Abstract
Introduction
Patients with Alzheimer’s disease (AD) show heterogeneity in profile of cognitive impairment. We aimed to identify cognitive subtypes in four large AD cohorts using a data-driven clustering approach.
Methods
We included probable AD dementia patients from the Amsterdam Dementia Cohort (n = 496), Alzheimer’s Disease Neuroimaging Initiative (n = 376), German Dementia Competence Network (n = 521), and University of California, San Francisco (n = 589). Neuropsychological data were clustered using nonnegative matrix factorization. We explored clinical and neurobiological characteristics of identified clusters.
Results
In each cohort, a two-clusters solution best fitted the data (cophenetic correlation >0.9): one cluster was memory-impaired and the other relatively memory spared. Pooled analyses showed that the memory-spared clusters (29%–52% of patients) were younger, more often apolipoprotein E (APOE) ε4 negative, and had more severe posterior atrophy compared with the memory-impaired clusters (all P < .05).
Conclusions
We could identify two robust cognitive clusters in four independent large cohorts with distinct clinical characteristics.
Keywords: Alzheimer’s disease, Cognition, Heterogeneity, Subtypes, Atypical, Neuropsychology
1. Introduction
Alzheimer’s disease (AD) dementia is characterized by progressive cognitive impairment in multiple cognitive domains, for example, memory, language, visuospatial and executive functioning, and attention. Typically, AD is characterized by early and prominent memory loss [1]. A minority of patients has a prominent and relatively focal cognitive presentation, such as logopenic-variant primary progressive aphasia, posterior cortical atrophy, or a behavioral/dysexecutive subtype [2–5]. Atypical variants have been associated with specific demographic, genetic, and neuroimaging/biomarker findings that are distinct from those of typical amnestic patients (e.g., age at onset, apolipoprotein E [APOE] genotype, distribution of cortical atrophy, hypometabolism, tau deposition, cerebrospinal fluid (CSF) biomarker concentrations, and pathologic findings) [6–10]. However, even patients who do not display a defined subtype also show a considerable variation in patterns of cognitive impairment. Earlier studies demonstrated the potential to capture cognitive heterogeneity in AD using a data-driven clustering approach [11–14]. Studies differed in sample size, clinical diagnosis of included patients, available neuropsychological (NP) test results, available neurobiological characteristics to compare clusters with, and clustering technique. This has resulted in different numbers of clusters, with different cognitive and neurobiological characteristics. Although those studies were clearly suggestive of variability in underlying pathologic mechanisms, it is difficult to generalize the findings, because they result from single studies that show considerable variability in patient population and methodological approaches.
In the present study, we aimed to identify cognitive subtypes and to study whether these subtypes could be replicated in three independent AD dementia cohorts. For the identification of cognitive AD subtypes, we used nonnegative matrix factorization (NMF) [15–18]. On the basis of the earlier descriptions of cognitive heterogeneity, we expected NMF to identify at least a cluster including patients with typical amnestic AD and one or more other clusters including patients with nonamnestic features [15–18].
2. Methods
2.1. Patients
We selected AD patients from four large cohorts: the Amsterdam Dementia Cohort (ADC), the Alzheimer’s Disease Neuroimaging Initiative (ADNI), the German Dementia Competence Network (DCN), and the University of California, San Francisco Memory and Aging Center research cohort (UCSF). Patients were selected based on (1) clinical diagnosis of probable AD dementia, (2) availability of NP test results, and (3) Mini–Mental State Examination (MMSE) score >16/30 [19]. In the ADC and UCSF cohort, patients with focal presentations logopenic-variant primary progressive aphasia, posterior cortical atrophy, and the behavioral/dysexecutive subtype of probable AD dementia were included, whereas such subjects were explicitly excluded from participation in the ADNI and DCN studies.
From the ADC we selected 496 patients with probable AD [20]. Patients visited the outpatient memory clinic of the VU University Alzheimer Center between 2008 and 2013. Standard dementia screening included for most patients medical history and medication use, physical and neurologic examination, extensive NP evaluation, screening laboratory tests, APOE genotyping, magnetic resonance imaging (MRI), and lumbar puncture (LP). In the ADC, level of education was defined according to a rating scale ranging from 1 (low, primary school not finished) to 7 (high, university degree) [21]. All participants provided written informed consent to use their clinical data for research purposes. The local ethical committee approved the study.
From the ADNI database (adni.loni.usc.edu) we selected 376 probable AD patients. Patients were recruited from more than 50 sites across the US and Canada (www.adni-info.org). Standard workup included medical history, physical and neurologic examination, extensive NP evaluation, screening laboratory tests, APOE genotyping, neuroimaging including MRI, and LP. For the present study, we used data of screening and baseline visits acquired for ADNI-1 or ADNI-2 between 2005 and 2013. All patients gave written informed consent at screening.
From the DCN cohort database (http://www.kompetenznetz-demenzen.de) we selected 521 probable AD patients [22]. The DCN is a collaboration of 14 specialized German memory clinics from university hospitals. All patients were offered a uniform dementia screening at first visit between 2003 and 2007, including medical history, physical and neurologic examination, extensive NP evaluation, screening laboratory tests, MRI scan, and LP. The DCN study protocol was approved by the institutional review boards of all participating study centers [22]. All patients, or their legal guardians, provided written informed consent.
From the UCSF research cohort we selected 589 probable AD patients [23]. Patients were either seen in the outpatient memory clinic or for a research assessment in the UCSF Alzheimer’s Disease Research Center. All patients were assessed at first visit between 1998 and 2013. Standardized dementia screening included medical history, physical and neurologic examination, NP evaluation, screening laboratory tests, APOE genotyping, and neuroimaging including MRI. A core screening NP battery was performed in both the clinical and research settings. All patients and informants provided written informed consent. Surrogate consent was accepted when patients lacked capacity to provide consent themselves. The local medical ethical committee approved the study.
2.2. NP tests
NP data included tests covering the major cognitive domains in each cohort, but the exact composition of NP test batteries differed across cohorts. NP tests included for analysis in this study are shown in Supplementary Table 1. The number of missing NP values differed across cohorts and within NP test batteries (on average 20% in ADC, 27% in ADNI, 1% in DCN, and 12% in UCSF). Main reasons for missingness are practical reasons unrelated to the data (random). In part of the cases, however, tests could not be finished because of cognitive impairment, whereas scoring differed across cohorts and between tests (i.e., assignment of missing value or minimum score). The clustering technique NMF does not allow for missing data or negative values. To reduce selection bias, we did not select patients based on completeness of data sets, but we completed the data sets using a multiple imputation approach that is commonly used as a reliable method to estimate missing data.
We imputed missing NP data using R package Multivariate Imputation by Chained Equations (MICE, version 2.25) [24,25]. MICE estimates missing NP values by predicting these values from the relationships with other NP variables. We also included predictors age, gender, MMSE, and when available education, duration of complaints, Alzheimer’s Disease Assessment Scale–Cognitive subscale, Cambridge Cognitive Examination, or Cognitive Dementia Rating sum of boxes in the imputation model. We ran MICE 50 times per cohort, resulting in 50 imputed data sets for each cohort. For further analyses, we included pooled measures over the 50 derived imputed data sets per cohort [26,27]. We inverted values when appropriate, so that for all tests lower scores reflect worse cognitive impairment. Next, the imputed NP data were normalized and scaled to include only positive values (0–1).
2.3. MRI characteristics
MRI characteristics were available for patients from the ADC, ADNI, and DCN cohorts.
For the ADC cohort, imaging data were obtained on a 1.5T or 3T scanner. Visual ratings of medial temporal lobe atrophy (range 0–4 [28]), posterior atrophy (range 0–3 [29]), and white matter hyperintensities (WMHs; range 0–3 [30]) were performed by an experienced neuroradiologist. For ADNI, a structural MRI 1.5T scan was performed on screening or baseline visit [31]. Image processing has been done with cortical reconstruction and volumetric segmentations using FreeSurfer 4.3 (surfer.nmr.mgh.harvard.edu/) [32]. Hippocampal and WMH volumes were downloaded from the ADNI website (ida.loni.usc.edu/) [33]. For the DCN cohort, MRI data were obtained from multiple 1.5T scanners with standardized MRI acquisition guidelines across centers [22,34]. Posterior atrophy and WMH were scored by experienced neuroradiologists using a visual rating scale in which higher scores reflect more severe abnormalities (range 0–3). Furthermore, hippocampal volumes were measured using the Functional MRI of the Brain (FMRIB) Integrated Registration and Segmentation Tool from the FMRIB Software Library package of tools [35,36]. For all atrophy measurements, we included the mean of left and right hemisphere.
2.4. APOE ε4 genotype
APOE ε4 genotype was available in the ADC (n = 448; 90%), ADNI (n = 196; 52%), DCN (n = 397; 76%), and UCSF (n = 175; 30%) cohorts. We dichotomized APOE ε4 genotype according to the presence or absence of one or more APOE ε4 alleles. APOE genotype was assessed using the Light-Cycler APOE mutation detection method (Roche Diagnostics GmbH, Mannheim, Germany) in the ADC. For ADNI, APOE alleles were genotyped using DNA extracted by Cogenics. For the DCN, APOE genotype was assessed using Qiagen blood isolation kit (Qiagen, Hilden, Germany). For UCSF, APOE genotype was conducted using a TaqMan Allelic Discrimination Assay on an ABI7900HT Fast Real-Time polymerase chain reaction system (Applied Biosystems, Foster City, CA, USA).
2.5. AD biomarkers
CSF markers amyloid β1–42 (Aβ1–42) and total tau (tau) were available for the ADC (n = 389; 79%), ADNI (n = 102; 27%), and DCN (n = 193; 37%). CSF biomarkers were assessed using Sandwich enzyme-linked immunosorbent assays (Fujirebio, Gent, Belgium) in the ADC and DCN [37], and Multiplex xMAP Luminex platform (Luminex Corp, Austin, TX) with Fujirebio immunoassay kit–based reagents (INNO-BIA Alzbio3; Fujirebio, Ghent, Belgium) in ADNI. CSF biomarkers were considered positive for AD when the tau/Aβ1–42 ratio was >0.52 in the ADC and DCN cohorts [38] and when >0.39 in the ADNI cohort [39]. Pittsburgh compound B positron emission tomography visual reading results (i.e., positive or negative) were available for the UCSF cohort (n = 52; 9%).
2.6. Statistical analysis
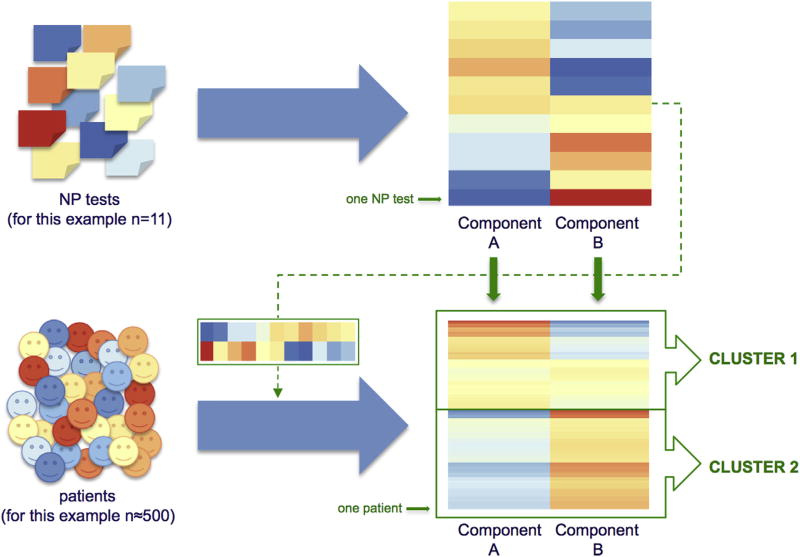
For statistical analyses, we used RStudio for Mac version 3.2.2 (Integrated Development for R. RStudio, Inc, Boston, MA, http://www.rstudio.com). Clustering was performed with the R package NMF (version 0.20.6) [40]. Clustering is a data-driven method to divide a heterogeneous set of objects (patients) in subgroups that are more homogeneous in terms of characteristics provided as input for the clustering (NP test results). NMF is a dual-clustering approach, meaning that clustering includes two parallel steps (illustrated in Fig. 1). First, NP test results are grouped together into NP profiles (“components”), and NP tests that determine these components can be identified by their component loading. Second, patients are grouped together based on the fit of their NP profile to the NP summary component. The optimal number of clusters is based on the most consistent assignment of patients to the identified cognitive profiles in the multiple runs of NMF. The stability of the cluster solution is assessed with the cophenetic correlation coefficient that measures how consistently tests and subjects are assigned to a given component, ranging between 0 and 1 (i.e., no stability to stable cluster solution). For each cohort, we determined the optimal number of clusters based on the highest cophenetic correlation coefficient for two to nine cluster solutions [15]. We used the “nonsmooth” NMF algorithm that introduces an intermediate smoothing matrix to enhance sparsity of the clusters [41].
Fig. 1.
Nonnegative matrix factorization is a dual-clustering approach, meaning that clustering includes two parallel steps. First, neuropsychological (NP) test results are grouped together into NP profiles (components), illustrated in the upper half of the figure, in which each row represents one NP test, and each column an identified NP component. The warmer the color, the higher the test loads to the component when test scores are high (relatively spared cognition); the colder the color, the lower the test loads to the component when test scores are high (relatively impaired cognition). The optimal number of components is based on the cophenetic correlation coefficient (for this example n = 2). Second, patients are grouped together (into “clusters”) based on the fit of their NP profile to the identified NP component, taking each test’s load to the component into account. This step is illustrated in the lower half of the figure, in which each row represents one patient. The warmer the color, the better the fit of patients’ NP profile to the NP profile of the identified component.
Characterization of identified clusters in terms of NP profile was based on the most strongly loading NP tests [17]. For characterization of identified clusters in terms of demographic, clinical, and neurobiological characteristics, we analyzed age, sex, education, disease duration reported by the patient, MMSE, APOE ε4 genotype, CSF biomarkers, MRI atrophy, and WMH measurements using χ2, t tests, or Kruskal-Wallis tests, where appropriate. These analyses were performed for each cohort separately. In addition, for each cluster we pooled the patient characteristics over the cohorts to compare them for the total sample. To this end we Z-transformed variables with different scales (i.e., education, CSF biomarkers, and MRI biomarkers) before pooling. When atrophy of the hippocampus and the posterior cortex was measured using a visual rating scale in which higher scores reflect more severe atrophy, the normalized scores were inverted so that higher scores reflect less atrophy in all cohorts and the pooled sample.
3. Results
3.1. Cohort characteristics
Characteristics of all cohorts are summarized in Table 1. On average, patients were 71 ± 9 years old, with the ADC being the youngest cohort. Fifty-four percent of patients were female, ranging from 44% (ADNI) to 60% (DCN). Patients were mildly to moderately demented, with an average MMSE score varying between cohorts from 22 to 24. Roughly two-thirds of patients were APOE ε4 carrier, ranging from 57% (UCSF) to 67% (ADC).
Table 1.
Demographical and neurobiological characteristics of study cohorts
| Characteristics | ADC (n = 496) | ADNI (n = 376) | DCN (n = 521) | UCSF (n = 589) | Pooled sample (n = 1.982) |
|---|---|---|---|---|---|
| Demographics | |||||
| Age (years) | 67 ± 8 | 75 ± 8 | 72 ± 8 | 71 ± 10 | 71 ± 9 |
| Female | 258 (52%) | 165 (44%) | 313 (60%) | 328 (56%) | 1064 (54%) |
| Education* | 5 (4–6) | 16 (13–18) | 11 (10–13) | 16 (14–18) | 0.00 ± 1.00 |
| Duration complaints (years) | 2 (2–4) | — | 2 (1–3) | — | 2 (1–4) |
| Global cognition | |||||
| MMSE | 22 ± 3 | 23 ± 2 | 23 ± 3 | 24 ± 4 | 23 ± 3 |
| APOE ε4 genotype | |||||
| APOE ε4 positive | 299 (67%) | 128 (65%) | 255 (64%) | 100 (57%) | 782 (64%) |
| AD biomarkers† | |||||
| AD biomarker available | 389 (79%) | 102 (27%) | 193 (37%) | 53 (9%) | 737 (37%) |
| AD biomarker positive | 358 (92%) | 89 (87%) | 164 (85%) | 52 (98%) | 654 (89%) |
| MRI | |||||
| Hippocampus‡ | 1.5 (1–2) | 2882 ± 511 | 2933 ± 473 | — | 0.00 ± 1.00 |
| Posterior cortex§ | 1 (1–2) | — | 1 (0–2) | — | 0.00 ± 1.00 |
| WMH | 1 (0–2) | 0.41 (0.16–1.25) | 0.5 (0–1) | — | 0.02 ± 1.01 |
Abbreviations: AD, Alzheimer’s disease; ADC, Amsterdam Dementia Cohort; ADNI, Alzheimer’s Disease Neuroimaging Initiative; APOE, apolipoprotein E; CSF, cerebrospinal fluid; DCN, German Dementia Competence Network; MMSE, Mini–Mental State Examination; MRI, magnetic resonance imaging; UCSF, University of California, San Francisco; WMH, white matter hyperintensity.
NOTE. Data are presented in the mean ± standard deviation, number (%), or median (2nd–4th quantile).
Education is given according to the Verhage scale (1–7, respectively, low-high education [21]) for ADC, years of education for ADNI, DCN, and UCSF, and normalized scores for the pooled sample.
AD biomarkers are available as CSF total tau/amyloid β1–42 (abnormal when >0.52 according to Duits et al. [38] in the ADC and DCN cohorts, and abnormal when >0.39 according to Shaw et al. [39] in the ADNI cohort), or as Pittsburgh compound B positron emission tomography positivity in the UCSF cohort.
Hippocampal atrophy is measured according to the medial temporal lobe atrophy (MTA) visual rating scale for the ADC (0–4, higher scores reflect more severe atrophy [28]), and hippocampal volumes in cubic millimeters for the ADNI and DCN, and z-scores (in which normalized MTA scores are inverted) for the pooled sample.
Posterior atrophy is scored using a visual rating scale for the ADC [29] and the DCN, inverted z-scores are given for the pooled sample. WMH are scored according to a visual rating scale for the ADC [30] and the DCN (0–3, higher scores reflect more sever pathology), WMH volumes for the ADNI [33] and z-scores are given for the pooled sample.
3.2. NMF clusters of cognitive subtypes
NMF is a dual-clustering approach; first, NP tests are grouped into components, and second patients are clustered based on the fit of their NP profiles to the profiles of the identified NP components, taking the load of each test to the component into account. NMF showed that within each cohort, the optimal number of clusters was two, as the solution with two clusters showed the strongest cophenetic correlation (>0.90).
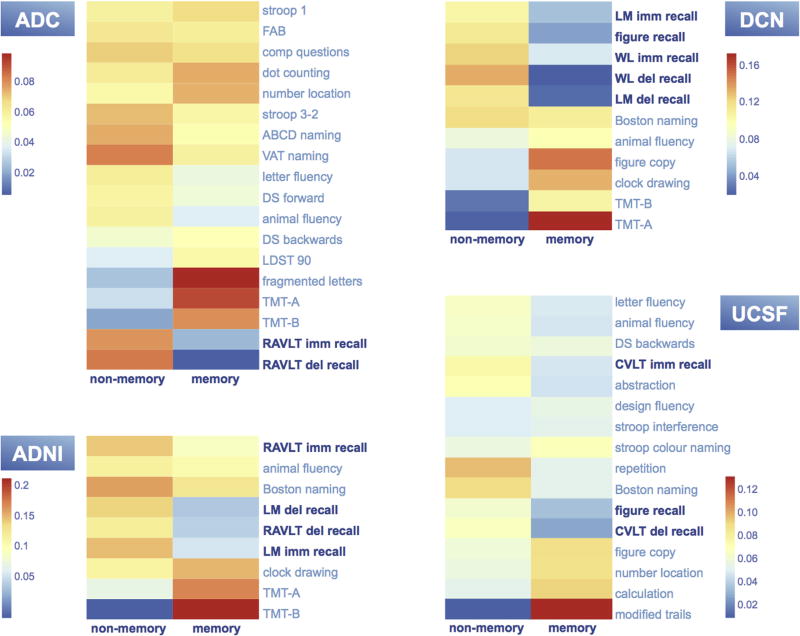
Results of the clustering of tests are shown in Fig. 2. In the ADC, one NP component mainly included memory tests Rey Auditory Verbal Learning Test (RAVLT) immediate and delayed recall. The other component included mainly nonmemory tests Trail Making Test (TMT)-A and TMT-B, and fragmented letters. In the ADNI cohort, one NP component mainly included memory tests logical memory immediate and delayed recall, and RAVLT delayed recall. The other component mainly included nonmemory tests TMT-A and TMT-B. In the DCN cohort, one NP component included mainly memory tests logical memory immediate and delayed recall, word list immediate and delayed recall of the Consortium to Establish a Registry for Alzheimer’s Disease (CERAD), and CERAD figure recall. The other component included mainly nonmemory tests TMT-A, TMT-B, clock drawing, and CERAD figure copy. In the UCSF cohort, one NP component included mainly memory tests figure recall and the California verbal learning test delayed recall. The other component included mainly nonmemory tests modified trails, design fluency, and stroop interference.
Fig. 2.
Memory tests (indicated in dark blue). Each row represents one neuropsychological test. The two columns represent the two found neuropsychological components. The warmer the color, the higher the test loads to the component when test scores are high (relatively spared cognition); the colder the color, the lower the test loads to the component when test scores are high (relatively impaired cognition). Interpretation: in each cohort, one component is associated with relative impairment of memory tests, therefore called the memory component. The other component is associated with relative impairment of nonmemory functions, therefore called the nonmemory component. In all cohorts, the optimal number of test clusters was two. Abbreviations: ABCD, Arizona Battery for Communication Disorders of Dementia; ADC, Amsterdam Dementia Cohort; ADNI, Alzheimer’s Disease Neuroimaging Initiative; comp questions, comparative questions; CERAD, Consortium to Establish a Registry for Alzheimer’s Disease; CVLT, California Verbal Learning Test; DCN, German Dementia Competence Network; DS, Digit Span; FAB, Frontal Assessment Battery; LDST, Letter Digit Substitution Test; LM, CERAD Logical Memory; RAVLT, Rey Auditory Verbal Learning Test; TMT, Trail Making Test; UCSF, University of California, San Francisco; VAT, Visual Association Test; WL, CERAD Word List.
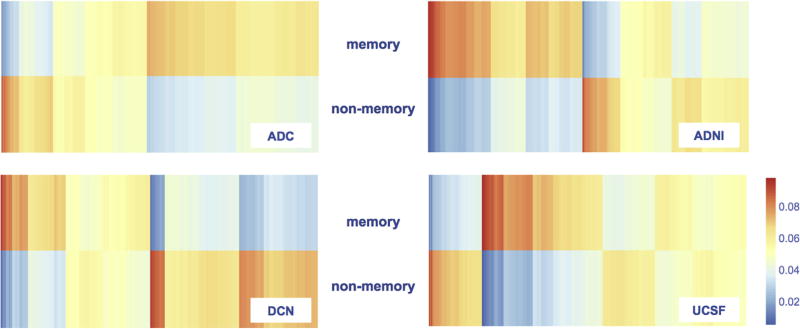
Patients were assigned to either of two clusters based on the fit of their NP test results to the memory or nonmemory component in each cohort (Fig. 3). Across all cohorts, the memory clusters included on average 60% of patients, ranging from 48% (ADNI) to 71% (ADC), and the nonmemory clusters included on average 40% of patients, ranging from 29% (ADC) to 52% (ADNI).
Fig. 3.
Patients were assigned to either the memory or nonmemory cluster based on the fit of their neuropsychological profile to the memory or nonmemory component (Fig. 2). Each column represents one patient. The warmer the color, the better the fit of patients’ neuropsychological profile to the neuropsychological profile of that component. Abbreviations: ADC, Amsterdam Dementia Cohort; ADNI, Alzheimer’s disease Neuroimaging Initiative; DCN, German Dementia Competence Network; UCSF, University of California, San Francisco.
3.3. Cluster characterization
We analyzed cluster characteristics in terms of demographic, clinical, and neurobiological characteristics. Associations for each cohort are shown in Table 2. In the ADC, patients in the nonmemory cluster were younger, had lower MMSE scores, and had more severe atrophy of the posterior cortex. In the ADNI cohort, nonmemory patients tended to be younger and less often APOE ε4 positive with relative hippocampal sparing, but these differences did not reach significance. In the DCN cohort, patients in the nonmemory cluster had more severe atrophy of the posterior cortex. In the UCSF cohort, the nonmemory cluster had lower MMSE scores and patients were less often APOE ε4 positive. There were no differences in WMHs (ADC, ADNI, DCN). When we analyzed pooled data, we found that across cohorts, patients in nonmemory clusters were younger, less educated, reported shorter disease duration, had lower MMSE scores, were less often APOE ε4 positive, had less severe hippocampal atrophy, and more severe atrophy of the posterior cortex than patients in memory clusters.
Table 2.
Demographic and neurobiological cluster characteristics
| ADC | ADNI | DCN | UCSF | Pooled sample | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||
| Characteristics | Memory (n = 352) (71%) |
Nonmemory (n = 144) (29%) |
Memory (n = 182) (48%) |
Nonmemory (n = 194) (52%) |
Memory (n = 335) (64%) |
Nonmemory (n = 186) (36%) |
Memory (n = 326) (55%) |
Nonmemory (n = 263) (45%) |
Memory (n = 1195) (60%) |
Nonmemory (n = 787) (40%) |
| Demographics | ||||||||||
| Age (years) | 67.7 ± 7.8 | 64.7 ± 8.4* | 76.2 ± 7.2 | 74.1 ± 8.3 | 72.3 ± 7.8 | 71.1 ± 9.0 | 72.1 ± 9.7 | 70.3 ± 11.0 | 71.5 ± 8.7 | 70.4 ± 9.9‡ |
| Female | 180 (51%) | 78 (54%) | 78 (43%) | 87 (45%) | 201 (60%) | 112 (60%) | 171 (52%) | 157 (60%) | 630 (53%) | 434 (55%) |
| Education | 0.03 ± 1.00 | −0.06 ± 1.01 | 0.01 ± 0.97 | −0.01 ± 1.03 | 0.03 ± 1.00 | −0.05 ± 1.00 | 0.1 ± 1.00 | −0.13 ± 0.99 | 0.04 ± 0.99 | −0.07 ± 1.01‡ |
| Duration complaints | 3.1 ± 2.2 | 2.7 ± 1.7 | — | — | 2.7 ± 2.3 | 2.4 ± 1.8 | — | — | 2.9 ± 2.3 | 2.5 ± 1.8‡ |
| Global cognition | ||||||||||
| MMSE | 22.6 ± 3.0 | 21.5 ± 3.2† | 23.5 ± 1.9 | 23.0 ± 2.3 | 23.4 ± 2.6 | 22.9 ± 3.0 | 24.4 ± 3.1 | 22.7 ± 3.8* | 23.5 ± 2.8 | 22.6 ± 3.2* |
| APOE ε4 genotype | ||||||||||
| APOE ε4 positive | 225 (70%) | 74 (59%) | 72 (67%) | 56 (63%) | 175 (67%) | 80 (58%) | 61 (66%) | 39 (48%)‡ | 533 (68%) | 249 (58%)* |
| AD biomarkers | ||||||||||
| AD biomarker available | 275 (78%) | 114 (79%) | 52 (29%) | 50 (26%) | 123 (38%) | 70 (38%) | 25 (8%) | 28 (11%) | 475 (40%) | 262 (33%) |
| AD biomarker positive | 253 (92%) | 105 (92%) | 45 (87%) | 44 (88%) | 106 (86%) | 58 (83%) | 24 (96%) | 28 (100%) | 423 (89%) | 231 (88%) |
| MRI | ||||||||||
| Hippocampus | −0.07 ± 1.00 | 0.18 ± 0.97 | −0.05 ± 1.01 | 0.05 ± 1.00 | −0.09 ± 0.95 | 0.17 ± 1.07 | — | — | −0.07 ± 1.00 | 0.12 ± 1.00† |
| Posterior cortex | 0.11 ± 0.94 | −0.29 ± 1.10† | — | — | 0.08 ± 0.98 | −0.15 ± 1.03‡ | — | — | 0.09 ± 0.96 | −0.21 ± 1.06* |
| WMH | 0.10 ± 0.01 | −0.07 ± 0.01 | 0.01 ± 0.32 | −0.01 ± 0.95 | −0.04 ± 0.96 | 0.07 ± 1.07 | — | — | 0.03 ± 0.01 | 0.00 ± 1.05 |
Abbreviations: AD, Alzheimer’s disease; ADC, Amsterdam Dementia Cohort; ADNI, Alzheimer’s Disease Neuroimaging Initiative; APOE, apolipoprotein E; CSF, cerebrospinal fluid; DCN, German Dementia Competence Network; MMSE, Mini–Mental State Examination; MRI, magnetic resonance imaging; UCSF, University of California, San Francisco; WMH, white matter hyperintensity.
NOTE. Data are presented in number (%) or mean ± standard deviation, also when not normally distributed enabling clearer comparison between clusters. P values are based on t tests, χ2, or Kruskal-Wallis analyses, where appropriate. Normalized values are given for education and MRI characteristics. AD biomarkers are available as CSF total tau/amyloid β1–42 (abnormal when >0.52 according to Duits et al. [38] in the ADC and DCN cohorts, and abnormal when >0.39 according to Shaw et al. [39] in the ADNI cohort), or as Pittsburgh compound B positron emission tomography positivity (in the UCSF cohort). When MRI atrophy characteristics were measured using a visual rating scale in which a higher score reflects more severe atrophy, results were inverted so that higher scores reflect less atrophy. Differences between memory and nonmemory clusters are shown in bold and are indicated as follows:
P ≤ .001,
P ≤ .01,
P ≤ .05.
Interpretation: the nonmemory clusters were younger (pooled sample, ADC), less educated (pooled sample), had shorter duration of complaints (pooled sample), lower MMSE scores (ADC, UCSF, pooled sample), were more often APOE ε4 negative (UCSF, pooled sample), had less hippocampal atrophy (pooled sample), but more severe atrophy of the posterior cortex (ADC, DCN, pooled sample) than the memory clusters.
3.4. Validation of cognitive clusters after stratification for disease severity
We tested whether disease severity influenced the cluster solutions by repeating NMF analyses in each cohort after stratifying for disease severity according to the cohort median MMSE score value. Results appeared to be robust across severity subgroups, with a memory cluster and nonmemory cluster appearing in both the mild and the moderately demented patient strata (Supplementary Figs. 1–4). Cluster characteristics in terms of demographic and neurobiological characteristics remained largely the same (Supplementary Tables 2 and 3). Differences between the memory and nonmemory clusters were most pronounced in the mildly demented stratum.
3.5. Validation of cognitive clusters in AD biomarker confirmed patients
Availability of AD biomarkers is given in Tables 1 and 2. On the basis of the χ2 analyses, no differences were found between the memory and nonmemory clusters in terms of AD biomarker positivity (P > .05). Only for the ADC, enough data were available to repeat data-driven NMF analyses in AD biomarker confirmed patients (n = 357 with CSF total tau/Aβ1–42 > 0.52 [38]). Results appeared to be consistent with findings of the total cohorts, with a memory component mainly including RAVLT immediate and delayed recall, and a nonmemory component mainly including TMT-A, TMT-B, fragmented letters, and letter digit substitution test, shown in Supplementary Fig. 5. The memory cluster included 73% of patients and the nonmemory cluster 27% (Supplementary Fig. 6). Consistent with characteristics of the total ADC, the nonmemory cluster of the biomarker confirmed subset was younger (64 ± 8 vs. 67 ± 8 years, P < .01), had lower MMSE scores (20.9 ± 3.1 vs. 22.7 ± 3.0, P < .001), and had more severe posterior atrophy (1.51 ± 0.82 vs. 1.13 ± 0.71, P < .05, rated according to a visual rating scale in which higher scores reflect more severe atrophy [29]) than the memory cluster. In addition, the nonmemory cluster was less often APOE ε4 positive than the memory cluster (61% vs. 74%, P < .05). Results are provided in Supplementary Table 4.
4. Discussion
Across four independent AD dementia cohorts, we robustly found two cognitive clusters using a data-driven dual-clustering approach. One cluster was characterized by more prominent memory impairment and the other cluster by more prominent impairment on nonmemory tests. These memory and nonmemory AD phenotypes were consistently found across cohorts, although these cohorts differed in their patient populations (e.g., age, disease severity, monocentre vs. multicentre, geographic location) and composition and extensiveness of NP test battery. Moreover, the clusters were associated with specific demographic, clinical, and neurobiological characteristics. These findings demonstrate the biological relevance of clinical heterogeneity in AD, as this may reflect variation in underlying disease mechanisms.
Of all included patients, 40% belonged to a nonmemory cluster. Compared with the memory clusters, nonmemory cluster patients were younger, less educated, more often APOE ε4 negative, had more severe posterior atrophy, and relatively spared hippocampi. In addition, patients assigned to the nonmemory clusters reported shorter disease duration, but they had on average lower MMSE scores. Possibly, nonmemory clusters are associated with a more aggressive disease progression [42,43]. Future studies should address the question whether cognitive subtypes are related to the rate of decline, and hence suitable as a putative prognostic marker.
Among the suggested disease mechanisms causing heterogeneity is the influence of copathologies, for example, vascular pathology. However, we did not find differences in severity of ischemic vascular pathology (WMH). Of note, cognitive heterogeneity is most prominent in early onset AD patients, where AD pathology is often pure and copathologies are less present [44]. Biomarker support for AD pathology was not available for each patient but the available data showed no difference in AD biomarker positivity between clusters, suggesting that misdiagnosis is not a major driver of our findings. Variation in disease mechanisms could also be sought in the origin and spreading of neurofibrillary tangles (typically characterized by origin in the entorhinal cortex, progressing through the hippocampus to the association cortex and finally to the cortex [45]) because the medial temporal lobe was relatively spared and the posterior cortex most prominently affected in the nonmemory phenotype. This idea fits with the hypothesis that early onset APOE ε4 negative AD patients are predisposed to vulnerability of cerebral networks beyond the medial temporal lobe [45,46]. This hypothesis coincides with an autopsy study that identified an AD subtype with relatively less tangles in the hippocampus that was associated with younger age at death, male sex, rapid disease progression, and more often focal cortical clinical syndromes [47]. However, this phenotype only tended to have less often an APOE ε4 positive genotype (P = .067). Probably, the APOE ε4 allele is not the only AD risk factor that modifies clinical heterogeneity; the effect of genetic risk factors on clinical AD phenotype is still poorly understood. An increasing number of promising genes, however, are discovered to be associated with higher or reduced risks for developing AD and with certain neuropathologic pathways [48].
A limitation of our study is that we did not have postmortem pathologic confirmation of patients, so we cannot rule out the possibility of misdiagnosis. We think, however, that misdiagnoses will not have driven our results, as in all cohorts diagnoses were made according to the careful application of clinical criteria, and repeating analyses in the CSF biomarker confirmed that subset of the ADC gave similar results.
Furthermore, the present study could be biased because we excluded patients that were already severely demented (i.e., MMSE score <17) at diagnosis. We think that this selection bias could have resulted in underrepresentation of the nonmemory phenotype, because atypical AD variants are less easily recognized as AD (especially at younger age, when other causes for cognitive complaints such as depression or burnout are more common) and therefore probably more often associated with patients’ and doctors’ delay, and delay because of initial misdiagnoses. In addition, nonmemory patients reported shorter disease duration, whereas MMSE scores were already lower, possibly because of faster disease progression before diagnosis, suggesting higher risk for more severe dementia at the time of diagnosis.
It could be argued that the substantial differences between cohorts in patient population (e.g., geographically, age, disease severity, and degree of cognitive heterogeneity within cohorts) and in extensiveness of NP test battery could be a limitation. However, we see this as a strong point of our study because we were able to replicate our finding of two robust clusters with their corresponding clinical characterization. This suggests that the clusters we identified are generalizable to other AD populations; an often-encountered limitation of data-driven methods to cluster patients is that of limited generalizability.
Nonmemory cluster patients had on average lower MMSE scores, and therefore we performed additional analyses to study whether clustering has been driven by disease severity. Repeating the clustering after stratification based on MMSE scores, memory and nonmemory clusters were identified in each stratified cohort as well. Cluster differences in terms of clinical characteristics were largely similar for the strata, albeit more pronounced in the mildly demented stratum. This suggests that clinical heterogeneity is more prominently present in early stages of AD.
Our results emphasize the presence of nonmemory phenotypes in AD. Being able to identify AD subtypes is important in a clinical setting for early and adequate diagnosis and personalized medicine. Also, cognitive profiling should be taken into account when including patients for clinical trials or when choosing cognitive outcomes to analyze the effect of an intervention. Furthermore, the existence of these clusters with similar patient characteristics across independent cohorts suggests that cognitive heterogeneity is caused by different disease mechanisms.
In conclusion, we found two robust AD subtypes using a data-driven clustering approach in four AD cohorts. Identified clusters were associated with distinct demographic, clinical, and neurobiological characteristics, emphasizing the presence of cognitive heterogeneity in AD and suggesting variation in underlying pathology.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: Alzheimer’s disease (AD) is characterized by cognitive heterogeneity. We searched PubMed for clinical and neurobiological heterogeneity in AD profiles and for data-driven approaches used to identify AD subtypes based on neuropsychological test scores. Several studies demonstrated the potential of clustering methods to identify cognitive AD subtypes. Identified subtypes showed distinct clinical characteristics. However, none of the previous studies tested the generalizability of the cluster solutions, because those results were based on single-cohort studies.
Interpretation: In four large AD cohorts, we consistently identified two cognitive clusters using nonnegative matrix factorization of neuropsychological test scores. In each cohort one cluster most prominently showed memory impairment and the other cluster was relatively memory spared. These clusters were associated with distinct clinical characteristics.
Future directions: Future research should aim to further study the underlying biological disease mechanisms that cause a nonmemory AD phenotype.
Acknowledgments
The VU University Alzheimer Center is supported by Alzheimer Nederland and Stichting VUmc Fonds. The clinical database structure of the Amsterdam Dementia Cohort (ADC) was developed with funding from Stichting Dioraphte.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health, Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc; Cogstate; Eisai Inc; Elan Pharmaceuticals, Inc; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc; Fujirebio; GE Healthcare; IXICO Ltd; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck &Co, Inc; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org/). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuroimaging at the University of Southern California.
The Dementia Competence Network (DCN) was supported by a grant from the German Federal Ministry of Education and Research (BMBF) (01GI0420). Additional funding related to randomized clinical trials was provided by Janssen-Cilag and Merz Pharmaceuticals. The latter funds were exclusively used for personnel, pharmaceuticals, blistering and shipment of medication, monitoring, and as capitation fees for recruiting centers.
Footnotes
Conflicts of Interest: N.M.E.S, B.M.T, T.K., S.W., M.W., B.I.C.-S., J.H.K., B.L.M., and W.M.F. report no conflicts of interest. F.B. serves as a consultant to Biogen-Idec, Janssen Alzheimer Immunotherapy, Bayer-Schering, Merck-Serono, Roche, Novartis, Genzyme, and Sanofi-aventis. He has received sponsoring from EU-H2020, NWO, SMSR, EU-FP7, TEVA, Novartis, and Toshiba; he is supported by NIHR-BRC-UCL and is a member of editorial boards of Radiology, Brain, Neuroradiology, MSJ, and Neurology. C.E.T. serves on the advisory board of Fujirebio and Roche, received research consumables from Euroimmun, IBL, Fujirebio, Invitrogen, and Mesoscale Discovery, and performed contract research for IBL, Shire, Boehringer, Roche, and Probiodrug; she has received grants from the European Commission, the Dutch Research Council (ZonMW), Association of Frontotemporal Dementia/Alzheimer’s Drug Discovery Foundation, ISAO, and the Alzheimer’s Drug Discovery Foundation. She received research consumables from Euroimmun, IBL, Fujirebio, Invitrogen, and Mesoscale Discovery, and performed contract research for IBL, Shire, Boehringer, Roche, and Probiodrug. She received lecture fee from Roche and Axon Neurosciences. J.K. reports no conflict of interest with the content of the present manuscript. He holds the following patents: Diagnosis of Alzheimer’s disease (WO 2004/092737 A1), Immunoglobulin-bound Ab-peptides and immunoglobulins-binding Ab-peptides in diagnosis and therapy of Alzheimer’s dementia (WO 2007/082750 A1), Large Aβ-peptide binding particles (LAPS) in diagnosis and therapy of Alzheimer’s dementia (EP 1 811304 A1, 2007), New formulations for diagnosis of Alzheimer’s disease (WO 2011/124376 A1), and Methods of differentially diagnosing dementias (EP 2095128B1, 2013). O.P. received consulting and lecture fees from Affiris, Piramal, Novartis, Lilly and Roche, and he received grant support from Lilly, Genentech, Lundbeck, Probiodrug and Takeda. G.D.R. receives research support from Avid Radiopharmaceuticals/Eli Lilly, GE Healthcare, and Piramal. He has received speaking honoraria or consulting fees from Eisai, Roche, Putnam, Lundbeck. P.S. has received grant support for the VU University Alzheimer Center from GE Healthcare, Nutricia Research, Piramal, and Merck. In the past 2 years he has received consultancy/speaker fees (paid to the institution) from Probiodrug, EIP Pharma, Sanofi, Novartis, Piramal, and GE Healthcare.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jalz.2017.03.002.
References
- 1.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorno-Tempini ML, Hillis AE, Weintraub S, Kertesz A, Mendez M, Cappa SF, et al. Classification of primary progressive aphasia and its variants. Neurology. 2011;76:1006–14. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson DF, Davis RJ, Snyder BD. Posterior cortical atrophy. Arch Neurol. 1988;45:789–93. doi: 10.1001/archneur.1988.00520310107024. [DOI] [PubMed] [Google Scholar]
- 4.Crutch SJ, Schott JM, Rabinovici GD, Boeve BF, Cappa SF, Dickerson BC, et al. Shining a light on posterior cortical atrophy. Alzheimers Dement. 2013;9:463–5. doi: 10.1016/j.jalz.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Ossenkoppele R, Pijnenburg YA, Perry DC, Cohn-Sheehy BI, Scheltens NM, Vogel JW, et al. The behavioural/dysexecutive variant of Alzheimer’s disease: clinical, neuroimaging and pathological features. Brain. 2015;138:2732–49. doi: 10.1093/brain/awv191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Der Flier WM, Schoonenboom SN, Pijnenburg YA, Fox NC, Scheltens P. The effect of APOE genotype on clinical phenotype in Alzheimer disease. Neurology. 2006;67:526–7. doi: 10.1212/01.wnl.0000228222.17111.2a. [DOI] [PubMed] [Google Scholar]
- 7.van der Vlies AE, Pijnenburg YA, Koene T, Klein M, Kok A, Scheltens P, et al. Cognitive impairment in Alzheimer’s disease is modified by APOE genotype. Dement Geriatr Cogn Disord. 2007;24:98–103. doi: 10.1159/000104467. [DOI] [PubMed] [Google Scholar]
- 8.Smits LL, Tijms BM, Benedictus MR, Koedam EL, Koene T, Reuling IE, et al. Regional atrophy is associated with impairment in distinct cognitive domains in Alzheimer’s disease. Alzheimers Dement. 2014;10:S299–305. doi: 10.1016/j.jalz.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Ossenkoppele R, Cohn-Sheehy BI, La Joie R, Vogel JW, Möller C, Lehmann M, et al. Atrophy patterns in early clinical stages across distinct phenotypes of Alzheimer’s disease. Hum Brain Mapp. 2015;36:4421–37. doi: 10.1002/hbm.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ossenkoppele R, Mattsson N, Teunissen CE, Barkhof F, Pijnenburg Y, Scheltens P, et al. Cerebrospinal fluid biomarkers and cerebral atrophy in distinct clinical variants of probable Alzheimer’s disease. Neurobiol Aging. 2015;36:2340–7. doi: 10.1016/j.neurobiolaging.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stopford CL, Snowden JS, Thompson JC, Neary D. Variability in cognitive presentation of Alzheimer’s disease. Cortex. 2008;44:185–95. doi: 10.1016/j.cortex.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Davidson JE, Irizarry MC, Bray BC, Wetten S, Galwey N, Gibson R, et al. An exploration of cognitive subgroups in Alzheimer’s disease. J Int Neuropsychol Soc. 2010;16:233–43. doi: 10.1017/S1355617709991160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Libon DJ, Drabick DA, Giovannetti T, Price CC, Bondi MW, Eppig J, et al. Neuropsychological syndromes associated with Alzheimer’s/vascular dementia: a latent class analysis. J Alzheimers Dis. 2014;42:999–1014. doi: 10.3233/JAD-132147. [DOI] [PubMed] [Google Scholar]
- 14.Scheltens NM, Galindo-Garre F, Pijnenburg YA, van der Vlies AE, Smits LL, Koene T, et al. The identification of cognitive subtypes in Alzheimer’s disease dementia using latent class analysis. J Neurol Neurosurg Psychiatr. 2016;87:235–43. doi: 10.1136/jnnp-2014-309582. [DOI] [PubMed] [Google Scholar]
- 15.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–9. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YX, Zhang YJ. Nonnegative matrix factorization: a comprehensive review. IEEE Trans Knowl Data Eng. 2013;25:1336–53. [Google Scholar]
- 17.Carmona-Saez P, Pascual-Marqui RD, Tirado F, Carazo JM, Pascual-Montano A. Biclustering of gene expression data by non-smooth nonnegative matrix factorization. BMC Bioinformatics. 2006;7:78. doi: 10.1186/1471-2105-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shokrollahi M, Krishnan S, Dopsa DD, Muir RT, Black SE, Swartz RH, et al. Nonnegative matrix factorization and sparse representation for the automated detection of periodic limb movements in sleep. Med Biol Eng Comput. 2016;54:1641–54. doi: 10.1007/s11517-015-1444-y. [DOI] [PubMed] [Google Scholar]
- 19.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state.” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–98. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 20.van der Flier WM, Pijnenburg YA, Prins N, Lemstra AW, Bouwman FH, Teunissen CE, et al. Optimizing patient care and research: the Amsterdam Dementia Cohort. J Alzheimers Dis. 2014;41:313–27. doi: 10.3233/JAD-132306. [DOI] [PubMed] [Google Scholar]
- 21.Verhage F. Intelligence and religious persuasion. Ned Tijdschr Psychol. 1964;19:247–54. [PubMed] [Google Scholar]
- 22.Kornhuber J, Schmidtke K, Frolich L, Perneczky R, Wolf S, Hampel H, et al. Early and differential diagnosis of dementia and mild cognitive impairment: design and cohort baseline characteristics of the German Dementia Competence Network. Dement Geriatr Cogn Disord. 2009;27:404–17. doi: 10.1159/000210388. [DOI] [PubMed] [Google Scholar]
- 23.Kramer JH, Jurik J, Sha SJ, Rankin KP, Rosen HJ, Johnson JK, et al. Distinctive neuropsychological patterns in frontotemporal dementia, semantic dementia, and Alzheimer disease. Cogn Behav Neurol. 2003;16:211–8. doi: 10.1097/00146965-200312000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Van Buuren S, Groothuis-Oudshoorn K. MICE: multivariate imputation by chained equations in R. J Stat Softw. 2011;45:1–67. [Google Scholar]
- 25.Van Buuren S. Flexible imputation of missing data. 20125245. Boca Raton, FL: CRC Press; 2012. [Google Scholar]
- 26.Rubin DB. Multiple imputation for nonresponse in surveys. Hoboken, NJ, USA: John Wiley & Sons; 2009. [Google Scholar]
- 27.Marshall A, Altman DG, Holder RL, Royston P. Combining estimates of interest in prognostic modelling studies after multiple imputation: current practice and guidelines. BMC Med Res Methodol. 2009;9:57. doi: 10.1186/1471-2288-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheltens P, Launer LJ, Barkhof F, Weinstein HC, van Gool WA. Visual assessment of medial temporal lobe atrophy on magnetic resonance imaging: interobserver reliability. J Neurol. 1995;242:557–60. doi: 10.1007/BF00868807. [DOI] [PubMed] [Google Scholar]
- 29.Koedam EL, Lehmann M, van der Flier WM, Scheltens P, Pijnenburg YA, Fox N, et al. Visual assessment of posterior atrophy development of a MRI rating scale. Eur Radiol. 2011;21:2618–25. doi: 10.1007/s00330-011-2205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fazekas F, Chawluk JB, Alavi A, Hurtig HI, Zimmerman RA. MR signal abnormalities at 1.5 T in Alzheimer’s dementia and normal aging. AJR Am J Roentgenol. 1987;149:351–6. doi: 10.2214/ajr.149.2.351. [DOI] [PubMed] [Google Scholar]
- 31.Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–91. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–81. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz CG, Fletcher E, DeCarli CS, Carmichael O. Fully-automated white matter hyperintensity detection with anatomical prior knowledge and without FLAIRs. Inf Process Med Imaging. 2009;21:239–51. doi: 10.1007/978-3-642-02498-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ewers M, Teipel SJ, Dietrich O, Schönberg SO, Jessen F, Heun R, et al. Multicenter assessment of reliability of cranial MRI. Neurobiol Aging. 2006;27:1051–9. doi: 10.1016/j.neurobiolaging.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 35.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–22. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–19. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 37.Lewczuk P, Kornhuber J, Wiltfang J. The German Competence Net Dementias: standard operating procedures for the neurochemical dementia diagnostics. J Neural Transm (Vienna) 2006;113:1075–80. doi: 10.1007/s00702-006-0511-9. [DOI] [PubMed] [Google Scholar]
- 38.Duits FH, Teunissen CE, Bouwman FH, Visser PJ, Mattsson N, Zetterberg H, et al. The cerebrospinal fluid “Alzheimer profile”: easily said, but what does it mean? Alzheimers Dement. 2014;10:713–723.e2. doi: 10.1016/j.jalz.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 39.Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–13. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pascual-Montano A, Carazo JM, Kochi K, Lehmann D, Pascual-Marqui RD. Nonsmooth nonnegative matrix factorization (nsNMF) IEEE Trans Pattern Anal Mach Intell. 2006;28:403–15. doi: 10.1109/TPAMI.2006.60. [DOI] [PubMed] [Google Scholar]
- 42.van der Vlies AE, Koedam EL, Pijnenburg YA, Twisk JW, Scheltens P, Van Der Flier WM. Most rapid cognitive decline in APOE epsilon4 negative Alzheimer’s disease with early onset. Psychol Med. 2009;39:1907–11. doi: 10.1017/S0033291709005492. [DOI] [PubMed] [Google Scholar]
- 43.Smits LL, Pijnenburg YA, van der Vlies AE, Koedam EL, Bouwman FH, Reuling IE, et al. Early onset APOE E4-negative Alzheimer’s disease patients show faster cognitive decline on nonmemory domains. Eur Neuropsychopharmacol. 2015;25:1010–7. doi: 10.1016/j.euroneuro.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 44.van der Flier WM. Clinical heterogeneity in familial Alzheimer’s disease. Lancet Neurol. 2016;15:1296–8. doi: 10.1016/S1474-4422(16)30275-7. [DOI] [PubMed] [Google Scholar]
- 45.Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 46.van der Flier WM, Pijnenburg YA, Fox NC, Scheltens P. Early-onset versus late-onset Alzheimer’s disease: the case of the missing APOE ε4 allele. Lancet Neurol. 2011;10:280–8. doi: 10.1016/S1474-4422(10)70306-9. [DOI] [PubMed] [Google Scholar]
- 47.Murray ME, Graff-Radford NR, Ross OA, Petersen RC, Duara R, Dickson DW. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: a retrospective study. Lancet Neurol. 2011;10:785–96. doi: 10.1016/S1474-4422(11)70156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.