Abstract
Macrophages are key participants in melanoma growth and survival. In general, macrophages can be classified as M1 or M2 activation phenotypes. Increasing evidence demonstrates that melanoma exosomes also facilitate tumor survival and metastasis. However, the role of melanoma exosomes in directly influencing macrophage function is poorly understood. Herein, we investigated the hypothesis that natural melanoma exosomes might directly influence macrophage polarization. To explore this hypothesis, ELISA, RT-qPCR, and macrophage functional studies were performed in vitro using an established source of melanoma exosomes (B16-F10). ELISA results for melanoma exosome induction of common M1 and M2 cytokines in RAW264.7 macrophages, revealed that melanoma exosomes do not polarize macrophages exclusively in the M1 or M2 direction. Melanoma exosomes induced the M1 and M2 representative cytokines TNF-α and IL-10 respectively. Further assessment, using an RT-qPCR array with RAW264.7 and primary macrophages, confirmed and extended the ELISA findings. Upregulation of markers common to both M1 and M2 polarization phenotypes included CCL22, IL-12B, IL-1β, IL-6, i-NOS, and TNF-α. The M2 cytokine TGF-β was upregulated in primary but not RAW264.7 macrophages. Pro-tumor functions have been attributed to each of these markers. Macrophage functional assays demonstrated a trend toward increased i-NOS (M1) to arginase (M2) activity. Collectively, the results provide the first evidence that melanoma exosomes can induce a mixed M1 and M2 pro-tumor macrophage activation phenotype.
Keywords: Melanoma, Exosomes, Macrophage, Polarization, M1/M2, Cytokine, Chemokine
INTRODUCTION
Exosomes are cell-derived extracellular nanovesicles, approximately 100 nm in diameter, depending on their cell of origin [1]. Similar to soluble mediators, tumor exosomes can promote tumor supportive processes. For example, melanoma derived exosomes can mediate immune suppression [2]. They can directly interact with and suppress natural killer and cytotoxic CD8+ lymphocytes or induce myeloid derived suppressor cells (MDSCs). MDSCs can anergize anti-tumor CD8+ lymphocytes, promote M2 macrophage (Mφ) polarization and recruit pro-tumor regulatory T cells.
Melanoma exosomes further support melanoma growth via pro-angiogenic functions. They directly induce endothelial proliferation, endothelial spheroid growth and endothelial spheroid sprouts in a dose-dependent manner in vitro [3]. Some pro-angiogenic and immunomodulatory factors found in melanoma exosomes include interleukin 6 (IL-6), vascular endothelial growth factor A (VEGF-A), and matrix metalloprotease 2 (MMP2) [4]. Melanoma exosomes have also been reported to re-program bone marrow progenitor cells toward a pro-vascular phenotype [5].
Previously, we discovered that melanoma exosomes naturally home to the subcapsular sinus (SCS) of lymph nodes [6, 7] and prepare them for tumor metastasis [6]. Induction of pro-angiogenic and inflammatory genes such as hypoxia inducible factor 1 alpha (HIF-1α) and tumor necrosis factor alpha (TNF-α) by melanoma exosomes in lymph nodes was observed [6]. Increased expression of these genes is suggestive of Mφ participation in melanoma exosome mediated preparation of pre-metastatic niches.
Macrophages are key participants in tumor pathogenesis. They can be divided into two general classes (M1 and M2) based on function [8]. M1 polarized Mφs possess anti-tumor functions whereas M2 tumor associated Mφs (TAMs) promote tumor growth [9]. Further subdivisions exist within the general M2 Mφ category [8]. Definitive M2 subclassification remains a work in progress given the overlap between polarization markers. Using a typical scheme, M2 Mφs can be divided into M2a, M2b, M2c, and M2d/TAM subclasses [8, 10]. In general, IL-10 production is common for all M2 subclasses [8].
To date, there have been minimal investigations into the direct influence of melanoma exosomes on Mφ function. Determining whether melanoma exosomes stimulate Mφ dependent pro-tumor processes will further our basic understanding of melanoma pathogenesis. In this study, we hypothesized that natural melanoma exosomes might directly influence Mφ polarization.
MATERIALS AND METHODS
Cell culture
B16-F10 melanoma cells (CRL-6475) and RAW264.7 mouse Mφs (TIB-71) were obtained directly from the American Type Culture Collection (ATCC). Primary C57BL/6 mouse bone marrow Mφs (C57-6030F) were obtained from Cell Biologics Inc. B16-F10 melanoma cells were cultured at 37°C in 90% Dulbecco’s modified Eagle’s medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS) and 5% CO2. RAW264.7 mouse Mφs were cultured at 37°C, and maintained in 90% DMEM with 10% heat-inactivated FBS and 5% CO2. Primary Mφs were cultured at 37°C, and maintained in complete Mφ medium (M3368, Cell Biologics Inc.) and 5% CO2.
Exosome isolation
Isolation and characterization of highly purified populations of B16-F10 mouse melanoma exosomes from cell culture, by means of differential centrifugation, have previously been established [3]. Briefly, to isolate exosomes, cells were grown to 70% confluence in three 300 cm2 flasks. Culture media was removed, and the cells were washed with DMEM. Cells were then cultured for 48 hrs in bovine exosome-free conditioned media. Conditioned culture media was prepared by subjecting normal culture media to overnight ultracentrifugation at 110,000g to remove bovine exosomes [11]. Cell culture media was harvested after 48 hrs, diluted 1:1 in 50 mM trehalose (cryoprotectant) PBS [12], and processed using differential centrifugation. Supernatants were collected, and the pellets were discarded, following 3,400g for 30 min to remove residual cells and debris, and 10,000g for 30 min to remove microparticles. Finally, the exosome pellet was collected after 110,000g for 1.5 hrs, and then washed at 110,000g for 1.5 hrs. Exosome protein concentrations were measured using a Pierce BCA protein assay (ThermoFisher Scientific). Exosomes were stored in 50 mM trehalose PBS until use [12].
Exosome Characterization
B16-F10 exosome size (hydrodynamic diameter) was determined in 1X PBS, using dynamic light scattering with a NanoBrook 90Plus (Brookhaven Instruments), according to established methods [3, 6, 12]. B16-F10 exosome density was determined by sucrose gradient centrifugation of fluorescently labeled exosomes as described previously [3]. Briefly, flotation of fluorescent red carbocyanine DiI (1.0 µM, ThermoFisher Scientific) labeled exosomes (400 µg of exosome protein) on a continuous sucrose gradient (2.0 – 0.25M sucrose, 20mM HEPES/NaOH, pH 7.4) was performed using a Beckman Coulter SW 41 rotor [3, 12]. The gradient was produced using a Gradient Master (Biocomp Instruments, Fredericton, NB, Canada) and was spun, after loading exosomes, for >20 h at 100,000g. Post centrifugation, 1 ml fractions were collected from the bottom up. The density of each fraction was calculated using a refractometer [3]. Two hundred microliters of each fraction was added to a black 96-well plate and DiI exosome fluorescence detected using a Tecan M200 infinite pro microplate reader according to established methods [3, 12]. Detection of the exosomal marker CD63 on B16-F10 exosomes (10 µg of exosome protein) was determined using an ExoELISA-ULTRA CD63 Kit (EXEL-ULTRA-CD63-1, System Biosciences) according to the manufacturer’s instructions. A CD63 standard, supplied with the kit, enabled quantification of CD63+ exosomes per µg of exosome protein.
Cell Treatments
RAW264.7 or primary Mφs (2500 per well of a 96 well plate) were grown for 24 hrs in their respective bovine exosome-free conditioned medias. Post 24 hrs of cell culture, media was replaced with fresh conditioned media containing either lipopolysaccharide (LPS), IL-4, B16-F10 melanoma exosomes, or a combination of LPS + exosomes, or IL-4 + exosomes for an additional 24 hrs. For LPS or IL-4 dosing of Mφs, 200 ng/ml LPS (E. coli, Sigma-Aldrich, L6529) or 40 ng/ml IL-4 (Sigma-Aldrich, SRP-3211) was used. These concentrations are based on previously established methods to induce M1 or M2 polarization in RAW264.7 cells [13]. For exosome dosing, a concentration of 0.01 mg/ml exosome protein, measured via BCA absorbance (ThermoFisher Scientific Inc.) was used. All treatments were performed in bovine exosome-free conditioned media.
Enzyme-linked immunosorbent assay (ELISA)
Following RAW264.7 cell treatments with LPS, IL-4, exosomes or a combination, cell supernatants were collected and processed using affymetrix eBioscience ready-set-go ELISA kits according to manufacturers’ instructions to detect RAW264.7 cell expression of IL-10 (cat# 88-7105), TGF-β (cat# 88-8350), TNF-α (cat# 88-7324) and IL-1β (cat# 88-7013-86). Cytokine levels were normalized to cell growth using PrestoBlue® (ThermoFisher Scientific) cell viability reagent. The 2-tailed Student's t test was used to determine statistically significant, P values for α = 0.05, differences in cytokine expression between treatment groups.
Reverse Transcriptase Quantitative PCR (RT-qPCR)
Following RAW264.7, or primary Mφ treatments with exosomes, cell culture media was removed and cells were washed in PBS (Sigma-Aldrich, cat# D8537). RNA was isolated using Qiagen’s miRNAeasy kit (cat# 217004) according to the manufacturer’s instructions. RNA quantity and quality were assessed using a Tecan M200 infinite pro microplate reader. For each sample, 1 µg of cellular RNA was converted to cDNA using Qiagen’s RT2 First Strand kit (cat#33041). Following conversion to cDNA, each sample was applied to a Qiagen Mouse Cancer Inflammation and Immunity Crosstalk RT2 Profiler PCR Array (PAMM-181Z). Quantitative PCR was performed using a StepOnePlus™ Real-Time PCR system (Applied Biosystems™). Array results, normalization gene selection, and statistics were determined using Qiagen’s online PCR array data analysis portal (http://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/rt2-profiler-pcr-arrays-data-analysis-center). Using Qiagen’s data analysis portal, the beta actin gene was selected as the best normalization gene across RAW 264.7 Mφ arrays, and (C-X-C) motif chemokine receptor 7 (CXCR7), VEGFA, and toll-like receptor 4 (TLR-4) were automatically selected as the best normalization genes across primary Mφ arrays.
Inducible nitric oxide synthase (i-NOS) and Arginase activity assays
To determine M1 or M2 RAW264.7, or primary Mφ function, following treatment with B16-F10 melanoma exosomes, assays to assess increased inducible nitric oxide synthase activity, indicative of M1 Mφ polarization, and increased arginase activity, indicative of M2 Mφ polarization, were performed. To assess i-NOS activity, a Nitric Oxide Synthase Detection System (Fluorometric, FCANOS1-1KT, Sigma-Aldrich) was used according to manufacturer’s instructions. Briefly, 25,000 cells per well of a 96 well plate were incubated for 24 hrs at 37°C in 200 µl of bovine exosome-free conditioned media containing mouse GM-CSF (0.1 mg/ml, PeproTech, 315-03)[14]. Post 24 hrs, cell culture supernatants were aspirated and wells were treated for an additional 24 hrs at 37°C with either LPS (200 ng/ml), washed exosomes (0.01 mg/ml in 50 mM Trehalose-PBS) or in an equivalent amount of 50 mM trehalose-PBS (vehicle-control) in 200 µl of fresh bovine exosome-free conditioned media containing mouse GM-CSF (0.1 mg/ml, PeproTech, 315-03) [14]. Subsequently, i-NOS activity was determined.
For assessment of arginase activity, an Arginase Activity Assay Kit (MAK112-1KT, Sigma-Aldrich) assay was performed according to manufacturer’s instructions. Briefly, 2 million cells per T-300 tissue culture treated flask were incubated for 24 hrs at 37°C in 25 ml of bovine exosome-free conditioned media containing mouse GM-CSF (0.1 mg/ml, PeproTech, 315-03)[14]. Post 24 hrs, cell culture supernatants were aspirated and flasks were treated with either LPS (200 ng/ml), washed exosomes (0.01 mg/ml in 50mM trehalose-PBS) or an equivalent amount of 50 mM trehalose-PBS (vehicle-control) in 25 ml of fresh bovine exosome-free conditioned media containing mouse GM-CSF (0.1 mg/ml, PeproTech, 315-03)[14] for 24 hrs at 37°C. Following treatment, cells were lysed from the flasks using lysis buffer containing 10 mM Tris-HCl pH 7.4, 1 µM pepstatin A, 1 µM leupeptin and 0.4% triton X-100. Arginase activity of the cell lysates was determined using the kit components.
Cell viability experiments using PrestoBlue Cell Viability Reagent (ThermoFisher Scientific) were performed as well to determine any influence of LPS or B16-F10 melanoma exosomes on Mφ growth. The same i-NOS and arginase activity kit parameters were used. For ease of comparison, i-NOS and arginase activity were normalized to cell viability by dividing activity by viability, determined using PrestoBlue Cell Viability Reagent, and setting the non-treatment (NT) control groups to 100%. The 2-tailed Student's t test was used to determine statistically significant, P values for α = 0.05, differences in i-NOS or arginase activity between treatment groups.
RESULTS
In this investigation we evaluated the ability of unmodified B16-F10 melanoma exosomes to directly influence Mφ function. Given the complexity of exosome signaling, we elected to use the mouse RAW264.7 cell line as our Mφ model to enable a degree of standardization between experiments. The RAW264.7 cell line is commonly employed for assessing the preliminary efficacy of vaccines, Mφ function and polarity [15–23].
Melanoma exosome characterization
We previously established that B16-F10 melanoma cells produce a homogenous population of exosomes as defined empirically [24], based on size and density [3]. B16-F10 melanoma exosomes were further characterized in the present study. Melanoma exosome size (diameter) was determined by dynamic light scattering to be 79 +/− 19 nm (n =30). This is consistent with previous B16-F10 melanoma exosome size reports [3, 6, 12]. B16-F10 exosome size is also well within the < 200 nm size range reported for exosomes in general [25]. B16-F10 exosomes isolated by sucrose density gradient centrifugation, had a density between 1.12 – 1.22 g/ml. This overlaps the density range we previously reported [3], as well as density ranges reported for other exosome types [26], and by melanoma exosome studies [27, 28]. Additional analysis using an ExoELISA-ULTRA CD63 kit (System Biosciences), demonstrated that the B16-F10 melanoma exosomes expressed the exosomal tetraspanin marker CD63. This corresponded to ~ 2×109 +/− 4×108 CD63+ exosomes per µg of exosomal protein (n = 3).
Melanoma exosomes induce M1 and M2 macrophage cytokines detected by ELISA
In the first set of experiments we sought to determine whether melanoma exosomes induce M1 or M2 cytokines in Mφs using cytokine ELISAs. LPS and IL-4 were selected as positive controls. Stimulation of RAW264.7 Mφs with lipopolysaccharide (LPS) or IL-4 induces M1 or M2 Mφ polarity respectfully [13]. Commonly used cytokine markers, indicative of Mφ polarization, include TNF-α and IL-1β for M1 or IL-10 and transforming growth factor beta (TGF-β) for M2 [29].
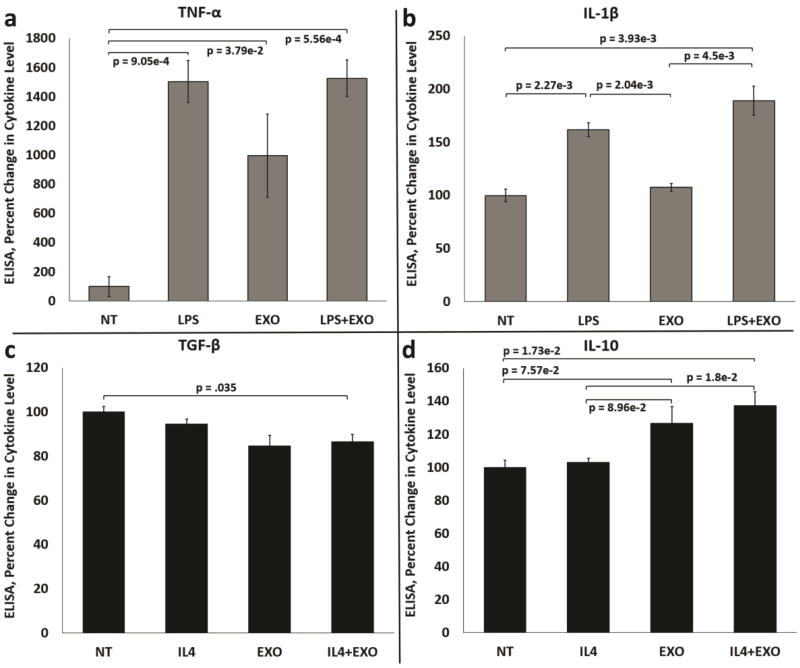
Treatment of Mφs with LPS or melanoma exosomes significantly increased the production of TNF-α (Fig. 1a). However, combination treatment using LPS + exosomes performed similarly to cells treated with LPS alone. Stimulation with LPS also significantly increased production of IL-1β (Fig. 1b). No significant influence on IL-1β production following exosome exposure alone was observed. However, treatment with combined LPS + exosomes trended toward more induction of IL-1β than LPS treatment alone.
Figure 1.
Melanoma exosomes stimulate production of mixed M1 and M2 cytokines by macrophages in cell culture. (a) Induction of the M1 cytokine TNF-α in control, LPS treated, exosome treated and LPS + exosome treated RAW264.7 Mφs. (b) Induction of the M1 cytokine IL-1β in control, LPS treated, exosome treated and LPS + exosome treated RAW264.7 Mφs. (c) Induction of the M2 cytokine TGF-β in control, IL-4 treated, exosome treated and IL-4 + exosome treated RAW264.7 Mφs. (d) Induction of the M2 cytokine IL-10 in control, IL-4 treated, exosome treated and IL-4 + exosome treated RAW264.7 Mφs. n = 3 independent experiments normalized to non-treated (NT) Mφs (percent change = 100%) using three pooled batches of melanoma exosomes. Error bars = S.E.M. Connecting bars denote a statistical comparison between two groups with p values listed above or below the connecting bars, p values < 0.05 were considered statistically significant.
Treatment of Mφs with IL-4 or melanoma exosomes resulted in no significant increase in TGF-β production by Mφs (Fig. 1c). Stimulation with IL-4 + exosomes resulted in decreased TGF-β. Treatment of Mφs with IL-4 resulted in no significant increase in IL-10 (Fig. 1d). However, exposure to melanoma exosomes significantly increased IL-10 production (Fig. 1d). Combination treatment with IL-4 + exosomes resulted in a significant increase in IL-10 produced versus treatment with IL-4 alone (Fig. 1d).
Melanoma exosomes upregulate M1 and M2 macrophage polarization markers identified on RT-qPCR array
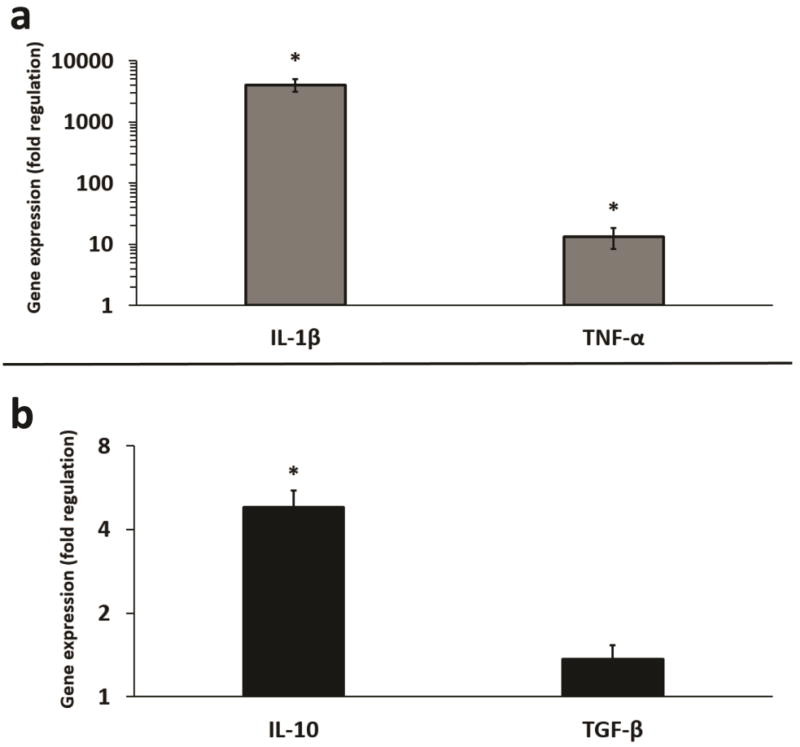
Given the minimal response of the RAW 264.7 Mφs to IL-4, as evidenced by cytokine ELISA, we sought to determine whether RT-qPCR might be more sensitive to detecting changes in cytokine induction. As shown, upregulation of the M1 cytokines IL-1β and TNF-α at the mRNA level, following treatment with LPS (Fig. 2a), was consistent with the ELISA results. Although, the extent of induction between the cytokine protein and mRNA levels differed. Significantly increased production of the M2 cytokine IL-10 at the mRNA level, following treatment with IL-4 (Fig. 2b), was more dramatic than the ELISA results. A modest, though insignificant increase in TGF-β expression, was observed at the mRNA level following stimulation with IL-4 as compared to the subtle decrease observed using ELISA.
Figure 2.
Fold regulation (RT-qPCR) in LPS and IL-4 induced gene expression of common M1 and M2 cytokines in macrophages. (a) LPS induces increased gene expression of IL-1β and TNF-α in RAW264.7 Mφs. (b) IL-4 induces increased gene expression of IL-10 and TGF-β in RAW264.7 Mφs. n = 3 replicate arrays, using three pooled independent batches of melanoma exosomes, and normalized to non-treated Mφs (fold change = 1). Error bars = S.E.M. * = p values < 0.05, and were considered statistically significant compared to non-treated Mφs.
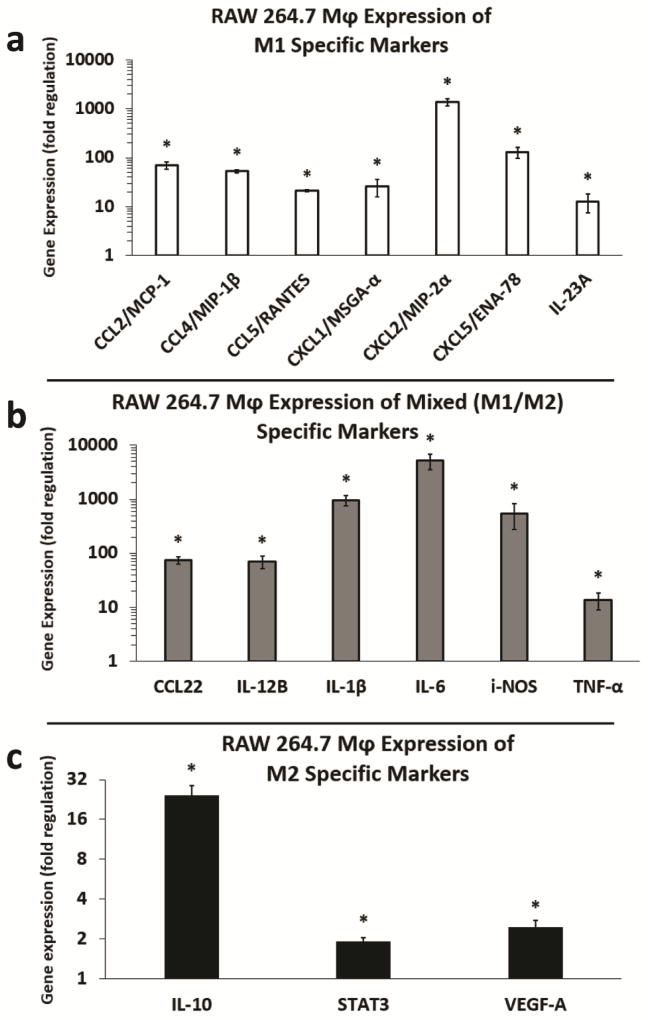
Based on the combined ELISA and RT-qPCR results assessing IL-10 and TGF-β induction by IL-4, RT-qPCR appeared to be more sensitive to detecting changes in cytokine expression in our system. Subsequently, using a cancer inflammation and immunity crosstalk RT-qPCR array, we assessed melanoma exosome mediated induction of additional M1 and M2 markers of RAW 264.7 Mφ polarization [8, 30]. Upregulated markers were divided into M1 specific, dual M1/M2 specific, or M2 polarization groups. Of the 84 inflammatory genes assessed on the array, 16 Mφ polarization markers were significantly upregulated with fold changes of approximately two or higher. As shown, melanoma exosomes induced the M1 markers (C-C motif) chemokine ligand 2 (CCL2), CCL4, CCL5, chemokine (C-X-C motif) ligand 1 (CXCL1), CXCL2, CXCL5, and IL-23A (Fig. 3a). Upregulation of markers common to both M1 and M2 polarization included CCL22, IL-12B, IL-1β, IL-6, i-NOS, and TNF-α (Fig. 3b). Melanoma exosome induction of M2 markers included IL-10, signal transducer and activator of transcription 3 (STAT3), and VEGF-A (Fig. 3c).
Figure 3.
Fold regulation (RT-qPCR) in M1 and M2 cytokine gene expression induced by melanoma exosomes in RAW264.7 macrophages. (a) Induction of M1 markers in RAW264.7 Mφs by melanoma exosomes. (b) Induction of mixed (M1/M2) markers in RAW264.7 Mφs by melanoma exosomes. (c) Induction of M2 markers in RAW264.7 Mφs by melanoma exosomes. n = 3 replicate arrays, using three pooled independent batches of melanoma exosomes, and normalized to non-treated Mφs (fold change = 1). Error bars = S.E.M. * = p values < 0.05, and were considered statistically significant compared to non-treated RAW264.7 Mφs.
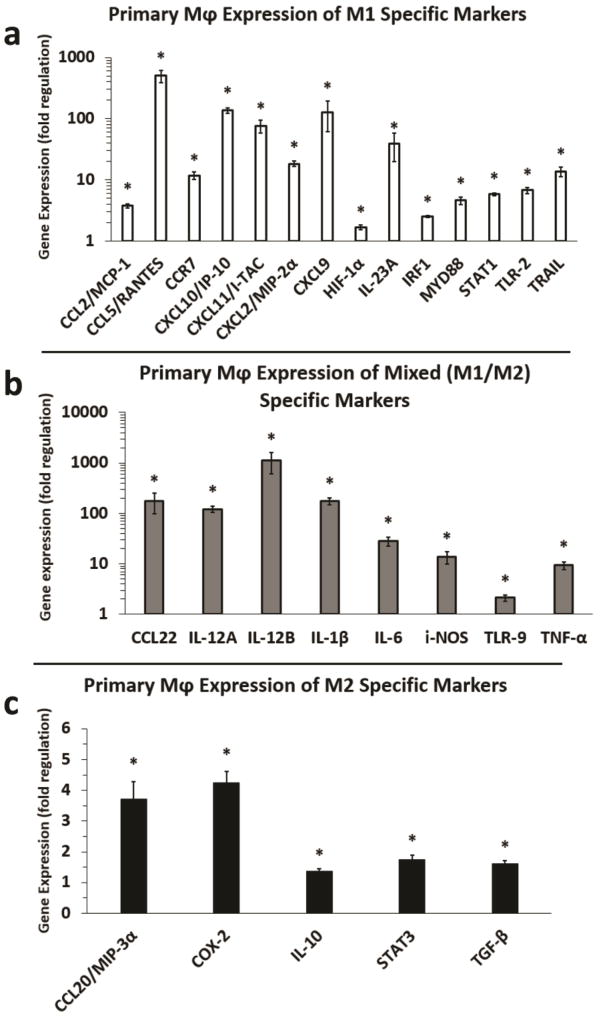
To confirm and extend the RAW 264.7 Mφ gene expression results, we assessed melanoma exosome mediated induction of M1 and M2 polarization markers in primary Mφs (Fig. 4). The results largely validated the RAW264.7 Mφ findings, but there were also a number of differences. As shown, melanoma exosomes induced fourteen M1 markers in primary Mφ compared to seven in RAW 264.7 Mφs. M1 markers included: CCL2, CCL5, (C-C motif) chemokine receptor type 7 (CCR7), CXCL2, CXCL9, CXCL10, CXCL11, IL-23A, HIF-1α, interferon regulatory factor 1 (IRF1), myeloid differentiation primary response 88 (MYD88), STAT1, toll-like receptor 2 (TLR-2), and TNF-related apoptosis-inducing ligand (TRAIL) (Fig. 4a). Upregulation of eight versus six markers common to both M1 and M2 polarization was observed for primary Mφs versus RAW 264.7 Mφs. These included CCL22, IL-12A, IL-12B, ILMelanoma 1β, IL-6, i-NOS, TLR-9, and TNF-α (Fig. 4b). Melanoma exosomes also induced five versus three M2 markers in primary Mφs compared to RAW 264.7 Mφs. This included CCL20, cyclooxygenase-2 (COX-2), IL-10, STAT3, and notably TGF-β (Fig. 4c), a key immunosuppressive cytokine, not upregulated in RAW 264.7 Mφs by melanoma exosomes. However, induction of the immunosuppressive cytokine IL-10, by melanoma exosomes in primary Mφs (Fig. 4c), was much less pronounced than in RAW 264.7 Mφs (Fig. 3c). Collectively, 7 M1, 6 mixed M1/2, and 3 M2 markers were upregulated in RAW 264.7 Mφs (Fig. 3). In contrast, 14 M1, 8 mixed M1/2, and 5 M2 markers were upregulated in primary Mφs (Fig. 4). Moreover, the majority of M1 versus M2 markers exhibited much greater increases in gene expression.
Figure 4.
Fold regulation (RT-qPCR) in M1 and M2 cytokine gene expression induced by melanoma exosomes in primary macrophages. (a) Induction of M1 markers in primary Mφs by melanoma exosomes. (b) Induction of mixed (M1/M2) markers in primary Mφs. (c) Induction of M2 markers in primary Mφs by melanoma exosomes. n = 3 replicate arrays, using three pooled independent batches of melanoma exosomes, and normalized to non-treated Mφs (fold change = 1). Error bars = S.E.M. * = p values < 0.05, and were considered statistically significant compared to non-treated primary Mφs.
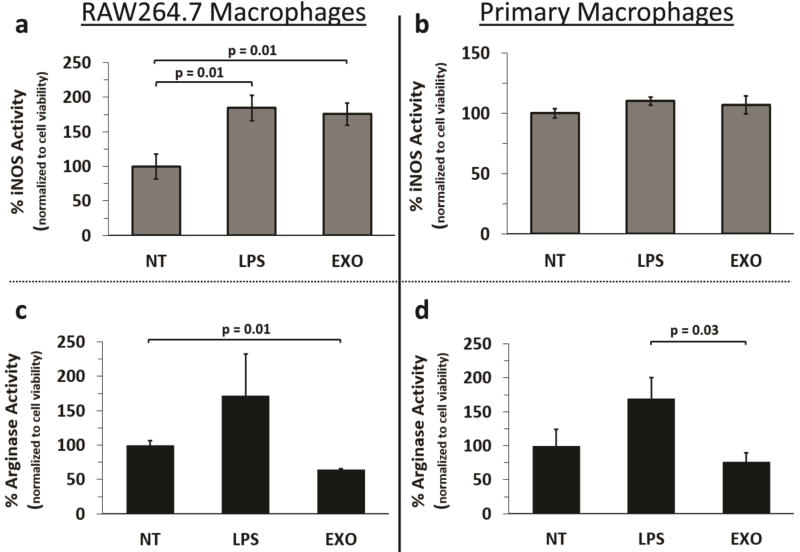
Macrophages treated with melanoma exosomes exhibit an increased ratio of i-NOS to arginase activity
Categorizing M1 and M2 Mφs using phenotypic surface markers is difficult given the potential for intermediates [29]. Alternatively, assessment of functional markers aids in the identification of M1 and M2 phenotypes [29]. These include upregulation of inducible nitrogen oxide synthase (i-NOS) for M1 and arginase 1 (Arg-1) for M2 respectively. Given the detected increased expression of i-NOS mRNA, common to both M1 and M2b Mφs [30], we assessed whether melanoma exosome induced Mφs exhibited M1 or M2 functions. As shown, treatment of RAW 264.7 Mφs with either melanoma exosomes or LPS (positive control) resulted in increased i-NOS activity (Fig. 5a). A similar, though not statistically significant, trend in increased i-NOS activity was observed for primary Mφs (Fig. 5b). No significant increase in arginase activity was observed following LPS treatment of Mφs (Fig. 5c, d). However, melanoma exosomes did produce a small decrease in RAW 264.7 Mφ arginase activity (Fig. 5c). A similar, though not statistically significant, trend in decreased arginase activity was observed for primary Mφs treated with melanoma exosomes (Fig. 5d).
Figure 5.
Assessment of macrophage M1 and M2 functional status following treatment with melanoma exosomes. (a) i-NOS activity detected for non-treated, LPS, or melanoma exosome treated RAW 264.7 Mφs (n = 5), (b) i-NOS activity detected for non-treated, LPS, or melanoma exosome treated primary Mφs (n = 9), (c) Arginase activity detected for non-treated, LPS, or melanoma exosome treated RAW264.7 Mφs (n = 3), (d) Arginase activity detected for non-treated, LPS, or melanoma exosome treated primary Mφs (n = 6). Error bars = S.E.M. Connecting bars denote a statistical comparison between two groups with p values listed above the connecting bars, p values < 0.05 were considered statistically significant.
DISCUSSION
M1 macrophage polarization factors upregulated by melanoma exosomes
Herein, we hypothesized that melanoma exosomes could directly stimulate Mφ polarization. Collectively our findings demonstrate that melanoma exosomes induce a mixed, ostensibly more M1 than M2 phenotype, characterized in part by increased expression of related signaling factors and cytokines including MyD88, TRAIL, IL-1β, IL-6 and TNF-α in primary Mφ (Table 2). Signaling through the MyD88 adaptor protein results in nuclear factor-κB (NF-κB) activation [31]. TRAIL can also activate NF-κB in TAMs. This results in increased expression of pro-inflammatory IL-1β, IL-6, TNF-α, and M1 polarization of TAMs [32]. Other signaling factors including IRF1 [33], and TLR-2 [34], upregulated in primary Mφs by melanoma exosomes, have recently been found to be associated with M1 polarization. However, while M1 polarization is typically associated with an anti-tumor response, many of the M1 polarization factors upregulated by melanoma exosomes in Mφs, including IL-6 and TNF-α, further discussed below, also possess melanoma supportive functions.
Table 2.
Melanoma exosomes induce mixed M1/M2 primary macrophage polarization. Representative markers of Mφ polarization, induced by B16-F10 melanoma exosomes, are sorted into their respective subclasses. Markers in bold are common to two or more Mφ subclasses.
| Primary Macrophage Polarization: |
M1 | M2a | M2b | M2c | M2d or TAM |
|---|---|---|---|---|---|
| Chemokines | CCL2 CCL5 CCL22 CCR7 CXCL2 CXCL9 CXCL10 CXCL11 | CCL22 | CCL20 | ||
| Cytokines | IL-1β IL-6 IL-12A IL-12B IL-23A TNF-α | IL-10 TGF-β | IL-10 IL-1β IL-6 TNF-α | IL-10 TGF-β | IL-10 IL-12A IL-12B TGF-β TNF-α |
| Signaling and Growth Factors | HIF-1α IRF1 MYD88 STAT1 TLR-2 TLR-9 TRAIL | STAT3 | COX-2 | COX-2 TLR-9 | |
| Metabolism | i-NOS | i-NOS |
Analysis of pcr array results revealed increased expression of a number of M1 chemokines by both RAW264.7 (Table 1) and primary Mφs (Table 2). Upregulated M1 chemokines include Mφ chemoattractant protein 1 (MCP-1)/CCL2, regulated on activation normal T cell expressed and secreted (RANTES)/CCL5, and Mφ inflammatory protein 2 alpha (MIP-2α)/CXCL2. Induction of the M1 chemokines MCP-1/CCL2 and MIP-2α/CXCL2 by exosomes derived from B16-F10 melanoma is consistent with a previous tumor exosome study. Marton et al. demonstrated that CCL2 and CXCL2 are also induced in RAW264.7 Mφs treated with exosomes derived from the B16-F1 melanoma variant [35].
Table 1.
Melanoma exosomes induce mixed M1/M2 macrophage polarization in RAW264.7 cells. Representative markers of Mφ polarization, induced by B16-F10 melanoma exosomes, are sorted into their respective subclasses. Markers in bold are common to two or more Mφ subclasses.
| RAW 264.7 Macrophage Polarization: |
M1 | M2a | M2b | M2c | M2d or TAM |
|---|---|---|---|---|---|
| Chemokines | CCL2 CCL4 CCL5 CCL22 CXCL1 CXCL2 CXCL5 | CCL22 | |||
| Cytokines | IL-1β IL-6 IL-12B IL-23A TNF-α | IL-10 | IL-10 IL-1β IL-6 TNF-α | IL-10 | IL-10 IL-12B TNF-α |
| Signaling and Growth Factors | STAT3 | VEGF-A | |||
| Metabolism | i-NOS | i-NOS |
M1 chemokines induced by melanoma exosomes are involved in the recruitment and activation of immune cell subsets. They also actively participate in immunological processes enhancing melanoma survival. MCP-1/CCL2 is particularly relevant to melanoma growth and metastasis. Within melanoma tumor microenvironments, MCP-1 produced by melanoma cells exhibits biphasic functionality [36]. Lower concentrations of MCP-1 can recruit a modest number of monocytes to non-tumorigenic melanoma cells resulting in tumorigenesis. In contrast, higher concentrations of MCP-1 greatly increase infiltration of anti-tumor monocytes/Mφs that destroy tumors. Induction of tumor growth by low levels of MCP-1 was shown to be mediated by monocyte derived TNF-α resulting in angiogenesis.
In other tumor models, macrophage inflammatory protein 1 beta (MIP-1β) /CCL4 (Table 1), and RANTES/CCL5 (Table 1, 2), were found to be produced by monocytic myeloid derived suppressor cells (MO-MDSCs) isolated from B16 melanoma [37]. MO-MDSCs were capable of recruiting regulatory T lymphocytes (Tregs) expressing C-C chemokine receptor 5 (CCR5) in vitro. Subsequent studies in vivo revealed that Tregs were recruited to tumors in response to increased concentrations of MIP-1β /CCL4, and CCL5 administered intratumorally. In contrast, impaired melanoma growth was observed in CCR5 deficient mice exhibiting decreased recruitment of intratumoral Tregs.
Mφ inflammatory protein 3 alpha (MIP-3α)/CCL20, produced by TAMS, was found to be upregulated in primary but not RAW264.7 Mφs. MIP-3α recruits pro-tumor Tregs [38], and increases tumor growth and metastasis [39].
Chemokine production can also influence recruitment of MDSCs to melanomas. Expression of HIF-1α, a pro-angiogenic M1 Mφ signaling factor [40, 41], in MDSCs is attenuated by the tumor suppressive activity of miR-155 [42]. In a recent study, increased HIF-1α activity in miR-155 deficient MDSCs was associated with increased expression of melanoma growth stimulating activity alpha (MSGA-α)/CXCL1 (Table 1), and other chemokines by MDSCs [42]. Expression of such chemokines facilitated recruitment of immunosuppressive and pro-angiogenic miR-155−/− MDSCs to tumors.
Interestingly, while CXCL1 was found to be upregulated by melanoma exosomes in RAW 264.7 Mφs but not primary Mφs, the reverse was true for HIF-1α. In a previous study, we demonstrated upregulation of HIF-1α in the lymph nodes of wild-type C57BL6 mice treated with B16-F10 melanoma exosomes [6]. Taken together, these findings begin to suggest that lymph node associated Mφs, possibly subcapsular sinus Mφs [2], produce HIF-1α in response to melanoma exosomes in lymph nodes.
Tumor associated neutrophils (TANs), in addition to other tumor infiltrating cells of myeloid lineage such as TAMs or MDSCs, facilitate tumor angiogenesis [43]. TANs can be recruited to tumors via CXCL2 gradients emanating from the tumor (high CXCL2 concentration) to the bone marrow (low CXCL2 concentration) [43]. This process is inhibited by interferon beta (IFN-β). Antibody blockade of CXCL2 in B16 melanoma bearing Ifnb1−/− mice results in decreased angiogenesis and decreased recruitment of TANs [43]. CXCL2 can also directly promote tumor angiogenesis via interaction with its receptor, CXCR2, on endothelial cells [43]. Epithelial neutrophil-activating peptide 78 (ENA-78)/CXCL5 (Table 1), also signals through CXCR2 and has been shown to promote tumor angiogenesis via endothelial cell activation [44].
In addition to chemokines, the M1 cytokine subunits, IL-23A (p19) and IL-12B (p40), were upregulated in RAW 264.7 (Table 1) and primary Mφs (Table 2) by melanoma exosomes. The heterodimeric cytokine IL-23, composed of p19 and p40 subunits, has been shown to oppose IL-12 activity. IL-23 is composed of the same p40 subunit associated with IL-12, which consist of p35 (IL-12A) and p40 (IL-12B) subunits. Combined IL-23 and IL-12 activities serve to maintain the equilibrium stage of cancer immunoediting [45]. IL-12 prevents cancer outgrowth whereas IL-23 promotes cancer persistence through suppression of innate and adaptive anti-tumor immune responses. The absence of significantly increased expression of the IL-12 p35 subunit in RAW 264.7 Mφs suggest that IL-23 rather than IL-12 was preferentially induced by the melanoma exosomes. In contrast, primary Mφs produced IL-12A, IL-12B, and IL-23A which may result in a degree of simultaneous expression of IL-12 and IL-23. This begins to suggest a novel role for melanoma exosomes in supporting the equilibrium stage of cancer. In addition to direct immunosuppressive effects, melanoma exosomes may mediate indirect immune suppression via induction of Mφ derived IL-23. The immunosuppressive effects of IL-23 may be further bolstered by melanoma exosome mediated induction of Mφ derived CCL22. CCL22 has been shown to recruit Tregs to melanoma [46]. Within tumors, Tregs suppress tumor immune surveillance.
Other M1 cytokines uniquely upregulated in primary but not RAW264.7 Mφs included chemokine receptor type 7 (CCR7), chemokine (C-X-C motif) ligand 9 (CXCL9), CXCL10, and CXCL11. CCR7 expression supports tumor metastasis by melanoma cells by enabling their recruitment to lymph nodes [47]. Mouse B16-F10 and human melanoma cells express CXCR3, the receptor for CXCL9, CXCL10, and CXCL11 [48]. CXCL9 and CXCL10 chemotaxis facilitates melanoma metastasis via transendothelial migration [49]. Interferon-inducible T-cell alpha chemoattractant (I-TAC)/CXCL11 also enables melanoma cell migration [50]. Further, pre-treatment of mouse lymph nodes with the immunopotentiator, complete Freud’s adjuvant, results in increased production of CXCL9 and CXCL10 in draining lymph nodes, and concomitant metastasis of B16-F10 melanoma cells to the nodes [48]. In the present study, the ability of melanoma exosomes to upregulate CXCL9 and CXCL10 in primary Mφs, coupled to our previous finding that B16-F10 melanoma cells preferentially home to melanoma exosome deposition sites in lymph nodes [6], supports a tumor exosome-dependent means of melanoma self-seeding of lymph nodes.
Dual (M1 and M2) specific macrophage polarization factors upregulated by melanoma exosomes
Assessment of markers common to both M1 and M2 Mφ polarization phenotypes, revealed that melanoma exosomes significantly increased the production of TNF-α in RAW264.7 and primary Mφs. TNF-α is known to participate in M1 Mφ mediated anti-tumor immunity. However, TNF-α also plays a role in promoting tumor angiogenesis [51]. TNF-α has also been shown to mediate vascular remodeling and lymphangiogenesis [52].
Results from the RAW264.7 Mφ RT-qPCR arrays largely corroborated the RAW264.7 Mφ ELISA data. However, in contrast to the ELISA results, the PCR data shows that exosomes significantly increased IL-1β mRNA synthesis. The discrepancy could be a result of ELISA timing, or assay sensitivity, with RT-qPCR being more sensitive, or may reflect undefined post-transcriptional regulation mechanisms requiring more investigation. Beyond its traditional role as a pro-inflammatory cytokine, IL-1β promotes tumor invasion [53] and angiogenesis via direct and indirect mechanisms including upregulation of endothelial VEGF and VEGF receptors [54]. This is consistent with the observation that induction of IL-1β mRNA by melanoma exosomes in RAW264.7 Mφ was also associated with increased VEGF mRNA.
Melanoma exosomes also upregulated IL-6 in RAW264.7 and primary Mφs. IL-6 is a pro-inflammatory cytokine that can also stimulate the growth of tumor cells and facilitate invasion and metastasis [55]. It is an important mediator of angiogenesis, possessing similar properties as VEGF-A such as promoting endothelial proliferation [55]. However, in contrast to VEGF-A, IL-6 stimulates vessel sprouting with defective pericyte coverage, potentially contributing to abnormal tumor vasculature.
Upregulation of TLR-9 by melanoma exosomes in primary Mφs is interesting, because TLR-9 activity is associated with M1 polarization in non-alcoholic steatohepatitis [56] and M2 polarization in cancer [57]. This finding further highlights the complexity associated with melanoma exosome-mediated influences on Mφ phenotype plasticity.
M2 specific macrophage polarization factors upregulated by melanoma exosomes
Detection of standard M2 cytokines induced at the mRNA level by melanoma exosomes revealed similar findings. The induction of the immunosuppressive cytokine transforming growth factor-beta (TGF-β) by melanoma exosomes in primary Mφs is similar to what has been reported concerning the ability of tumor microvesicles to impair CD14+ monocyte differentiation into dendritic cells [58]. Valenti et al. demonstrated that the impaired monocytes produced increased levels of TGF-β and suppressed activated T-lymphocyte proliferation and cytotoxic functions. Further, a similar population of CD14+ cells was found in the circulation of melanoma patients.
Induction of the M2a marker VEGF-A [8] in RAW 264.7 Mφs, and the M2d marker STAT3 [30] in RAW 264.7 and primary Mφs by melanoma exosomes, further supports a degree of polarization toward an M2-like phenotype. IL-10/STAT3 signaling induces M2 TAMs in hypoxic melanoma core microenvironments associated with increased expression of HIF-1α and VEGF-A [59]. The STAT3 transcription factor induces anti-apoptotic, angiogenic, immunosuppressive and metastatic gene expression profiles conducive to melanoma growth and survival [60]. In contrast, the M1 signaling factor STAT1 [30], antagonizes these same processes. Upregulation of STAT1 mRNA was detected only in primary Mφs, possibly in response to increased STAT3 signaling.
COX-2 expression was upregulated in primary (Fig. 4) but not RAW 264.7 (Fig. 3) Mφs. Increased COX-2 expression is associated with M2b [61], and M2d/TAM [62] Mφ phenotypes. Inhibition of COX-2 in M2 TAMs polarizes them to an M1 phenotype [63].
IL-10 was expressed in RAW264.7 Mφs, and in primary Mφs to a lesser degree. In general, IL-10 is produced by all M2 Mφ subclasses [8]. IL-10, when released by type 2 helper T cells, also polarizes Mφs toward an immunosuppressive M2 phenotype in melanoma microenvironments [64].
A number of previous studies suggest a complicated relationship between the M2 cytokine IL-10 and the M1 cytokine TNF-α. TNF-α can induce IL-10 expression [65], or alternatively, IL-10 can suppress TNF-α [66]. Melanoma cells themselves can also produce TNF-α, IL-10 and IL-1β cytokines [67]. The ability of melanoma exosomes to induce these same cytokines in Mφs highlights the complexity of tumor resilience. A further implication is that melanoma can directly produce tumor supportive cytokines locally, and/or indirectly induce them via exosome-mediated stimulation of Mφs remotely, in pre-metastatic niches for example. TNF-α can also increase HIF-1α production by Mφs [68]. This is consistent with what we previously reported showing increased expression of TNF-α and HIF-1α in lymph nodes by melanoma exosomes [6].
Melanoma exosome induction of mixed M1 and M2 macrophage polarization
Melanoma cells can overexpress i-NOS [69]. Pro-tumor functions attributed to i-NOS include promoting angiogenesis, and melanoma cell proliferation via nitrosylation of tuberous sclerosis complex [69]. Conceivably, i-NOS pathway associated mRNAs, miRNAs, or signaling molecules present in melanoma cells may be relayed to Mφs via melanoma exosomes. Exosomal “shuttle” mRNA has been shown to be transferrable and functional in target cells [70].
Decreased arginase activity in the context of increased i-NOS activity, and increased i-NOS mRNA expression was observed for RAW 264.7 Mφs. A similar, more modest trend was observed for primary Mφs. Taken together, these data support melanoma exosome-mediated induction of a more M1 functional phenotype, especially for RAW 264.7 Mφs. This is consistent with the RT-qPCR array findings demonstrating increased gene expression of M1 versus M2 polarization markers (Table 1, 2). A quick calculation reveals an M1:M2 marker ratio of ~ 2.3 for RAW 264.7 Mφs and ~ 2.8 for primary Mφs. This shows that both RAW 264.7 and primary Mφs were predominantly M1 polarized by melanoma exosomes. Nevertheless, a smaller degree of M2 polarization is also apparent, particularly given the observed upregulation of mixed M1/M2 markers (Fig. 3b, 4b).
The M2 component of RAW 264.7 Mφ polarization is most similar to an M2b phenotype, which is characterized by IL-10 expression in the context of typical M1 representative cytokines including TNF-α, IL-1β, IL-6 and i-NOS activity [30]. The M2b response is traditionally associated with humoral immunity [30]. In contrast, the M2 component of primary Mφ polarization is most similar to an M2d/TAM phenotype, given the additional upregulation of TGF-β, COX-2, and TLR-9. The ability of melanoma exosomes to skew primary Mφ polarization toward a TAM-like phenotype is consistent with melanoma exosome-mediated preparation of tumor microenvironments [6]. It is possible that prolonged exposure to melanoma exosomes may further increase the M2 polarization observed. Conceivably, such a scenario may occur in the context of sustained production of locally high concentrations of melanoma exosomes in tumor bearing or draining lymph nodes [71]. Further, Mφ type and/or pre-existing M1 or M2 subtype polarization, may also dictate the extent to which melanoma exosomes can influence Mφ polarization [71]. Future studies will be necessary to tease apart the exosomal cargo (proteins, mRNA, miRNA, lipids etc.) responsible for mediating the complex pattern of Mφ polarization observed. Undoubtedly, the effect likely requires the activity of multiple exosomal components working in concert.
Conclusions
The ability of melanoma exosomes to influence Mφ polarity is consistent with what has been reported for other tumor exosome types [72–77]. However, to the best of our knowledge, our studies provide the first evidence that natural, unmodified melanoma exosomes are able to directly polarize Mφs toward a “mixed” M1/M2 phenotype. The implication of the finding is that it enables melanoma exosome tuned Mφs to maintain a certain degree of flexibility and adaptability in terms of facilitating tumor survival and resistance to biologic therapies. This “mixed” phenotype would be expected to promote a variety of pro-tumor functions. Based on the identified polarization factors, such pro-tumor functions may include: 1. stimulating TAM polarization, tumor growth, and metastasis, 2. recruiting immunosuppressive cell types (Tregs, MDSCs, TAMs and TANs), 3. supporting the cancer immune equilibrium stage, 4. facilitating tumor “self-seeding” of lymph nodes, angiogenesis and lymphangiogenesis, and 5. promoting immune suppression. Ultimately, this has important ramifications for our understanding of melanoma exosome mediated tumor pathogenesis and the development of new immunotherapies for melanoma.
Supplementary Material
Highlights.
Anti-tumor (M1) and pro-tumor (M2) macrophages influence melanoma survival.
Melanoma exosomes also facilitate tumor survival.
Melanoma exosomes might directly influence M1 or M2 macrophage polarization.
Melanoma exosomes induce a mixed M1/M2 pro-tumor macrophage activation phenotype.
Acknowledgments
This research was supported by the NCI R25 grant, University of Louisville Cancer Education Program NIH/NCI (R25-CA134283), the School of Medicine Summer Research Scholar Program, and NIH/NIGMS (R21-GM107894). This work was also supported in part by a grant from the University of Louisville School of Medicine, and a pilot project funded through the NIH 1P30GM106396 Molecular Targets Phase III CoBRE grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The authors declare no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
J. L. Hood and G. T. Bardi wrote and edited the manuscript. J. L. Hood developed the experimental concepts and hypotheses and analyzed data. G. T. Bardi and M. A. Smith performed experiments and analyzed data.
References
- 1.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-'t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, Hochberg F. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of extracellular vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hood JL. The association of exosomes with lymph nodes. Seminars in cell & developmental biology. 2017;67:29–38. doi: 10.1016/j.semcdb.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Hood JL, Pan H, Lanza GM, Wickline SA. Paracrine induction of endothelium by tumor exosomes. Lab Invest. 2009;89(11):1317–28. doi: 10.1038/labinvest.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekstrom EJ, Bergenfelz C, von Bulow V, Serifler F, Carlemalm E, Jonsson G, Andersson T, Leandersson K. WNT5A induces release of exosomes containing pro-angiogenic and immunosuppressive factors from malignant melanoma cells. Molecular cancer. 2014;13:88. doi: 10.1186/1476-4598-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature medicine. 2012;18(6):883–91. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer research. 2011;71(11):3792–801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 7.Hu L, Wickline SA, Hood JL. Magnetic resonance imaging of melanoma exosomes in lymph nodes. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2015;74:266–71. doi: 10.1002/mrm.25376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roszer T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediators of inflammation. 2015;2015:816460. doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen JL, Mohamadzadeh M. Macrophages and chemokines as mediators of angiogenesis. Frontiers in physiology. 2013;4:159. doi: 10.3389/fphys.2013.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duluc D, Corvaisier M, Blanchard S, Catala L, Descamps P, Gamelin E, Ponsoda S, Delneste Y, Hebbar M, Jeannin P. Interferon-gamma reverses the immunosuppressive and protumoral properties and prevents the generation of human tumor-associated macrophages. International journal of cancer. 2009;125(2):367–73. doi: 10.1002/ijc.24401. [DOI] [PubMed] [Google Scholar]
- 11.Thery C, Clayton A, Amigorena S, Raposo G. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Current Protocols in Cell Biology. 2006:3.22.1–3.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 12.Hood JL, Scott MJ, Wickline SA. Maximizing exosome colloidal stability following electroporation. Anal Biochem. 2014;448:41–9. doi: 10.1016/j.ab.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barth KA, Waterfield JD, Brunette DM. The effect of surface roughness on RAW 264.7 macrophage phenotype. Journal of biomedical materials research. Part A. 2013;101(9):2679–88. doi: 10.1002/jbm.a.34562. [DOI] [PubMed] [Google Scholar]
- 14.Na YR, Jung D, Gu GJ, Seok SH. GM-CSF Grown Bone Marrow Derived Cells Are Composed of Phenotypically Different Dendritic Cells and Macrophages. Molecules and cells. 2016;39(10):734–741. doi: 10.14348/molcells.2016.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar V, Gour JK, Singh N, Bajpai S, Singh RK. Leishmania donovani-specific 25- and 28-kDa urinary proteins activate macrophage effector functions, lymphocyte proliferation and Th1 cytokines production. Parasitology research. 2013;112(4):1427–35. doi: 10.1007/s00436-013-3272-z. [DOI] [PubMed] [Google Scholar]
- 16.Huang HN, Rajanbabu V, Pan CY, Chan YL, Wu CJ, Chen JY. A cancer vaccine based on the marine antimicrobial peptide pardaxin (GE33) for control of bladderassociated tumors. Biomaterials. 2013;34(38):10151–9. doi: 10.1016/j.biomaterials.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 17.Fan Q, Lu M, Xia ZY, Bao L. Mycobacterium tuberculosis MPT64 stimulates the activation of murine macrophage modulated by IFN-gamma. European review for medical and pharmacological sciences. 2013;17(24):3296–305. [PubMed] [Google Scholar]
- 18.Sun T, Yan X, Guo W, Zhao D. Evaluation of cytotoxicity and immune modulatory activities of soyasaponin Ab: an in vitro and in vivo study. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2014;21(13):1759–66. doi: 10.1016/j.phymed.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Devi KS, Sahoo B, Behera B, Maiti TK. Nanoparticle and polysaccharide conjugate: a potential candidate vaccine to improve immunological stimuli. International journal of biological macromolecules. 2015;72:1254–64. doi: 10.1016/j.ijbiomac.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H, Zhang X, Chen X, Li Y, Ke Z, Tang T, Chai H, Guo AM, Chen H, Yang J. Isoliquiritigenin, a flavonoid from licorice, blocks M2 macrophage polarization in colitis-associated tumorigenesis through downregulating PGE2 and IL-6. Toxicology and applied pharmacology. 2014;279(3):311–21. doi: 10.1016/j.taap.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Chen M, Zhang J, Hu F, Liu S, Zhou Z. Metformin affects the features of a human hepatocellular cell line (HepG2) by regulating macrophage polarization in a co-culture microenviroment. Diabetes/metabolism research and reviews. 2015;31(8):781–9. doi: 10.1002/dmrr.2761. [DOI] [PubMed] [Google Scholar]
- 22.Gao S, Zhou J, Liu N, Wang L, Gao Q, Wu Y, Zhao Q, Liu P, Wang S, Liu Y, Guo N, Shen Y, Wu Y, Yuan Z. Curcumin induces M2 macrophage polarization by secretion IL-4 and/or IL-13. Journal of molecular and cellular cardiology. 2015;85:131–9. doi: 10.1016/j.yjmcc.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Liu Y, Gu J, Wang Y, Liu L, Zhang P, Li Y. Endostatin inhibits the growth and migration of 4T1 mouse breast cancer cells by skewing macrophage polarity toward the M1 phenotype. Cancer immunology, immunotherapy : CII. 2016;65(6):677–88. doi: 10.1007/s00262-016-1824-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. Journal of extracellular vesicles. 2013;2 doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupert DL, Claudio V, Lasser C, Bally M. Methods for the physical characterization and quantification of extracellular vesicles in biological samples. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbagen.2016.07.028. [DOI] [PubMed] [Google Scholar]
- 26.Hood JL, Wickline SA. A systematic approach to exosome-based translational nanomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4(4):458–67. doi: 10.1002/wnan.1174. [DOI] [PubMed] [Google Scholar]
- 27.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nature medicine. 2001;7(3):297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 28.Riteau B, Faure F, Menier C, Viel S, Carosella ED, Amigorena S, Rouas-Freiss N. Exosomes bearing HLA-G are released by melanoma cells. Hum Immunol. 2003;64(11):1064–72. doi: 10.1016/j.humimm.2003.08.344. [DOI] [PubMed] [Google Scholar]
- 29.Italiani P, Boraschi D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Frontiers in immunology. 2014;5:514. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foey A. Macrophage Polarisation: A collaboration of Differentiation, Activation and Pre-Programming? J Clin Cell Immunol. 2015;6(1) [Google Scholar]
- 31.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1–m2 polarization balance. Frontiers in immunology. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J, Wang D, Liu D, Liu M, Ge Y, Jiang M, Liu Y, Zheng D. Tumor necrosis factor-related apoptosis-inducing ligand induces the expression of proinflammatory cytokines in macrophages and re-educates tumor-associated macrophages to an antitumor phenotype. Mol Biol Cell. 2015;26(18):3178–89. doi: 10.1091/mbc.E15-04-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie C, Liu C, Wu B, Lin Y, Ma T, Xiong H, Wang Q, Li Z, Ma C, Tu Z. Effects of IRF1 and IFN-beta interaction on the M1 polarization of macrophages and its antitumor function. Int J Mol Med. 2016;38(1):148–60. doi: 10.3892/ijmm.2016.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni W, Zhang Q, Liu G, Wang F, Yuan H, Guo Y, Zhang X, Xie F, Li Q, Tai G. Escherichia coli maltose-binding protein activates mouse peritoneal macrophages and induces M1 polarization via TLR2/4 in vivo and in vitro. Int Immunopharmacol. 2014;21(1):171–80. doi: 10.1016/j.intimp.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 35.Marton A, Vizler C, Kusz E, Temesfoi V, Szathmary Z, Nagy K, Szegletes Z, Varo G, Siklos L, Katona RL, Tubak V, Howard OM, Duda E, Minarovits J, Nagy K, Buzas K. Melanoma cell-derived exosomes alter macrophage and dendritic cell functions in vitro. Immunology letters. 2012;148(1):34–8. doi: 10.1016/j.imlet.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Nesbit M, Schaider H, Miller TH, Herlyn M. Low-level monocyte chemoattractant protein-1 stimulation of monocytes leads to tumor formation in nontumorigenic melanoma cells. Journal of immunology. 2001;166(11):6483–90. doi: 10.4049/jimmunol.166.11.6483. [DOI] [PubMed] [Google Scholar]
- 37.Schlecker E, Stojanovic A, Eisen C, Quack C, Falk CS, Umansky V, Cerwenka A. Tumor-infiltrating monocytic myeloid-derived suppressor cells mediate CCR5-dependent recruitment of regulatory T cells favoring tumor growth. Journal of immunology. 2012;189(12):5602–11. doi: 10.4049/jimmunol.1201018. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Zhang N, Li Q, Zhang W, Ke F, Leng Q, Wang H, Chen J, Wang H. Tumor-associated macrophages recruit CCR6+ regulatory T cells and promote the development of colorectal cancer via enhancing CCL20 production in mice. PloS one. 2011;6(4):e19495. doi: 10.1371/journal.pone.0019495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B, Jia Y, Ma J, Wu S, Jiang H, Cao Y, Sun X, Yin X, Yan S, Shang M, Mao A. Tumor-associated macrophage-derived CCL20 enhances the growth and metastasis of pancreatic cancer. Acta Biochim Biophys Sin (Shanghai) 2016;48(12):1067–1074. doi: 10.1093/abbs/gmw101. [DOI] [PubMed] [Google Scholar]
- 40.Weigert A, Brune B. Nitric oxide, apoptosis and macrophage polarization during tumor progression. Nitric Oxide. 2008;19(2):95–102. doi: 10.1016/j.niox.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 41.Aarup A, Pedersen TX, Junker N, Christoffersen C, Bartels ED, Madsen M, Nielsen CH, Nielsen LB. Hypoxia-Inducible Factor-1alpha Expression in Macrophages Promotes Development of Atherosclerosis. Arteriosclerosis, thrombosis, and vascular biology. 2016;36(9):1782–90. doi: 10.1161/ATVBAHA.116.307830. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Yu F, Jia X, Iwanowycz S, Wang Y, Huang S, Ai W, Fan D. MicroRNA-155 deficiency enhances the recruitment and functions of myeloid-derived suppressor cells in tumor microenvironment and promotes solid tumor growth. Int J Cancer. 2015;136(6):E602–13. doi: 10.1002/ijc.29151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jablonska J, Wu CF, Andzinski L, Leschner S, Weiss S. CXCR2-mediated tumor-associated neutrophil recruitment is regulated by IFN-beta. Int J Cancer. 2014;134(6):1346–58. doi: 10.1002/ijc.28551. [DOI] [PubMed] [Google Scholar]
- 44.Li A, King J, Moro A, Sugi MD, Dawson DW, Kaplan J, Li G, Lu X, Strieter RM, Burdick M, Go VL, Reber HA, Eibl G, Hines OJ. Overexpression of CXCL5 is associated with poor survival in patients with pancreatic cancer. The American journal of pathology. 2011;178(3):1340–9. doi: 10.1016/j.ajpath.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teng MW, Swann JB, Koebel CM, Schreiber RD, Smyth MJ. Immune-mediated dormancy: an equilibrium with cancer. Journal of leukocyte biology. 2008;84(4):988–93. doi: 10.1189/jlb.1107774. [DOI] [PubMed] [Google Scholar]
- 46.Klarquist J, Tobin K, Farhangi Oskuei P, Henning SW, Fernandez MF, Dellacecca ER, Navarro FC, Eby JM, Chatterjee S, Mehrotra S, Clark JI, Le Poole IC. Ccl22 Diverts T Regulatory Cells and Controls the Growth of Melanoma. Cancer research. 2016;76(21):6230–6240. doi: 10.1158/0008-5472.CAN-16-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lazennec G, Richmond A. Chemokines and chemokine receptors: new insights into cancer-related inflammation. Trends in molecular medicine. 2010;16(3):133–44. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawada K, Sonoshita M, Sakashita H, Takabayashi A, Yamaoka Y, Manabe T, Inaba K, Minato N, Oshima M, Taketo MM. Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer research. 2004;64(11):4010–7. doi: 10.1158/0008-5472.CAN-03-1757. [DOI] [PubMed] [Google Scholar]
- 49.Amatschek S, Lucas R, Eger A, Pflueger M, Hundsberger H, Knoll C, Grosse-Kracht S, Schuett W, Koszik F, Maurer D, Wiesner C. CXCL9 induces chemotaxis, chemorepulsion and endothelial barrier disruption through CXCR3-mediated activation of melanoma cells. British journal of cancer. 2011;104(3):469–79. doi: 10.1038/sj.bjc.6606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee E, Choi SY, Bin BH, Kim NH, Kim KH, Choi DH, Han J, Choi H, Lee AY, Lee TR, Cho EG. Interferon-inducible T-cell alpha chemoattractant (ITAC) induces the melanocytic migration and hypopigmentation through destabilizing p53 via histone deacetylase 5: a possible role of ITAC in pigment-related disorders. The British journal of dermatology. 2017;176(1):127–137. doi: 10.1111/bjd.14878. [DOI] [PubMed] [Google Scholar]
- 51.Kim NH, Jung HJ, Shibasaki F, Kwon HJ. NBBA, a synthetic small molecule, inhibits TNF-alpha-induced angiogenesis by suppressing the NF-kappaB signaling pathway. Biochemical and biophysical research communications. 2010;391(3):1500–5. doi: 10.1016/j.bbrc.2009.12.101. [DOI] [PubMed] [Google Scholar]
- 52.Baluk P, Yao LC, Feng J, Romano T, Jung SS, Schreiter JL, Yan L, Shealy DJ, McDonald DM. TNF-alpha drives remodeling of blood vessels and lymphatics in sustained airway inflammation in mice. The Journal of clinical investigation. 2009;119(10):2954–64. doi: 10.1172/JCI37626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krelin Y, Voronov E, Dotan S, Elkabets M, Reich E, Fogel M, Huszar M, Iwakura Y, Segal S, Dinarello CA, Apte RN. Interleukin-1beta-driven inflammation promotes the development and invasiveness of chemical carcinogen-induced tumors. Cancer research. 2007;67(3):1062–71. doi: 10.1158/0008-5472.CAN-06-2956. [DOI] [PubMed] [Google Scholar]
- 54.Voronov E, Carmi Y, Apte RN. The role IL-1 in tumor-mediated angiogenesis. Frontiers in physiology. 2014;5:114. doi: 10.3389/fphys.2014.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopinathan G, Milagre C, Pearce OM, Reynolds LE, Hodivala-Dilke K, Leinster DA, Zhong H, Hollingsworth RE, Thompson R, Whiteford JR, Balkwill F. Interleukin-6 Stimulates Defective Angiogenesis. Cancer research. 2015;75(15):3098–107. doi: 10.1158/0008-5472.CAN-15-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mridha AR, Haczeyni F, Yeh MM, Haigh WG, Ioannou GN, Barn V, Ajamieh H, Adams L, Hamdorf JM, Teoh NC, Farrell GC. TLR9 is up-regulated in human and murine NASH: pivotal role in inflammatory recruitment and cell survival. Clinical science (London, England : 1979) 2017;131(16):2145–2159. doi: 10.1042/CS20160838. [DOI] [PubMed] [Google Scholar]
- 57.Sanchez-Reyes K, Bravo-Cuellar A, Hernandez-Flores G, Lerma-Diaz JM, Jave-Suarez LF, Gomez-Lomeli P, de Celis R, Aguilar-Lemarroy A, Dominguez-Rodriguez JR, Ortiz-Lazareno PC. Cervical cancer cell supernatants induce a phenotypic switch from U937-derived macrophage-activated M1 state into M2-like suppressor phenotype with change in Toll-like receptor profile. Biomed Res Int. 2014;2014:683068. doi: 10.1155/2014/683068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valenti R, Huber V, Filipazzi P, Pilla L, Sovena G, Villa A, Corbelli A, Fais S, Parmiani G, Rivoltini L. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-beta-mediated suppressive activity on T lymphocytes. Cancer research. 2006;66(18):9290–8. doi: 10.1158/0008-5472.CAN-06-1819. [DOI] [PubMed] [Google Scholar]
- 59.Goswami KK, Sarkar M, Ghosh S, Saha A, Ghosh T, Guha I, Barik S, Banerjee S, Roy S, Bose A, Dasgupta P, Baral R. Neem leaf glycoprotein regulates function of tumor associated M2 macrophages in hypoxic tumor core: Critical role of IL-10/STAT3 signaling. Molecular immunology. 2016;80:1–10. doi: 10.1016/j.molimm.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 60.Nicholas C, Lesinski GB. The Jak-STAT Signal Transduction Pathway in Melanoma. In: Tanaka DY, editor. Breakthroughs in Melanoma Research, InTech. 2011. [Google Scholar]
- 61.Kudlik G, Hegyi B, Czibula A, Monostori E, Buday L, Uher F. Mesenchymal stem cells promote macrophage polarization toward M2b-like cells. Exp Cell Res. 2016;348(1):36–45. doi: 10.1016/j.yexcr.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 62.Li H, Yang B, Huang J, Lin Y, Xiang T, Wan J, Chouaib S, Ren G. Cyclooxygenase-2 in tumor-associated macrophages promotes breast cancer cell survival by triggering a positive-feedback loop between macrophages and cancer cells. Oncotarget. 2015;6(30):29637–50. doi: 10.18632/oncotarget.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakanishi Y, Nakatsuji M, Seno H, Ishizu S, Akitake-Kawano R, Kanda K, Ueo T, Komekado H, Kawada M, Minami M, Chiba T. COX-2 inhibition alters the phenotype of tumor-associated macrophages from M2 to M1 in ApcMin/+ mouse polyps. Carcinogenesis. 2011;32(9):1333–9. doi: 10.1093/carcin/bgr128. [DOI] [PubMed] [Google Scholar]
- 64.Ilkovitch D, Lopez DM. Immune modulation by melanoma-derived factors. Experimental dermatology. 2008;17(12):977–85. doi: 10.1111/j.1600-0625.2008.00779.x. [DOI] [PubMed] [Google Scholar]
- 65.Wanidworanun C, Strober W. Predominant role of tumor necrosis factor-alpha in human monocyte IL-10 synthesis. Journal of immunology. 1993;151(12):6853–61. [PubMed] [Google Scholar]
- 66.Armstrong L, Jordan N, Millar A. Interleukin 10 (IL-10) regulation of tumour necrosis factor alpha (TNF-alpha) from human alveolar macrophages and peripheral blood monocytes. Thorax. 1996;51(2):143–9. doi: 10.1136/thx.51.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kruger-Krasagakes S, Krasagakis K, Garbe C, Diamantstein T. Production of cytokines by human melanoma cells and melanocytes. Recent results in cancer research. Fortschritte der Krebsforschung. Progres dans les recherches sur le cancer. 1995;139:155–68. doi: 10.1007/978-3-642-78771-3_11. [DOI] [PubMed] [Google Scholar]
- 68.Palazon A, Goldrath AW, Nizet V, Johnson RS. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41(4):518–28. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez-Rivera E, Jayaraman P, Parikh F, Davies MA, Ekmekcioglu S, Izadmehr S, Milton DR, Chipuk JE, Grimm EA, Estrada Y, Aguirre-Ghiso J, Sikora AG. Inducible nitric oxide synthase drives mTOR pathway activation and proliferation of human melanoma by reversible nitrosylation of TSC2. Cancer research. 2014;74(4):1067–78. doi: 10.1158/0008-5472.CAN-13-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9(6):654–U72. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 71.Hood JL. Melanoma exosome induction of endothelial cell GM-CSF in pre-metastatic lymph nodes may result in different M1 and M2 macrophage mediated angiogenic processes. Medical hypotheses. 2016;94:118–22. doi: 10.1016/j.mehy.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soki FN, Koh AJ, Jones JD, Kim YW, Dai J, Keller ET, Pienta KJ, Atabai K, Roca H, McCauley LK. Polarization of prostate cancer-associated macrophages is induced by milk fat globule-EGF factor 8 (MFG-E8)-mediated efferocytosis. The Journal of biological chemistry. 2014;289(35):24560–72. doi: 10.1074/jbc.M114.571620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trivedi M, Talekar M, Shah P, Ouyang Q, Amiji M. Modification of tumor cell exosome content by transfection with wt-p53 and microRNA-125b expressing plasmid DNA and its effect on macrophage polarization. Oncogenesis. 2016;5(8):e250. doi: 10.1038/oncsis.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Su MJ, Aldawsari H, Amiji M. Pancreatic Cancer Cell Exosome-Mediated Macrophage Reprogramming and the Role of MicroRNAs 155 and 125b2 Transfection using Nanoparticle Delivery Systems. Sci Rep. 2016;6:30110. doi: 10.1038/srep30110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ying X, Wu Q, Wu X, Zhu Q, Wang X, Jiang L, Chen X, Wang X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. 2016;7(28):43076–43087. doi: 10.18632/oncotarget.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, Iguchi T, Ito S, Eguchi H, Ochiya T, Yanaga K, Miyano S, Mimori K. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017;8(45):78598–78613. doi: 10.18632/oncotarget.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen X, Ying X, Wang X, Wu X, Zhu Q. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol Rep. 2017;38(1):522–528. doi: 10.3892/or.2017.5697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.