Summary
Bacterial resistance mechanisms must cope with transient fast-changing conditions. These systems are often repressed in absence of the drug and it is unclear how their regulation can provide a quick response when challenged. Here we focus on the tet operon, which provides resistance to tetracycline through efflux pump TetA. We show that, somewhat counter-intuitively, prompt expression of TetA repressor TetR is key for cellular survival upon abrupt drug exposure. Tracking individual cells upon exposure, we find that differences in the rate of TetR elevation result in three distinct cell fates: recovery (high rate), death due to excess of TetA (intermediate rate) and death from the drug (low rate). A surge of TetR expression optimizes the response by allowing sensitive detection of both the initial rise and the later decline of intracellular drug, avoiding an undesirable overshoot in TetA expression. These results show how regulatory circuits of resistance genes have evolved for optimized dynamics.
Introduction
The expression of an antibiotic resistance gene is often regulated by the concentration of its corresponding antibiotic (Depardieu et al., 2007). Many antibiotic resistance mechanisms are repressed by a dedicated transcription factor, which guarantees expression only in the presence of the drug. Such regulation avoids costly expression when the drug is absent and, in presence of the drug, tunes expression according to drug concentration to optimize cost and benefit (Batchelor et al., 2004; Dekel and Alon, 2005). Beyond regulating the gene to its optimal level at steady antibiotic concentrations, regulation should also cope with transient conditions (Alon, 2007), since the cell begins the response from a drug-susceptible state. Indeed, the choice of regulatory architecture of a response mechanism was shown to be determined not only by static requirements of stability, demand and robustness, but also by requirements for dynamic responsiveness (Savageau, 1998; Wall et al., 2004) (Guet et al., 2005; Le et al., 2006). However, it is unclear to what extent the regulation of a single antibiotic resistance gene can optimize cell survival both statically in steady drug environments and dynamically during abrupt changes in drug concentrations.
Here we focus on the tet operon, which provides resistance against tetracycline (a translation inhibitor) in Escherichia coli, to understand the role of regulation upon an abrupt increase in drug concentration. The expression of tetracycline specific efflux pump TetA (Fernández and Hancock, 2012) is tightly repressed by the transcription factor TetR (Ramos et al., 2005) (Figures 1A and S1A). In the presence of tetracycline, TetR binds the drug and greatly diminishes the affinity for its operators (Lederer et al., 1995), releasing expression of TetA (Muthukrishnan et al., 2012). TetA then actively transports tetracycline out of the cytoplasm in a process involving an ion exchange that disrupts the membrane potential (Bertrand and Lenski, 1989; Eckert and Beck, 1989), thereby posing a trade-off between resistance to tetracycline and the adverse effects of TetA expression. TetR also represses itself, which is known to speed up response times (Camas et al., 2006; Rosenfeld et al., 2002), reduce noise (Foglierini et al., 2006) and increase the input dynamic range (Madar et al., 2011; Nevozhay et al., 2009), and when TetR is induced upon exposure it can cause TetA levels to drop (Le et al., 2006). The tet operon therefore provides an ideal system to study dynamic regulation – it is tightly repressed, well characterized and controls a gene that carries a significant cost (Berens and Hillen, 2003; Meier et al., 1988; Palmer et al., 2010; Sigler et al., 2000; Wu and Rao, 2010).
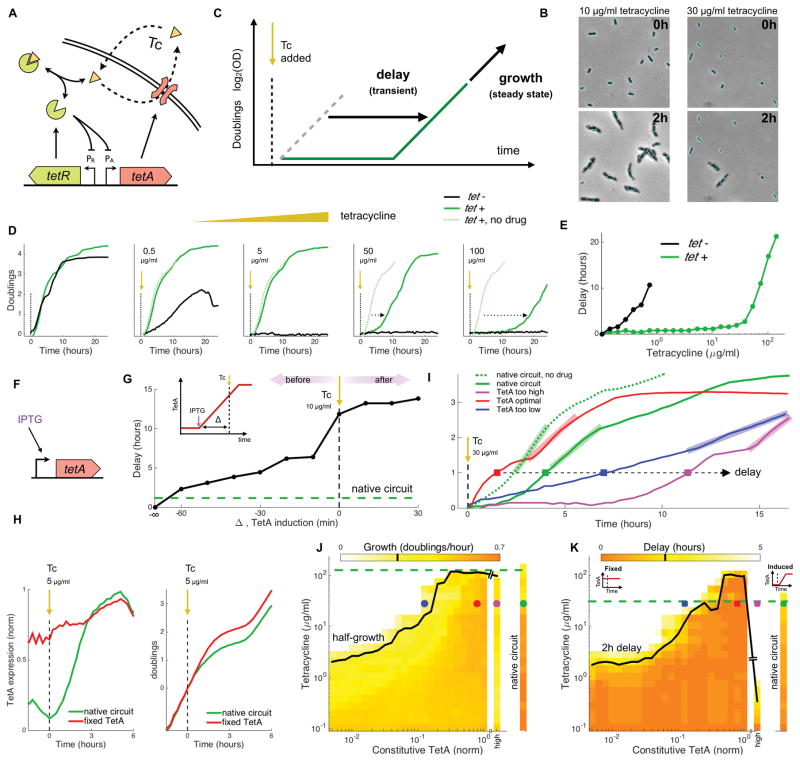
Figure 1. Optimal response to tetracycline requires fast expression of TetA with minimal overshoot.
(A) tet resistance mechanism: tetracycline (Tc), a translation inhibitor, diffuses across the cell membrane and binds transcription repressor TetR, which becomes inactive and releases expression of both TetR and TetA. Efflux pump TetA exports tetracycline out of the cell in an ion exchange that disrupts the membrane potential. (B) Imaging of cells carrying the native resistance mechanism upon exposure to 10 and 30μg/ml Tc. The contour of cells at the time of exposure is overlaid on an image taken two hours later to highlight cell growth. (C) In experiments with in liquid cultures exposed to a step increase in drug concentration, we measure population-level performance during the transient following exposure by the delay in the time taken to reach one doubling that is introduced by the addition of drug during mid-log phase. Performance in the steady state is measured by the maximum growth over one doubling at any point after exposure. (D) Growth of sensitive strains and strains carrying the native tet operon upon exposure to step increases of different tetracycline concentrations (at time zero). Dotted arrows indicate the delay in the time taken to reach one doubling in comparison to growth in the absence of drug. (E) Delays in the time taken to reach one doubling as a function of tetracycline concentration for liquid cultures of sensitive cells (black) and cells with the tet operon (green). (F) We compare the native tet circuit to synthetic strains where TetA is expressed from an inducible promoter. (G) Increasing delays in the recovery of growth in liquid cultures as TetA is synthetically induced at closer times to the exposure to 10μg/ml Tc (inset), loosing the advantage of early expression. If TetA is induced at the same time or after exposure to tetracycline, very long delays are observed. A culture of cells carrying the native circuit (green) shows a short delay in recovery, despite only inducing TetA upon exposure. (H) TetA expression and growth in cells carrying the native circuit (green) and a strain with fixed TetA expression (red) upon exposure to 5μg/ml Tc. The synthetic strain has the advantage of expressing TetA in advance. (I) Growth in liquid cultures of cells carrying the native circuit and of cells with different levels of constitutive TetA expression, measured following exposure to 30μg/ml Tc. Bars indicate growth in the steady state and squares indicate the delay in reaching one doubling following exposure. (J) Steady-state growth and (K) Delays in the recovery of liquid cultures of cells carrying the native circuit and of cells expressing a range of TetA constitutively, following exposure to a gradient of tetracycline. Tetracycline concentrations where growth is reduced in half and where delays reach 2 hours are indicated in black for constitutive TetA expression and green for the native circuit. Points obtained from the curves in (I) are also indicated. See also Figure S1.
Results
Dynamics of TetA induction is critical for cell survival upon drug exposure
The native regulation of TetA provides resistance to an abrupt increase in tetracycline up to a critical concentration, beyond which growth and survival of the cell are compromised. We focus here on experiments where tetracycline is added suddenly to a growing population. We started by examining how a population carrying the native resistance mechanism responds to such step-like increase in drug concentration. Sudden (and sustained) exposure to tetracycline during mid-log phase shows no impact on population growth for concentrations of up to 100 times larger than the minimal inhibitory concentration for wild-type cells (IC50, measured under constant conditions; Figure S1B and S1C). At even higher concentrations, growth defects start showing in the transient following exposure, as a fraction of the cells is arrested (Deris et al., 2013) (Figure 1B and 1D). At the population level, an initial pause in growth is observed, followed by recovery when a growing population rises above the background of arrested cells. The speed of recovery following exposure to tetracycline, accounting for both cell death and slow growth, can be measured as the delay in the time it takes to achieve one doubling due to the presence of drug (Figure 1C). Both the speed of recovery from the transient and the steady-state growth rate decline as a function of drug concentration (Figure 1E and S1B-S1C). We next asked how these transient and steady state behaviors depend on the regulation of TetA.
Comparing the native tet circuit to engineered strains with synthetically controlled TetA expression, we found that optimal TetA expression requires a quick response with minimal overshoot. Expression of TetA is strongly repressed in the absence of drug (Berens and Hillen, 2003), so a temporary deficiency of TetA immediately following exposure can result in high drug levels inhibiting translation, causing the arrest of cells before optimal TetA levels are reached. In order to study TetA induction, we engineered an alternative strain where TetA is expressed by a chemically inducible promoter (using lac analog isopropyl β-D-1-thiogalactopyranoside, IPTG, Figure 1F). We induced TetA expression in liquid cultures of these alternative cells at different intervals in relation to a moderate step-like increase in tetracycline concentration. The addition of drug has no effect in cells where TetA is induced from inoculation (cells carrying the native circuit, in comparison, show a small delay in recovery). However, as TetA is induced closer to the time of exposure slower recovery is observed (Figures 1G and S1E). Since growth defects can arise from the loss of early expression, cells that already constitutively express TetA prior to drug exposure have an advantage over the slower response of the native circuit, where TetA is only induced when the drug is already present (Koutsolioutsou et al., 2005, Figures 1H and S1F–S1G). In order to compare fixed TetA expression with the native regulatory circuit, we pre-treated the alternative cells to a range of IPTG prior to exposure to steps of different tetracycline concentrations, searching for an expression level that optimizes tetracycline resistance (Figures 1I and S1H). While no fixed TetA expression levels improved on the steady-state growth of the native circuit (Figure 1J), constitutive expression of optimal levels of TetA can beat the performance of the native circuit during the transient, showing faster recovery at high drug concentrations (Figure 1K). However, further increase of TetA expression using a high-copy plasmid proved to be deficient, particularly during the transient, as cells showed lower steady-state growth rates and slow recovery following exposure, consistent with high expression of TetA permeabilizing the cell membrane in the presence of the drug (Bertrand and Lenski, 1989; Eckert and Beck, 1989). Therefore, the native circuit is able to optimize TetA levels in the steady state, but cannot induce TetA fast enough during the transient to achieve the ideal step-response efficiency, where optimal levels are reached immediately. The risk of exceeding safe levels limits the ability to arbitrarily increase the speed of TetA induction, so TetR repression could play an important role in curbing TetA expression.
Increased expression of the TetR repressor improves cell survival
To understand the role of TetR expression in cell survival, we engineered a strain where TetA is expressed from its native promoter but TetR is expressed constitutively from an IPTG-inducible promoter, bypassing the negative feedback loop in its regulation (Figure 2A). Fluorescent proteins expressed from matching promoters (PA-mCherry and Plac-GFPmut3) allowed tracking of TetA and TetR expression. Liquid cultures of these cells were grown in a range of IPTG concentrations, with or without tetracycline, and expression levels were measured in steady growth far after the introduction of the drug (Figures 2B and S2A). In the absence of tetracycline, increasing TetR expression greatly decreased TetA levels; this is consistent with TetR’s biochemical function as a repressor of TetA transcription and shows that TetR regulation can provide a wide dynamic range. In this condition, the native circuit expresses just enough TetR to keep TetA levels low. In the presence of tetracycline, higher levels of TetR are needed for TetA repression, shifting the regulation curve and setting a new steady state for the native circuit expression at higher TetR and TetA levels (Figure 2B). Therefore, expression of TetR keeps TetA at very low levels prior to drug exposure. We next asked if cells with such strong TetA repression can survive an abrupt exposure to tetracycline.
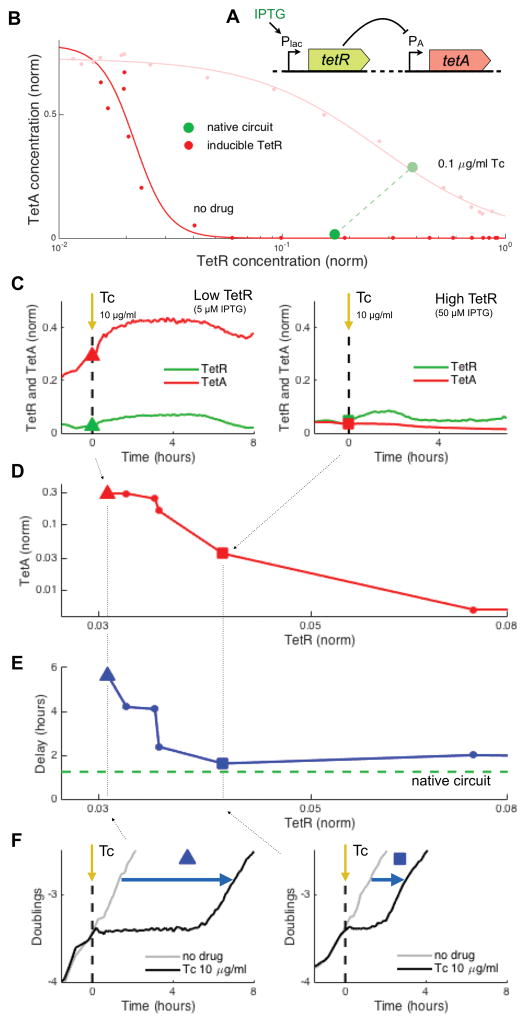
Figure 2. Increased constitutive expression of TetR shortens recovery, despite repressing TetA.
(A) We compare the native tet circuit to a synthetic strain where TetR is expressed from an inducible promoter and TetA is expressed from its native promoter. (B) Dynamic range of TetA expression provided by TetR regulation, obtained by constitutively expressing TetR at different levels, in the absence and in the presence of tetracycline. Green circles indicate the expression levels of the native circuit under the same conditions. (C) Gene expression in liquid cultures during exposure to 10μg/ml Tc, for low and high levels of constitutive expression of TetR. (D) TetA and TetR expression levels measured at the time of exposure for a range of TetR expressions. Points from (C) are indicated. (E) Delay in the recovery from drug exposure for the same cultures in (D), compared to the delay observed for the native circuit (green). (F) Growth curves for the points indicated in (C–E), showing a reduced delay in recovery for increased TetR expression. See also Figure S2.
Counter-intuitively, we find that upon a step increase in drug concentration, cells with increased TetR expression were more resilient despite their lower TetA expression. When cell populations with a range of TetR expression levels were exposed to a step increase in tetracycline, populations with higher TetR expression recovered growth earlier, despite decreased levels of TetA (Figures 2C–2F and S2B). In the absence of TetR expression, the cell culture shows long delays in recovery, consistent with the delays observed in high constitutive TetA expression, suggesting that the native TetA promoter can lead to dangerously high expression levels when left unsuppressed. Synthetically increasing TetR expression reduces the expression level of TetA, which improves cell survival upon encountering the antibiotic. Although high TetR expression could result in increased delays in TetA induction (Bertrand et al., 1984), this deficit is at least partially mitigated by excess TetR binding and sequestering drug molecules (Figure S2C). The advantage conferred to cells by TetR, through both drug sequestration and avoiding dangerously high TetA levels, suggests that the fate of individual cells upon drug exposure may result from cell-to-cell differences in its expression.
The rate of TetR elevation upon drug exposure dictates single-cell fates
Tracking of individual cells upon drug exposure showed three distinct growth and expression patterns: Recovered, Moribund and Arrested (R, M and A; Figure 3B). To follow physiological changes in single cells responding to a step increase in drug concentrations (Rosenfeld et al., 2005), we developed a microfluidic device that incorporates mother-machine single cell trapping (Wang et al., 2010) with on-chip valving for fast media switching (Figure 3A). We tracked the growth of individual cells carrying the native tet operon and a two-color reporter plasmid indicating TetR and TetA expression (PR-GFPmut3 and PA-mCherry, Figure S3). Following an abrupt exposure to tetracycline at the IC50 concentration, some of the cells returned to growth (Recovered) while others eventually stopped growing (Figure 3C–3E). There were two distinct subgroups among cells that stopped growing: while some cells arrested growth shortly after exposure with little expression (Arrested), others stopped growing only after an initial slow growth phase showing very high TetA expression levels (Moribund). Although ultimately cells either live or die, the dynamical response reveals trajectories to three distinct fates.
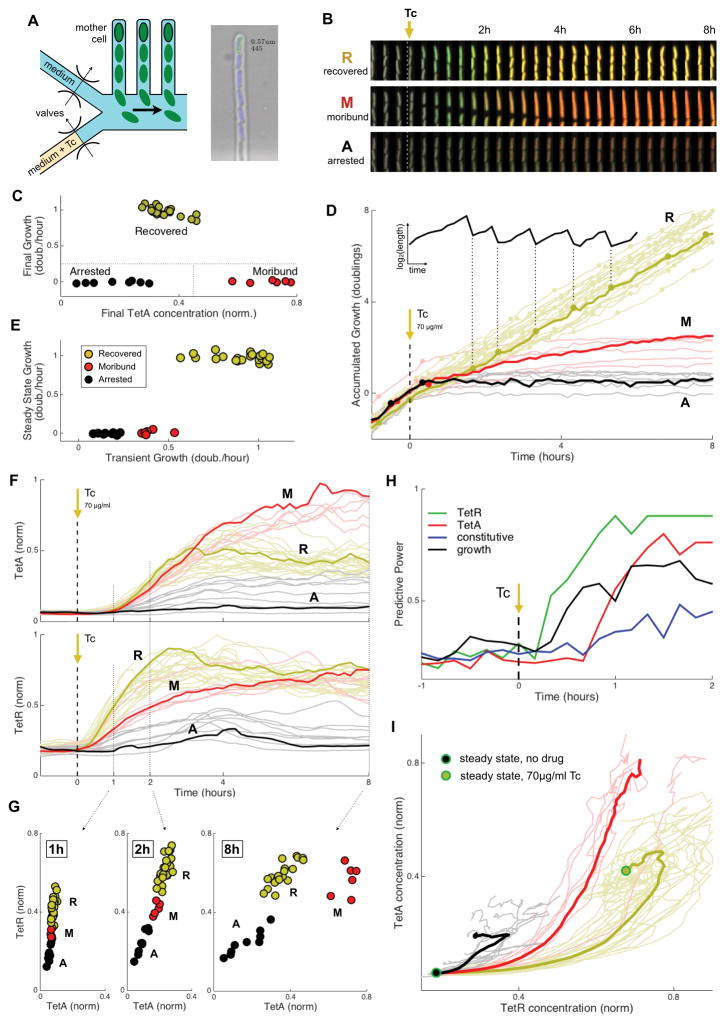
Figure 3. Abrupt exposure of single-cells to tetracycline shows three distinct fates: Recovered, Moribund and Arrested.
(A) Design of the microfluidic device, which places single cells in fixed locations as they go through division cycles. A valving layer allows on-chip rapid switching of media. Right: analysis of a microscopy image obtained from a channel. Cells carrying the native resistance mechanism with fluorescent reporters were exposed to a step increase of 70μg/ml Tc (IC50). (B) Time course of individual cells representing the three different cell fates observed: Recovered, Moribund and Arrested. (C) Cell fates were defined according to final growth rate and final TetA expression, with cells clustering into three very distinct groups. (D) Accumulated growth in cell length (shown in inset) for all cells observed, colored by cell fate and with division events indicated. (E) Average growth rates during transient (first 4h after exposure) and steady state (6h to 10h), showing three distinct growth patterns. (F) Expression of TetA (top) and TetR (bottom), with cell fate indicated by color. (G) TetR and TetA concentrations at three time points after exposure to tetracycline, showing the distinct expression pattern of each cell fate. (H) Predictive power in determining cell fate during early dynamics for growth rate, versus expression of TetR, TetA or a control constitutive promoter. TetR expression predicts cell fate within the first hour following exposure, before predictions from growth (TetR predictability may even precede TetA, though this measurement may be influenced by the different maturation times of the corresponding fluorescent proteins, 6.5 versus 15 minutes, respectively; Megerle et al., 2008; Shaner et al., 2004). (I) Expression trajectories in the TetR and TetA concentration space following drug exposure, departing from the no-drug steady state and colored by cell fate. Bold lines indicate the average trajectory of each cell fate. See also Figure S3.
We next asked whether these different cell fates were dictated by the expression of either TetR or TetA at the onset of the drug. We followed the fluorescent reporters for TetR and TetA in individual cells during the dynamical response and separated the cells according to their fate (Figure 3F). Recovered cells showed the fastest TetR expression, leading to intermediate levels of TetA. Moribund cells showed slower expression of TetR, resulting in high expression of TetA. Arrested cells stopped growing quickly, before any significant expression of either protein (Figure S3). Proper gene regulation is important in this dynamical context; recovered cells follow a particular trajectory to the final steady state after drug exposure, while Moribund and Arrested cells are diverted and fail to reach the optimized expression (Figure 3I). To understand whether early gene expression can predict cell fate, we consider the fate of the cells as a function of the expression of TetR and TetA at different times following drug exposure (Figure 3G). Within the first two hours cells can already be separated by fate according to their TetR expression. Quantitatively, we used an information gain approach to calculate the prediction power of gene expression in the outcome of individual cells (Ross Quinlan, 1993). Cell fates can be reliably predicted from TetR expression already within the first hour of the response, while cells are still growing (Figure 3H). In fact, at these early times, TetR expression is even a better predictor of cell fate than the growth rate itself. While TetA is ultimately responsible for drug export, high expression of its repressor TetR at the onset of drug response dictates individual cell survival.
Surge of TetR expression upon drug exposure: while low TetR levels release TetA expression quickly, high TetR levels are required for its timely repression
To understand the dependence of cell fate on TetR expression, we considered a simple dynamical model of drug concentration and gene regulation. Our model describes the accumulation of intracellular tetracycline due to diffusion into the cell, export by TetA and dilution due to cell growth (Figure 4A). Within the cell, TetR binds tetracycline in chemical equilibrium, thus sequestering and thereby inactivating a fraction of the intracellular drug. We assume that TetR and TetA are expressed upon drug exposure at fixed rates αR and αA, until the concentration of free (unbound) TetR crosses above a given threshold, at which point it turns off TetA expression (here we consider an on/off regulation for a simpler illustration, but a more gradual regulation yields qualitatively similar results; Figure S4A). Motivated by our experimental results, we assume that cells can die not only from excess of intracellular tetracycline, but also from expressing too high levels of TetA during response to the drug (Arrested and Moribund, Figure 4B). For a range of TetR and TetA expression rates, we simulated the model in a range of external tetracycline concentrations and defined resistance as the maximal concentration for which the concentrations of neither intracellular free tetracycline nor TetA exceed their limits (Figure 4C). For a given TetR expression rate, there is an optimal TetA expression that maximizes resistance (Figure 4C, white square). Assuming that the native circuit operates with optimized TetA expression, we find that even small changes in TetR expression can lead to the three different fates: For the given TetR expression rate, cells will survive (Recovered), but slightly reducing TetR expression fails to shut down TetA expression in time, causing death due to high expression levels (Moribund). Reducing expression even further diminishes tetracycline sequestration by TetR, causing death from excess of the drug (Arrested; Figures 3F, and 4D). The model thus helps explain the dependence of cell fate on TetR expression, as seen in the single cell measurements.
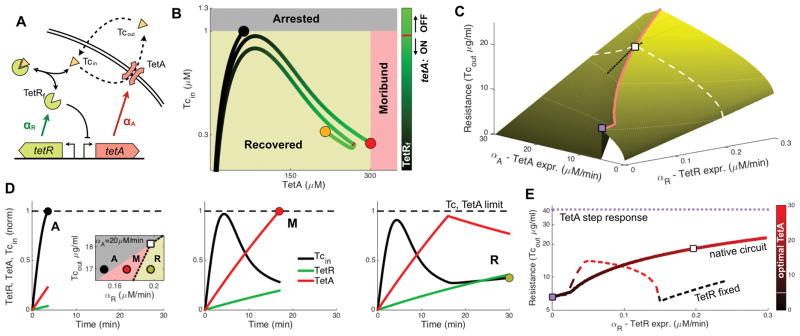
Figure 4. Mathematical model shows that fast expression of TetR is required for timely induction and repression of TetA.
(A) Mathematical model tracking intracellular drug concentration, taking into account diffusion of tetracycline across the membrane, dilution due to growth, export of tetracycline by TetA and binding of tetracycline and TetR. We model expression in the native circuit by both TetR and TetA being induced upon exposure, with TetA being repressed when the concentration of free (unbound) TetR crosses a given threshold (10nM). (B) Trajectories for different TetR expression rates in the space of TetA and free intracellular tetracycline concentrations, showing the upper limits on both TetA and tetracycline. (C) Maximum tetracycline concentration resisted by the native circuit for different values of TetR and TetA expressions. Each given TetR expression rate is optimized by a corresponding TetA expression rate (red line). The white square indicates the optimal TetA expression for a fixed TetR expression rate (white dashed line). The purple square shows the maximal resistance obtained with TetA induced in the absence of regulation by TetR. The black line shows the region depicted in the inset in (D). (D) Inset: cell fates for different values of Tcout and TetR expression. All three fates are possible within a small range of TetR expression. Time courses of intracellular concentrations of TetR, TetA and Tcin for the different points in the inset, showing the three cell fates observed experimentally. Tcin and TetA are normalized by their upper limits, and TetR is normalized by its optimized steady state concentration. (E) Resistance with optimized TetA expression as a function of TetR expression. The purple line indicates the ideal scenario of a step response (or constitutive expression) of TetA at its limit, while the purple square indicates the resistance obtained when TetA is induced upon exposure a the rate resulting in expression at the limit (maximal constitutive rate). The dashed line represents resistance when TetR is expressed constitutively instead of being induced. See also Figure S4.
The model also explains the limitations of constitutive TetR and TetA expressions, and shows that the native tet circuit increases the speed of the response by using a dynamic regulation where low levels of TetR regulate the induction of TetA and subsequently high levels regulate its repression. Ideally the cell would respond to drug exposure with a step increase in the concentration of TetA to optimal values (Figure 4E, step-response). To approach that theoretical limit, cells need to express TetA fast, yet in the absence of TetR repression this strong expression would overshoot above the maximal tolerated limit (Figure 4, maximum constitutive TetA expression). If fixed expression of TetR is used to provide this repression, a cell can withstand higher rates of TetA expression, and drug resistance increases with TetR expression initially (Figure 4E and S4B–S4C). However, if TetR levels are too high upon exposure, TetA initiation is delayed, decreasing resistance. Therefore, low TetR levels are needed at the time of exposure for sensitivity to the presence of the drug and quick initiation of TetA, while high levels of TetR are important later for timely repression of TetA (and possibly for increasing resistance through drug sequestration; Figure S4D). The native circuit enjoys both of these benefits: it triggers TetA when TetR concentration is low, but it then quickly expresses TetR to be able to repress TetA in time to avoid expression above the limit. This strategy allows prompt expression of TetA at much faster rates than in the absence of regulation, resulting in higher resistance (Figure 4E).
Discussion
The dynamical behavior of antibiotic resistance mechanisms is critical to cell fate. The tet operon has evolved sophisticated regulation, with multiple overlapping promoters and operators, to optimize not only steady state concentrations, but also the dynamics of the response. In this dynamical context, the repressor of the system, TetR, has a critical role both in releasing expression of TetA upon exposure and in timing a window for strong expression. Fluctuations in the rate of TetR expression can therefore dictate ultimate cell fate. Fast TetR expression provides a strong feedback that increases the responsiveness of the tet operon, a system where both under- and overdamping of the response can cause the arrest of cell growth. It will be interesting to see if this circuit design functions in other resistance genes and organisms where cell survival depends on its ability to respond promptly and accurately to the drug (Miyashiro and Goulian, 2007; Shin et al., 2006). Multiple regulated resistance mechanisms share the structure of the tet operon of a divergently expressed repressor that also regulates itself (Grkovic et al., 2006; Li et al., 2015). Other systems use alternative architectures to achieve fast responses, such as the mar operon in E. coli, which combines a positive and a negative feedback loop in its regulation (Rodrigo et al., 2016). Following single-cell dynamics of these systems during the transients following drug exposure can help uncover specific vulnerabilities of cellular responses to antibiotics.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to Lead Contact Roy Kishony at the Technion - Israel Institute of Technology (rkishony@technion.ac.il).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Media, drugs and strains
All experiments were conducted in M63 minimal medium (2g/l (NH4)2SO4, 13.6g/l KH2PO4, 0.5mg/l FeSO4·7H2O) supplemented with 0.2% glucose, 0.01% casamino acids, 1mM MgSO4 and 1.5mM thiamine. Tetracycline and IPTG solutions were freshly made from powder stocks (Sigma) and filter-sterilized before each experiment. All strains were derived from E. coli K-12 strain MG1655 rph+ ΔLacIZYA. For assays with inducible TetA expression at higher levels, TetA was PCR-amplified with a proofreading polymerase (phusion), cloned into pZS*32 (low copy) and ASKA (high copy) vectors by Gibson Assembly (NEB), sequenced and transformed using a TSS protocol. Chromosomal inducible TetA expression (used in Figures 1G and S1F–S1G for comparison of expression levels), the native tet resistance mechanism from the Tn10 transposon and its pLac-TetR variation were ordered from Genewiz in a pIT3-CH integrating plasmid and integrated in the chromosome at site HKO22. Matching fluorescent reporters (GFPmut3 and mCherry), plus CFP in a constitutive promoter, were also ordered from Genewiz in a pZS1 plasmid and transformed using TSS. For the microfluidic assay, cell motility was hindered by a deletion of flagellum motor gene motA, introduced by P1 transduction.
METHOD DETAILS
Growth rate assays
Overnight cultures were diluted 5000-fold and grown on an automated robotic system (Hudson) at 30°C with rapid shaking in 96-well microtiter plates (Costar) containing 150μl medium per well. Optical density (OD, absorbance at 600nm) and fluorescence (when needed) were recorded by a plate reader (EnVision, Perkin-Elmer) at short intervals for at least 24h, with background subtracted. Two-dimensional concentration gradients of tetracycline and IPTG were set up over multiple 96-well plates. Tetracycline was added either when OD was first detected above background or in the middle of log-phase, when a reading of fluorescence was needed. Maximum growth rates were calculated using Matlab by linear regression of log2(OD) over one doubling during exponential growth. Gene expression in liquid culture assays was defined as fluorescence/OD. TetA expression from plasmids was estimated by measuring fluorescence from a cell expressing YFP from the same vector, grown across the same gradient of IPTG. All relevant data was included in this study and made available online (Mendeley Data).
Single-cell imaging
An overnight culture was diluted 100-fold and incubated at 37°C for 2h (in IPTG if appropriate). Then tetracycline was added and 5μl of culture was applied onto a 1.5% low-melting agarose pad (Sigma) containing final concentrations of both IPTG and tetracycline. The agarose pad was then placed onto a coverslip slide (VWR) for imaging. Images were obtained at 30°C with a Nikon Ti inverted microscope with an incubation chamber and a Hamamatsu Orca-ER camera, an automated stage (Prior), and a Lumencor SOLA fluorescent illumination system.
Single-cell assay in the microfluidic device
E. coli cells were grown for 2h from an overnight culture and injected into the feeding channels of the device. The chip was mounted on a custom-machined platform and cells were forced into the cell channels by centrifugation. The microfluidic device was mounted on the same microscope described above immediately after loading. Image acquisition was performed using Nikon NIS-Elements software. Exposures were done at very low illumination intensities with 4×4 binning. Cells were allowed to equilibrate in the device for several hours before imaging.
Growth and expression in single cells
We followed 40 cells containing the native tet resistance mechanism in the microfluidic device, recording cell sizes and gene expression every 10min over 15h. Cells were grown in M63 medium until growth was stabilized, then exposed to 70μg/ml of tetracycline. Gene expression was measured using a plasmid expressing fluorescent reporters GFPmut3 (6.5min maturation time) and mCherry (15min maturation time) from the native promoters of TetR and TetA, respectively. All data analysis was based on a custom Matlab image-processing pipeline. For each image, the top cell in each channel was identified and their length and mean fluorescence intensity was calculated. Cell division events were identified by looking for instances where a cell’s length dropped to less than 60% of its previous value. Growth rate was measured by taking the derivative of log2(cell length) (Figure S3).
Microfluidic device fabrication
The device consists of a flow layer and a control layer, placed about 50μm above and used for valving. When pressure is applied to a channel in the control layers it expands, collapsing the flow channels below and stopping its flow. The master molds for these two layers were fabricated by ultraviolet photolithography using standard methods. Shipley or SU-8 (Microchem) photoresist was applied to a silicon wafer by spin coating to appropriate thickness (corresponding to the channel height) and patterns were then created by exposing the uncured photoresist to ultraviolet light through custom quartz masks (Toppan). Dimethyl siloxane monomer (Sylgard 184) with 1:20 curing agent was spin coated onto the flow layer, while the control layer was molded using dimethyl siloxane monomer with 1:7 curing agent. After baking both layers at 65°C for 35min, the two layers were aligned and baked together overnight at 65°C for bonding (due to the difference in curing agent ratio). The following day holes were introduced using a biopsy punch to connect the feeding channels to the external tubing, and individual chips were cut and bonded onto KOH-cleaned cover slips using oxygen plasma treatment. Media was pumped through the device using a custom-built system that uses air pressure to push media through the tubing: a control board (SwitchAndSense) controls airflow through solenoid valves, applying pressure both to the media containers and the valving layer of the device.
Mathematical model for intracellular drug accumulation
The flux of a drug freely diffusing through a membrane with a constant Ki is Ki (X – x), where X and x are the extracellular and intracellular concentrations of drug, respectively. If the extracellular drug concentration is fixed, a cell with a constant growth rate λ will accumulate intracellular drug as
The action of efflux pump TetA is modeled using Michaellis-Menten kinetics, where a(t) is TetA concentration and KA and KA are the maximum rate and Michaellis constant associated with TetA. Binding of TetR r(t) and tetracycline x(t) is considered in equilibrium rf + xf ⇌ c with a dissociation constant Kb (subscript f denotes free form, and c is the inactive complex formed by TetR and tetracycline, r = rf + c and x = xf + c ). Since only unbound tetracycline can diffuse or be exported out of the cell, the equation describing the accumulation of total intracellular tetracycline is
with x = xf [1 + r / (Kb + xf)] obtained from the equilibrium. Given TetR and TetA expressions r(t) and a(t) (fixed rates with binary repression of TetA, as described in the main text) we numerically integrate the solution x(t) in discrete time steps for a given concentration of extracellular tetracycline. We simulated the system only up to the point where the cell has reversed the influx of drug and subsequently shut down expression of TetA, since longer simulations would relate to the steady state and would depend on the self-regulation of TetR, absent in the model. We chose a threshold of 10nM of free TetR for the repression of TetA, which affects the extent of sequestration of tetracycline by TetR (Figure S4). The constants used in the numerical simulation were estimated from data or obtained from the literature (McMurry et al., 1980; Sigler et al., 2000; Takahashi et al., 1986; Table S1).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative analysis as reported in the results and figure legends were calculated with custom scripts written in MATLAB.
Predictive power of an attribute
At the end of an experiment with N cells the outcomes are divided into N1 recovered cells, N2 moribund cells and N3 arrested cells (N1 + N2 + N3 = N), and the total information entropy of this system is
At time t, for given values xi and xj of an attribute x of the cell (TetR expression, for instance), we divide the population into three subpopulations: NA cells where x ≤ xi, NB cells where xi < x ≤ xj and NC cells where x > xj. Considering the final outcome, the information entropy within each subpopulation at time t is
The information gain of splitting of the original population at xi and xj is then defined as
If each subpopulation at time t shows roughly the same distribution of outcomes as the final population, then SA ≈ SB ≈ SC ≈ S0 and G ≈ 0. However, if the population at time t is split along xi and xj values that separate between recovered, moribund and arrested cells, then SA ≈ SB ≈ SC ≈ 0 and G ≈ 1. We define the predictive power of attribute x at time t as the gain obtained from the best split of the population, at xi and xj values that maximize G.
DATA AND SOFTWARE AVAILABILITY
Software
MATLAB code used for analysis is available upon request.
Data Resources
Growth curves of the liquid-culture assays are available as supplemental data file Data S1. Imaging files of the single-cell assay in the microfluidic device are available as supplemental data file Data S2. Both files can be accessed in Mendeley Data: https://data.mendeley.com/datasets/fbwd37ynp8/draft?a=e505a88d-b8f3-4100-a9b0-67f3567ad37b.
Supplementary Material
Related to Figure 3
Cells carrying the native tet resistance mechanism were exposed to a step increase of 70μg/ml tetracycline at time zero. Fluorescent reporters with matching promoters were used to measure expression of efflux pump TetA (red, mCherry) and its repressor TetR (green, GFP) during exposure. Imaging was done in a microfluidic device that places single cells in fixed locations as they go through division cycles. A valving layer allows on-chip rapid switching of media.
Acknowledgments
We thank N. Lord and R. Chait for help with designing the microfluidic device. We thank U. Alon for comments and insights. R.K. acknowledges the support of the European Research Council Seventh Framework Programme (ERC grant no. 281891), the National Institutes of Health (grant no. R01GM081617) and the Israeli Centers of Research Excellence I-CORE Program (ISF grant no. 152/11).
Footnotes
Author Contributions
All authors designed the research. D.S. constructed the microfluidic device, performed the experiments and developed the mathematical model. All authors analyzed the data, D.S. and R.K. wrote the paper.
References
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Batchelor E, Silhavy TJ, Goulian M. Continuous Control in Bacterial Regulatory Circuits. J Bacteriol. 2004;186:7618–7625. doi: 10.1128/JB.186.22.7618-7625.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens C, Hillen W. Gene regulation by tetracyclines. Eur J Biochem. 2003;270:3109–3121. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- Bertrand KP, Lenski RE. Effects of Carriage and Expression of the Tn10 Tetracycline-Resistance Operon on the Fitness of Escherichia coli K12. Mol Biol Evol. 1989;6:213–225. doi: 10.1093/oxfordjournals.molbev.a040545. [DOI] [PubMed] [Google Scholar]
- Bertrand KP, Postle K, Wray LV, Reznikoff WS. Construction of a Single-Copy Promoter Vector and Its Use in Analysis of Regulation of the Transposon Tn10 Tetracycline Resistance Determinant. J Bacteriol. 1984;158:910–919. doi: 10.1128/jb.158.3.910-919.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camas FM, Blázquez J, Poyatos JF. Autogenous and nonautogenous control of response. Proc Natl Acad Sci. 2006;103:12718–12723. doi: 10.1073/pnas.0602119103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. Modes and Modulations of Antibiotic Resistance Gene Expression. Clin Microbiol Rev. 2007;20:79–114. doi: 10.1128/CMR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deris JB, Kim M, Zhang Z, Okano H, Hermsen R, Groisman A, Hwa T. The Innate Growth Bistability and Fitness Landscapes of Antibiotic Resistant Bacteria. Science. 2013;342:1237435. doi: 10.1126/science.1237435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert B, Beck CF. Overproduction of Transposon Tn10-Encoded Tetracycline Resistance Protein Results in Cell Death and Loss of Membrane Potential. J Bacteriol. 1989;171:3557–3559. doi: 10.1128/jb.171.6.3557-3559.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L, Hancock REW. Adaptive and Mutational Resistance: Role of Porins and Efflux Pumps in Drug Resistance. Clin Microbiol Rev. 2012;25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foglierini M, Serrano L, Dublanche Y, Michalodimitrakis K, Ku N. Noise in transcription negative feedback loops: simulation and experimental analysis. Mol Syst Biol. 2006:1–12. doi: 10.1038/msb4100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grkovic S, Brown MH, Skurray RA, Repressor A, Subtilis B. Regulation of Bacterial Drug Export Systems. Microbiol Mol Biol Rev. 2006;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guet C, Dittmar K, Emonet T, Pan T, Cluzel P, Le TT. Real-time RNA profiling within a single bacterium. Proc Natl Acad Sci. 2005;102:9160–9164. doi: 10.1073/pnas.0503311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsolioutsou A, Peña-Llopis S, Demple B. Constitutive soxR Mutations Contribute to Multiple-Antibiotic Resistance in Clinical Escherichia coli Isolates. Antimicrob Agents Chemoterapy. 2005;49:2746–2752. doi: 10.1128/AAC.49.7.2746-2752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Emonet T, Harlepp S, Guet C, Cluzel P. Dynamical Determinants of Drug-Inducible Gene Expression in a Single Bacterium. Biophys J. 2006;90:3315–3321. doi: 10.1529/biophysj.105.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer T, Takahashi M, Hillen W. Thermodynamic Analysis of Tetracycline-Mediated Induction of Tet Repressor by a Quantitative Methylation Protection Assay. Anal Biochem. 1995;196:190–196. doi: 10.1006/abio.1995.0006. [DOI] [PubMed] [Google Scholar]
- Li X, Plésiat P, Nikaido H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin Microbiol Rev. 2015;28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madar D, Dekel E, Bren A, Alon U. Negative auto-regulation increases the input dynamic-range of the arabinose system of Escherichia coli. BMC Syst Biol. 2011;5:111. doi: 10.1186/1752-0509-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L, Petrucci RE, Levy SB. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megerle JA, Fritz G, Gerland U, Jung K, Ra JO. Timing and Dynamics of Single Cell Gene Expression in the Arabinose Utilization System. Biophys J. 2008;95 doi: 10.1529/biophysj.107.127191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Wray LV, Hillen W. Differential regulation of the resistance genes tetA and tetR by the tandem tet operators O1 and O2. EMBO J. 1988;7:567–572. doi: 10.1002/j.1460-2075.1988.tb02846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro T, Goulian M. Stimulus-dependent differential regulation in the Escherichia coli PhoQ – PhoP system. Proc Natl Acad Sci. 2007;104:16305–16310. doi: 10.1073/pnas.0700025104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan A, Kandhavelu M, Lloyd-price J, Kudasov F, Chowdhury S, Yli-harja O, Ribeiro AS. Dynamics of transcription driven by the tetA promoter, one event at a time, in live Escherichia coli cells. Nucleic Acids Res. 2012;40:8472–8483. doi: 10.1093/nar/gks583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevozhay D, Adams RM, Murphy KF. Negative autoregulation linearizes the dose – response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci. 2009;106:5123–5128. doi: 10.1073/pnas.0809901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AC, Angelino E, Kishony R. Chemical decay of an antibiotic inverts selection for resistance. Nat Chem Biol. 2010;6:105–107. doi: 10.1038/nchembio.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Tera W, Brennan R, Tobes R. The TetR Family of Transcriptional Repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo G, Bajic D, Elola I, Poyatos JF. Antagonistic autoregulation speeds up a homogeneous response in Escherichia coli. Sci Rep. 2016:1–10. doi: 10.1038/srep36196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U. Negative Autoregulation Speeds the Response Times of Transcription Networks. J Mol Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene Regulation at the Single-Cell Level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Ross Quinlan J. C4.5: Programs for Machine Learning. San Mateo, CA: Morgan Kaufmann; 1993. [Google Scholar]
- Savageau MA. Quantitative Development of the Theory. 1998. Demand Theory of Gene Regulation. I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shin D, Lee E, Huang H, Groisman EA. A Positive Feedback Loop Promotes Transcription Surge That Jump-Starts Salmonella Virulence Circuit. Science. 2006;314:1607–1609. doi: 10.1126/science.1134930. [DOI] [PubMed] [Google Scholar]
- Sigler A, Schubert P, Hillen W, Niederweis M. Permeation of tetracyclines through membranes of liposomes and Escherichia coli. Eur J Biochem. 2000;267:527–534. doi: 10.1046/j.1432-1327.2000.01026.x. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Altschmied L, Hillen W. Kinetic and Equilibrium Characterization of the Tet Repressor-Tetracycline complex by Fluorescence Measurements. J Mol Biol. 1986;187:341–348. doi: 10.1016/0022-2836(86)90437-7. [DOI] [PubMed] [Google Scholar]
- Wall ME, Hlavacek WS, Savageau MA. Design of Gene Circuits: Lessons from Bacteria. Nat Rev Genet. 2004;5:34–42. doi: 10.1038/nrg1244. [DOI] [PubMed] [Google Scholar]
- Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A. Robust Growth of Escherichia coli. Curr Biol. 2010;20:1099–1103. doi: 10.1016/j.cub.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Rao CV. The role of configuration and coupling in autoregulatory gene circuits. Mol Microbiol. 2010;75:513–527. doi: 10.1111/j.1365-2958.2009.07011.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 3
Cells carrying the native tet resistance mechanism were exposed to a step increase of 70μg/ml tetracycline at time zero. Fluorescent reporters with matching promoters were used to measure expression of efflux pump TetA (red, mCherry) and its repressor TetR (green, GFP) during exposure. Imaging was done in a microfluidic device that places single cells in fixed locations as they go through division cycles. A valving layer allows on-chip rapid switching of media.