Abstract
The spirochetes Borrelia (Borreliella) burgdorferi and Borrelia hermsii, the etiologic agents of Lyme disease and relapsing fever, respectively, cycle in nature between an arthropod vector and a vertebrate host. They have extraordinarily unusual genomes that are highly segmented and predominantly linear. The genetic analyses of Lyme disease spirochetes have become increasingly more sophisticated, while the age of genetic investigation in the relapsing fever spirochetes is just dawning. Molecular tools available for B. burgdorferi and related species range from simple selectable markers and gene reporters to state-of-the-art inducible gene expression systems that function in the animal model and high-throughput mutagenesis methodologies, despite nearly overwhelming experimental obstacles. This armamentarium has empowered borreliologists to build a formidable genetic understanding of the cellular physiology of the spirochete and the molecular pathogenesis of Lyme disease.
1 Introduction
The maxim spirochetes do it differently has proved repeatedly to be true, from ultrastructure to motility to gene regulation (Charon and Goldstein 2002; Charon et al. 2012; Johnson 1977; Samuels and Radolf 2009; Samuels 2011), yet a few exceptions exist; one of these rule-proving exceptions regards genetically manipulating Borrelia spp. Except for the labyrinthine tactics required to grow these serpentine bacteria in culture and handle their Gordian genomes, which are exceedingly segmented and largely linear (Brisson et al. 2012; Fraser et al. 1997), the transformation and complementation of Borrelia (Borreliella) burgdorferi (and related species that cause Lyme disease) and Borrelia hermsii (and related species that cause relapsing fever) are not unlike that of model microorganisms (Battisti et al. 2008; Samuels 1995).
The awesome power of reverse genetics has been tapped to dissect the microbial physiology of B. burgdorferi and, to a lesser extent, B. afzelii, B. garinii, and B. hermsii as well as the molecular pathogenesis of Lyme disease and, more recently, relapsing fever (Brisson et al. 2012; Groshong and Blevins 2014; Raffel et al. 2014; Rosa et al. 2005). One good reason for the success of the genetic approach is the availability of robust animal models (Barthold et al. 2010); these experimental systems mimic the natural course of disease as well as the two-host enzootic cycles that include a vertebrate and an arthropod vector, usually a tick (Caimano et al. 2016; Piesman and Schwan 2010; Radolf et al. 2012). The genetic toolbox constructed since Borrelia was first transformed (Samuels et al. 1994a) includes genome sequences (Di et al. 2014; Fraser et al. 1997), selectable markers and gene reporters (see Sect. 3), shuttle vectors (see Sect. 6), transposon mutagenesis (see Sect. 7), inducible promoters (see Sect. 5), and a counterselectable marker (see Sect. 8). Borreliogists have mutated a multitude of genes and now molecularly probe the Borrelia lifestyle encompassing all things from metabolism to transcriptional regulation to host interactions.
2 Transformation
B. burgdorferi was first transformed in the early 1990s with an allele of the endogenous gyrB gene, encoding the B subunit of DNA gyrase, that confers resistance to coumarin antibiotics (Samuels et al. 1994a). The electrotransformation protocol is similar to that used for many bacteria (Nickoloff 1995), but low transformation efficiency and retention of the native suite of plasmids continues to challenge borreliologists. The basic protocol has remained relatively unchanged: competent cells are prepared by removing salts, purified DNA substrate is added, and a high-voltage electric current is applied to the cells. Electroporation transiently compromises the integrity of the outer membrane, visualized by electron microscopy to reveal dark-stained patches that likely represent breaches in the membrane (Fig. 1) (Samuels and Garon 1997). The membrane rapidly recovers (within 30 min) when the cells are placed in growth medium following electroporation. Substantial improvements in transformation efficiency have been wrought by increasing the amount of substrate DNA to tens of micrograms (Eggers et al. 2002; Elias et al. 2002; Hübner et al. 2001; Stewart et al. 2001; Tilly et al. 2000) and transforming with modified DNA (Chen et al. 2008; Jewett et al. 2007).
Fig. 1.
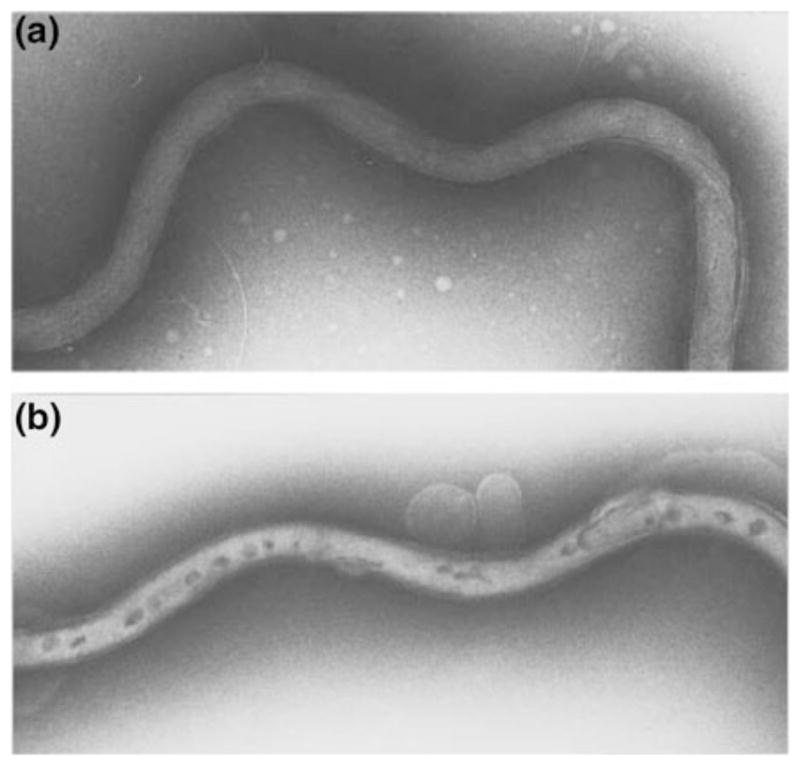
B. burgdorferi before (a) and after (b) electroporation as visualized by negative staining and transmission electron microscopy. Note the darkly stained regions that are thought to be transient pores generated by the electric pulse that serve as a passageway for DNA to enter the spirochete. Reprinted with permission from Samuels and Garon (1997)
2.1 Competent Cells and Electroporation
Successful preparation of B. burgdorferi competent cells requires growth to mid- to late-log phase; cell density is probably the most vital parameter (Samuels et al. 1994a; Tilly et al. 2000). Certain low-passage and mutant strains are refractory to transformation due to inordinate clumping during competent cell preparation (Hübner et al. 2001; Samuels 1995; Tilly et al. 2000). Salts are removed from the growth medium by a series of cold, sterile washes and the Borrelia cells are resuspended in a glycerol-sucrose solution. Arcing will occur during electroporation if salts are not effectively removed, which will kill the vast majority of cells and, obviously, prevent transformation. Electrotransformation is most efficient with fresh competent cells, but cells can be stored at −80 °C and slow-thawed on ice, although there is about a 50% decrease in efficiency (Samuels 1995; Samuels and Garon 1997).
2.2 DNA Source, Form and Modification
Many different substrates have been transformed into B. burgdorferi, from short oligonucleotides (Samuels and Garon 1997) to an entire native 28-kb linear plasmid (Grimm et al. 2004a). The original allelic exchange transformation used PCR products to mutate the gyrB gene and confer antibiotic resistance (Samuels et al. 1994a). More recently, linear segments of cloned DNA containing a selectable marker are typically used for allelic exchange to generate mutants. In addition, circular suicide plasmids have been integrated into the genome by recombination (Stevenson et al. 1998). An important advance in the Borrelia genetic toolbox was the development of circular shuttle vectors that autonomously replicate in both B. burgdorferi and Escherichia coli (Eggers et al. 2002; Sartakova et al. 2000; Stewart et al. 2001). These plasmid vectors enhanced the number and sophistication of genetic experiments in B. burgdorferi, including trans-complementation, ectopic gene expression, and inducible gene expression. Linear shuttle vectors based on lp25, lp28-1, and lp17 have also been built using a similar strategy to the circular shuttle vector, although they have not been used frequently (Beaurepaire and Chaconas 2005; Stewart et al. 2003).
The low frequency of transformation is likely due to restriction-modification systems (Chen et al. 2008; Kawabata et al. 2004; Lawrenz et al. 2002; Rego et al. 2011). B. burgdorferi putatively possesses two type-IV restriction-modification enzymes: BBE02 on lp25 and BBQ67 on lp56 (Kawabata et al. 2004; Lawrenz et al. 2002). The gene products of bbe02 and bbq67 are predicted to methylate certain adenines and cleave foreign DNA lacking this modification. The presence of these two plasmids is correlated with low transformation efficiency of shuttle vectors isolated from E. coli. Transformants often lose lp25 and transformation efficiency increases about 40-fold in bbe02 mutants (Kawabata et al. 2004). To complicate matters, lp25 also carries genes essential for vertebrate and tick infectivity (Grimm et al. 2004a; Labandeira-Rey and Skare 2001; Purser and Norris 2000; Purser et al. 2003; Revel et al. 2005; Strother and de Silva 2005). Thus, low-passage, infectious strains are more difficult to transform than high-passage, attenuated strains. BBE02 is not required for in vitro cultivation, so replacing bbe02 with a selectable marker both increased transformation efficiency and provided a selection for lp25, allowing transformants to retain infectivity (Chan et al. 2015; Gilbert et al. 2007; Kasumba et al. 2015; Rego et al. 2011). Clever tactics to subvert the restriction-modification systems have been developed over the years. Transformation efficiency can be increased by first transforming the DNA into a strain carrying only one of the putative restriction-modification systems and using this partially methylated DNA for subsequent transformation (Jewett et al. 2007). Alternatively, in vitro CpG methylation of substrate DNA increases transformation efficiency (Chen et al. 2008). Both approaches have facilitated genetic manipulation of B. burgdorferi.
2.3 Relapsing Fever Borrelia
B. hermsii, one of the species of Borrelia that causes relapsing fever in humans (Barbour and Guo 2010), has also been successfully transformed (Battisti et al. 2008), but the history of genetic manipulation is considerably younger. The protocol for preparation of competent cells, electrotransformation and selection are basically the same as for B. burgdorferi. The notable changes include using large amounts of DNA for transformation (25 μg) and selecting for transformants in antibiotics for up to a week in batch before plating to isolate clones (Battisti et al. 2008; Fine et al. 2011; Raffel et al. 2014). The shuttle vector pBhSV2, based on the B. burgdorferi shuttle vector pBSV2, functions in B. hermsii, but only the antibiotics gentamicin and kanamycin have been effective for selection (Battisti et al. 2008). So far, three genes have been mutated: vtp, which encodes variable tick protein, an outer surface lipoprotein synthesized when the spirochete is resident in its tick vector (Battisti et al. 2008; Raffel et al. 2014), vmp, which is the expression locus for the genes encoding the variable major proteins that are synthesized in the mammalian host and allow the spirochete to evade the immune response (Raffel et al. 2014), and dam, which encodes DNA adenine methyltransferase (James et al. 2016). In addition, the green fluorescent protein (GFP) gene has been expressed in B. hermsii (Fine et al. 2011) and its cousin Borrelia turicatae (Krishnavajhala et al. 2017). With these advances in hand, we are beginning to see the dawn of the age of reverse genetics in relapsing fever spirochetes.
3 Selectable Markers and Assayable Reporters
Selectable markers allow for the preferential isolation of genetically altered microorganisms, including B. burgdorferi. Antibiotic resistance markers have proved to be remarkably utilitarian tools for negative selection, the inhibition of the unaltered parental cells. B. burgdorferi is susceptible to many antibiotics and the genes conferring resistance to antibiotics such as streptomycin, kanamycin, gentamicin, and erythromycin are paramount for genetic studies. The antibiotics ampicillin and tetracycline are used to treat patients with Lyme disease, so their use in genetic experiments is discouraged.
The challenges, both in cost and time, in culturing B. burgdorferi to study gene expression have made reporter genes also incredibly useful tools. Reporter gene products provide both a convenient readout to measure transcriptional regulation when fused to promoters and a signal for visualization of tagged proteins or entire cells in vitro and in vivo. Advances in imaging of live mice and ticks infected with B. burgdorferi have provided an unprecedented vista of the interaction of the spirochete with the host (Dunham-Ems et al. 2009; Moriarty et al. 2008).
3.1 Native Antibiotic Resistance
The first selectable marker used for genetic manipulation in B. burgdorferi was an allele of the gyrB gene conferring resistance to the coumarin antibiotics (Samuels et al. 1994a, b; Samuels 2006). A synthetic version of gyrB designed to minimize recombination into the native gyrB locus on the chromosome was used briefly as a selectable marker (Elias et al. 2003). Unfortunately, the gyrB mutations also affect DNA supercoiling in B. burgdorferi (Samuels and Garon 1993), causing pleiotropic effects such as increased expression of groEL, encoding a chaperonin (Alverson and Samuels 2002) and ospC, encoding an outer surface lipoprotein (Alverson et al. 2003; Drecktrah et al. 2013; Yang et al. 2005), which spurred the exploitation of antibiotic resistance markers from other microorganisms.
3.2 Foreign Antibiotic Resistance Genes
The efficiency of genetic manipulation of B. burgdorferi was enhanced when new selectable markers were developed by fusing endogenous promoters to nonnative antibiotic resistance genes (Bono et al. 2000). The strong, constitutive promoters of the flagellin and basal body rod genes flaB and flgB, respectively, were chosen to drive gene expression (Bono et al. 2000; Ge et al. 1997; Sohaskey et al. 1997). These promoters were first fused to the aphI gene from Tn903, resulting in flaBp-aphI and flgBp-aphI, which mediate resistance to kanamycin (Bono et al. 2000). Other markers were constructed soon after by the same strategy and conferred resistance to streptomycin using aadA from Shigella flexneri plasmid R100 (Frank et al. 2003) and resistance to gentamicin using aacC1 from Tn1696 (Elias et al. 2003). These antibiotic resistance markers function in both E. coli and B. burgdorferi, making them versatile and practical tools for genetic studies. Not all antibiotic resistance cassettes function in B. burgdorferi, including cat and pac, the genes encoding resistance to chloramphenicol and puromycin, respectively (Elias et al. 2003). The cat gene encodes a chloramphenicol acetyl transferase, but the acetylation of chloramphenicol is reversed by esterases present in the B. burgdorferi growth medium (Sohaskey and Barbour 1999), which might be overcome by modifying the medium. Some foreign antibiotic resistance genes driven by their own promoters function in B. burgdorferi, such as erm conferring erythromycin resistance (Sartakova et al. 2000). Others include aph(3′)-IIIa conferring kanamycin resistance, aad9 conferring spectinomycin resistance, linA’ conferring lincomycin resistance, and aac(3)-VIa conferring sisomycin and gentamicin resistance, but they have not been widely used (Sartakova et al. 2003). Use of the erm gene has been restricted by some Institutional Biosafety Committees because erythromycin can be used to treat Lyme disease patients (Terekhova et al. 2002). Spectinomycin is not efficient at selecting for transformants containing the aadA (and probably the aad9) gene because B. burgdorferi readily becomes resistant to this antibiotic (Criswell et al. 2006). Lincomycin has not been utilized for selection in B. burgdorferi, but could be adapted to expand the toolbox for genetic manipulation. While the early years entailed plodding development of methodologies for genetic manipulation, borreliologists have refined their experimental approaches and now almost exclusively employ selectable markers conferring antibiotic resistance to kanamycin, streptomycin and gentamicin.
3.3 Mutant Isolation
There are two techniques for plating and selecting transformed B. burgdorferi. One is semi-solid agarose plating where transformed cells are allowed to recover overnight before plating in growth medium mixed with molten agarose (Kurtti et al. 1987; Rosa and Hogan 1992; Samuels et al. 1994a). B. burgdorferi is mixed with the molten agarose-growth medium containing the antibiotic so the colonies form within the mixture, not on the surface. After incubation for 5 days to 2 months in a CO2 incubator, individual colonies appear as small white clouds in the plate and are then expanded in liquid culture to be screened for the desired genetic alteration. The second method is dilution plating in which the transformed cells are diluted in 100 ml growth medium and then distributed in four 96-well plates following overnight recovery (Yang et al. 2004). Plates are incubated in a CO2 incubator until the medium in individual wells begins to turn from red to yellow, as metabolizing B. burgdorferi excretes lactic acid that is indicated by phenol red in the medium. Positive wells are expanded in liquid culture and screened.
3.4 Reporter Genes
The first reporter used in B. burgdorferi was the cat gene (Sohaskey et al. 1997). Although the gene product does not confer resistance to chloramphenicol in B. burgdorferi, the reporter provided some of the first assays of promoter activity (Alverson et al. 2003; Sohaskey et al. 1997, 1999), just as the age of spirochete genomics was dawning (Fraser et al. 1997). More recently, the gene for β-galactosidase, lacZ, fused to the ospC promoter, was used to study the regulation of ospC expression in both liquid and semi-solid media (Hayes et al. 2010, 2014; Sarkar et al. 2011).
The luciferase (luc) gene from the firefly Photinus pyralis has been adapted as a transcriptional reporter to assay gene expression from endogenous promoters (Blevins et al. 2007; Ouyang et al. 2010). The luc reporter also provided the means to follow B. burgdorferi infection in vivo by bioluminescence imaging of infected mice (Chan et al. 2015; Hyde et al. 2011). B. burgdorferi infection and dissemination of mutant strains lacking the fibronectin-binding protein BBK32 or the decorin-binding proteins DbpA and DbpB (Hyde et al. 2011) were assayed as well as the pathogenokinetics of ospC expression (Skare et al. 2016). Recently, a dual luc reporter system has been successfully implemented to study gene expression in the tick vector. The second luc reporter, from Renilla reniformis, allows for normalization to B. burgdorferi number, while the P. pyralis luc gene product can then accurately measure transcriptional activity (Adams et al. 2017a). The luc reporter systems developed for in vivo work provide a more convenient and lower cost system to follow gene expression in the tick-mouse model.
The convenience and versatility of GFP has made it a popular reporter for studies of B. burgdorferi gene expression (Babb et al. 2004; Carroll et al. 2003; Eggers et al. 2002, 2004) and served to demonstrate the functionality of an inducible gene system (Whetstine et al. 2009). The GFP reporter has the advantage of allowing the analyses of single cells, which has highlighted the heterogeneity of B. burgdorferi populations (Srivastava and de Silva 2008). Additionally, the yellow fluorescent protein and cyan fluorescent protein have both been used as reporters (Eggers et al. 2002). The monomeric red fluorescent protein has proved useful for studying lipoprotein secretion to the cell surface (Schulze and Zückert 2006; Schulze et al. 2010) and was a crucial component of a FACS-based protein localization screen (Kumru et al. 2010). The GFP marker could not be directly visualized in these studies, as fluorescence was compromised in the periplasm (Schulze and Zückert 2006). Constitutively expressed gfp has allowed for visualization of B. burgdorferi in live-cell imaging studies. A strain of B. burgdorferi containing a plasmid expressing gfp from the flaB promoter allowed for exquisite examination of the spirochete’s interaction with the mouse vasculature in vivo by live-cell intravital microscopy (Moriarty et al. 2008). Stable integration of a constitutively expressed gfp driven by the flaB promoter on cp26 has served as a fluorescent tag of B. burgdorferi to allow in vivo studies on morphology and population dynamics in ticks (Dunham-Ems et al. 2009).
Finally, GFP has been expressed in the relapsing fever agents B. hermsii (Fine et al. 2011) and B. turicatae (Krishnavajhala et al. 2017), and used to follow the persistence of the latter in the Ornithodoros turicatae tick midgut and salivary glands (Krishnavajhala et al. 2017). Visualization of GFP-relapsing fever Borrelia in vivo should aid in describing the spirochete lifestyle in both the tick vector and mammalian hosts of these understudied Borrelia species.
4 Mutagenesis
Using reverse genetics to reveal the function of a gene entails generating mutations in the gene, which is often suspected of producing a particular phenotype, and isolating the resulting mutants. Although transduction has been demonstrated in Borrelia (Eggers et al. 2001, 2016), mutagenesis currently relies exclusively on transformation (see Sect. 2) and selection with an antibiotic resistance marker (see Sect. 3). Besides pleiotropy and polar effects, a crucial caveat is off-target repercussions of genetic manipulation, which is particularly problematic in B. burgdorferi due to plasmid loss from the segmented genome (Kawabata et al. 2004; Labandeira-Rey and Skare 2001; Lawrenz et al. 2002; Purser and Norris 2000); therefore, mutant phenotypes should be confirmed by complementation or genetic reconstitution (see Sect. 6).
4.1 Gene Inactivation by Homologous Recombination
Almost every mutant in Borrelia has been generated by allelic exchange, requiring recombination of a synthetic DNA construct into the genome. The other mechanisms for mutagenesis, albeit not commonly utilized, are displacement of entire plasmids (Grimm et al. 2004a) and deletion of large segments of plasmids (Beaurepaire and Chaconas 2005; Bestor et al. 2010; Chaconas et al. 2001). Both B. burgdorferi (and other Lyme disease species) and B. hermsii (and other relapsing fever species) have antigenic variation systems that entail intra-genomic recombination (Nordstrand et al. 2000; Stoenner et al. 1982; Zhang et al. 1997) and a RecA homolog (Liveris et al. 2004; Putteet-Driver et al. 2004) that are likely hijacked to incorporate foreign DNA. VlsE antigenic variation in B. burgdorferi requires the branch migrase RuvAB (Dresser et al. 2009; Lin et al. 2009), which is also implicated in homologous recombination during genetic manipulation.
One curious phenomenon that was observed during allelic exchange (Brisson et al. 2012; Knight et al. 2000) and also documented during VlsE variation (Coutte et al. 2009) is the presence of mosaic recombination products, which implies that the crossover hops back and forth between the donor and recipient DNA strands, so the recombination machinery skips short stretches of sequence. This intermittent recombination generates further diversity during antigenic variation (Brisson et al. 2012; Coutte et al. 2009), but the caveat for genetic manipulation is that not all the expected changes may be present, so mutant and flanking sequences should be experimentally confirmed.
4.2 Site-Directed Mutagenesis
There have been only a handful of studies in which the sequence of a gene or regulatory element in its endogenous genomic location was specifically and subtly altered (as opposed to grossly deleting or inserting a gene) in B. burgdorferi. In the original report of transformation, a single codon of the gyrB gene was mutated to confer resistance to the antibiotic coumermycin A1 (Samuels et al. 1994a; Samuels 2006). Several other genes have since been surgically manipulated in situ.
The C-terminal domain of the A subunit of DNA gyrase is uniquely synthesized as an independent DNA-binding protein termed Gac in B. burgdorferi (Knight and Samuels 1999). In order to disrupt gac expression without affecting the essential gyrA gene in which gac is embedded, the ribosome-binding site, the start codon and the second methionine codon of gac were mutated, which abrogated Gac production, although the mutant phenotype was disappointingly suppressed (Knight et al. 2000).
Rrp2 is an enhancer-binding protein and two-component system response regulator (Fraser et al. 1997; Samuels 2011). Exhaustive efforts failed to generate a null rrp2 mutant, suggesting that Rrp2 had an essential function. Yang et al. (2003) changed a single residue in the putative activation domain, which eliminated expression of rpoS and the RpoS regulon, establishing the Rrp2-RpoN-RpoS pathway paradigm for gene regulation during the enzootic cycle (Burtnick et al. 2007; Caimano et al. 2007; Hübner et al. 2001; Samuels 2011), but retained the essential, and still unidentified, function of Rrp2.
OspC is a transmission factor required to establish B. burgdorferi infection in the vertebrate host (Dunham-Ems et al. 2012; Fingerle et al. 2007; Gilbert et al. 2007; Gilmore and Piesman 2000; Grimm et al. 2004b; Pal et al. 2004; Stewart et al. 2006; Tilly et al. 2006), but its molecular function has not been fully elucidated. Marconi and colleagues have mutated both a presumptive ligand-binding site (Earnhart et al. 2010) and a cysteine residue involved in quaternary structure (Earnhart et al. 2011): infectivity was attenuated when the binding site was mutated, although the specific ligand has not been defined, and oligomerization was prevented by eradicating the disulfide bond.
The regulation of ospC expression has also been probed by site-directed mutagenesis. OspC levels increase dramatically during transmission from the tick to the vertebrate (Fingerle et al. 2002; Gilmore and Piesman 2000; Leuba-Garcia et al. 1998; Montgomery et al. 1996; Ohnishi et al. 2001; Rathinavelu and de Silva 2001; Schwan et al. 1995; Schwan and Piesman 2000), but OspC synthesis is turned off after the vertebrate infection is established in order for the spirochete to evade adaptive immunity (Crother et al. 2004; Liang et al. 2002a, b; 2004; Tilly et al. 2006). We (Drecktrah et al. 2013) introduced site-directed mutations in situ into an inverted repeat that constitutes part of an ospC operator (Xu et al. 2007) and demonstrated that impairing the intra-complementarity of the inverted repeat reduced induction of ospC expression in response to environmental factors in vitro.
Site-directed mutations have also been incorporated in trans into the genome by transforming a mutated gene or regulatory sequence on a shuttle vector into a null background or fused to a reporter, respectively. Targets of these plasmid-borne dissections include the ospC operator/promoter (Eggers et al. 2004; Yang et al. 2005), the ospE and ospF promoters (Eggers et al. 2006), ospA (Schulze and Zückert 2006), ospC (Kumru et al. 2011a, b), p13 (Kumru et al. 2011a), bb0646 (Shaw et al. 2012), and bbd18 (Hayes et al. 2014).
4.3 Promoter Fusion
Borrelia promoters have been fused to selectable markers (see Sect. 3.2), reporters (see Sect. 3.4), and transposons (see Sect. 7) as well as genes of interest to effect constitutive, high-level expression, often as an alternative approach when attempts to disrupt the gene fail. One of the first applications was the fusion of the flaB promoter to the ospC ORF, which corroborated that constitutive synthesis of OspC during vertebrate infection prevented B. burgdorferi from avoiding the immune response (Xu et al. 2006). In a more recent study, Tilly et al. (2013) swapped the promoters and ORFs for three outer membrane lipoprotein genes, including ospC and vlsE, revealing that OspC and VlsE have similar in vivo functions.
4.4 Cre-lox and Telomere-Mediated Recombination
Perhaps the most conspicuous genetic method missing from the Borrelia armamentarium is markerless mutagenesis, which is somewhat surprising because Bestor et al. (2010) have developed an elegant Cre-lox recombination system that functions in B. burgdorferi. Two loxP sites were inserted several kilobase pairs apart into one of the linear plasmids and the Cre recombinase was introduced on a shuttle vector, which resulted in the deletion of a substantial swath of genes (Bestor et al. 2010). Chaconas et al. (2001) fashioned a similar system utilizing a replicated telomere that is inserted into a linear plasmid and triggers a telomere resolvase-mediated deletion between the inserted telomere and the end of the linear plasmid opposite the origin of replication (Beaurepaire and Chaconas 2005). Genetic borreliology stoically awaits a tractable system to make markerless mutations.
5 Inducible Gene Expression
The ability to artificially regulate gene expression in B. burgdorferi has been immensely valuable for probing the function of essential genes and for assaying the kinetic requirements of gene expression in vivo (Samuels 2011). Failure to generate a null mutant after exhaustive attempts suggests that the targeted gene is essential (Bandy et al. 2014; Chu et al. 2016; Dunn et al. 2015; Lenhart et al. 2012; Liang et al. 2010). In these instances, a conditional mutant, generated by replacing the endogenous promoter with an inducible promoter, allows for gradual reduction of gene expression, which is not possible with a null mutation. Essentiality is demonstrated if growth ceases in the absence of gene expression. In addition, the phenotype associated with decreased expression may provide clues to the function of the gene. Inducible systems can also be harnessed to experimentally overcome a growth phenotype even if the gene is not essential. For example, changes in morphology, metabolism or gene expression may be challenging or laborious to assess in a null mutant, but may be expedited using a conditional mutant where expression is induced for outgrowth and then stopped by removing the inducer.
5.1 lac System
A couple of strategies for inducible gene expression that have been workhorses in other bacteria have been adapted for B. burgdorferi. The first was based on the lac operon of E. coli where the repressor LacI binds to operators to repress gene expression (Jacob and Monod 1961). Repression is relieved by an inducer, such as isopropyl β-D-1-thiogalactopyranoside (IPTG), which binds LacI and causes an allosteric change that inhibits binding to the operators. In B. burgdorferi, the strong flgB promoter (Ge et al. 1997) fused to lacI was inserted in bbe02 on lp25, resulting in high levels of LacI production (Gilbert et al. 2007). The inducible flac promoter was fashioned by integrating an optimized operator, lacOid (Oehler et al. 1994), into the flgB promoter; the hybrid flacp is then fused to any gene (Gilbert et al. 2007). In vitro, IPTG induced OspC production in a flacp-ospC strain (Gilbert et al. 2007) and RpoS production in a flacp-rpoS strain (Caimano et al. 2007; Gilbert et al. 2007). Furthermore, using a mouse model in which IPTG was administered in the drinking water, OspC was required immediately upon injection of B. burgdorferi into the mouse as well as during the early stages of murine infection (Gilbert et al. 2007). flacp has also been fused to bosR, encoding a transcriptional regulator (Hyde et al. 2010), bamA and tamB, encoding the outer membrane protein transport machinery (Iqbal et al. 2016; Lenhart and Akins 2010), and ftsH, encoding a membrane protein protease (Chu et al. 2016).
Another inducible system based on the lac operon consists of the flaB promoter fused to lacI and the hybrid T5/lac promoter pQE30 from plasmid QE30 (Blevins et al. 2007). IPTG induced expression of several genes fused to pQE30, including bptA, encoding an outer surface lipoprotein required for tick persistence (Blevins et al. 2007), rrp2 (Groshong et al. 2012; Ouyang et al. 2014a), csrA, encoding an RNA-binding protein (Ouyang et al. 2014b), dhhP, encoding a cyclic-di-AMP phosphodiesterase (Ye et al. 2014), and resT, encoding the telomere resolvase (Bandy et al. 2014).
5.2 tet System
In addition to the lac-based gene regulation in B. burgdorferi, there have been two other inducible systems incorporating tetracycline resistance regulation from the E. coli transposon Tn10 (Cabello et al. 2006; Whetstine et al. 2009). The TetR repressor binds to the tet operator and the tetracycline analog anhydrotetracycline induces gene expression. In one version, the flaB promoter was fused to tetR and the tet operator was combined with the B. burgdorferi ospA promoter forming the hybrid Post (Whetstine et al. 2009), which was fused to gfp (Whetstine et al. 2009) as well as bpaB and ebfC, which encode small DNA-binding proteins (Jutras et al. 2012a, b). In the other tet system in B. burgdorferi, tetR was also driven by the flaB promoter and expression of gfp or bmpA, encoding an outer membrane protein, was controlled by fusing tet operators to the bmpA promoter (Cabello et al. 2006).
Thus, a few variations on inducible gene regulation have been developed for B. burgdorferi, although the lacI systems are more widely used and, notably, function in vivo in an animal model (Chu et al. 2016; Gilbert et al. 2007).
6 Complementation
The fulfillment of molecular Koch’s postulates assigning a function to a gene product in a microbe requires complementation of an isogenic mutant (Falkow 1988). In bacteria, including B. burgdorferi, synthetically constructed mutants are typically complemented either in cis by genetically restoring the wild-type allele to the native locus or in trans by introducing the functional gene to a heterologous locus, often on a plasmid vector. Each approach has strengths and weaknesses.
6.1 Obstacles
The barriers to B. burgdorferi transformation, especially the restriction-modification systems (the bbe02 and bbq56 gene products) and loss of genomic elements, some of which are essential for host infection, are particularly pertinent to complementation. In addition to the aforementioned tactics of bbe02 disruption and DNA methylation, various methods facilitate transformation of infectious mutant strains for complementation. A bbe02 mutant has been deployed as the parental strain, in both wild-type and mutant bbq56 backgrounds (Chan et al. 2015; Gilbert et al. 2007; Kasumba et al. 2015; Rego et al. 2011). This modification serves double duty by increasing transformation efficiency and furnishing a selection for lp25, which is readily lost during in vitro cultivation, but essential for host infection (Grimm et al. 2004a; Labandeira-Rey and Skare 2001; Purser and Norris 2000; Purser et al. 2003; Revel et al. 2005; Strother and de Silva 2005). Another method employed to ease complementation utilizes the pncA gene from lp25, which is necessary and sufficient to maintain mouse infectivity, carried on a shuttle vector, along with the complementing gene, and transformed into a strain that lacks lp25 (and lp56) (Seshu et al. 2006; Xu et al. 2005). This approach allows for vertebrate host studies, but not tick experiments as undefined genes from lp25, besides pncA, are necessary for persistence in the tick (Grimm et al. 2005; Revel et al. 2005; Strother and de Silva 2005). More recently, a suicide plasmid that recombines both the complementing gene and a selectable marker into the bbe02 locus on lp25 has been established that is competent for in vivo experiments in the tick-mouse model (Kasumba et al. 2015). Thus, what was once an impediment to genetic manipulation and animal studies, a restriction-modification system on a plasmid unstable in vitro but required in vivo, has been exploited to study the role of gene products in B. burgdorferi.
6.2 Trans-complementation and Shuttle Vectors
B. burgdorferi sequences essential for plasmid partitioning and replication have been incorporated to construct several shuttle vectors that originally made complementation experiments possible. pBSV2 was the first, and likely remains the most popular, shuttle vector; it contains a 3.3-kb segment from the endogenous plasmid cp9 and is capable of autonomous replication in both E. coli and B. burgdorferi (Stewart et al. 2001). This region contains three open reading frames of a paralogous gene family (PGF) that is not homologous to any known replication or partitioning genes, but is found in many of the B. burgdorferi circular and linear plasmids (Casjens et al. 2000; Chaconas and Norris 2013; Zückert and Meyer 1996). In all, there are five different members in the PGF, with different combinations present on B. burgdorferi plasmids (Beaurepaire and Chaconas 2005; Chaconas and Norris 2013; Eggers et al. 2002; Zückert and Meyer 1996). pBSV2 (Stewart et al. 2001), and its derivatives containing alternative selectable markers (Elias et al. 2003; Frank et al. 2003), as well as the pCE320 series derived from cp32 (Eggers et al. 2002) are stably maintained both in vitro and in vivo. A number of shuttle vectors have been constructed using the PGFs from lp25, lp28-1, lp17, a cp32, and cp26 (Beaurepaire and Chaconas 2005; Byram et al. 2004; Eggers et al. 2002; Stewart et al. 2003; Tilly et al. 2012). Interestingly, all of these shuttle vectors are incompatible with their native plasmid from which the partitioning region has been cloned, as evidenced by transformation displacing the endogenous plasmid (Chaconas and Norris 2013), except cp26, which contains other essential genetic elements (Bandy et al. 2014; Byram et al. 2004).
Broad-host-range plasmids also have served as shuttle vectors to complement B. burgdorferi mutants, and to ectopically express genes. These plasmids can be replicated and maintained in a number of different bacterial hosts. The plasmid pJRS525 and related pGK12, both from Lactococcus lactis, have been used to complement B. burgdorferi mutants and express gfp, respectively (Hübner et al. 2001; Sartakova et al. 2000). pGK12 is less stable, without selection, and present at lower copy number than the more widely used pBSV2, potentially practical properties for plasmid curing and matching gene dosage during complementation (Sartakova et al. 2000).
6.3 Genetic Reconstitution
The other way to complement a mutant in B. burgdorferi is genetic reconstitution. This strategy involves allelic exchange at the original mutated locus to restore the wild-type gene (Hübner et al. 2001; Tilly et al. 2001). The restriction-modification systems do not pose a hurdle with this method as linear, non-autonomously replicating DNA substrates are used, but loss of native plasmids during this second transformation remains a concern. The advantage of genetic reconstitution is that returning the wild-type gene to its native locus restores the copy number, regulation, supercoiling state, and any small noncoding RNAs that may have been disrupted in the mutant. Disadvantages of genetic reconstitution include additional manipulation of the targeted loci, which may cause polar effects, thereby confounding comparison of the mutant and complemented strain phenotypes. Genetic reconstitution in a separate noncoding region of the genome may alleviate some of these concerns, but we now know that B. burgdorferi has an extensive, and incompletely characterized, sRNA transcriptome that must be considered when altering any region of the genome that is considered “non-coding” (Adams et al. 2017b; Arnold et al. 2016; Lybecker and Samuels 2017; Popitsch et al. 2017).
6.4 Caveats and Considerations
Occasionally, despite a plethora of painstaking attempts, complementing a B. burgdorferi mutant is simply not successful (Stewart et al. 2008). The reason is rarely clear, but some mutants have severe growth phenotypes, including retarded replication, elongated morphology and “clumping,” which adversely affect the preparation of competent cells or efficient electrotransformation and recovery. In the case where a mutant is unable to be complemented, the last best course to assign a function to a mutated gene is to generate multiple independent clones from separate transformations. If all the clones have the same phenotype, then this is stronger evidence than relying on the phenotype of a single mutant. This line of reasoning is obviously not as robust as complementation of the mutant, but decreases the chance that an unrelated second-site mutation in the genome is responsible for the phenotype, as that change is unlikely to occur in two independent mutants, particularly if two different selectable markers are used. There are several analyses that should be undertaken if complementation does not restore the wild-type phenotype (Bugrysheva et al. 2005). Of course, the plasmid profile should be determined following all genetic manipulations, and transformants lacking an intact genome should not be further studied. Second, an alternative complementation construction (in trans or genetic reconstitution) should be attempted as both methods have their limitations. Third, the recombinant DNA in the mutant should be recovered from the complemented strain and sequenced to determine if any changes have occurred during genetic manipulation. Inadvertent mutations could be introduced during subcloning in E. coli or in B. burgdorferi. If the sequence is correct, then the level of gene expression should be compared between the complemented strain and the isogenic wild type by qRT-PCR or Northern blot analyses. Northern blot analysis of the mRNA has the advantage of detecting differential transcript processing between the two strains, which may affect stability or translation. A different complementation strategy would be prudent if the levels or kinetics of expression are significantly different between the complemented and wild-type strains. Finally, if all else fails, then polar effects from the mutation, second-site mutations and disruption of small noncoding RNAs should be considered. If the expression of adjacent genes has been altered in either the mutant or complemented strain, then another strategy such as site-directed mutagenesis may be warranted. Recently, B. burgdorferi has been found to have an extensive collection of noncoding sRNAs, nearly all of unknown function (Adams et al. 2017b; Arnold et al. 2016; Lybecker and Samuels 2017; Popitsch et al. 2017). These include antisense, intergenic, and intragenic (within a gene on the coding strand) RNAs. Mutant construction and complementation may alter these previously unappreciated sRNAs, resulting in failed complementation or confounding data interpretation. These genetic elements should be considered when designing both mutant and complementation strategies. Finally, the effect of extraneous second-site mutations on complementation may finally be possible to address as the cost of sequencing entire bacterial genomes continues to decrease. This may be especially applicable to the small genome (*1.5 Mb) of B. burgdorferi (Fraser et al. 1997). While there remain numerous challenges for genetically manipulating B. burgdorferi, particularly during complementation, there is now truly light at the end of the tunnel.
7 Transposon Mutagenesis and High-Throughput Genetics
Transposon mutagenesis is undoubtedly an exceedingly powerful genetic manipulation tool (Lin et al. 2014; Morozova et al. 2005; Stewart et al. 2004; Stewart and Rosa 2008). The most popular transposon system employed in B. burgdorferi entails a hyperactive allele of Himar1, a mariner-family transposable element from Haematobia irritans, fused to a strong, constitutive promoter and carried on a suicide vector; also included are the inverted terminal repeats that serve as binding sites for the transposon and flank a ColE1 origin as well as a selectable marker (Stewart et al. 2004; Stewart and Rosa 2008). The genome-wide mutagenesis technology has led to the generation of a signature-tagged mutant (STM) library in an infectious background (Botkin et al. 2006; Lin et al. 2012), the application of transposon sequencing (Tn-seq) (Troy et al. 2013, 2016), and the identification of scores of genes involved in diverse aspects of spirochete physiology as well as disease pathogenesis (Botkin et al. 2006; Khajanchi et al. 2016; Lin et al. 2009, 2012, 2015; Liu et al. 2009; Morozova et al. 2005; Ramsey et al. 2017; Stewart et al. 2004; Troy et al. 2013, 2016). A notable caveat is that STM and Tn-seq libraries may include mutants that either carry second-site mutations or are missing genomic elements; again, the segmented nature of the genome is a challenge for high-throughput methodologies in Borrelia. Prudence dictates that clean mutants be constructed before a function is ascribed to a gene. For example, the tour de force implementation of in vivo expression technology, another high-throughput genetic screen, inadvertently identified a red herring due to the loss of a plasmid (Ellis et al. 2014), although the system has proved insightful in defining an infectivity-associated sRNA transcriptome in the spirochete (Adams et al. 2017b).
8 Counterselection
Counterselectable markers (CSM) select for the loss of a DNA sequence (Reyrat et al. 1998) and can provide another avenue to generate markerless mutations. CSM generally confer susceptibility to a substance that is lethal to an organism. This scenario requires that the parental strain in the absence of the CSM is resistant to the substance used for negative selection, which limits employing counterselection.
Counterselection has been demonstrated using the rpsL gene, encoding the S12 ribosomal protein, in the spirochetes Leptospira biflexa (Picardeau et al. 2001) and B. burgdorferi (Drecktrah et al. 2010). A mutant rpsL gene located on the chromosome confers resistance to the antibiotic streptomycin (Criswell et al. 2006). The presence of a second, wild-type allele of rpsL in trans renders the merodiploid B. burgdorferi susceptible to streptomycin; although the mechanism has not been fully defined, susceptibility is dominant because translational errors by the sensitive ribosomes (Karimi and Ehrenberg 1994, 1996) will poison the cell. The addition of streptomycin was shown to select for the loss of the plasmid carrying the wild-type rpsL gene that confers susceptibility, thus providing the first functioning CSM in Borrelia (Drecktrah et al. 2010). Additionally, mutations to the parC gene, encoding a subunit of topoisomerase IV, confer resistance to fluoroquinolones in B. burgdorferi (Galbraith et al. 2005), providing another potential CSM.
9 Surrogate Genetics
Many barriers to genetic manipulation of B. burgdorferi, including low transformation efficiency, loss of endogenous plasmids during transformation, low GC content, slow growth, and expensive, complex media, have made surrogate genetics using E. coli an attractive and productive method to study spirochete gene function. Numerous B. burgdorferi genes have been heterologously expressed in E. coli mutants to complement the phenotype in order to define the gene products: grpE and dnaJ (Tilly et al. 1993), guaA (Margolis et al. 1994), lon (Cloud et al. 1997; Coleman et al. 2009), gac (Knight and Samuels 1999), oppA (Lin et al. 2001), pncA (Purser et al. 2003), recA (Liveris et al. 2004; Putteet-Driver et al. 2004), rpoS (Eggers et al. 2004), relBbu (Bugrysheva et al. 2005), and hfq (Lybecker et al. 2010). Conversely, the E. coli hfq gene has been heterologously expressed in a B. burgdorferi hfq mutant and partially complemented the phenotype, providing evidence that the spirochete has an Hfq-like RNA chaperone (Lybecker et al. 2010). Several endogenous B. burgdorferi promoters are capable of driving gene expression in E. coli. In fact, determining the consensus sequences for the sigma factors RpoS and RpoN was facilitated by studies in E. coli (Alverson et al. 2003; Burtnick et al. 2007; Caimano et al. 2004; Eggers et al. 2004, 2006). In addition, key experiments characterizing the promoter for the operon encoding the decorin-binding proteins (Ouyang et al. 2010) and describing the transcriptional regulator BosR (Boylan et al. 2003) were performed in E. coli.
Curiously, B. burgdorferi itself has served as a surrogate to heterologously express genes from less genetically tractable spirochetes, such as Treponema pallidum, the etiological agent of syphilis. T. pallidum lacks systems for both genetic manipulation and in vitro culture. The T. pallidum gene encoding the putative lipoprotein TP0435 was expressed in B. burgdorferi and shown to mediate adherence to host cells (Chan et al. 2016). tp0435 expression was driven by its endogenous promoter, demonstrating that a T. pallidum promoter functions in B. burgdorferi (Chan et al. 2016). Using a similar system, the T. pallidum gene tp0751, encoding pallilysin, was also expressed in B. burgdorferi to complement an adhesin-attenuated mutant and restore interactions with the mouse vasculature in vivo (Kao et al. 2017). The promise of these surrogate systems to study surface-exposed adhesins provides a significant step toward the goal of developing T. pallidum vaccine candidates. B. burgdorferi, which has its own experimental hurdles, can, surprisingly, serve as a valuable tool to facilitate studies in other genetic-less members of the spirochete phylum.
10 Conclusions
The post-genomic era has ushered in powerful genetic approaches that, combined with robust animal models, have elevated spirochetology from its relegated plight as a microbiological curiosity into the limelight. The disciplines of Borrelia microbiology and Lyme disease pathogenesis have come to depend on genetic manipulation, and the study of relapsing fever spirochetes is not too far behind. Mutagenesis in these recalcitrant bacteria is still not for the faint-hearted, but most molecular laboratories are proficient at reverse genetics, although ignoring several considerations and caveats remains perilous. Certain genetic tricks, notably markerless mutations, are still absent from the borreliologist’s quiver, but this just requires some attention and a bit of technical elbow grease. The state-of-the-art technologies for inducible gene expression in vivo and intravital microscopy are destined to reveal incredible insights into how these wily microbes interact with their hosts and are maintained in nature.
Acknowledgments
We thank Ben Adler for thoughtful reading of the manuscript and Darrin Akins, Jim Battisti, Melissa Caimano, Sherwood Casjens, George Chaconas, Christian Eggers, Frank Gherardini, Mike Gilbert, Laura Hall, Chris Li, Meghan Lybecker, Rich Marconi, Motaleb, Steve Norris, Justin Radolf, Sandy Raffel, Patti Rosa, Tom Schwan, Jon Skare, Phil Stewart, Kit Tilly, and Frank Yang for useful discussions about the genetic manipulation of Borrelia. Our laboratory is supported by National Institutes of Health grant AI051486 (to D.S.S.).
References
- Adams PP, Flores Avile C, Jewett MW. A dual luciferase reporter system for B. burgdorferi measures transcriptional activity during tick-pathogen interactions. Front Cell Infect Microbiol. 2017a;7:225. doi: 10.3389/fcimb.2017.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PP, Flores Avile C, Popitsch N, Bilusic I, Schroeder R, Lybecker M, Jewett MW. In vivo expression technology and 5′ end mapping of the Borrelia burgdorferi transcriptome identify novel RNAs expressed during mammalian infection. Nucleic Acids Res. 2017b;45:775–792. doi: 10.1093/nar/gkw1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson J, Samuels DS. groEL expression in gyrB mutants of Borrelia burgdorferi. J Bacteriol. 2002;184:6069–6072. doi: 10.1128/JB.183.21.6069-6072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson J, Bundle SF, Sohaskey CD, Lybecker MC, Samuels DS. Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi. Mol Microbiol. 2003;48:1665–1677. doi: 10.1046/j.1365-2958.2003.03537.x. [DOI] [PubMed] [Google Scholar]
- Arnold WK, Savage CR, Brissette CA, Seshu J, Livny J, Stevenson B. RNA-seq of Borrelia burgdorferi in multiple phases of growth reveals insights into the dynamics of gene expression, transcriptome architecture, and noncoding RNAs. PLoS ONE. 2016;11:e0164165. doi: 10.1371/journal.pone.0164165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb K, McAlister JD, Miller JC, Stevenson B. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J Bacteriol. 2004;186:2745–2756. doi: 10.1128/JB.186.9.2745-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandy NJ, Salman-Dilgimen A, Chaconas G. Construction and characterization of a Borrelia burgdorferi strain with conditional expression of the essential telomere resolvase, ResT. J Bacteriol. 2014;196:2396–2404. doi: 10.1128/JB.01435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Guo BP. Pathogenesis of relapsing fever. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular biology, host interaction and pathogenesis. Norfolk, UK: Caister Academic Press; 2010. pp. 333–357. [Google Scholar]
- Barthold SW, Cadavid D, Philipp MT. Animal models of borreliosis. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular biology, host interaction and pathogenesis. Norfolk, UK: Caister Academic Press; 2010. pp. 359–411. [Google Scholar]
- Battisti JM, Raffel SJ, Schwan TG. A system for site-specific genetic manipulation of the relapsing fever spirochete Borrelia hermsii. In: DeLeo FR, Otto M, editors. Bacterial pathogenesis: Methods and protocols. Totowa, New Jersey: Humana Press; 2008. pp. 69–84. [DOI] [PubMed] [Google Scholar]
- Beaurepaire C, Chaconas G. Mapping of essential replication functions of the linear plasmid lp17 of B. burgdorferi by targeted deletion walking. Mol Microbiol. 2005;57:132–142. doi: 10.1111/j.1365-2958.2005.04688.x. [DOI] [PubMed] [Google Scholar]
- Bestor A, Stewart PE, Jewett MW, Sarkar A, Tilly K, Rosa PA. Use of the Cre-lox recombination system to investigate the lp54 gene requirement in the infectious cycle of Borrelia burgdorferi. Infect Immun. 2010;78:2397–2407. doi: 10.1128/IAI.01059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins JS, Revel AT, Smith AH, Bachlani GN, Norgard MV. Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl Environ Microbiol. 2007;73:1501–1513. doi: 10.1128/AEM.02454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JL, Elias AF, Kupko JJ, III, Stevenson B, Tilly K, Rosa P. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botkin DJ, Abbott AN, Stewart PE, Rosa PA, Kawabata H, Watanabe H, Norris SJ. Identification of potential virulence determinants by Himar1 transposition of infectious Borrelia burgdorferi B31. Infect Immun. 2006;74:6690–6699. doi: 10.1128/IAI.00993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JA, Posey JE, Gherardini FC. Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc Natl Acad Sci USA. 2003;100:11684–11689. doi: 10.1073/pnas.2032956100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D, Drecktrah D, Eggers CH, Samuels DS. Genetics of Borrelia burgdorferi. Annu Rev Genet. 2012;46:515–536. doi: 10.1146/annurev-genet-011112-112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva JV, Bryksin AV, Godfrey HP, Cabello FC. Borrelia burgdorferi rel is responsible for generation of guanosine-3′-diphosphate-5′-triphosphate and growth control. Infect Immun. 2005;73:4972–4981. doi: 10.1128/IAI.73.8.4972-4981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, Hoover TR, Gherardini FC. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol Microbiol. 2007;65:277–293. doi: 10.1111/j.1365-2958.2007.05813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byram R, Stewart PE, Rosa P. The essential nature of the ubiquitous 26-kilobase circular replicon of Borrelia burgdorferi. J Bacteriol. 2004;186:3561–3569. doi: 10.1128/JB.186.11.3561-3569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello FC, Dubytska L, Bryksin AV, Bugrysheva JV, Godfrey HP. Genetic studies of the Borrelia burgdorferi bmp gene family. In: Cabello FC, Hulinska D, Godfrey HP, editors. Molecular Biology of Spirochetes. Amsterdam, Netherlands: IOS Press; 2006. pp. 235–249. [Google Scholar]
- Caimano MJ, Eggers CH, Hazlett KRO, Radolf JD. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect Immun. 2004;72:6433–6445. doi: 10.1128/IAI.72.11.6433-6445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Drecktrah D, Kung F, Samuels DS. Interaction of the Lyme disease spirochete with its tick vector. Cell Microbiol. 2016;18:919–927. doi: 10.1111/cmi.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JA, Stewart PE, Rosa P, Elias AF, Garon CF. An enhanced GFP reporter system to monitor gene expression in Borrelia burgdorferi. Microbiology. 2003;149:1819–1828. doi: 10.1099/mic.0.26165-0. [DOI] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- Chaconas G, Norris SJ. Peaceful coexistence amongst Borrelia plasmids: getting by with a little help from their friends? Plasmid. 2013;70:161–167. doi: 10.1016/j.plasmid.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G, Stewart PE, Tilly K, Bono JL, Rosa P. Telomere resolution in the Lyme disease spirochete. EMBO J. 2001;20:3229–3237. doi: 10.1093/emboj/20.12.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Alter L, Barthold SW, Parveen N. Disruption of bbe02 by insertion of a luciferase gene increases transformation efficiency of Borrelia burgdorferi and allows live imaging in Lyme disease susceptible C3H mice. PLoS ONE. 2015;10:e0129532. doi: 10.1371/journal.pone.0129532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Nasereddin T, Alter L, Centurion-Lara A, Giacani L, Parveen N. Treponema pallidum lipoprotein TP0435 expressed in Borrelia burgdorferi produces multiple surface/periplasmic isoforms and mediates adherence. Sci Rep. 2016;6:25593. doi: 10.1038/srep25593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet. 2002;36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol. 2012;66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Fischer JR, Benoit VM, Dufour NP, Youderian P, Leong JM. In vitro CpG methylation increases the transformation efficiency of Borrelia burgdorferi strains harboring the endogenous linear plasmid lp56. J Bacteriol. 2008;190:7885–7891. doi: 10.1128/JB.00324-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C-Y, Stewart PE, Bestor A, Hansen B, Lin T, Gao L, Norris SJ, Rosa PA. Function of the Borrelia burgdorferi FtsH homolog is essential for viability both in vitro and in vivo and independent of HflK/C. MBio. 2016;7:e00404–16. doi: 10.1128/mBio.00404-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud JL, Marconi RT, Eggers CH, Garon CF, Tilly K, Samuels DS. Cloning and expression of the Borrelia burgdorferi lon gene. Gene. 1997;194:137–141. doi: 10.1016/s0378-1119(97)00196-0. [DOI] [PubMed] [Google Scholar]
- Coleman JL, Katona LI, Kuhlow C, Toledo A, Okan NA, Tokarz R, Benach JL. Evidence that two ATP-dependent (Lon) proteases in Borrelia burgdorferi serve different functions. PLoS Pathog. 2009;5:e1000676. doi: 10.1371/journal.ppat.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutte L, Botkin DJ, Gao L, Norris SJ. Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection. PLoS Pathog. 2009;5:e1000293. doi: 10.1371/journal.ppat.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criswell D, Tobiason VL, Lodmell JS, Samuels DS. Mutations conferring aminoglycoside and spectinomycin resistance in Borrelia burgdorferi. Antimicrob Agents Chemother. 2006;50:445–452. doi: 10.1128/AAC.50.2.445-452.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crother TR, Champion CI, Whitelegge JP, Aguilera R, Wu X-Y, Blanco DR, Miller JN, Lovett MA. Temporal analysis of the antigenic composition of Borrelia burgdorferi during infection in rabbit skin. Infect Immun. 2004;72:5063–5072. doi: 10.1128/IAI.72.9.5063-5072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di L, Pagan PE, Packer D, Martin CL, Akther S, Ramrattan G, Mongodin EF, Fraser CM, Schutzer SE, Luft BJ, Casjens SR, Qiu W-G. BorreliaBase: a phylogeny-centered browser of Borrelia genomes. BMC Bioinf. 2014;15:233. doi: 10.1186/1471-2105-15-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Douglas JM, Samuels DS. Use of rpsL as a counterselectable marker in Borrelia burgdorferi. Appl Environ Microbiol. 2010;76:985–987. doi: 10.1128/AEM.02172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Hall LS, Hoon-Hanks LL, Samuels DS. An inverted repeat in the ospC operator is required for induction in Borrelia burgdorferi. PLoS ONE. 2013;8:e68799. doi: 10.1371/journal.pone.0068799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser AR, Hardy PO, Chaconas G. Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase. PLoS Pathog. 2009;5:e1000680. doi: 10.1371/journal.ppat.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Ems SM, Caimano MJ, Pal U, Wolgemuth CW, Eggers CH, Balic A, Radolf JD. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J Clin Invest. 2009;119:3652–3665. doi: 10.1172/JCI39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Ems SM, Caimano MJ, Eggers CH, Radolf JD. Borrelia burgdorferi requires the alternative sigma factor RpoS for dissemination within the vector during tick-to-mammal transmission. PLoS Pathog. 2012;8:e1002532. doi: 10.1371/journal.ppat.1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JP, Kenedy MR, Iqbal H, Akins DR. Characterization of the β-barrel assembly machine accessory lipoproteins from Borrelia burgdorferi. BMC Microbiol. 2015;15:70. doi: 10.1186/s12866-015-0411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnhart CG, LeBlanc DV, Alix KE, Desrosiers DC, Radolf JD, Marconi RT. Identification of residues within ligand-binding domain 1 (LBD1) of the Borrelia burgdorferi OspC protein required for function in the mammalian environment. Mol Microbiol. 2010;76:393–408. doi: 10.1111/j.1365-2958.2010.07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnhart CG, Rhodes DV, Marconi RT. Disulfide-mediated oligomer formation in Borrelia burgdorferi outer surface protein C, a critical virulence factor and potential Lyme disease vaccine candidate. Clin Vaccine Immunol. 2011;18:901–906. doi: 10.1128/CVI.05004-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Kimmel BJ, Bono JL, Elias A, Rosa P, Samuels DS. Transduction by ϕBB-1, a bacteriophage of Borrelia burgdorferi. J Bacteriol. 2001;183:4771–4778. doi: 10.1128/JB.183.16.4771-4778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, Radolf JD. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for expression of fluorescent reporters in the Lyme disease spirochaete. Mol Microbiol. 2002;43:281–296. doi: 10.1046/j.1365-2958.2002.02758.x. [DOI] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Radolf JD. Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J Bacteriol. 2004;186:7390–7402. doi: 10.1128/JB.186.21.7390-7402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Radolf JD. Sigma factor selectivity in Borrelia burgdorferi: RpoS recognition of the ospE/ospF/elp promoters is dependent on the sequence of the –10 region. Mol Microbiol. 2006;59:1859–1875. doi: 10.1111/j.1365-2958.2006.05066.x. [DOI] [PubMed] [Google Scholar]
- Eggers CH, Gray CM, Preisig AM, Glenn DM, Pereira J, Ayers RW, Alshahrani M, Acabbo C, Becker MR, Bruenn KN, Cheung T, Jendras TM, Shepley AB, Moeller JT. Phage-mediated horizontal gene transfer of both prophage and heterologous DNA by ϕBB-1, a bacteriophage of Borrelia burgdorferi. Pathog Dis. 2016;74:ftw107. doi: 10.1093/femspd/ftw107. [DOI] [PubMed] [Google Scholar]
- Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, Bono JL, Akins DR, Radolf JD, Schwan TG, Rosa P. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun. 2002;70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Bono JL, Kupko JJ, Stewart PE, 3rd, Krum JG, Rosa PA. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol. 2003;6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- Ellis TC, Jain S, Linowski AK, Rike K, Bestor A, Rosa PA, Halpern M, Kurhanewicz S, Jewett MW. Correction: In vivo expression technology identifies a novel virulence factor critical for Borrelia burgdorferi persistence in mice. PLoS Pathog. 2014;10:e1004260. doi: 10.1371/journal.ppat.1004260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkow S. Molecular Koch’s postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10(Suppl 2):S274–276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- Fine LM, Earnhart CG, Marconi RT. Genetic transformation of the relapsing fever spirochete Borrelia hermsii: stable integration and expression of green fluorescent protein from linear plasmid 200. J Bacteriol. 2011;193:3241–3245. doi: 10.1128/JB.05037-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerle V, Rauser S, Hammer B, Kahl O, Heimerl C, Schulte-Spechtel U, Gern L, Wilske B. Dynamics of dissemination and outer surface protein expression of different European Borrelia burgdorferi sensu lato strains in artificially infected Ixodes ricinus nymphs. J Clin Microbiol. 2002;40:1456–1463. doi: 10.1128/JCM.40.4.1456-1463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerle V, Goettner G, Gern L, Wilske B, Schulte-Spechtel U. Complementation of a Borrelia afzelii OspC mutant highlights the crucial role of OspC for dissemination of Borrelia afzelii in Ixodes ricinus. Int J Med Microbiol. 2007;297:97–107. doi: 10.1016/j.ijmm.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. aadA confers streptomycin-resistance in Borrelia burgdorferi. J Bacteriol. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb J-F, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quakenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MK, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Genomic sequence of a Lyme disease spirochete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Galbraith KM, Ng AC, Eggers BJ, Kuchel CR, Eggers CH, Samuels DS. parC mutations in fluoroquinolone-resistant Borrelia burgdorferi. Antimicrob Agents Chemother. 2005;49:4354–4357. doi: 10.1128/AAC.49.10.4354-4357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Old IG, Saint Girons I, Charon NW. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus σ70 promoter. J Bacteriol. 1997;179:2289–2299. doi: 10.1128/jb.179.7.2289-2299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MA, Morton EA, Bundle SF, Samuels DS. Artificial regulation of ospC expression in Borrelia burgdorferi. Mol Microbiol. 2007;63:1259–1273. doi: 10.1111/j.1365-2958.2007.05593.x. [DOI] [PubMed] [Google Scholar]
- Gilmore RD, Jr, Piesman J. Inhibition of Borrelia burgdorferi migration from the midgut to the salivary glands following feeding by ticks on OspC-immunized mice. Infect Immun. 2000;68:411–414. doi: 10.1128/iai.68.1.411-414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Eggers CH, Caimano MJ, Tilly K, Stewart PE, Elias AF, Radolf JD, Rosa PA. Experimental assessment of the roles of linear plasmids lp25 and lp28-1 of Borrelia burgdorferi throughout the infectious cycle. Infect Immun. 2004a;72:5938–5946. doi: 10.1128/IAI.72.10.5938-5946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, Schwan TG, Policastro PF, Elias AF, Rosa PA. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc Natl Acad Sci USA. 2004b;101:3142–3147. doi: 10.1073/pnas.0306845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Bueschel DM, Fisher MA, Policastro PF, Gherardini FC, Schwan TG, Rosa PA. Defining plasmids required by Borrelia burgdorferi for colonization of tick vector Ixodes scapularis (Acari: Ixodidae) J Med Entomol. 2005;42:676–684. doi: 10.1093/jmedent/42.4.676. [DOI] [PubMed] [Google Scholar]
- Groshong AM, Blevins JS. Insights into the biology of Borrelia burgdorferi gained through the application of molecular genetics. Adv Appl Microbiol. 2014;86:41–143. doi: 10.1016/B978-0-12-800262-9.00002-0. [DOI] [PubMed] [Google Scholar]
- Groshong AM, Gibbons NE, Yang XF, Blevins JS. Rrp2, a prokaryotic enhancer-like binding protein, is essential for viability of Borrelia burgdorferi. J Bacteriol. 2012;194:3336–3342. doi: 10.1128/JB.00253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BM, Jewett MW, Rosa PA. lacZ reporter system for use in Borrelia burgdorferi. Appl Environ Microbiol. 2010;76:7407–7412. doi: 10.1128/AEM.01389-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BM, Dulebohn DP, Sarkar A, Tilly K, Bestor A, Ambroggio X, Rosa PA. Regulatory protein BBD18 of the Lyme disease spirochete: essential role during tick acquisition? MBio. 2014;5:e01017–14. doi: 10.1128/mBio.01017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci USA. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Shaw DK, Smith R, III, Trzeciakowski JP, Skare JT. Characterization of a conditional bosR mutant in Borrelia burgdorferi. Infect Immun. 2010;78:265–274. doi: 10.1128/IAI.01018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Weening EH, Chang M, Trzeciakowski JP, Höök M, Cirillo JD, Skare JT. Bioluminescent imaging of Borrelia burgdorferi in vivo demonstrates that the fibronectin-binding protein BBK32 is required for optimal infectivity. Mol Microbiol. 2011;82:99–113. doi: 10.1111/j.1365-2958.2011.07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal H, Kenedy MR, Lybecker M, Akins DR. The TamB ortholog of Borrelia burgdorferi interacts with the β-barrel assembly machine (BAM) complex protein BamA. Mol Microbiol. 2016;102:757–774. doi: 10.1111/mmi.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- James AE, Rogovskyy AS, Crowley MA, Bankhead T. Characterization of a DNA adenine methyltransferase gene of Borrelia hermsii and its dispensability for murine infection and persistence. PLoS ONE. 2016;11:e0155798. doi: 10.1371/journal.pone.0155798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, VanRaden M, Gherardini F, Rosa PA. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol Microbiol. 2007;64:1358–1374. doi: 10.1111/j.1365-2958.2007.05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC. The spirochetes. Annu Rev Microbiol. 1977;31:89–106. doi: 10.1146/annurev.mi.31.100177.000513. [DOI] [PubMed] [Google Scholar]
- Jutras BL, Bowman A, Brissette CA, Adams CA, Verma A, Chenail AM, Stevenson B. EbfC (YbaB) is a new type of bacterial nucleoid-associated protein and a global regulator of gene expression in the Lyme disease spirochete. J Bacteriol. 2012a;194:3395–3406. doi: 10.1128/JB.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Verma A, Adams CA, Brissette CA, Burns LH, Whetstine CR, Bowman A, Chenail AM, Zückert WR, Stevenson B. BpaB and EbfC DNA-binding proteins regulate production of the Lyme disease spirochete’s infection-associated Erp surface proteins. J Bacteriol. 2012b;194:778–786. doi: 10.1128/JB.06394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao W-CA, Pětrošová H, Ebady R, Lithgow KV, Rojas P, Zhang Y, Kim Y-E, Kim Y-R, Odisho T, Gupta N, Moter A, Cameron CE, Moriarty TJ. Identification of Tp0751 (pallilysin) as a Treponema pallidum vascular adhesin by heterologous expression in the Lyme disease spirochete. Sci Rep. 2017;7:1538. doi: 10.1038/s41598-017-01589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Ehrenberg M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur J Biochem. 1994;226:355–360. doi: 10.1111/j.1432-1033.1994.tb20059.x. [DOI] [PubMed] [Google Scholar]
- Karimi R, Ehrenberg M. Dissociation rates of peptidyl-tRNA from the P-site of E. coli ribosomes. EMBO J. 1996;15:1149–1154. [PMC free article] [PubMed] [Google Scholar]
- Kasumba IN, Bestor A, Tilly K, Rosa PA. Use of an endogenous plasmid locus for stable in trans complementation in Borrelia burgdorferi. Appl Environ Microbiol. 2015;81:1038–1046. doi: 10.1128/AEM.03657-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata H, Norris SJ, Watanabe H. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect Immun. 2004;72:7147–7754. doi: 10.1128/IAI.72.12.7147-7154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajanchi BK, Odeh E, Gao L, Jacobs MB, Philipp MT, Lin T, Norris SJ. Phosphoenolpyruvate phosphotransferase system components modulate gene transcription and virulence of Borrelia burgdorferi. Infect Immun. 2016;84:754–764. doi: 10.1128/IAI.00917-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Samuels DS. Natural synthesis of a DNA-binding protein from the C-terminal domain of DNA gyrase A in Borrelia burgdorferi. EMBO J. 1999;18:4875–4881. doi: 10.1093/emboj/18.17.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Kimmel BJ, Eggers CH, Samuels DS. Disruption of the Borrelia burgdorferi gac gene, encoding the naturally synthesized GyrA C-terminal domain. J Bacteriol. 2000;182:2048–2051. doi: 10.1128/jb.182.7.2048-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnavajhala A, Wilder HK, Boyle WK, Damania A, Thornton JA, Pérez de León AA, Teel PD, Lopez JE. Imaging of Borrelia turicatae producing the green fluorescent protein reveals persistent colonization of the Ornithodoros turicata midgut and salivary glands from nymphal acquisition through transmission. Appl Environ Microbiol. 2017;83:e02503–16. doi: 10.1128/AEM.02503-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumru OS, Schulze RJ, Slusser JG, Zückert WR. Development and validation of a FACS-based lipoprotein localization screen in the Lyme disease spirochete Borrelia burgdorferi. BMC Microbiol. 2010;10:277. doi: 10.1186/1471-2180-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumru OS, Bunikis I, Sorokina I, Bergström S, Zückert WR. Specificity and role of the Borrelia burgdorferi CtpA protease in outer membrane protein processing. J Bacteriol. 2011a;193:5759–5765. doi: 10.1128/JB.05622-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumru OS, Schulze RJ, Rodnin MV, Ladokhin AS, Zückert WR. Surface localization determinants of Borrelia OspC/Vsp family lipoproteins. J Bacteriol. 2011b;193:2814–2825. doi: 10.1128/JB.00015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtti TJ, Munderloh UG, Johnson RC, Ahlstrand GG. Colony formation and morphology in Borrelia burgdorferi. J Clin Microbiol. 1987;25:2054–2058. doi: 10.1128/jcm.25.11.2054-2058.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M, Skare JT. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun. 2001;69:446–455. doi: 10.1128/IAI.69.1.446-455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenz MB, Kawabata H, Purser JE, Norris SJ. Decreased electroporation efficiency in Borrelia burgdorferi containing linear plasmids lp25 and lp56: impact on transformation of infectious B. burgdorferi. Infect Immun. 2002;70:4798–4804. doi: 10.1128/IAI.70.9.4798-4804.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart TR, Akins DR. Borrelia burgdorferi locus BB0795 encodes a BamA orthologue required for growth and efficient localization of outer membrane proteins. Mol Microbiol. 2010;75:692–709. doi: 10.1111/j.1365-2958.2009.07015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart TR, Kenedy MR, Yang X, Pal U, Akins DR. BB0324 and BB0028 are constituents of the Borrelia burgdorferi β-barrel assembly machine (BAM) complex. BMC Microbiol. 2012;12:60. doi: 10.1186/1471-2180-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuba-Garcia S, Martinez R, Gern L. Expression of outer surface proteins A and C of Borrelia afzelii in Ixodes ricinus ticks and in the skin of mice. Zentralbl Bakteriol. 1998;287:475–484. doi: 10.1016/s0934-8840(98)80187-4. [DOI] [PubMed] [Google Scholar]
- Liang FT, Jacobs MB, Bowers LC, Philipp MT. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J Exp Med. 2002a;195:415–422. doi: 10.1084/jem.20011870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FT, Nelson FK, Fikrig E. Molecular adaptation of Borrelia burgdorferi in the murine host. J Exp Med. 2002b;196:275–280. doi: 10.1084/jem.20020770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FT, Yan J, Mbow ML, Sviat SL, Gilmore RD, Mamula M, Fikrig E. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect Immun. 2004;72:5759–5767. doi: 10.1128/IAI.72.10.5759-5767.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FT, Xu Q, Sikdar R, Xiao Y, Cox JS, Doerrler WT. BB0250 of Borrelia burgdorferi is a conserved and essential inner membrane protein required for cell division. J Bacteriol. 2010;192:6105–6115. doi: 10.1128/JB.00571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Short SA, Eskildsen M, Klempner MS, Hu LT. Functional testing of putative oligopeptide permease (Opp) proteins of Borrelia burgdorferi: a complementation model in opp− Escherichia coli. Biochim Biophys Acta. 2001;1499:222–231. doi: 10.1016/s0167-4889(00)00121-x. [DOI] [PubMed] [Google Scholar]
- Lin T, Gao L, Edmondson DG, Jacobs MB, Philipp MT, Norris SJ. Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi. PLoS Pathog. 2009;5:e1000679. doi: 10.1371/journal.ppat.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, Chaconas G, Philipp MT, Norris SJ. Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PLoS ONE. 2012;7:e47532. doi: 10.1371/journal.pone.0047532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Troy EB, Hu LT, Gao L, Norris SJ. Transposon mutagenesis as an approach to improved understanding of Borrelia pathogenesis and biology. Front Cell Infect Microbiol. 2014;4:63. doi: 10.3389/fcimb.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Gao L, Zhao X, Liu J, Norris SJ. Mutations in the Borrelia burgdorferi flagellar type III secretion system genes fliH and fliI profoundly affect spirochete flagellar assembly, morphology, motility, structure, and cell division. MBio. 2015;6:e00579–15. doi: 10.1128/mBio.00579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liveris D, Mulay V, Schwartz I. Functional properties of Borrelia burgdorferi recA. J Bacteriol. 2004;186:2275–2280. doi: 10.1128/JB.186.8.2275-2280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, Abel CA, Feig AL, Samuels DS. Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2010;78:622–635. doi: 10.1111/j.1365-2958.2010.07374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, Samuels DS. Small RNAs of Borrelia burgdorferi: characterizing functional regulators in a sea of sRNAs. Yale J Biol Med. 2017;90:317–323. [PMC free article] [PubMed] [Google Scholar]
- Margolis N, Hogan D, Tilly K, Rosa PA. Plasmid location of Borrelia purine biosynthesis gene homologs. J Bacteriol. 1994;176:6427–6432. doi: 10.1128/jb.176.21.6427-6432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RR, Malawista SE, Feen KJ, Bockenstedt LK. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J Exp Med. 1996;183:261–269. doi: 10.1084/jem.183.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty TJ, Norman MU, Colarusso P, Bankhead T, Kubes P, Chaconas G. Real-time high resolution 3D imaging of the Lyme disease spirochete adhering to and escaping from the vasculature of a living host. PLoS Pathog. 2008;4:e1000090. doi: 10.1371/journal.ppat.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova OV, Dubytska LP, Ivanova LB, Moreno CX, Bryksin AV, Sartakova ML, Dobrikova EY, Godfrey HP, Cabello FC. Genetic and physiological characterization of 23S rRNA and ftsJ mutants of Borrelia burgdorferi isolated by mariner transposition. Gene. 2005;357:63–72. doi: 10.1016/j.gene.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Nickoloff JA. Electroporation protocols for microorganisms. In: Walker JM, editor. Methods in Molecular Biology. Totowa, New Jersey: Humana Press; 1995. p. 372. [Google Scholar]
- Nordstrand A, Barbour AG, Bergström S. Borrelia pathogenesis research in the post-genomic and post-vaccine era. Curr Opin Microbiol. 2000;3:86–92. doi: 10.1016/s1369-5274(99)00056-9. [DOI] [PubMed] [Google Scholar]
- Oehler S, Amouyal M, Kolkhof P, von Wilcken-Bergmann B, Müller-Hill B. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 1994;13:3348–3355. doi: 10.1002/j.1460-2075.1994.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi J, Piesman J, de Silva AM. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc Natl Acad Sci USA. 2001;98:670–675. doi: 10.1073/pnas.98.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Haq S, Norgard MV. Analysis of the dbpBA upstream regulatory region controlled by RpoS in Borrelia burgdorferi. J Bacteriol. 2010;192:1965–1974. doi: 10.1128/JB.01616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Zhou J, Norgard MV. Synthesis of RpoS is dependent on a putative enhancer binding protein Rrp2 in Borrelia burgdorferi. PLoS ONE. 2014a;9:e96917. doi: 10.1371/journal.pone.0096917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Zhou J, Norgard MV. CsrA (BB0184) is not involved in activation of the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. Infect Immun. 2014b;82:1511–1522. doi: 10.1128/IAI.01555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, Norgard MV, Fikrig E. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113:220–230. doi: 10.1172/JCI19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardeau M, Brenot A, Saint Girons I. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol Microbiol. 2001;40:189–199. doi: 10.1046/j.1365-2958.2001.02374.x. [DOI] [PubMed] [Google Scholar]
- Piesman J, Schwan TG. Ecology of borreliae and their arthropod vectors. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular biology, host interaction and pathogenesis. Norfolk, UK: Caister Academic Press; 2010. pp. 251–278. [Google Scholar]
- Popitsch N, Bilusic I, Rescheneder P, Schroeder R, Lybecker M. Temperature-dependent sRNA transcriptome of the Lyme disease spirochete. BMC Genom. 2017;18:28. doi: 10.1186/s12864-016-3398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser JE, Norris SJ. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2000;97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol. 2003;48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- Putteet-Driver AD, Zhong J, Barbour AG. Transgenic expression of RecA of the spirochetes Borrelia burgdorferi and Borrelia hermsii in Escherichia coli revealed differences in DNA repair and recombination phenotypes. J Bacteriol. 2004;186:2266–2274. doi: 10.1128/JB.186.8.2266-2274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffel SJ, Battisti JM, Fischer RJ, Schwan TG. Inactivation of genes for antigenic variation in the relapsing fever spirochete Borrelia hermsii reduces infectivity in mice and transmission by ticks. PLoS Pathog. 2014;10:e1004056. doi: 10.1371/journal.ppat.1004056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey ME, Hyde JA, Medina-Perez DN, Lin T, Gao L, Lundt ME, Li X, Norris SJ, Skare JT, Hu LT. A high-throughput genetic screen identifies previously uncharacterized Borrelia burgdorferi genes important for resistance against reactive oxygen and nitrogen species. PLoS Pathog. 2017;13:e1006225. doi: 10.1371/journal.ppat.1006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinavelu S, de Silva AM. Purification and characterization of Borrelia burgdorferi from feeding nymphal ticks (Ixodes scapularis) Infect Immun. 2001;69:3536–3541. doi: 10.1128/IAI.69.6.3536-3541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rego ROM, Bestor A, Rosa PA. Defining the plasmid-borne restriction-modification systems of the Lyme disease spirochete Borrelia burgdorferi. J Bacteriol. 2011;193:1161–1171. doi: 10.1128/JB.01176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel AT, Blevins JS, Almazán C, Neil L, Kocan KM, de la Fuente J, Hagman KE, Norgard MV. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc Natl Acad Sci USA. 2005;102:6972–6977. doi: 10.1073/pnas.0502565102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyrat J-M, Pelicic V, Gicquel B, Rappuoli R. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect Immun. 1998;66:4011–4017. doi: 10.1128/iai.66.9.4011-4017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa PA, Hogan DM. Colony formation by Borrelia burgdorferi in solid medium: clonal analysis of osp locus variants. In: Munderloh UG, Kurtti TJ, editors. First international conference on tick-borne pathogens at the host-vector interface: an agenda for research. St. Paul University of Minnesota; 1992. pp. 95–103. [Google Scholar]
- Rosa PA, Tilly K, Stewart PE. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat Rev Microbiol. 2005;3:129–143. doi: 10.1038/nrmicro1086. [DOI] [PubMed] [Google Scholar]
- Samuels DS. Electrotransformation of the spirochete Borrelia burgdorferi. In: Nickoloff JA, editor. Electroporation protocols for microorganisms. Totowa, New Jersey: Humana Press; 1995. pp. 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS. Antibiotic resistance in Borrelia burgdorferi: applications for genetic manipulation and implications for evolution. In: Cabello FC, Hulinska D, Godfrey HP, editors. Molecular biology of spirochetes. Amsterdam, Netherlands: IOS Press; 2006. pp. 56–70. [Google Scholar]
- Samuels DS. Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol. 2011;65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- Samuels DS, Garon CF. Coumermycin A1 inhibits growth and induces relaxation of supercoiled plasmids in Borrelia burgdorferi, the Lyme disease agent. Antimicrob Agents Chemother. 1993;37:46–50. doi: 10.1128/aac.37.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS, Garon CF. Oligonucleotide-mediated genetic transformation of Borrelia burgdorferi. Microbiology. 1997;143:519–522. doi: 10.1099/00221287-143-2-519. [DOI] [PubMed] [Google Scholar]
- Samuels DS, Mach KE, Garon CF. Genetic transformation of the Lyme disease agent Borrelia burgdorferi with coumarin-resistant gyrB. J Bacteriol. 1994a;176:6045–6049. doi: 10.1128/jb.176.19.6045-6049.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS, Radolf JD. Who is the BosR around here anyway? Mol Microbiol. 2009;74:1295–1299. doi: 10.1111/j.1365-2958.2009.06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS, Marconi RT, Huang WM, Garon CF. gyrB mutations in coumermycin A1-resistant Borrelia burgdorferi. J Bacteriol. 1994b;176:3072–3075. doi: 10.1128/jb.176.10.3072-3075.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Hayes BM, Dulebohn DP, Rosa PA. Regulation of the virulence determinant OspC by bbd18 on linear plasmid lp17 of Borrelia burgdorferi. J Bacteriol. 2011;193:5365–5373. doi: 10.1128/JB.01496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartakova M, Dobrikova E, Cabello FC. Development of an extrachromosomal cloning vector system for use in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2000;97:4850–4855. doi: 10.1073/pnas.080068797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartakova ML, Dobrikova EY, Terekhova DA, Devis R, Bugrysheva JV, Morozova OV, Godfrey HP, Cabello FC. Novel antibiotic-resistance markers in pGK12-derived vectors for Borrelia burgdorferi. Gene. 2003;303:131–137. doi: 10.1016/s0378-1119(02)01146-0. [DOI] [PubMed] [Google Scholar]
- Schulze RJ, Zückert WR. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol Microbiol. 2006;59:1473–1484. doi: 10.1111/j.1365-2958.2006.05039.x. [DOI] [PubMed] [Google Scholar]
- Schulze RJ, Chen S, Kumru OS, Zückert WR. Translocation of Borrelia burgdorferi surface lipoprotein OspA through the outer membrane requires an unfolded conformation and can initiate at the C-terminus. Mol Microbiol. 2010;76:1266–1278. doi: 10.1111/j.1365-2958.2010.07172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, Höök M, Skare JT. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol Microbiol. 2006;59:1591–1601. doi: 10.1111/j.1365-2958.2005.05042.x. [DOI] [PubMed] [Google Scholar]
- Shaw DK, Hyde JA, Skare JT. The BB0646 protein demonstrates lipase and haemolytic activity associated with Borrelia burgdorferi, the aetiological agent of Lyme disease. Mol Microbiol. 2012;83:319–334. doi: 10.1111/j.1365-2958.2011.07932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare JT, Shaw DK, Trzeciakowski JP, Hyde JA. In vivo imaging demonstrates that Borrelia burgdorferi ospC is uniquely expressed temporally and spatially throughout experimental infection. PLoS ONE. 2016;11:e0162501. doi: 10.1371/journal.pone.0162501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey CD, Barbour AG. Esterases in serum-containing growth media counteract chloramphenicol acetyltransferase activity in vitro. Antimicrob Agents Chemother. 1999;43:655–660. doi: 10.1128/aac.43.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey CD, Arnold C, Barbour AG. Analysis of promoters in Borrelia burgdorferi by use of a transiently expressed reporter gene. J Bacteriol. 1997;179:6837–6842. doi: 10.1128/jb.179.21.6837-6842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey CD, Zückert WR, Barbour AG. The extended promoters for two outer membrane lipoprotein genes of Borrelia spp. uniquely include a T-rich region. Mol Microbiol. 1999;33:41–51. doi: 10.1046/j.1365-2958.1999.01443.x. [DOI] [PubMed] [Google Scholar]
- Srivastava SY, de Silva AM. Reciprocal expression of ospA and ospC in single cells of Borrelia burgdorferi. J Bacteriol. 2008;190:3429–3433. doi: 10.1128/JB.00085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Bono JL, Elias A, Tilly K, Rosa P. Transformation of the Lyme disease spirochete Borrelia burgdorferi with heterologous DNA. J Bacteriol. 1998;180:4850–4855. doi: 10.1128/jb.180.18.4850-4855.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Rosa PA. Transposon mutagenesis of the Lyme disease agent Borrelia burgdorferi. In: DeLeo FR, Otto M, editors. Bacterial pathogenesis: methods and protocols. Totowa, New Jersey: Humana Press; 2008. pp. 85–95. [DOI] [PubMed] [Google Scholar]
- Stewart PE, Chaconas G, Rosa P. Conservation of plasmid maintenance functions between linear and circular plasmids in Borrelia burgdorferi. J Bacteriol. 2003;185:3202–3209. doi: 10.1128/JB.185.10.3202-3209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P, Thalken R, Bono J, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol. 2001;39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- Stewart PE, Wang X, Bueschel DM, Clifton DR, Grimm D, Tilly K, Carroll JA, Weis JJ, Rosa PA. Delineating the requirement for the Borrelia burgdorferi virulence factor OspC in the mammalian host. Infect Immun. 2006;74:3547–3553. doi: 10.1128/IAI.00158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Bestor A, Cullen JN, Rosa PA. A tightly regulated surface protein of Borrelia burgdorferi is not essential to the mouse-tick infectious cycle. Infect Immun. 2008;76:1970–1978. doi: 10.1128/IAI.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Hoff J, Fischer E, Krum JG, Rosa PA. Genome-wide transposon mutagenesis of Borrelia burgdorferi for identification of phenotypic mutants. Appl Environ Microbiol. 2004;70:5973–5979. doi: 10.1128/AEM.70.10.5973-5979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoenner HG, Dodd T, Larsen C. Antigenic variation of Borrelia hermsii. J Exp Med. 1982;156:1297–1311. doi: 10.1084/jem.156.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strother KO, de Silva A. Role of Borrelia burgdorferi linear plasmid 25 in infection of Ixodes scapularis ticks. J Bacteriol. 2005;187:5776–5781. doi: 10.1128/JB.187.16.5776-5781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terekhova D, Sartakova ML, Wormser GP, Schwartz I, Cabello FC. Erythromycin resistance in Borrelia burgdorferi. Antimicrob Agents Chemother. 2002;46:3637–3640. doi: 10.1128/AAC.46.11.3637-3640.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Hauser R, Campbell J, Ostheimer GJ. Isolation of dnaJ, dnaK, and grpE homologues from Borrelia burgdorferi and complementation of Escherichia coli mutants. Mol Microbiol. 1993;7:359–369. doi: 10.1111/j.1365-2958.1993.tb01128.x. [DOI] [PubMed] [Google Scholar]
- Tilly K, Elias AF, Bono JL, Stewart P, Rosa P. DNA exchange and insertional inactivation in spirochetes. J Mol Microbiol Biotechnol. 2000;2:433–442. [PubMed] [Google Scholar]
- Tilly K, Elias AF, Errett J, Fischer E, Iyer R, Schwartz I, Bono JL, Rosa P. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J Bacteriol. 2001;183:5544–5553. doi: 10.1128/JB.183.19.5544-5553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, Byram R, Dorward D, Vanraden MJ, Stewart P, Rosa P. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun. 2006;74:3554–3564. doi: 10.1128/IAI.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Checroun C, Rosa PA. Requirements for Borrelia burgdorferi plasmid maintenance. Plasmid. 2012;68:1–12. doi: 10.1016/j.plasmid.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Bestor A, Rosa PA. Lipoprotein succession in Borrelia burgdorferi: similar but distinct roles for OspC and VlsE at different stages of mammalian infection. Mol Microbiol. 2013;89:216–227. doi: 10.1111/mmi.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy EB, Lin T, Gao L, Lazinski DW, Camilli A, Norris SJ, Hu LT. Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing. Infect Immun. 2013;81:2347–2457. doi: 10.1128/IAI.00266-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy EB, Lin T, Gao L, Lazinski DW, Lundt M, Camilli A, Norris SJ, Hu LT. Global Tn-seq analysis of carbohydrate utilization and vertebrate infectivity of Borrelia burgdorferi. Mol Microbiol. 2016;101:1003–1023. doi: 10.1111/mmi.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine CR, Slusser JG, Zückert WR. Development of a single-plasmid-based regulatable gene expression system for Borrelia burgdorferi. Appl Environ Microbiol. 2009;75:6553–6558. doi: 10.1128/AEM.02825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Seemanapalli SV, Lomax L, McShan K, Li X, Fikrig E, Liang FT. Association of linear plasmid 28-1 with an arthritic phenotype of Borrelia burgdorferi. Infect Immun. 2005;73:7208–7215. doi: 10.1128/IAI.73.11.7208-7215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Seemanapalli SV, McShan K, Liang FT. Constitutive expression of outer surface protein C diminishes the ability of Borrelia burgdorferi to evade specific humoral immunity. Infect Immun. 2006;74:5177–5184. doi: 10.1128/IAI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, McShan K, Liang FT. Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi. Mol Microbiol. 2007;64:220–231. doi: 10.1111/j.1365-2958.2007.05636.x. [DOI] [PubMed] [Google Scholar]
- Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2003;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med. 2004;199:641–648. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Lybecker MC, Pal U, Alani SM, Blevins J, Revel AT, Samuels DS, Norgard MV. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J Bacteriol. 2005;187:4822–4829. doi: 10.1128/JB.187.14.4822-4829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Zhang J-J, Fang X, Lawlis GB, Troxell B, Zhou Y, Gomelsky M, Lou Y, Yang XF. DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect Immun. 2014;82:1840–1849. doi: 10.1128/IAI.00030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-R, Hardham JM, Barbour AG, Norris SJ. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- Zückert WR, Meyer J. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


