2015.
Figure 2. Francisella polysaccharide locus comparisons.
Operon cluster of polysaccharide synthesis genes in F. tularensis strain SchuS4 (top), F. novicida-like strain TCH2015 (middle), and F. novicida strain U112T (bottom). Capsule biosynthesis genes FTT0794, FTT0795, and FTT0796, while absent in non-F. tularensis strains, are curiously preserved in TCH2015 with 99% identity. In strain U112T, these genes are replaced with two unrelated genes encoding KpsC and a hypothetical protein. Gene numbers for strains SchuS4 and U112T correspond to NCBI gene accession numbers, and to RAST gene numbers for strain TCH2015.
1.3.3. TCH2015 shares variable oxidase activity with F. novicida strains
TCH2015 tested positive for oxidase activity, which was initially the only contraindication against F. tularensis. In the literature, F. tularensis strains are consistently oxidase-negative, while F. philomiragia gives a strong oxidase positive result (Table 2). A newly proposed species, F. opportunistica, was also found to be oxidase positive. In contrast, F. novicida strains give inconsistent oxidase test results, with only three out of at least 10 previously reported as oxidase positive (Whitehouse et al., 2012). Genomes of these three oxidase-positive F. novicida isolates have not been fully sequenced. In addition to this strain-to-strain variation observed, intra-strain variation is also observed, as only two of three laboratories reported an oxidase positive result for TCH2015 (Figure S3). An alternative explanation is that different oxidase activity tests under different conditions produced disparate results.
Finding no evidence among the extra gene content to explain the oxidase activity in TCH2015, we then compared metabolic reconstructions of F. philomiragia 25017 to F. opportunistica MA06-7296 for all proteins within the respiration subsystem using the SEED framework (theseed.org). We hypothesized that the respiration subsystem for two oxidase-positive Francisella organisms would include the common proteins required for this activity (Table S5). We also individually compared these two strains to F. tularensis SchuS4, an oxidase-negative strain (Tables S6 and S7). In both comparisons, the single functional protein lacking in SchuS4 among terminal cytochrome oxidases was that of a putative cytochrome bd-II subunit I (appC). Further examination revealed a G243A substitution in SchuS4 resulting in an early stop codon 20% into the protein (Figure 3). This mutation is present in all type A and type B strains examined, with type B containing an additional stop codon 26 amino acids earlier. Francisella AppC shares 45% amino acid homology with E. coli AppC (Sturr et al., 1996), and is annotated as a pseudogene in strain SchuS4. Therefore, one possible explanation for the oxidase negative phenotype of F. tularensis is that only in strains where both app subunits are predicted as functional does oxidase activity occur. In other cases, however, appBC is not sufficient for oxidase activity, as appC in TCH2015 has 100% sequence homology with F. novicida strains known (or presumed) to be oxidase-negative. Non-coding regulatory sequences and environmental cues might contribute to the irregular oxidase activity observed in F. novicida and F. novicida-like strains. In future analyses, matching biochemical evidence with genomic support could grant a richer understanding of these organisms than either would alone.
Figure 3. Protein alignment of AppC among Francisella strains.
MacVector-generated alignment shows the presence of early stop codons in virulent F. tularensis strains (SchuS4, OSU18, and FSC200) compared to F. novicida strains (AZ06-7470 and U112T), F. uliginis (TX07-7310), F. philomiragia (ATCC 25017), F. opportunistica (MA06-7296), or clinical isolate TCH2015. Strains are ordered according to oxidase activity indicated in the right column. Several avirulent strains are oxidase negative even though they do not contain disruptions in appBC.
1.4. DISCUSSION
Epidemiological investigations of F. novicida and F. novicida-like infections reveal a strong association with salt-water environments (Petersen et al., 2009). While the mode of transmission in this case was not determined, the patient was active in water sports at the beach as well as canals around his home, suggesting water as the likely source of infection. Due to the finding of an F. novicida-like isolate and a history of neutropenia in infancy, a comprehensive immunologic evaluation was performed, and the patient was found to have decreased natural killer cell function (NK Lytic Units at 0.2; reference range > 2.6). We hypothesize that decreased natural killer cell function left the patient particularly susceptible to opportunistic infection. Further workup is in progress. Our whole-genome sequencing results reveal that TCH2015 most closely resembles F. novicida (or F. tularensis subsp. novicida), as determined by ANI comparisons, phylogenetic analyses, a single copy of the FPI, a 144-bp insert of the pdpD gene, as well as clinical presentation and biochemical test results. Even so, TCH2015 has several unique genotypic and phenotypic traits compared to other F. novicida strains, leading to a more conservative designation of TCH2015 as F. novicida-like.
The biochemical profile of TCH2015 led us to focus on the oxidase activity of F. novicida strains, which yield inconsistent results to this important clinical test. Initially, this result was the only contraindication against virulent F. tularensis, warranting further biochemical studies to determine underlying mechanisms of the variable oxidase activity observed in F. novicida strains. The G243A substitution in appC found in virulent F. tularensis strains is yet another distinguishing factor identified by WGS. Interestingly, in an F. novicida transposon-mutagenesis library, appB and appC mutants displayed a growth reduction of 3–7 logs in human-derived U937 macrophages and D. melanogaster-derived S2 cells (Asare and Kwaik, 2010, 2011). AppC mutants also displayed trafficking defects in U937 cells. A separate study identified appC in a negative selection screen in C57BL/6 mouse lung infections after 48 hours (Peng and Monack, 2010).
Challacombe et al. recently challenged current dogma of Francisella classification and proposed five functional loci as distinguishing features for Francisella species based on a comparative genomics study with 31 complete genomes (see section 1.1). TCH2015 contains three of these loci, one of which has not been identified in any other F. novicida or F. novicida-like strain to date (Sjödin et al., 2012; see section 1.3.2), further blurring the line between which genomic features distinguish virulent Francisella species from avirulent ones. The contribution of these genes to TCH2015 is not known, and it should be noted that two other LPS loci highlighted by Challacombe et al. were not found in TCH2015 (FTT1188 and FTT1453c, 54c, and 58c). While one or the other has been found in some F. novicida strains, only virulent F. tularensis strains possess both loci. Less surprising was the presence of SpeADE and AguAB, which, in addition to being present in F. tularensis, were previously described in detail in Australian clinical isolate 3523 and are also found in F. novicida strains AL97-2214, D9876, F6168, and TX07-6608. Siddaramappa et al. (2011) speculate that F. hispaniensis 3523 and F. novicida-like Fx1 represent strains transitioning from an environmental state to a pathogenic one, as may be the case for TCH2015. Alternatively, TCH2015 may represent an ancestral lineage at the junction of F. novicida and F. tularensis divergence which maintained or acquired partial sequences, possibly due to unique ecological pressure.
This study illustrates the current challenges pertaining to Francisella diagnostics and classification and exemplifies the utility of WGS in resolving identities of clinical isolates with atypical characteristics. Currently, determining the disease-causing potential for Francisella organisms isolated in clinical laboratories is challenging, as the field has outgrown current virulence marker identification methods. In our case—which we surmise is true for other clinical laboratories—the automated identification methods typically used do not include Francisella in the database system out of concern for laboratory exposure, creating an additional barrier to diagnosis and classification. Moreover, the high degree of sequence similarity in Francisella 16S rRNA causes more clinically relevant features to be overlooked. Discrepancies between genomic and biochemical data further open the door to misclassification of a virulent strain as avirulent, with potentially severe consequences. In the reverse scenario, valuable time and resources are wasted, and laboratory personnel undergo unnecessary monitoring and prophylaxis (Brett et al., 2012; cdc.gov).
Our results echo the conclusion of Challacombe et al. that correct classification will remain a challenge until the abundance of isolates with finished genomes increases but also emphasize the need to explore stratifications that do not rely solely on “presence/absence” genotypes (Sjödin et al., 2012). For instance, pseudogene content (Figure S4). Other useful stratifications may exist that we cannot currently appreciate. To our knowledge, this is the first report of a Francisella strain isolated from Guatemala. It is not known whether TCH2015 represents an emerging strain or is part of an underappreciated Francisella clade. The continued effort to sequence clinical and environmental isolates across more diverse sampling sites will lead to a greater understanding of the factors that contribute to Francisella evolution and virulence. Insights provided by studies like these may also be useful for the development of new strategies to reliably detect, classify, and monitor virulence of emerging strains.
Supplementary Material
Highlights.
Determining the disease-causing potential for Francisella isolates is ambiguous.
Whole-genome sequencing was performed on a clinical F. novicida-like isolate.
TCH2015 most closely resembles F. novicida, but possesses additional gene content.
First report of the FTT0794-0796 polysaccharide locus in a non-F. tularensis strain.
Explores possible cause of inconsistent oxidase test results by F. novicida strains.
Acknowledgments
1.6. FUNDING INFORMATION
This work was supported by the National Institutes of Health [Grant numbers U54 AI057156, U54 HG003273], and generous funds gifted by the Alkek Foundation. Special thanks to Dr. Nadim Ajami for comments to the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
1.5. ACCESSION NUMBER
TCH2015 has been deposited in GenBank under the accession number CP021490.
References
- Asare R, Kwaik YA. Molecular complexity orchestrates modulation of phagosome biogenesis and escape to the cytosol of macrophages by Francisella tularensis. Environ Microbiol. 2010;12:2559–2586. doi: 10.1111/j.1462-2920.2010.02229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare R, Kwaik YA. Exploitation of host cell biology and evasion of immunity by Francisella tularensis. Front Microbiol. 2011;1:145. doi: 10.3389/fmicb.2010.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins LM, Holder ME, Ajami NJ, Metcalf GA, Weissenberger GM, Wang M, Vee V, Han Y, Muzny DM, Gibbs RA, Petrosino JF. High-quality draft genome sequence of Francisella tularensis subsp. holarctica strain OR96-0246. Genome Announc. 2015;3:1997–1998. doi: 10.1128/genomeA.00898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacterio.net. accessed October 21st, 2017 from http://www.bacterio.net/francisella.html.
- Banada P, Deshpande S, Chakravorty S, Russo R, Occi J, Meister G, Jones KJ, Gelhaus CH, Valderas MW, Jones M, Connell N, Alland D. Sensitive detection of Francisella tularensis directly from whole blood by use of the GeneXpert System. J Clin Microbiol. 2017;55:291–301. doi: 10.1128/JCM.01126-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara AB, Champion AE, Wang X, Berg G, Apicella MA, McLendon M, Azadi P, Snyder DS, Inzana TJ. Isolation and mutagenesis of a capsule-like complex (CLC) from Francisella tularensis, and contribution of the CLC to F. tularensis virulence in mice. PLoS One. 2011;6:e19003. doi: 10.1371/journal.pone.0019003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett M, Doppalapudi A, Respicio-Kingry LB, Myers D, Husband B, Pollard K, Mead P, Petersen JM, Whitener CJ. Francisella novicida bacteremia after a near-drowning accident. J Clin Microbiol. 2012;50:2826–2829. doi: 10.1128/JCM.00995-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett ME, Respicio-Kingry LB, Yendell S, Ratard R, Hand J, Balsamo G, Scott-Waldron C, O’Neal C, Kidwell D, Yockey B, Singh P, Carpenter J, Hill V, Petersen JM, Mead P. Outbreak of Francisella novicida bacteremia among inmates at a Louisiana correctional facility. Clin Infect Dis. 2014;59:826–833. doi: 10.1093/cid/ciu430. [DOI] [PubMed] [Google Scholar]
- Busse HJ, Huber B, Anda P, Escudero R, Scholz HC, Seibold E, Splettstoesser WD, Kämpfer P. Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis – response to Johansson et al. Int J Syst Evol Microbiol. 2010;60:1718–1720. doi: 10.1099/00207713-60-8-1718. [DOI] [PubMed] [Google Scholar]
- Challacombe JF, Petersen JM, Gallegos-Graves LV, Hodge D, Pillai S, Kuske CR. Whole genome relationships among Francisella bacteria of diverse origin define new species and provide specific regions for detection. Appl Environ Microbiol. 2016;83:e02589–16. doi: 10.1128/AEM.02589-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarridge JE, Raich TJ, Sjöstedt A, Sandström G, Darouiche RO, Shawar RM, Georghiou PR, Osting C, Vo L. Characterization of two unusual clinically significant Francisella strains. J Clin Microbiol. 1996;34:1995–2000. doi: 10.1128/jcm.34.8.1995-2000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay CD, Son S, Gunn JS, Schlesinger LS. Evasion of complement-mediated lysis and complement C3 deposition are regulated by Francisella tularensis lipopolysaccharide O antigen. J Immunol. 2008;181:5568–5578. doi: 10.4049/jimmunol.181.8.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DSMZ.de. accessed October 21st, 2017 from https://www.dsmz.de/microorganisms/pnu/bacterial_nomenclature_info_mm.php?genus=Francisella&show_genus_info=1.
- Forsman M, Sandström G, Sjöstedt A. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int J Syst Bacteriol. 1994;44:38–46. doi: 10.1099/00207713-44-1-38. [DOI] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Gunnell MK, Lovelace CD, Satterfield BA, Moore EA, Neill KLO, Robison RA. A multiplex real-time PCR assay for the detection and differentiation of Francisella tularensis subspecies. J Med Microbiol. 2012;61:1525–1531. doi: 10.1099/jmm.0.046631-0. [DOI] [PubMed] [Google Scholar]
- Hollis DG, Weaver RE, Steigerwalt AG, Wenger JD, Moss CW, Brenner DJ. Francisella philomiragia comb. nov. (Formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (Formerly Francisella novicida) associated with human disease. J Clin Microbiol. 1989;27:1601–1608. doi: 10.1128/jcm.27.7.1601-1608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber B, Escudero R, Busse HJ, Seibold E, Scholz HC, Anda P, Kämpfer P, Splettstoesser WD. Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al. 1955) Olsufiev et al. 1959 as Francisella tularensis subsp. novicida comb. nov. and emended description of the genus Francisella. Int J Syst Evol Microbiol. 2010;60:1887–1896. doi: 10.1099/ijs.0.015941-0. [DOI] [PubMed] [Google Scholar]
- Johansson A, Celli J, Conlan W, Elkins KL, Forsman M, Keim PS, Larsson P, Manoil C, Nano FE, Petersen JM, Sjöstedt A. Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis. Int J Syst Evol Microbiol. 2010;60:1717–1718. doi: 10.1099/ijs.0.022830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Daligault HE, Davenport KW, Coyne SR, Frey KG, Koroleva GI, Broomall SM, Bishop-Lilly KA, Bruce DC, Chertkov O, Freitas T, Jaissle J, Ladner JT, Rosenzweig CN, Gibbons HS, Palacios GF, Redden CL, Xu Y, Minogue TD, Chain PS. Genome sequencing of 18 Francisella strains to aid in assay development and testing. Genome Announc. 2015;3:2–3. doi: 10.1128/genomeA.00147-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingry LC, Petersen JM. Comparative review of Francisella tularensis and Francisella novicida. Front Cell Infect Microbiol. 2014;4:35. doi: 10.3389/fcimb.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones S, Marra M. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson C, Wicht W, Jellison W. A new organism resembling P. tularensis isolated from water. Public Heal Rep. 1955;70:253–258. [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Oyston PCF, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Hälltorp G, Johansson D, Isherwood KE, Karp PD, Larsson E, Liu Y, Michell S, Prior J, Prior R, Malfatti S, Sjöstedt A, Svensson K, Thompson N, Vergez L, Wagg JK, Wren BW, Lindler LE, Andersson SGE, Forsman M, Titball RW. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–9. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- Larsson P, Elfsmark D, Svensson K, Wikström P, Forsman M, Brettin T, Keim P, Johansson A. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 2009;5:e1000472. doi: 10.1371/journal.ppat.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima R, Escudero R, Molina DM, Rodriguez-Vargas M, Randall A, Jasinskas A, Pablo J, Felgner PL, AuCoin DP, Anda P, Davies DH. Towards development of improved serodiagnostics for tularemia by use of Francisella tularensis proteome microarrays. J Clin Microbiol. 2016;54:1755–1765. doi: 10.1128/JCM.02784-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KKM, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, Elkins KL. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano FE, Schmerk C. The Francisella pathogenicity island. Ann N Y Acad Sci. 2007;1105:122–37. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- NCBI Prokaryotic Genome Annotation Pipeline. doi: 10.1093/nar/gkw569. accessed October 26th, 2017 from https://www.ncbi.nlm.nih.gov/genome/annotation_prok. [DOI] [PMC free article] [PubMed]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacific Biosciences. Protocols accessed October 26th, 2017 from http://www.pacb.com/wp-content/uploads/Procedure-Checklist-Preparing-SMRTbell-Libraries-PacBio-Barcoded-Adapters-Multiplex-SMRT-Sequencing.pdf and http://www.pacb.com/wp-content/uploads/2015/09/SMRT-Analysis-Software-Installation-v2.3.0.pdf.
- Peng K, Monack DM. Indoleamine 2,3-dioxygenase 1 is a lung-specific innate immune defense mechanism that inhibits growth of Francisella tularensis tryptophan auxotrophs. Infect Immun. 2010;78:2723–2733. doi: 10.1128/IAI.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JM, Carlson J, Yockey B, Pillai S, Kuske C, Garbalena G, Pottumarthy S, Chalcraft L. Direct isolation of Francisella spp. from environmental samples. Lett Appl Microbiol. 2009;48:663–667. doi: 10.1111/j.1472-765X.2009.02589.x. [DOI] [PubMed] [Google Scholar]
- SEED Viewer version 2.0. can be accessed from http://rast.nmpdr.org/seedviewer.cgi.
- Segata N, Börnigen D, Morgan XC, Huttenhower C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun. 2013;4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Select Agents and Toxins Exclusions. accessed October 26th, 2017 from https://www.selectagents.gov/exclusions-hhs.html#francisella.
- Siddaramappa S, Challacombe JF, Petersen JM, Pillai S, Hogg G, Kuske CR. Common ancestry and novel genetic traits of Francisella novicida-like isolates from North America and Australia as revealed by comparative genomic analyses. Appl Environ Microbiol. 2011;77:5110–5122. doi: 10.1128/AEM.00337-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin A, Svensson K, Ohrman C, Ahlinder J, Lindgren P, Duodu S, Johansson A, Colquhoun DJ, Larsson P, Forsman M. Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. BMC Genomics. 2012;13:268. doi: 10.1186/1471-2164-13-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöstedt AB. Family III. Francisellaceae fam. nov. In: Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. 2nd. Springer; USA: 2005. pp. 199–210. (Vol II. Part B. The gammaproteobacteria). [DOI] [Google Scholar]
- Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturr MG, Krulwich TA, Hicks DB. Purification of a cytochrome bd terminal oxidase encoded by the Escherichia coli app locus from a Δcyo Δcyd strain complemented by genes from Bacillus firmus OF4. J Bacteriol. 1996;178:1742–1749. doi: 10.1128/jb.178.6.1742-1749.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RM, Twine SM, Fulton KM, Tessier L, Kilmury SLN, Ding W, Harmer N, Michell SL, Oyston PCF, Titball RW, Prior JL. Glycosylation of DsbA in Francisella tularensis subsp. tularensis. J Bacteriol. 2011;193:5498–509. doi: 10.1128/JB.00438-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipp MJ, Davis JM, Lum G, De Boer J, Zhou Y, Bearden SW, Petersen JM, Chu MC, Hogg G. Characterization of a novicida-like subspecies of Francisella tularensis isolated in Australia. J Med Microbiol. 2003;52:839–842. doi: 10.1099/jmm.0.05245-0. [DOI] [PubMed] [Google Scholar]
- Whitehouse CA, Kesterson KE, Duncan DD, Eshoo MW, Wolcott M. Identification and characterization of Francisella species from natural warm springs in Utah, USA. Lett Appl Microbiol. 2012;54:313–324. doi: 10.1111/j.1472-765X.2012.03214.x. [DOI] [PubMed] [Google Scholar]
- Zarrella TM, Singh A, Bitsaktsis C, Rahman T, Sahay B, Feustel PJ, Gosselin EJ, Sellati TJ, Hazlett KRO. Host-adaptation of Francisella tularensis alters the bacterium’s surface-carbohydrates to hinder effectors of innate and adaptive immunity. PLoS One. 2011;6:e22335. doi: 10.1371/journal.pone.0022335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


