Abstract
An outbreak of multi-drug resistant (MDR) tuberculosis (TB) has been reported on Daru Island, Papua New Guinea. Mycobacterium tuberculosis strains driving this outbreak and the temporal accrual of drug resistance mutations have not been described. Whole genome sequencing of 100 of 165 clinical isolates referred from Daru General Hospital to the Supranational reference laboratory, Brisbane, during 2012–2015 revealed that 95 belonged to a single modern Beijing sub-lineage strain. Molecular dating suggested acquisition of streptomycin and isoniazid resistance in the 1960s, with potentially enhanced virulence mediated by an mycP1 mutation. The Beijing sub-lineage strain demonstrated a high degree of co-resistance between isoniazid and ethionamide (80/95; 84.2 %) attributed to an inhA promoter mutation combined with inhA and ndh coding mutations. Multi-drug resistance, observed in 78/95 samples, emerged with the acquisition of a typical rpoB mutation together with a compensatory rpoC mutation in the 1980s. There was independent acquisition of fluoroquinolone and aminoglycoside resistance, and evidence of local transmission of extensively drug resistant (XDR) strains from 2009. These findings underline the importance of whole genome sequencing in informing an effective public health response to MDR/XDR TB.
Keywords: genomics, tuberculosis, mycobacteria, evolution, drug resistance
Data Summary
1. The Illumina sequencing reads generated and analysed during the current study are available in the NCBI under the project file number PRJNA385247, https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA385247
2. All scripts and command lines used for generation of structural variants are available, https://github.com/arnoldbaino/Daru_scripts
3. The file used for molecular clock analysis is available, https://figshare.com/s/73cfe6062e657419e084
Impact Statement.
The Papua New Guinea government declared a multi-drug-resistant (MDR) tuberculosis (TB) emergency on Daru Island following excessive case numbers in recent years. Clinical experience suggested that most cases were epidemiologically linked, but to date limited molecular epidemiology studies have been performed. We utilized whole genome sequencing (WGS) to refine our understanding of the epidemiology and acquisition of drug resistance. We confirmed that the MDR TB outbreak on Daru Island is being driven by transmission of a modern Beijing sub-lineage strain. Molecular clock analysis of the dominant strain identified a long evolutionary history with four circulating clades. Clade C was dominant and included the majority of MDR strains, as well as a small cluster of recently evolved extensively drug-resistant (XDR) strains. Detailed characterization of resistance-conferring mutations provided novel insight into the underlying mechanisms of drug resistance and helped to elucidate putative compensatory or virulence genes. The insight gained by WGS provides added urgency to contain the spread of MDR and XDR TB transmission on Daru Island and beyond. It emphasized the need for early accurate diagnosis and increased access to new agents and regimens to improve patient outcomes.
Introduction
Globally, an estimated 10.4 million cases of tuberculosis (TB) occurred in 2015 and TB caused by Mycobacterium tuberculosis (MTB) was the leading cause of death from a single infectious agent [1]. The emergence and spread of drug-resistant MTB strains pose a major challenge to global TB prevention efforts [2]. Multi-drug-resistant (MDR) TB, which is resistant to at least isoniazid and rifampicin, accounted for an estimated 480 000 new cases and 250 000 deaths in 2015 [1]. Additional resistance to fluoroquinolones and second-line injectables defines extensively drug-resistant (XDR) TB [3]. In MTB, drug resistance occurs mainly due to the accumulation of chromosomal resistance-conferring mutations without evidence of lateral gene transfer [4]. The emergence of drug resistance is dependent on the rate of acquisition of resistance-conferring mutations and the frequency with which these drug-resistant strains are transmitted, especially in communities with inadequate treatment or poor adherence to treatment [5, 6]. Compensatory mutations that limit the fitness cost imposed by drug resistance may enhance the clonal spread of the most successful drug-resistant strains [6–8].
Genotyping of MTB using the 24-locus mycobacterial interspersed repetitive unit (MIRU-24) remains widely used as a primary approach to strain characterization [9]. However, MIRU-24 interrogates small genomic regions that are susceptible to homoplasy and offers sub-optimal discriminatory power [10]. The discriminatory power of MIRU-24 is of particular concern with Beijing lineage strains [11]. Whole genome sequencing (WGS) offers optimal resolution to explore local transmission dynamics while also revealing the molecular mechanisms and evolution of drug resistance [6, 12, 13].
Papua New Guinea (PNG) has an estimated TB incidence of 432 per 100 000 population [1]. However, this reported incidence may be an underestimate because most rural areas are isolated with limited access to proper health services, and there are inaccuracies in census data and variations in population growth [14]. Daru Island (Fig. 1) is a 14.7-km2 island with a population of 16 714 [15, 16] and is one of the major hotspots for TB outbreaks [17]. The case notification rate in 2016 was estimated to be 2901 per 100 000 population (based on 485 cases notified) [15]. A recent national MDR TB survey showed that Daru Island had the highest concentration of MDR TB cases in PNG [17], with estimates of 0.76 % (125 cases) of the population diagnosed with drug-resistant TB in 2016 [15]. However, the MTB strains driving the epidemic on Daru Island, as well as the associated drug resistance mutations and temporal accrual of these mutations have not been described. Surveillance of underlying drug resistance mutations, in addition to routine phenotypic susceptibility testing, is important for optimizing TB treatment. In particular, it will help caution the appropriate use of the latest as well as re-purposed antibiotics to maximize their effectiveness. Understanding the evolution of drug resistance mutations is important for evaluating the strengths and weaknesses of public health responses.
Fig. 1.
Map of Papua New Guinea illustrating the study location of Daru Island (inset). Daru Town is the capital of South Fly district, Western Province of Papua New Guinea. The original map was obtained fromhttp://d-maps.com
Methods
We performed a retrospective assessment of all clinical isolates referred from Daru General Hospital to the Supra-National Referral Laboratory (SRL) in Brisbane, Australia, over a 29.5-month period from 1 October 2012 to 15 March 2015, which had a positive M. tuberculosis culture. Clinical specimens are routinely referred to SRL for culture and drug sensitivity testing. The criterion for referral is detection of rifampicin resistance by Xpert MTB/RIF assay (Cephid) on concurrent samples or at the individual discretion of a clinician.
MIRU-24 and WGS were performed to understand the genetic diversity of circulating strains, reveal the mutations associated with drug resistance and explore the evolution of these mutations.
Study setting and specimen selection
Daru Town, located on Daru Island, is the provincial capital of PNG’s Western Province and has an estimated population of around 16 714 [16]. Daru General Hospital provides healthcare services to residents of Daru Island and the surrounding mainland communities in South Fly District.
Study procedures
DNA was extracted from cultures with confluent growth on Löwenstien-Jensen (LJ) slopes using a Spin column High pure PCR template prep kit (Roche Diagnostics). MIRU-24 genotyping was performed as per standard protocols [18], and GeneMapper version 4.0 (Applied Biosystems) was used for fragment analysis.
Phenotypic drug susceptibility to the first-line drugs rifampicin (1.0 µg ml−1), isoniazid (0.1 µg ml−1, low-level; 0.4 µg ml−1, high-level), streptomycin (1.0 µg ml−1), ethambutol (5.0 µg ml−1) and pyrazinamide (100 µg ml−1) was performed based on the proportion method using the automated Bactec Mycobacterial Growth Indicator Tube (MGIT) 960 system (Becton Dickinson). For MDR TB isolates, susceptibility to the second-line drugs amikacin (1.0 µg ml−1), capreomycin (2.5 µg ml−1), kanamycin (2.5 µg ml−1), ethionamide (5.0 µg ml−1), ofloxacin (2.0 µg ml−1), p-aminosalicylic acid (4.0 µg ml−1) and cycloserine (50 µg ml−1) was determined using the MGIT system [19, 20].
DNA for WGS was isolated using a lysozyme-phenol chloroform-based method and DNA was purified using a QIAamp DNA mini prep kit (Qiagen). Paired end libraries were prepared using an Illumina Nextera XT DNA library preparation kit and sequenced using the Illumina MiSeq sequencing platform at Westmead Institute for Medical Research and Australia Genome Research Facility (Sydney, Australia). Raw reads were used to derive octal codes using in silico SpolPred [21] and spoligotypes were inferred using the international database (SpolDB4) [22]. Paired end reads were checked for quality using FastQC v0.11.2 [23], and trimmomatic v0.27 [24] was used to remove low-quality base pairs (Phred score <30) especially at 3′ ends. Trimmed reads were mapped to the H37Rv reference genome (GenBank: NC_000962.3) using BWA-mem with default settings [25]. The mean reference coverage was 98.4 % (range 96.4–99.8 %) and mean high-quality base coverage was 70.7× (range 25–182×).
GATK UnifiedGenotyper was used to call SNPs and small indels [26]. SNPs and small indels with at least 10× read depth, 80 % allele frequency and with at least 10 bp difference between neighbouring SNPs/indels were retained. Indels of greater than four mutations were excluded from the analysis unless occurring within a known drug resistance-associated gene. High-quality SNPs and indels were annotated using SnpEff v4.1 [27] and those in repetitive regions such as proline glutamine/proline proline glutamate family genes (PE/PPE) were excluded from analysis. We characterized mutations in known genes (including regulatory mutations) that confer resistance to rifampicin, isoniazid, ethambutol, streptomycin, pyrazinamide, fluoroquinolones, amikacin, capreomycin, kanamycin, ethionamide, p-aminosalicyclic acid, cycloserine, bedaquiline, linezolid and delamanid according to a literature review (Table S1, available in the online version of this article).
Allelic diversity, phylogeny and molecular dating
The clonal structure from MIRU-24 was inferred using a minimum spanning tree algorithm implemented in Bionumerics v6.7 (Applied Maths). Data sets from two previous independent WGS studies, PRJEB7281 and PRJEB2358 [28], were used as MTB global representatives in phylogenetic analysis.
Concatenated SNP alignment was used to construct a maximum-likelihood phylogenetic tree using RAxML v7.4.2, the GTRCAT model with 1000 bootstrap replicates [29] and visualized using FigTree v1.4.2.
Molecular dating of the Beijing outbreak cluster was first performed by Beauti using an alignment containing both invariable and variable sites as an input file for beast v1.8.2 [30]. Base substitution was modelled using the Hasegawa-Kishino-Yano (HKY) or General Time Reversible (GTR) model with an estimated base frequency and a gamma distribution among-site rate variation with four rate categories. The lognormal relaxed clock (uncorrelated) model which assumes independent mutation rates on different branches was used [12, 13, 31, 32]. The tree was calibrated using date of sample collection as tip dates for each genome, specified in years before the present.
We used a uniform prior distribution for all the trees using a mean mutation rate of 0.35 SNP/genome/year [12, 33–35] and compared the model performance under the different demographic models by calculating the Bayes factors from marginal likelihood estimates obtained from path sampling/stepping stone sampling [36]. For each analysis, two independent runs of 50 million steps using the Markov chain Monte Carlo (MCMC) method were performed, discarding 10 % burn-in and drawing samples every 5000 steps. Three independent runs of the best model were performed for consistency. Tracer was used to examine Markov chain convergence, adequate mixing, chain length and effective sample size (ESS >200). TreeAnnotator was used to obtain the best supported topology under the maximum clade credibility method.
A constant demographic model that used GTR substitution and gamma distribution at four categories was found to be the best fit since it had a better marginal likelihood estimate compared to the other models (Table S2). The overall mutation rate was estimated to be 0.36 SNP/genome/year [95 % highest posterior density (HPD) 0.24–0.48], which is consistent with other published reports [33, 37, 38].
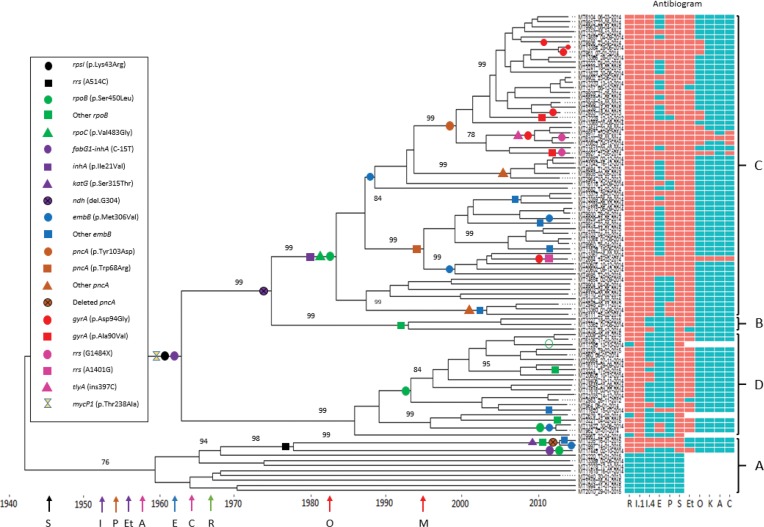
Resistance-conferring mutations were parsimoniously mapped on the phylogenetic tree (considering no reversion) and divergence time was used to infer the likely timing of drug resistance acquisition. The number of strains that shared known resistance-conferring SNPs from the tree nodes (Fig. 3) was enumerated to infer transmitted drug resistance-conferring versus non-shared drug resistance-conferring SNPs (tree branches) to infer acquired drug resistance [6, 13, 39]. SeqTrack was used to create transmission networks among the clades. A Fisher’s exact test in R statistical package was used to assess the association of observed drug resistance mutations and MDR/XDR phenotypes.
Fig. 3.
Phylogeny of Beijing sub-lineage strains with dated acquisition of drug resistance mutations. With four clades (A–D) recognizable, coloured shapes representing fixed resistance-conferring mutations for a TB drug or class of TB drug and putative mutations (ndh and mycP1) mapped on to tree branches parsimoniously assuming no reversion. Small red circle, gyrA (p.Asp94Ala); open green circle, no rpoB mutation. On the x-axis are coloured arrows representing the year of drug discovery: black, S (streptomycin, 1946); green, R (rifampicin, 1966); purple, I (isoniazid, 1952) and Et (ethionamide, 1956); blue, E (ethambutol, 1961); brown, P (pyrazinamide, 1954); red, O (ofloxacin, 1982) and M (moxifloxacin, 1996); pink, A (amikacin, 1957) and C (capreomycin, 1963) while antigiobram represents in vitro susceptibility: red squares, resistant; blue squares, susceptible. Bootstrap support values are shown where they exceed 70 %.
Results
Global phylogeny
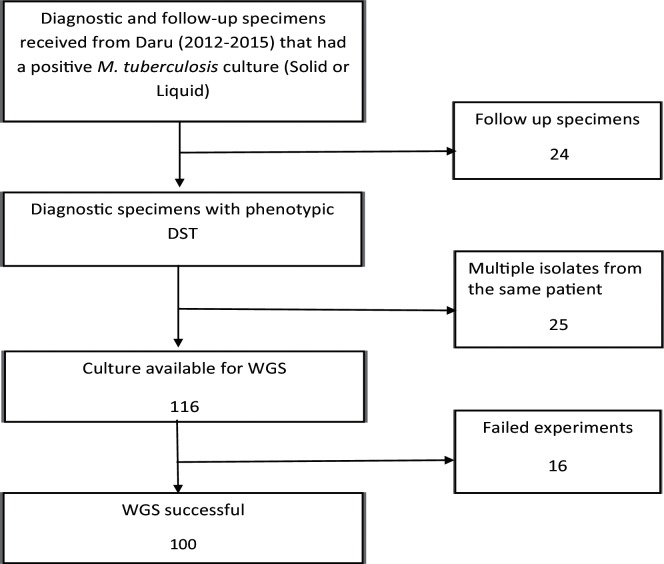
We characterized 100 out of 165 isolates collected during 2012–2015 from Daru, PNG, by MIRU-24 and WGS (Fig. 2). MIRU-24 analysis revealed that 95 isolates formed a single dominant cluster (Fig. S1). Analysis of SNPs derived from WGS revealed all the strains in the dominant cluster formed a monophyletic clade in the East-Asian lineage (sensu stricto modern Beijing lineage), while all the remaining strains belonged to the Euro-American lineage (Fig. S2). According to the classification of Coll et al. [40], the modern Beijing lineage was revealed to be Beijing sub-lineage 2.2.1.1 (Table S3). The Beijing sub-lineage had a median of 23 pairwise differing SNPs between samples (range 0–62), highlighting its limited genetic diversity (Fig. S3). This lineage was separated by at least 32 SNPs from the nearest neighbouring modern Beijing genome included in the representative phylogenetic tree (Table S4).
Fig. 2.
Workflow to derive clinical isolates included in the study collected between 1 October 2012 and 15 March 2015. PNG, Papua New Guinea; SRL, Supra-National Reference Laboratory; DST, drug susceptibility test; WGS, whole genome sequencing.
Molecular dating and drug resistance
Table 1 summarizes known and new putative drug resistance mutations detected within the Beijing sub-lineage, which were parsimoniously mapped to a molecular clock phylogenetic tree that identified four clades: A (n=11), B (n=3), C (n=60) and D (n=21) (Fig. 3). Strains in clade A were the most distantly related and accumulated the fewest drug resistance mutations. Clade C demonstrated massive clonal expansion and included the majority of the MDR strains (54 MDR and six XDR). Ten SNPs, including a mutation in mycP1 (p.Thr238Ala), differentiated clades B, C and D from clade A (Table S5; polymorphisms that are specific to Beijing lineage outbreak clusters clade B, C and D).
Table 1. Drug resistance mutations and phenotypic drug resistance observed among Beijing sub-lineage strains.
Aminoglycosides: A, amikacin; K, kanamycin; C, capreomycin. Isoniazid, all samples with phenotypic resistance had one of the mutations listed; ethambutol, 27/54 samples with one of the listed mutations were phenotypically susceptible. Known mutations to bedaquiline, delamanid, linezolid and clofazimine genes were not identified.
| TB drug | Gene | Mutation | Phenotypically resistant (n) | Phenotypically susceptible (n) | Critical concentration (µg/ml) |
|---|---|---|---|---|---|
| Rifampicin | rpoB | Ser450Leu* | 78 | 0 | 1 |
| Asp435Val | 2 | 0 | |||
| Asp435Tyr | 2 | 0 | |||
| His445Arg | 1 | 0 | |||
| Ile480Val* | 1 | 0 | |||
| His445Asp | 1 | 0 | |||
| rpoC | Val483Gly | 60 | 0 | ||
| lle491Thr | 1 | 0 | |||
| rpoA | Val183Gly | 1 | 0 | ||
| Isoniazid | inhA | Ile21Val | 60 | 0 | 0.4 |
| fabG1-inhA | C15T | 65 | 0 | 0.4 | |
| 20 | 0 | 0.1 | |||
| katG | Ser315Thr | 2 | 0 | 0.4 | |
| ndh | del, G304† | 61 | 0 | 0.4 | |
| 2 | 0 | 0.1 | |||
| Ethambutol | embB | Met306Val* | 23 | 21 | 5 |
| Met306Ile | 1 | 1 | |||
| Gly406Ala | 0 | 1 | |||
| Gln497Arg* | 2 | 5 | |||
| Pyrazinamide | pncA | Tyr103Asp | 27 | 1 | 100 |
| Trp68Arg | 17 | 0 | |||
| Thr135Pro | 1 | 0 | |||
| Gln10Pro | 3 | 0 | |||
| Asp12Glu | 5 | 0 | |||
| Del, gene† | 2 | 0 | |||
| Streptomycin | rpsL | Lys43Arg | 84 | 0 | 8 |
| rrs | A514C | 3 | 0 | ||
| Ethionamide | fabG1-inhA | C15T | 80 | 2 | 10 |
| Quinolones | gyrA | Ala90Val | 2 | 0 | 2.5 |
| Asp94Ala | 1 | 0 | |||
| Asp94Gly | 8 | 0 | |||
| Aminoglycosides | rrs | A1401G | 1 (A,K,C) | 0 | 4 |
| G1484A | 1 (K,C) | 1 (A) | |||
| G1484T | 1 (A,K,C) | 0 | |||
| tlyA | ins397C | 3 (C) | 1 (C) |
*Two dual mutations (Ile480Val and Ser450Val, and Gln497Arg and Met306Val).
†Putative mutations.
Strains in clades B, C and D demonstrated universal streptomycin resistance conferred by an ancestral rpsL (p.Lys43Arg) mutation acquired in the 1960s. The strain ancestral to clades B, C and D also acquired an inhA promoter mutation (fabG1-inhA, C-15T) associated with low-level isoniazid resistance in the 1960s. The same inhA promoter mutation occurred independently in a single clade A strain, while some (3/11) clade A strains acquired streptomycin resistance conferred by an rrs (A514C) mutation. Clade C displayed universal high-level isoniazid resistance potentially conferred by an intragenic inhA mutation (p.Ile21Val) acquired in the 1980s. Occasional high-level isoniazid resistance in clade B (1/3) and clade D (2/21) strains was not associated with an inhA coding mutation while a katG (p.Ser315Thr) mutation was detected in only two clade A strains.
Of the 84 isoniazid-resistant strains tested for ethionamide resistance, 80 (95.2 %) were ethionamide co-resistant (comprising 64/67 high-level and 16/17 low-level isoniazid-resistant strains). Of the four remaining ethionamide-susceptible strains resistant to isoniazid, two have a katG mutation but no inhA coding or promoter mutations. All strains with co-resistance to at least low-level isoniazid and ethionamide had a fabG1-inhA mutation. We explored the co-occurring mutations observed in these strains (Table 2). All strains with co-occurring fabG1-inhA, inhA (p.Ile21Val) and ndh (del.G304) mutations were ethionamide-resistant, while 2/3 with both fabG1-inhA and ndh mutations were ethionamide-resistant. No other mutations associated with ethionamide resistance, such as ethA or ethR, were observed.
Table 2. Association of isoniazid and ethionamide phenotypes and common genes involved in resistance among Beijing sub-lineage strains.
INH, isoniazid; ETH, ethionamide. A total of 64/67 strains with high-level INH resistance were ethionamide-resistant. Note that three strains (fabG–inhA only) with low-level isoniazid resistance were not tested against ethionamide as they were not MDR.
| Gene combinations* | Total | High-level INH resistant | Low-level INH resistant | Ethionamide resistant | Ethionamide susceptible |
|---|---|---|---|---|---|
| fabG1-inhA+inhA+ndh | 60 | 60 | 0 | 60 | 0 |
| fabG1-inhA+ndh only | 3 | 1 | 2 | 2 | 1† |
| fabG1-inhA only | 22 | 4 | 18 | 18 | 1 |
| katG only | 2 | 2 | 0 | 0 | 2 |
| inhA only | 0 | 0 | 0 | 0 | 0 |
| ethA only | 0 | 0 | 0 | 0 | 0 |
| ethr only | 0 | 0 | 0 | 0 | 0 |
*Observed mutations are in Table 1.
†High-level resistance to INH.
Multi-drug resistance was first acquired by clade C in the 1980s with acquisition of a frequently encountered rpoB mutation (p.Ser450Leu), together with a compensatory rpoC mutation (p.Val483Gly). The ancestral strain in clade B acquired a different rpoB mutation (p.His445Arg) in the 1990s without a compensatory mutation. One rifampicin-resistant clade A strain acquired a different rpoC compensatory mutation (p.Ile491Thr). The majority of clade D strains (17/21) also acquired the p.Ser450Leu rpoB mutation, but at a later time point and only one strain had a compensatory rpoA (p.Val183Gly) mutation. One rifampicin-resistant clade D strain had two rpoB mutations (p.Ser450Leu and p.Ile480Val), as confirmed by sequence reads spanning both mutations (Fig. S4). One rifampicin-susceptible isolate in clade D was found to be part of a rifampicin-resistant clone carrying a common rpoB (pSer450Leu) mutation although we did not observe this mutation in this isolate despite adequate sequence coverage (Fig. S5).
There was good correlation between genotypic mutations and phenotypic drug susceptibility (Table 1). However, only 50 % (27/54) of the strains with putative ethambutol resistance-conferring mutations were phenotypically resistant at the critical concentration (5.0 mg l−1); MIC testing to encompass lower concentrations of ethambutol was not performed. The embB (p.Met306Val) mutation first occurred in clade C during the 1980s with independent acquisition at later time points in clades A, C and D. Another embB mutation (p.Gln497Arg) occurred independently on two occasions in clade C.
The majority of clade C (53/60; 88.3 %) strains had pyrazinamide resistance mutations. The earliest mutation pncA (p.Trp68Arg) was acquired in the 1990s. The only two pyrazinamide-resistant clade A strains shared a 3990-bp genomic deletion (position 2 287 064–2 291 054, Fig. S6) spanning four genes including pncA.
XDR TB genotypes
Fluoroquinolone resistance due to mutations in the gyrA gene was detected in 11/60 (18 %) clade C strains; six were XDR TB (Table 1). A single four-member XDR clone emerged around 2009, characterized by an additional capreomycin resistance mutation (tlyA, insertion C.397). One XDR strain with phenotypic resistance to amikacin, kanamycin and capreomycin had both rrs (G1484T) and tlyA (insertion 397C) mutations, while another with an rrs (G1484A) mutation was phenotypically only resistant to kanamycin and capreomycin (Fig. S7). We utilized the arrangement of drug resistance mutations on the tree (Fig. 3) together with transmission network analysis of the large clades C and D (Fig. S8) to determine whether MDR/XDR TB emerged because of acquired resistance or direct transmission. Transmitted drug resistance to isoniazid, rifampicin, ethambutol and streptomycin was evident in clades B, C and D (P<0.0001, Table 3). From transmission network analysis, six and four distinct clusters (2–4 isolates per cluster) that are tightly linked (three or fewer SNP differences between samples within a cluster) were identified in clades C and D, respectively (Fig. S8). For the remaining isolates within the clades, 23 clade C strains (included four XDR) and six clade D strains were within 3–5 SNP differences to at least one other isolate reminiscent of transmission.
Table 3. Frequency of transmitted (cluster) and acquired (independent) drug resistance mutations identified in different clades of the Beijing sub-lineage strains inferred from Fig. 3.
| Clade A | Clade B | Clade C | Clade D | |||||
|---|---|---|---|---|---|---|---|---|
| Independent | Cluster | Cluster | Independent | Cluster | Independent | Cluster | ||
| Locus | rpoB | 1 | 2 | 3 | 0 | 60 | 3 | 16 |
| fabG1-inhA | 1 | 0 | 3 | 0 | 60 | 0 | 21 | |
| inhA | 0 | 0 | 0 | 0 | 60 | 0 | 0 | |
| katG | 0 | 2 | 0 | 0 | 0 | 0 | 0 | |
| rpsl | 0 | 3 | 3 | 0 | 60 | 0 | 21 | |
| embB | 2 | 0 | 0 | 3 | 46 | 1 | 2 | |
| pncA | 0 | 2 | 0 | 0 | 53 | 0 | 0 | |
| gyrA | 0 | 0 | 0 | 7 | 4 | 0 | 0 | |
| rrs | 0 | 0 | 0 | 3 | 0 | 0 | 0 | |
| tlyA | 0 | 0 | 0 | 0 | 4 | 0 | 0 | |
Discussion
We provide the first insight into the molecular basis of drug resistance and genealogy of the dominant strains circulating in a high-prevalence setting on Daru Island in the Western Province, PNG. Our findings indicate that the large number of MDR TB cases is driven by the transmission of a highly drug-resistant cluster of a modern Beijing lineage strain. Studies from other parts of PNG have demonstrated dominance of Euro-American lineage strains, [20, 41] although the contribution of Beijing sub-lineage strains may have been underestimated. We cannot comment on the geographical spread of the Beijing sub-lineage, but phylogenetic analysis combined with detailed molecular dating suggests that it has been in local circulation since the 1940s and first acquired drug resistance mutations in the 1960s. The identification of four different clades with distinct evolutionary trajectories suggests a ‘permissive’ environment for MTB strains to acquire and spread drug resistance within the study setting.
Clade C was the most successful clade, acquiring resistance to all four first-line drugs, and demonstrated clonal spread of both MDR and XDR strains. Phylogenetic analysis indicated that isoniazid and streptomycin resistance were acquired by strains ancestral to clades B, C and D. This is consistent with the use of non-rifampicin-containing regimens in the 1960s that were highly reliant on isoniazid and streptomycin [42]. Similar analysis in Russia and South Africa where large MDR outbreaks have been recorded also indicated that streptomycin and isoniazid resistance were acquired before the introduction of rifampicin-containing regimens [6, 12]. These settings mostly reported high-level isoniazid resistance due to katG mutations. In a recent multinational study, katG (p.Ser315Thr) was identified as the harbinger mutation that is most frequently associated with future MDR occurrence and risks subsequent clonal spread [39].
Other settings with strains possessing the signature fabG1-inhA (C-15T) mutation that confers low-level isoniazid resistance with ethionamide co-resistance [43] have been observed in South Africa [44] and Portugal [45], although the Lisbon strain was of the LAM family. Phenotypic drug susceptibility results of clade C strains demonstrated universal high-level isoniazid resistance, which might be explained by the accumulation of an additional inhA (p.Ile21Val) mutation. The double accumulation of mutations may confer high-level isoniazid resistance without the fitness cost associated with katG mutations, supporting successful clonal expansion and acquisition of additional resistance-conferring mutations to other drugs [46]. An additional consideration is the possibility that the ndh frame shift mutation (deletion of Glu102fs) observed only in clades B and C may also have contributed, potentially as a compensatory mutation, because SNPs in the ndh gene have been associated with increased intracellular NADH/NAD+ ratios, and hence competitive inhibition of activated isoniazid [47]. With the recognized description of mutations that confer resistance to isoniazid and ethionamide, we anticipate that introduction of a short course regimen for MDR TB in this setting may be ineffective. This is because the dominant drug-resistant strain has underlying high-level resistance to two of six regimen drugs (isoniazid and proethionamide-ethionamide analogue) due to coexistence of inhA promoter and inhA gene mutations.
The selective advantage of clade C might have been enhanced further by the acquisition of a classic rpoB (p.Ser450Leu) rifampicin resistance determining mutation together with a compensatory rpoC (p.Val483Gly) mutation that abrogates its negative fitness effects [7]. Similar to previous observations, [48] one strain had two rpoB mutations, of which p.Ile480Val, which lies outside the 81-bp rifampicin resistance determining region, may represent a compensatory mutation. The single strain within a clade D rpoB (p.Ser450Leu) cluster, in which we were unable to detect an rpoB mutation (75× sequence depth) even though all neighbouring strains had the same mutation, may have been due to reversion that might have occurred after a transient mutator phenotype. Schmalstieg et al. attributed this ‘unstable or transient state’ event to be caused by sub-therapeutic drug exposures and effects of efflux pumps especially in the first steps of acquiring higher level resistance [49]. This has never been observed in rifampicin resistance and further investigation into the functionality of this aspect will help to determine its effects on the development of resistance and transmission.
An effective host immune response is critical to protect individuals against TB and to limit ongoing transmission within communities. We identified a putative non-synonymous mycp1 (p.Thr238Ala) mutation that is ancestral to clades B, C and D. Mouse models used to identify MTB ESX-1 (ESAT-6 secretion system 1) substrates and their effects on host cells found mycP1 proteins to be essential for early replication in macrophages and contribution to virulence by allowing escape of mycobacteria from the phagosome into the cytosol of infected macrophages [50, 51]. Site-directed mutagenesis that inactivated mycP1 increased the expression of ESAT-6. Altered host immune responses effected by the putative mycp1 (p.Thr238Ala) mutation may have increased the virulence and transmissibility of the Beijing sub-lineage strain because the observed mutation is within the active site [52], but this remains speculative as clade B demonstrated limited clonal expansion.
Previous studies have found a strong correlation between MDR TB and resistance to pyrazinamide or ethambutol, which highlights the problem of drug resistance amplification with continued use of first-line treatment regimens in patients with undiagnosed MDR TB [53, 54]. Our study provides evidence that streptomycin resistance in this setting is long standing and, as such, probably provided little protection against the development of resistance to first-line oral drugs when used in the World Health Organization (WHO)-endorsed retreatment regimen, since it is potentially compromised by prior treatment.
There was good correlation between genotypic and phenotypic drug resistance profiles, except for ethambutol. This reflects the complex genetic basis of ethambutol resistance, but also emphasizes the highly variable results achieved with phenotypic drug susceptibility testing [13]. Safi et al. demonstrated the epistatic nature of ethambutol resistance by establishing how mutations in embB are often accompanied by polymorphisms in other genes such as nuoD and Rv3806c and hence lead to progressive increases in the critical concentration [55]. We could not identify any of the accompanying mutations previously identified, but hypothesize that a reduction in the critical concentration used may improve the correlation between resistance phenotype and genotype.
The manifestation of transmitted MDR TB and minimal transmission of XDR TB highlights a problem with preventing transmissions. There is need for rapid access to reliable drug susceptibility testing (DST) and increased timely access to new and repurposed agents such as bedaquiline, delamanid and linezolid for effective management of MDR/XDR TB. Controlled clinical trials performed in other endemic settings such as Russia [56] before roll out of regimens with newer drug agents are required for establishment of the minimum number of drugs and duration of treatment that constitute an effective regimen. No putative resistance mutations were detected which have been implicated in resistance to newer agents. Mutations at rrs position 1484 may affect aminoglycoside susceptibility differently, since one XDR sample with an rrs (G1484T) mutation had phenotypic amikacin resistance, while another with an rrs (G1484A) mutation was susceptible to amikacin. In a systematic review of studies that assessed the use of second-line injectables, the rrs (G1484T) mutation was described as a specific predictor of injectable drug resistance but not the rrs (G1484A) mutation [57]. The Hain MTBDRsl line probe assay recently endorsed by WHO for rapid detection of second-line drug resistance has a probe for rrs G1484T but may detect G1484A as a failure of wild-type binding [58] and hence incorrectly infer resistance to all second-line injectables, which could lead to an incorrect choice of treatment.
Our study is limited by its retrospective nature and the fact that the clinical isolates tested failed to capture the whole population with MDR TB on Daru Island. Although we sequenced all available isolates, unreliable transport of specimens to the reference laboratory resulted in an incomplete sampling of MDR TB during the study period, leading to a modest sample size. Due to the modest sample size and short study period we chose to use a fixed mutation rate, which may have reduced the accuracy of evolutionary time point estimates. Another limitation is the use of sample collection date rather than date of diagnosis, although exclusion of follow-up samples should limit the impact of this on evolutionary analysis. Despite these limitations, our study represents the largest drug resistance collection from PNG described so far and provides an unbiased view of the evolution of MDR TB on Daru Island. The estimated dates of divergence are biologically plausible and are temporally consistent with respect to introduction of anti-TB drugs on the global market. Unfortunately a detailed description of when specific drugs were first used in the study setting was unavailable, although culture and drug susceptibility testing were started in 2010. Finally, we lacked clinical and epidemiological information to enhance our genomic analysis. This information is critical to identify individuals and population characteristics that facilitate ongoing transmission of drug-resistant strains.
WGS confirmed both recent transmission of drug-resistant TB and repeated acquisition of drug resistance mutations, driven by a modern Beijing sub-lineage strain. A major concern is that further spread of the strain to adjacent geographical areas, including the capital city, Port Moresby, may amplify transmission within PNG. There is urgent need for improved early detection of drug-resistant TB cases with linkage to effective care programmes in order to limit drug resistance amplification and terminate ongoing transmissions.
Data bibliography
NCBI under project file number PRJNA385247 (2017)
NCBI under project file number PRJEB7281 (2014)
NCBI under project file number PRJEB2358 (2011)
Funding information
This study was funded by the National Health and Medical Research Council, Australia (grant APP1044986), and AusAid (grant 01 HHISP-2013–0015). L.J.M.C. is an NHMRC Career Development Fellow (APP1130084).
Acknowledgement
We thank the Papua New Guinea National Tuberculosis Program, Provincial Health Staff (Western Province) and staff of Queensland Mycobacterium Reference Laboratory for their assistance. Devika Ganesamoorthy, Derek Benson, Nour Ben Zakour and Alex Outhred are thanked for their assistance with data analysis. Associate Professor Vitali Sintchenko is thanked for his leadership in pursuing WGS strategies in Australia as outlined in NHMRC grant APP1044986. We thank Professor Matthew Cooper for discussions and interpretation of the results. This research was supported by the use of the NeCTAR Research Cloud, by QCIF and by the University of Queensland's Research Computing Centre (RCC). The NeCTAR Research Cloud is a collaborative Australian research platform supported by the National Collaborative Research Infrastructure Strategy.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The study was approved by institutional review boards of the University of Queensland, Australia (study number 2015000572), and PNG Medical Research Advisory Committee (study number 16.42), and all the experiments were performed in accordance with relevant guidelines and regulations.
Supplementary Data
Footnotes
Abbreviations: TB, tuberculosis; MTB, Mycobacterium tuberculosis TB; MDR, multi-drug resistant; XDR, extensively drug-resistant; WGS, whole genome sequencing; PNG, Papua New Guinea; MIRU-24, 24-locus mycobacterial interspersed repetitive unit; SRL, Supra-National Referral Laboratory.
Data statement: All supporting data, code and protocols have been provided within the article or through supplementary data files. Five supplementary tables and eight supplementary figures are available in the online version of this article.
References
- 1.World Health Organization., Global Tuberculosis Programme . Geneva: Global Tuberculosis Programme; Global tuberculosis control: WHO report; p. 15. [Google Scholar]
- 2.Abubakar I, Zignol M, Falzon D, Raviglione M, Ditiu L, et al. Drug-resistant tuberculosis: time for visionary political leadership. Lancet Infect Dis. 2013;13:529–539. doi: 10.1016/S1473-3099(13)70030-6. [DOI] [PubMed] [Google Scholar]
- 3.Trauner A, Borrell S, Reither K, Gagneux S. Evolution of drug resistance in tuberculosis: recent progress and implications for diagnosis and therapy. Drugs. 2014;74:1063–1072. doi: 10.1007/s40265-014-0248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int J Tuberc Lung Dis. 2015;19:1276–1289. doi: 10.5588/ijtld.15.0389. [DOI] [PubMed] [Google Scholar]
- 5.Ragonnet R, Trauer JM, Denholm JT, Marais BJ, McBryde ES. High rates of multidrug-resistant and rifampicin-resistant tuberculosis among re-treatment cases: where do they come from? BMC Infect Dis. 2017;17:36. doi: 10.1186/s12879-016-2171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comas I, Borrell S, Roetzer A, Rose G, Malla B, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller B, Chihota VN, Pillay M, Klopper M, Streicher EM, et al. Programmatically selected multidrug-resistant strains drive the emergence of extensively drug-resistant tuberculosis in South Africa. PLoS One. 2013;8:e70919. doi: 10.1371/journal.pone.0070919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanekom M, van der Spuy GD, Gey van Pittius NC, McEvoy CR, Hoek KG, et al. Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J Clin Microbiol. 2008;46:3338–3345. doi: 10.1128/JCM.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One. 2009;4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurjav U, Outhred AC, Jelfs P, McCallum N, Wang Q, et al. Whole genome sequencing demonstrates limited transmission within identified Mycobacterium tuberculosis clusters in New South Wales, Australia. PLoS One. 2016;11:e0163612. doi: 10.1371/journal.pone.0163612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen KA, Abeel T, Manson McGuire A, Desjardins CA, Munsamy V, et al. Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med. 2015;12:e1001880. doi: 10.1371/journal.pmed.1001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eldholm V, Monteserin J, Rieux A, Lopez B, Sobkowiak B, et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun. 2015;6:7119. doi: 10.1038/ncomms8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross GB, Coles K, Nikpour M, Moore OA, Denholm J, et al. TB incidence and characteristics in the remote gulf province of Papua New Guinea: a prospective study. BMC Infect Dis. 2014;14:93. doi: 10.1186/1471-2334-14-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Western Province TB Program Report Western Province TB Program Report. 2016.
- 16.Final Figures Papua New Guinea National Population and Housing Census 2011 [press release] 2011.
- 17.Aia P, Kal M, Lavu E, John LN, Johnson K, et al. The burden of drug-resistant tuberculosis in Papua New Guinea: results of a large population-based survey. PLoS One. 2016;11:e0149806. doi: 10.1371/journal.pone.0149806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krüüner A, Yates MD, Drobniewski FA. Evaluation of MGIT 960-based antimicrobial testing and determination of critical concentrations of first- and second-line antimicrobial drugs with drug-resistant clinical strains of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:811–818. doi: 10.1128/JCM.44.3.811-818.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballif M, Harino P, Ley S, Coscolla M, Niemann S, et al. Drug resistance-conferring mutations in Mycobacterium tuberculosis from Madang, Papua New Guinea. BMC Microbiol. 2012;12:191. doi: 10.1186/1471-2180-12-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coll F, Mallard K, Preston MD, Bentley S, Parkhill J, et al. SpolPred: rapid and accurate prediction of Mycobacterium tuberculosis spoligotypes from short genomic sequences. Bioinformatics. 2012;28:2991–2993. doi: 10.1093/bioinformatics/bts544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demay C, Liens B, Burguière T, Hill V, Couvin D, et al. SITVITWEB-a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12:755–766. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 23.FastQC: a quality control tool for high throughput sequence data Available online at: 2010. www.bioinformatics.babraham.ac.uk/projects/fastqc [Internet]
- 24.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013:1303.3997v1 [Google Scholar]
- 26.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerra-Assunção JA, Crampin AC, Houben RM, Mzembe T, Mallard K, et al. Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. Elife. 2015;4 doi: 10.7554/eLife.05166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 30.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47:242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo T, Comas I, Luo D, Lu B, Wu J, et al. Southern East Asian origin and coexpansion of Mycobacterium tuberculosis Beijing family with Han Chinese. Proc Natl Acad Sci USA. 2015;112:8136–8141. doi: 10.1073/pnas.1424063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bryant JM, Schürch AC, van Deutekom H, Harris SR, de Beer JL, et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis. 2013;13:110. doi: 10.1186/1471-2334-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baele G, Li WL, Drummond AJ, Suchard MA, Lemey P. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Mol Biol Evol. 2013;30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43:482–486. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merker M, Kohl TA, Roetzer A, Truebe L, Richter E, et al. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One. 2013;8:e82551. doi: 10.1371/journal.pone.0082551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manson AL, Cohen KA, Abeel T, Desjardins CA, Armstrong DT, et al. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat Genet. 2017;49:395–402. doi: 10.1038/ng.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ley SD, Harino P, Vanuga K, Kamus R, Carter R, et al. Diversity of Mycobacterium tuberculosis and drug resistance in different provinces of Papua New Guinea. BMC Microbiol. 2014;14:307. doi: 10.1186/s12866-014-0307-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ley SD, Riley I, Beck HP. Tuberculosis in Papua New Guinea: from yesterday until today. Microbes Infect. 2014;16:607–614. doi: 10.1016/j.micinf.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Domínguez J, Boettger EC, Cirillo D, Cobelens F, Eisenach KD, et al. Clinical implications of molecular drug resistance testing for Mycobacterium tuberculosis: a TBNET/RESIST-TB consensus statement. Int J Tuberc Lung Dis. 2016;20:24–42. doi: 10.5588/ijtld.15.0221. [DOI] [PubMed] [Google Scholar]
- 44.Marais BJ, Victor TC, Hesseling AC, Barnard M, Jordaan A, et al. Beijing and Haarlem genotypes are overrepresented among children with drug-resistant tuberculosis in the Western Cape Province of South Africa. J Clin Microbiol. 2006;44:3539–3543. doi: 10.1128/JCM.01291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machado D, Perdigão J, Ramos J, Couto I, Portugal I, et al. High-level resistance to isoniazid and ethionamide in multidrug-resistant Mycobacterium tuberculosis of the Lisboa family is associated with inhA double mutations. J Antimicrob Chemother. 2013;68:1728–1732. doi: 10.1093/jac/dkt090. [DOI] [PubMed] [Google Scholar]
- 46.Ogbunugafor CB, Wylie CS, Diakite I, Weinreich DM, Hartl DL. Adaptive landscape by environment interactions dictate evolutionary dynamics in models of drug resistance. PLoS Comput Biol. 2016;12:e1004710. doi: 10.1371/journal.pcbi.1004710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee AS, Teo AS, Wong SY. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2001;45:2157–2159. doi: 10.1128/AAC.45.7.2157-2159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramaswamy SV, Dou SJ, Rendon A, Yang Z, Cave MD, et al. Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J Med Microbiol. 2004;53:107–113. doi: 10.1099/jmm.0.05343-0. [DOI] [PubMed] [Google Scholar]
- 49.Schmalstieg AM, Srivastava S, Belkaya S, Deshpande D, Meek C, et al. The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrob Agents Chemother. 2012;56:4806–4815. doi: 10.1128/AAC.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, et al. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe. 2010;7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solomonson M, Huesgen PF, Wasney GA, Watanabe N, Gruninger RJ, et al. Structure of the mycosin-1 protease from the mycobacterial ESX-1 protein type VII secretion system. J Biol Chem. 2013;288:17782–17790. doi: 10.1074/jbc.M113.462036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoek KG, Schaaf HS, Gey van Pittius NC, van Helden PD, Warren RM. Resistance to pyrazinamide and ethambutol compromises MDR/XDR-TB treatment. S Afr Med J. 2009;99:785–787. [PubMed] [Google Scholar]
- 54.Zhao LL, Sun Q, Liu HC, Wu XC, Xiao TY, et al. Analysis of embCAB mutations associated with ethambutol resistance in multidrug-resistant Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother. 2015;59:2045–2050. doi: 10.1128/AAC.04933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Safi H, Lingaraju S, Amin A, Kim S, Jones M, et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat Genet. 2013;45:1190–1197. doi: 10.1038/ng.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borisov SE, Dheda K, Enwerem M, Romero Leyet R, D'Ambrosio L, et al. Effectiveness and safety of bedaquiline-containing regimens in the treatment of MDR- and XDR-TB: a multicentre study. Eur Respir J. 2017;49:1700387. doi: 10.1183/13993003.00387-2017. [DOI] [PubMed] [Google Scholar]
- 57.Georghiou SB, Magana M, Garfein RS, Catanzaro DG, Catanzaro A, et al. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One. 2012;7:e33275. doi: 10.1371/journal.pone.0033275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.World Health Organization The use of molecular line probe assays for the detection of resistance to second-line anti-tuberculosis drugs. 2016. Geneva: Switzerland.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.