Abstract
Amyloid fibrils are highly ordered protein aggregates associated with more than 40 human diseases. The exact conditions in which the fibrils are grown determine many types of reported fibril polymorphism, including different twist patterns. Twist-based polymorphs display unique mechanical properties in vitro and the relevance of twist polymorphism in amyloid diseases has been suggested. We present transmission electron microscopy images of Aβ42-derived (amyloid β) fibrils, which are associated with Alzheimer’s disease, demonstrating the presence of twist variability even within a single long fibril. To better understand the molecular underpinnings of twist polymorphism, we present a structural and thermodynamics analysis of molecular dynamics simulations of the twisting of β-sheet protofilaments of a well-characterized cross-β model: the GNNQQNY peptide from the yeast prion Sup35. The results show that a protofilament model of GNNQQNY is able to adopt twist angles from −11 degrees on the left-handed side to +8 degrees on the right-handed side in response to various external conditions while keeping an unchanged peptide structure. The potential of mean force (PMF) of this cross-β structure upon twisting revealed that only ~2 kBT per peptide are needed to stabilize a straight conformation with respect to the left-handed free energy minimum. The PMF also shows that the canonical structural core of β-sheets, i.e. the hydrogen-bonded backbone β-strands, favours the straight conformation. However, the concerted effects of the side chains contribute to twisting, which provides a rationale to correlate polypeptide sequence, environmental growth conditions and number of protofilaments in a fibril with twist polymorphisms.
Graphical Abstract
Introduction
Amyloid fibrils are elongated protein aggregates characterized by a cross-β sheet structure. Amyloid fibrils can originate from more than 40 different peptides or proteins,1 which share no primary structure homology. Amyloids are associated with up to 40 human disease syndromes, ranging from common neurodegenerative disorders such as Alzheimer’s disease (AD), to rare disease syndromes like Familial Amyloid Polyneuropathy. 2,3 Amyloid fibrils can also be functional4,5 and display certain mechanical properties that make them potentially attractive for use as artificial biomaterials. 6–8
By formal pathological definition, amyloid deposits are filamentous structures of variable length that deposit in body tissues and fluoresce a green–yellow colour when stained with the dye Congo red and viewed between crossed polarizers1. Moreover, the fibrils must display the ‘cross-β’ diffraction pattern caused by the cross-β-sheet motif when irradiated with X-rays 9.
The ability to form amyloid-like structures is a property shared by many polypeptide sequences given that the right conditions (pH, T, salt concentration, etc.) are met.10 This observation strongly suggests the involvement of a feature common to all polypeptides independent of their sequence. The primary building blocks of amyloid fibrils are extended β-sheet structures assembled from numerous peptide monomers, known as protofilaments, which are stabilized primarily by backbone-backbone hydrogen bond interactions. 3,6,11–19 A protofilament often consists of two extended β-sheets with interlocked side chains forming so-called steric zippers. The β-sheets, which consist of stacked β-strands, run along the long axis of the filament and the β-strands run perpendicular to the filament. This arrangement results in the characteristic cross-β signature of amyloid-like structures shown by X-ray diffraction. 20–23
Mature amyloid fibrils, which generally consist of several filaments twisted into a cable-like structure, typically show a high degree of heterogeneity or polymorphism that determines their mechanical properties, but also pathology. 9,24–33 While the backbone provides the structural framework for the cross-β structure,14,34–36 side chain properties and their interactions will dictate the peptide conformation within the protofilament as well as the interactions between them.37,38 Amyloid fibrils derived from peptides or proteins of the same sequence display profound polymorphic variations. In these cases, the conditions of fibril growth define the structure of the mature fibril.38–41 Certain conditions even lead to the co-existence of different fibril polymorphs, suggesting that these polymorphs can be very close in energy.42,43 One key type of polymorphism (sometimes known as supramolecular polymorphism) relates specifically to differences in the arrangement of protofilaments in the mature fibril, even while the individual protofilaments remain virtually identical in their internal structure. 44–47
Protofilament twisting (pitch angle), which can occur without significant peptide conformational changes, defines the appearance of protofilaments and mature amyloid fibrils. While Atomic Force Microscopy (AFM) experiments and modelling approaches allow one to assess some mechanical properties of fibrils, such as shear and bending,48 only a few theoretical studies have derived phenomenological models that link the pitch of fibril twists observed in experimental studies to some other structural properties, such as the width of the fibril and some elastic term describing the shear and interaction between protofilaments often extracted from experiments.49,50
Here we focus on amyloid fibril twist polymorphism. We first analyse transmission electron microscopy (TEM) images to show that variable pitch may be observed in vitro on a single fibril of Aβ42. Next, we use the Sup35 peptide GNNQQNY from the prion protein as a model system to explore this phenomenon in more detail. High-resolution structures of this peptide in an amyloid-like, but microcrystalline assembly11,51 and structural data of actual amyloid-like fibrils52–54 showed that GNNQQNY and derivatives form structurally related fibrils and microcrystals. We studied the properties of this amyloid model system and its cross-β structure to rationalize the existence of twisting polymorphisms. We performed molecular dynamics (MD) simulations of full atomistic models and developed an innovative protocol to measure the free energy associated with protofilament twisting. Our results provide structural and thermodynamic insights into the origin and extent of fibril twist polymorphism.
Results
Twisting structure in Alzheimer’s disease Aβ42 fibril
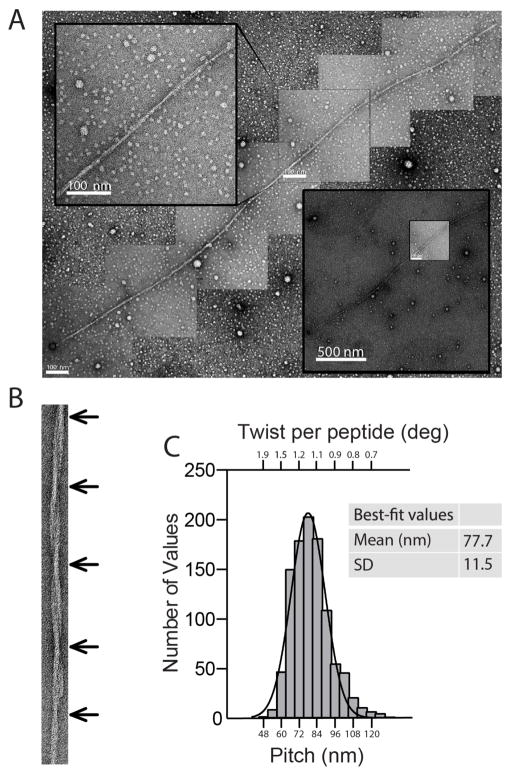
In vitro structural studies have shown that environmental conditions such as temperature, growth conditions (i.e., quiescent versus shaking or stirring) and pH have a dramatic impact on the twisting structure of Aβ fibrils. 28,55 To demonstrate the twisting of Aβ42 fibrils, we incubated Aβ42 peptide under quiescent conditions in sodium borate phosphate, pH 8.6. TEM images revealed a flexible and flat, ribbon-like fibril with a 12-nm width and twisting period between 50 and 125 nm with a mean at 77.7 nm and a standard deviation of 11.5 nm (Figure 1). This image suggests that the degree of fibril twist may vary within a single fibril. A similar observation was reported for an 11-residue fragment of the protein transthyretin.45 Note that different incubation conditions resulted in rigid and rope-like Aβ42 fibrils characterized by different twisting structure and abundant branching (data not shown).
Figure 1.
Twisting structure of an Alzheimer’s disease Aβ42 fibril prepared in vitro. A. Representative negative staining transmission electron microscopy images of a long Aβ42 amyloid fibril. The bigger image is a composite reconstruction obtained by arranging high magnification (scale bar = 100 nm) images. A lower magnification of the same fibril is shown in the lower right panel (scale bar = 500 nm). The upper left panel shows a fibril segment (scale bar = 100 nm) where the twisting is visible. B. Representative negative staining transmission electron microscopy image of a Aβ42 fibril. The arrows indicate the twist points. C. Normal distribution of the distance between twists in a Aβ42 fibril (arrows in B). Mean = 77.7 nm and SD = 11.5 nm. Data are expressed as number of values (tot n = 1012) per bin (bin size = 6 nm). The twist per peptide for a given fibril pitch was obtained as twist=180 deg/(pitch/0.5 nm), where 0.5 nm is the expected distance between strands in a sheet. See Figure S1 for more details.
At present the Aβ42 protofilament, like other clinically-relevant amyloidogenic peptides and proteins, is too intricate to study extensively using all-atom MD simulations. However, an amyloid-forming heptapeptide derived from the yeast prion Sup35 provides a satisfactory model system to address, for example, the factors that contribute to protofilament twisting polymorphisms.
Twist-polymorphism of the GNNQQNY cross-β structure
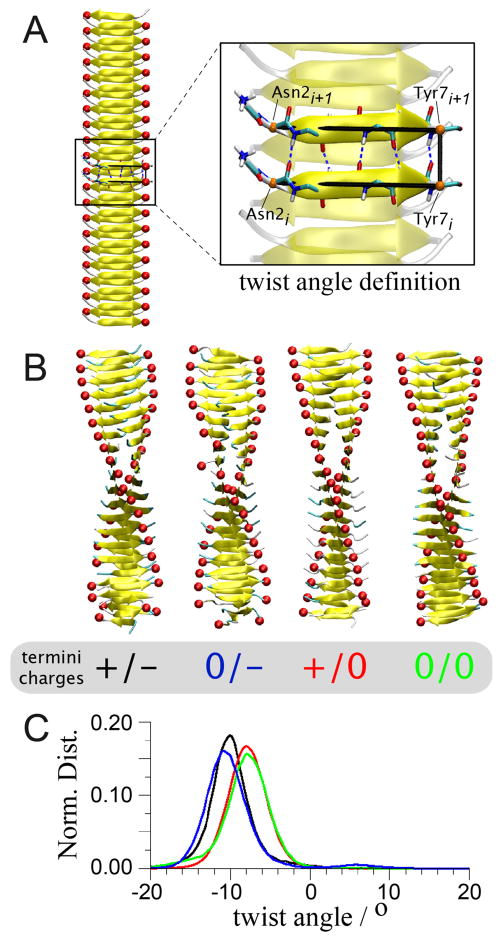
To explore twist polymorphism of the cross-β structure of the GNNQQNY peptide, a model consisting of two β-sheets with 20 peptides each (Figure 2) was simulated in monomeric, dimeric and tetrameric forms. Here, and elsewhere in the manuscript we will reserve the terms mono-, di-, and tetramer(ic) for multiples of a cross-β filament that is itself assembled from two interacting β-sheets (Figure 1). The peptide’s termini charges and the mode of interaction between the cross-β filament building blocks in the polymeric structures were varied. Initial conformations obey the lattice parameters and space group reported for the crystal.11 The twist angle of the cross-β structures, as defined in Figure 2, is used to follow the evolution of the structure.
Figure 2.
Twist polymorphism in isolated cross-β filament structures. A. Definition of the individual twist angle of our model structure shown on a straight conformation of the cross-β. The angles between the vectors Asn2-Tyr7 (Cα) of two consecutive peptides, i and i+1, are used; in other words, the torsion angle around Tyr7i-Tyr7i+1 (Cα) B. Equilibrated conformations of the cross-β structure free in solution obtained by a 15 ns MD simulation started from the straight conformation found in a microcrystal environment (shown in A). Four configurations of the termini charges were studied (+/−, 0/−, +/0 and 0/0) where the charges for the N- and C-terminus, respectively, are indicated. Red spheres are placed on the C-terminus of each peptide to guide the eye in discerning the cross-β structure twist. C. Normalized distributions of the fibril twist angle (colour code indicated in B).
Monomeric cross-β structures
Starting from the straight conformation, which is stable in a microcrystal environment,11,56 the cross-β model structure readily adopts a steady left-handed twist.42,56,57 Our simulations show that this is true regardless of the charge states of the peptide’s termini (Figure 2). The termini’s charge configuration, however, defines the extent of the twist of the structure. While the presence of positive charges on the N-terminus does not affect the twist of the structure, the presence of negative charges on the C-terminus increases the twist angle by ~20% as compared to the both-neutral (0/0) and charged N-termini (+/0) structures (from −8 to −10 degrees). Most informative, these effects of the N- and C-terminus charges are independent from each other. Since strands are parallel within a sheet while the sheets are antiparallel to each other, these charge effects are dominated by repulsive intra-sheet electrostatic interactions as opposed to attractive inter-sheet interactions. As noted in the Methods, the experimental ssNMR studies of the microcrystal structures 11,51–53 indicate that the N-termini are protonated and positively charged, while the C-termini are negatively charged (likely reflecting a shift in the C-terminal pKa in the microcrystalline environment due to the proximity of the oppositely charged N-terminus in comparison a situation in solution).
Polymeric cross-β structures
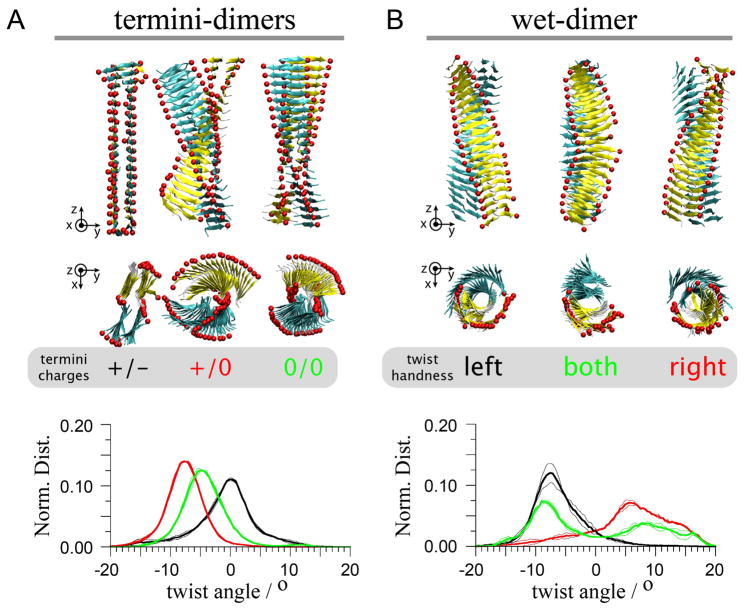
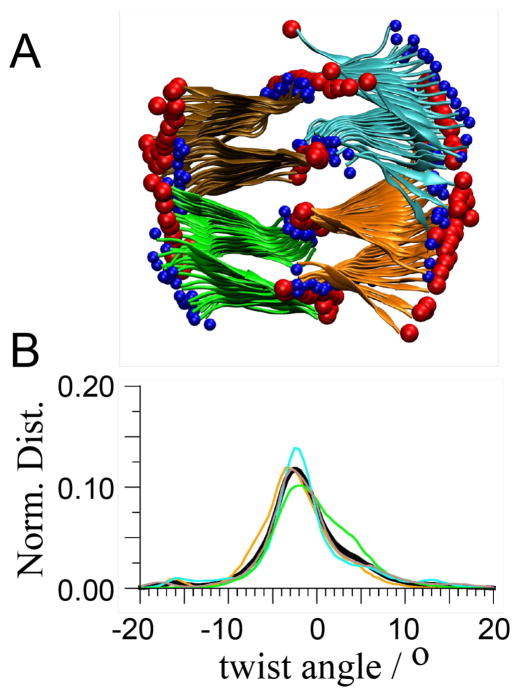
The possible effects of interactions between cross-β structures on the structure of the monomer were addressed by studying systems in which dimers of cross-β structures interact in two distinct manners (Figure 3). In one case the cross-β structures interact through their termini (termini–dimer), and in the other through the so-called “wet” interface (wet-dimer), defined as the cross-β structures interacting through their face exposed to the solvent when isolated. 11 A tetramer was also built (Figure 4). In that case both termini- and wet-dimer interfaces are present in the structure, partially recovering the supramolecular interactions present in the crystal lattice. Starting from these interacting straight cross-β structures, placed at distances as found in the microcrystalline environment,11 the systems were relaxed until they reached a stable and long-lived conformation. The final structures of the polymeric assemblies differ systematically as shown in Figure 3 and 4.
Figure 3.
Twist polymorphism in dimers of cross-β structures. Equilibrated conformations of cross-β structure dimers interacting through their termini (A, termini-dimers) or through their wet interface (B, wet-dimer) are shown. The wet interface is the surface of the sheets facing the solvent in the monomer of the cross-β structure.10 Structures were obtained from at least 15 ns MD simulations free in solution. All simulations were started from the straight conformation of the crystal structure (Figure 2A) and inter-monomers distances were taken from the crystal parameters.10 Three different charge configurations of the termini are shown for the termini-dimers (A), and three different conformations obtained for the wet-dimers in the same termini conditions (B). In each case a side (top panel) and a top (middle panel) view are shown together with the normalized distributions of twist angles (bottom panel). The colour code of the normalized distributions is as indicated in the grey areas. Coloured spheres are placed on the C-termini to guide the eye in following the twist of the cross-β structures.
Figure 4.
Morphology of a tetramer of cross-β structures. A. Cartoon representation of the resulting conformation of the tetramer after a 30 ns MD simulation free in solution and started from the straight conformation found in the crystal10. Both termini were charged (+/−). To guide the eye of the reader, red spheres are placed on the C-termini and smaller blue spheres on the N-termini. Each cross-β unit has a different colour. B. Normalized distribution of the twist angle of each monomer obtained with the last 20 ns of the simulation. The colour code is in accordance with panel A.
In the case of the termini-dimers, three configurations of the termini charges were probed: both charged (+/−), both neutral (0,0) and with the N-terminus of the peptide (only) charged (+/0). The three configurations relaxed to a different conformation of the cross-β monomers and of their assembly (Figure 3). Most notably, in the +/− configuration the cross-β structures maintained a straight conformation (twist angle=0). The formation of a twist in the two monomers (each a cross-β pair of β-sheets) would impair the energetically favourable charge-charge interactions between the termini of the two protofilaments. Although it cannot be completely excluded that the system is kinetically trapped in a local minimum, these interactions are strong enough to counter balance the intrinsic propensity of the monomers to twist (see above). This system was simulated six times longer than the time needed by the other systems to reach a stable twist. These charge-charge interactions have been reported previously and were suggested to stabilize the straight conformation observed in the crystalline environment.56
In the 0/0 configuration of the termini-dimer, the water molecules initially found at the termini interface were rapidly expelled leading to more intricate contacts between the termini of the two protofilaments when compared with the +/− configuration (Figure 3A). The interacting protofilaments adopted a −5 degrees twist angle indicating that the contacts between neutral termini restrain the twisting compared to the similarly charged isolated monomer that adopted a −8 degrees twist angle (Figure 2). In the case +/0, the monomers in the complex adopt the same left-handed twist as found in the isolated filament. In all cases the two interacting cross-β structures adopted similar conformations (see Figure 3A).
The wet-dimer relaxed into three symmetrical, but distinct and stable conformations. The three conformations appeared despite the use of identical starting conformations and termini configuration (+/−). In one case the cross-β structures adopt a fully left-handed twist, in a second one a fully right-handed twist, and in a third both right- and left-handed sections are present and divide the dimer into two segments of equal size (Figure 3B). At this point it is not clear precisely which energetic factors guided the system to its final conformation, but we observed a strong sensitivity of the conformation to the care given to the preparation of the interface and in particular to the water. The fully left-handed conformation, more likely candidate in view of the monomeric and termini-dimer simulations, is obtained only if the interfacial water molecules that solvate the wet interface of the experimental structure are included in the starting structure. Half and full right-handed sections were obtained when the wet interface lacked the experimental water molecules. Although no further attempt was made to elucidate these observations, they underline the sensitivity of the structure to the protofilament-protofilament interactions and the presence of solvent.
In the case of the tetramer with interfacial water included, the straight conformation (as found in a microcrystal 11) relaxed into a stable structure in which each monomer adopted a small, but non-zero, left-handed twist angle of −2.5° (Figure 4). This value of the twist angle when compared to the ones described above indicates that there is a compensation of effect due to the presence of both termini- and wet-interfaces in the structure. While the termini-interface stabilizes the straight conformation, the wet-interface favours the formation of a small twist.
The larger twist observed in the monomeric structure (−10°, Figure 2) compared to the wet-dimer (−8°, Figure 3B) is in line with the changes reported by Berryman et al.42 and Knowles et al.50, and predicted by Aggeli et al.50 for systems with increasing numbers of interacting protofilaments. The termini-dimer (0°, Figure 3A) compared to the tetramer (−2.5°, Figure 4) does not follow the trend. The absence of twist in the termini-dimer suggests that interactions among protofilaments mimic an increase of the peptide length through the termini interaction, thereby favouring a straight conformation.36 In contrast, the side chain interactions at the wet interface of the tetramer push the system to a slightly twisted conformation.
Thermodynamics of twisting a cross-β structure
The cross-β structure free energy minimum is left-handed
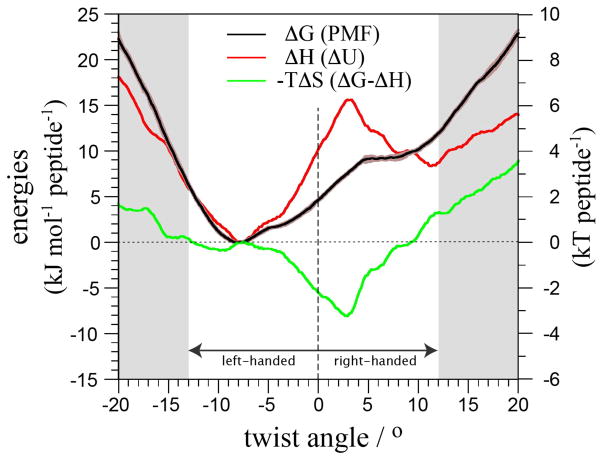
The PMF of the isolated cross-β structure with neutralized termini as a function of its twist angle (Figure 5) undeniably shows that the free energy minimum of the solvated cross-β spine has a left-handed twist angle of about 8°. This is in agreement with previous simulation results from us56 and others groups42,43,50,57 and contrast with the straight conformation observed in a microcrystal environment.11,51 The free energy profile has a v-shape with a higher slope (2 kJ/mol/peptide per degree) towards larger left-handed twist values than when reducing the twist and going to a right-handed twist structure (1 kJ/mol/peptide per degree). The profile indicates the presence of a metastable state for right-handed twist in the range of 5–9 degrees. In the following we focus our interpretation on the values of the twist angle of the cross-β structure within the range −13 to +12°. Beyond these limits the cross-β structure showed clear signs of irregularity and looses its integrity (see Movie in SI).
Figure 5.
Thermodynamics of twisting a cross-β structure. Relative free energy (ΔG=PMF), enthalpy (ΔH=ΔU) and entropy (−TΔS=ΔG−ΔH) of the cross-β structure in solution as a function of its twist angle. The twist angle is defined by the average of the 38 individual dihedrals (Figure 2A). The grey areas indicate the limit of the cross-β structural integrity. For convenience, the energies are expressed per peptide.
The cross-β structure twist is enthalpy-driven
The enthalpy of the system (ΔH) was determined from the total potential energy of the system (ΔU) (Figure 5). The strong similarity of ΔH with ΔG around the ΔG minimum demonstrates that the twist of the cross-β structure is enthalpy-driven. The v-shape of ΔH, however, brakes down as the twist angle of the cross-β structure exceeds +3 degrees, at which point ΔH decreases again until the twist angle reaches +12 degrees where ΔH starts increasing again. This change occurs close to the plateau in the free energy profile and suggests that the metastable state is also of enthalpic origin, i.e., both left- and right-handed structures are at a minimum of ΔH.
The entropy contribution (−TΔS) to the system’s free energy (ΔG) was estimated from the difference between ΔG and the enthalpy of the system (ΔH) (Figure 5). It shows that −TΔS has only a limited contribution for structures with twist angles more negative than −3°. In contrast it contributes significantly to stabilizing the cross-β structure in the range of twist angles between −3 and +7/8°. The gain in entropy compensates for the increase of enthalpy with a maximum contribution at +3 degrees where the enthalpy of the system is also at its maximum.
Contributions to the enthalpy of the system
The terms in the system’s enthalpy relevant to the twist of the cross-β structure were identified. Bonded terms might be discarded since they do not contribute significantly to the system’s enthalpy in the range of twist angle (−13 to +12°) relevant to our analysis. It is only for large values of the twist angle (<−10 and > 11°) that they become relevant and then mainly reveal that the peptide is under stress under these extreme twist angles (Figure S2).
Protein-protein non-bonded interactions shape the systems’ enthalpy
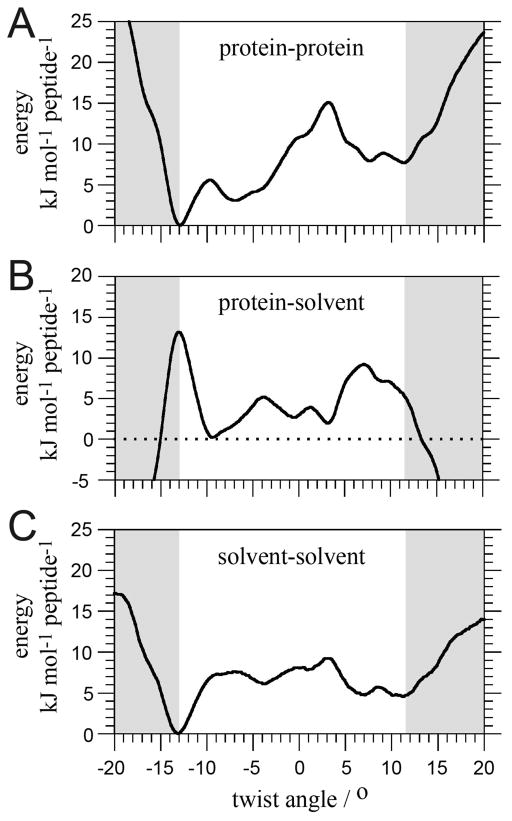
The non-bonded interactions (sum of electrostatic and van der Waals terms) clearly shape the enthalpy of the system upon moderate twisting (Figure 6) with the protein-protein interactions being the major contribution for twist angles between −10 and +10°. They stabilize both left- and right-handed twist conformations. Outside that region, large contributions from protein-protein, protein-solvent, and solvent-solvent interactions compensate each other. Although they are difficult to interpret in detail, the increasing role of solvent interactions reflects the loss of integrity of the structure.
Figure 6.
Decomposition of the protein and solvent non-bonded contributions to the enthalpy of the cross-β filament system shown in Figure 5A, ΔH (red in Figure 5). ΔH= protein-protein (A) + protein-solvent (B) + solvent-solvent (C) + bonded-terms. Bonded terms are shown in Figure S1. For convenience, the energies are expressed per peptide.
Peptide backbone and side chain contributions
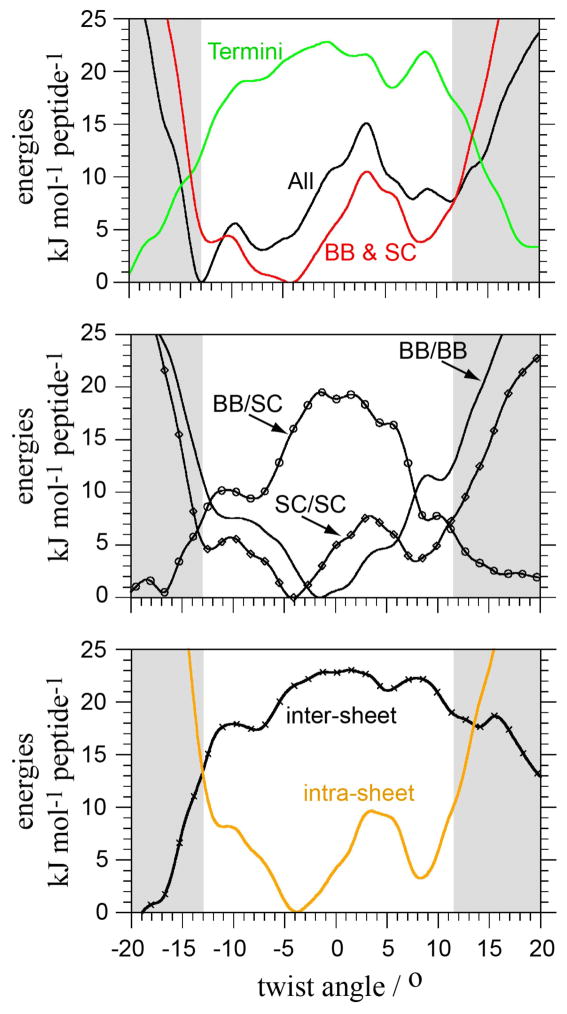
A further decomposition of the protein-protein non-bonded interactions reveals that backbone (BB)/BB non-bonded interactions have a v-shape and stabilize an almost straight conformation of the cross-β structure. In contrast, BB/side chain (SC) interactions favour twisted forms (Figure 7). Importantly, SC/SC interactions stabilize both left- (−4°) and right-handed (+8°) twists of the cross-β structure with a preference for the left-handed form, thereby shaping up the double well of the enthalpy profile as defined by the sum of BB and SC contributions. The termini contribution becomes significant only for large twist angle and tends to stabilize higher twisted forms.
Figure 7.
Decomposition of the backbone (BB), side chain (SC) and termini contributions to the protein-protein non-bonded interactions shown in Figure 6. protein-protein = All = Termini + BB & SC. BB & SC = BB/BB + BB/SC + SC/SC = inter-sheet + intra-sheet. For convenience, the energies are expressed per peptide.
Inter- and intra-sheet interactions
The determination of the intra- and inter-sheet contributions of BB and SC interactions indicate that intra-sheet terms govern the potential energy of the system as indicated by the presence of both left- and right-handed twist minima. The inter-sheet interactions in the isolated cross-β structure tend to stabilize larger twists and left-handed ones in the range of stable protofilament. The double well shape of intra-sheet interactions reflects SC/SC interactions at the dry interface (Figure S3).
Structural changes of the cross-β structure accompanying the twist
Although significant changes of the morphology of the cross-β structure are observed in the range of twist angles studied (Figures 2, 3) the peptide itself undergoes only very little conformational change. Thus, we observe a preservation of the basic conformation of the peptide as found in the microcrystalline environment11,51 within the range of twist for which the cross-β structure is stable. The phi and psi angles of the peptide backbone show monotonous evolution upon twisting of the protofilament (Figure S4) resulting in relatively small changes of each residue. These changes, however, add up to an overall backbone twist of 50 degrees (from +20 to −30) over the length of the peptide in the range of twist angle keeping the integrity of cross-β structure (Figure S5A). The cross-angle between the two sheets in the cross-β structure shows that the two sheets stay parallel for twist angle of the structure around a straight conformation (Figure S5B). It is only when the twist angle exceeds 5 degrees on both right- and left-handed directions that the sheets twist one relative to the other. The solvent accessible surface area (Figure S5C) of the cross-β structure decreases slightly with its twist, in line with a recent analysis of protein structures showing that the twist of β-sheets in proteins has a tendency to minimize SASA.58
Discussion
The pathophysiological hallmark of AD is the appearance of AD plaques in brain tissue. The plaques consist of Aβ42 (and other related peptides including Aβ40) amyloid fibrils. Biophysical investigations of Aβ-derived amyloids often indicate the presence of several polymorphs – internal structure (β-sheet formation and side-chain packing)38, fibril diameter and shape (e.g., ribbon-like or rope-like), branching and amount and frequency of twisting events9,59 contribute to fibril polymorphism. Importantly, a close correlation between structural polymorphisms and disease phenotype has been suggested31,60.
Twist-polymorphism can be demonstrated in filaments and fibrils prepared in vitro under the same conditions. Fibrils with the same morphology experience varying pitches falling into a quasi normal distribution. Our aim was to understand the energetics that underlie fibril twisting polymorphism. Many studies have modelled Aβ42 using all-atom MD simulations, 61–67 however, for the purpose of investigating the twist of fibrils we choose a more simple model system. The yeast prion Sup35 is amenable for our study since the relevant model heptapeptide GNNQQNY forms canonical filaments and fibrils in vitro, and is small enough for comprehensive MD studies. Furthermore, relevant structural data are available to establish initial conditions for MD simulations (Figures 2–4) and extensive experimental studies have been performed on crystal and fibril assemblies.
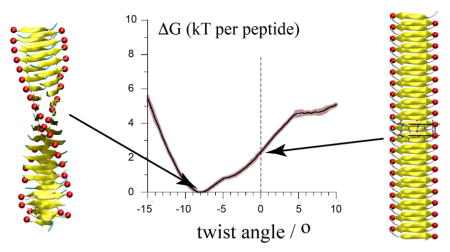
The free energy profile of twisting the cross-β structure of GNNQQNY (a pair of β-sheets) confirmed that its free energy minimum in solution is a left-handed twist (−8°) conformation42,43,50,56,57 and showed that it is enthalpy driven (Figure 5). The decomposition of the potential energy of the system further revealed that protein-protein interactions shape up the enthalpy, that the polypeptide backbone favours the straight conformation and that side chains are determinant in stabilizing the twist of the cross-β structure (Figure 5–7). These observations are in line with the consensus views of β-sheet structures. 14,34,36,68–70 For the Aβ it was suggested that the twist improved side chains packing between neighbouring β-sheets.61
The PMF also revealed that protofilaments with a −13 to +10° twist angle are accessible within 2–4 kT/peptide (Figure 5), making the protofilament quite flexible. Interestingly, the straight conformation found in the microcrystalline environment11,51 is not more than 2 kT/peptide away from the twisted free energy minimum (Figure 5). Our simulations show that the necessary stabilization energy may be provided by the interaction between two protofilaments through their termini (Figure 3A) as found in the microcrystalline environment. 11,51
Although the GNNQQNY protofilament is quite flexible when considered per peptide, collectively, it sits in a deep free energy minimum and is therefore a very stable structure. The systems started from a straight conformation indeed equilibrate very fast (<100 ps) to their respective final form.56 Our ability to twist a protofilament in contrast to breaking it relied on the application of forces homogeneously along the structure, on each peptide. In the determination of the PMF presented in Figure 5, we applied a restraint per peptide-pair in order to mimic what we assume occurs naturally when protofilaments interact. In experiments, forces may not be homogeneously distributed resulting in local accumulation of strain leading to defects, breakage and cracks as often seen in AFM and EM images. It may also occur that a single fibril feels different strains (of any type) along its length thereby twisting to different extents within its own length without affecting the overall morphology of the fibril (Figure 1).
In terms of the typically multimeric protofilament structures, our simulations show that the systems composed of several copies of a cross-β structure all adopted a different twist angle than that observed for the isolated cross-β structure. This observation strongly supports the notion that the mode of interaction between the cross-β structures is determinant for the final conformation of a multimeric system (Figure 2–4). Protofilaments will easily adapt to external forces such as the ones resulting from interactions with other protofilaments in fibrils but also changes of termini charges –a mimic of pH change. This idea, that a conserved cross-β filament structure can self-assemble in different ways (e.g. with different extents of twisting), is reminiscent of experimentally observed supramolecular polymorphism. In this case, amyloidogenic proteins such as tau or huntingtin exon 1 form different fiber polymorphs that upon closer inspection appear to be based on a different way of assembling essentially the same cross-β filament structure. 44–47
It is, however, not clear at this point if and how the changes in the twist of the cross-β structure reported here may account for the reported polymorphic aggregation by GNNQQNY in an experimental setting. This one peptide forms both monoclinic and orthorhombic crystals, in which the cross-β spines are straight.11,51 At higher concentrations, but otherwise identical conditions, amyloid-like fibrils form, which are characteristically assembled from three distinct but reproducible peptide conformers.52,53 The GNNQQNY fibrils are ribbon-like in appearance, with striations thought to reflect cross-β filaments. Presumably it is twisting of the filaments that prevents their growth into a three-dimensional crystalline state. Although we show the possibility of variable twists in a single long fibril in the case of Aβ42 (Figure 1), our computational experiments suggest that the conformation of the peptide is conserved along a protofilament, at least within the range of protofilament twists explored.
To perform our simulations of GNNQQNY peptides we have built protofilament models starting from the straight arrangement as found in the untwisted microcrystal structure and interpreted the degree of twist they acquired due to the combination of their intrinsic propensity to twist and external factors (mimic of pH change and interaction with other protofilaments). The effects are clear: namely, the interactions between protofilaments determine the twist observed in individual protofilaments, without requiring a significant change in the local monomer structures within the fibrils. Thus, once formed, a particular filament structure appears to be quite robust even when submitted to varying external forces or interactions.
This raises an interesting question that relates to the factors that dictate the fundamental structure of the filaments: the nucleation of amyloid structure. The structure of the initial nucleus encodes a certain degree of spontaneous twist that will be reflected in the final assembly. External conditions that favour the formation of twisted nucleating structures would trigger the formation of long twisted fibrils with a couple of interacting protofilaments. Conditions favouring the formation of untwisted β-sheets (e.g., termini-dimer in Figure 3) would allow the three-dimensional stacking of sheets into large straight assemblies as observed in microcrystals. In other words the nucleation of twisted protofilaments would prevent the formation of microcrystals by disfavouring protofilament interactions. Both paths are likely to be close in energy enabling them to occur simultaneously or to compete at specific growth conditions. 54,71
In prior ssNMR studies,52,53 GNNQQNY fibrils contain three different conformers that are closely interacting (<1 nm of each other) within a co-assembled and complex fibril structure that is distinct from that in the microcrystalline state.11,51,56,57 Each protofilament featured an internal polymorphism that is distinct from the type of supramolecular polymorphism examined here, but was for instance also reported for the SNNFGAILSS fragment from hIAPP.13 The three-fold ssNMR signature of the GNNQQNY filaments was found to be reproduced faithfully even when different types of fibril polymorphs were seen by EM.71 This is consistent with our current finding that an identical filament base structure is flexible enough to partake in different types of supramolecular architectures, potentially enabled by variations in the protofilament twists, without sufficient local structural change to see an observable effect by ssNMR. This is reminiscent of a recent study of differently curved HIV capsid assemblies, which examined by ssNMR that the local structural changes associated with obvious difference in capsid curvature.72 Thus, supramolecular structure and local monomer conformations are clearly related, but their interplay appears to be subject to various subtleties that may not be immediately apparent. It appears to be a combination of both the nucleating structure and the interfilament interactions that are needed to fully explain the final fibril architecture, the protofilament twists, fibril stability, and its functional characteristics.
Conclusion
We have demonstrated the twist-flexibility of a model of a cross-β structure of the GNNQQNY peptide from the yeast prion Sup35 and its adaptation to the external forces resulting from the environment (pH or quaternary interactions). This feature of cross-β filaments should help characterizing further the mechanical properties of amyloid-like fibrils and stimulate new experiments to estimate the extent of its contribution to different systems. Notably, the degree of twist of a mature fibril must affect its mechanical properties and could be considered as a tuning feature in the development of functionalized bio-materials73 assuming one could develop a strategy to control the fibril twist. Given the apparent role of the side chains in dictating the twisting, our data would point to compounds interacting with the side chains or termini of amyloidogenic peptides to be good starting point. It would also be important to determine the impact of fibrils’ twist in their pathology,31 in which case the development of drugs affecting the twist could be pharmacologically relevant.
Methods
Amyloid Aβ42 fibril preparation
Human amyolid β peptide (Aβ42), which is a proteolysis product of amyloid precursor polypeptide (APP) was obtained as a synthetic peptide (American Peptide) and solubilized in HFIP at 1μg/μl, dried and stored at −80 °C. On the day of the experiment, the peptide was reconstituted in 2mM NaOH to 1μg/μl, dried and diluted in 10 mM sodium borate phosphate buffer, pH8.6. Aβ42 (10μM) was incubated at 37 °C for 24 h and successively placed in a volume of 5μl onto carbon film 200-mesh copper TEM grids, rinsed with ddH2O and counterstained with 1% aqueous uranyl acetate solution. Samples were viewed with a JEOL 1400 Plus Transmission Electron Microscope and images acquired with a Gatan 2K × 2K digital camera.
Models of the GNNQQNY cross-β structure
The structure of the peptide GNNQQNY was taken from the Protein Data Bank (entry 1YJP11). The various systems simulated were constructed, as in our previous study,56 respecting the lattice parameters and space group reported for the microcrystal.11 A basic unit containing a pair of peptides related by a 21 screw axis was built and replicated 20 times along the axis of the fibril to form a two-stranded β–sheet structural model of a cross-β protofilament. To generate multi-layered starting structures, this system was replicated along the a and/or c axes.
Simulation details
Each system simulated was placed in a periodic rectangular box. The minimal distance between the solute and the box edges was at least 1 nm. The systems were solvated in water and energy minimized. The protonation state of titratable groups in the peptide was as expected at pH 7.0 except for the simulations performed with neutralized termini in which case the NH3+ and COO− groups at the termini were replaced by NH2 and COOH, respectively. Note that none of the side chains are actually titratable. The range of pH in which most experiments on the GNNQQNY peptide are performed, pH 2–3,11,51–53 would correspond to a situation where the N-terminus is positively charged and is near the pKa of the C-terminus.74 Referencing published NMR studies of pH-dependent protonation,75,76 we interpret published MAS ssNMR chemical shifts52 of the microcrystals to indicate a positively charged N-terminus and a negatively charged C-terminus. In the calculation of the PMF we used a peptide with both termini neutral. The charge configuration of the termini is noted +/−, 0/−, +/0 and 0,0 in which +/0/− indicate the charge carried by the N/ and C/- terminus, respectively.
All simulations were performed using the GROMACS simulation package.77 The peptides were modelled using the GROMOS 43a1 force field78 and the water molecules using the SPC model.79 The solute and solvent were weakly coupled80 independently to an external temperature bath at 300 K using a Berendsen thermostat with relaxation time of 0.1 ps. The pressure was weakly coupled80 to an external bath at 1 bar using a relaxation time of 1.0 ps. Covalent bonds within the peptide were constrained using the LINCS algorithm.81 The SETTLE algorithm82 was used to constrain the solvent. Non-bonded interactions were calculated using a twin-range cutoff of 0.8/1.4 nm. The charge-group pair list was updated every five integration steps (Δt=2 fs). To adjust for the truncation of electrostatic interactions beyond the long-range cutoff a Reaction-Field correction83 was applied (εRF=78).
Potential of mean force (PMF) of the cross-β structure upon twisting
PMF values were calculated using an umbrella sampling approach84 where the relative orientation of consecutive peptides was controlled by an additional dihedral angle harmonic potential (Figure 2A). This extra potential was applied to the 19 pairs of peptides along each β-sheet resulting in 38 biasing potentials used simultaneously. The angles were constrained to values from −20 to +20 degrees with 0.5 degree increments resulting in 81 umbrella windows; kθ was set to 104 kJ mol−1 rad−2 (3.05 kJ mol−1 deg−2). A small angle increment was used to assure a sufficient overlap between windows, which is challenging due to the effective 38 dimensions of the restrains. Each umbrella window was simulated for 5 ns, starting from a conformation of the cross-β structure close to the target value. The last 4 ns of simulation for each window were used to calculate the PMF. The error estimates on the final PMF were obtained through a bootstrap procedure in which each umbrella simulation was decomposed into four 1 ns windows. One hundred bootstrapping cycles were performed.
Once the PMF was resolved in the 38 dimensional dihedral space, the probability distribution was projected onto a reduced two-dimensional space, x0 and x1, where x0 is the average dihedral angle (twist angle as defined below) of the 38 individual angles and x1 is the root mean square deviation of the individual dihedral angles from the average dihedral angle of a conformation. The final PMF, w(x0), was obtained by integrating the 2D-PMF on the RMSD dimension from 0 to 1 degree. More details for the unbiasing of the umbrella simulations are provided in our earlier implementation85 of Kumar86,87 and Souaille and Roux88 algorithms and in the SI.
Twist angle
The twist angle of a cross-β structure was defined as the average of the dihedral angle between the vectors Cα/Asn2–Cα/Tyr7 of neighbouring peptides within the same β–sheet. Left-handed angles were given a negative sign (Figure 2).
Energetic analysis
The enthalpy of the different systems was monitored and decomposed into a series of terms as defined in the GROMOS 43a1 force field.78 To account for the contribution of the N- and C-termini, interactions involving the atoms of the terminal -NH3+ (-NH2) and -COO− (-COOH) groups were considered separately. Although the approach used to determine the enthalpy and entropy of the system is not rigorously precise, it has been previously shown to provide good qualitative and quantitative estimations of their contributions.89 The different energy terms were shifted along the z-axis to assure that the relevant part of all the curves shows in the plot so that to allow their comparison. Therefore the energies reported in Figures 5, 6 and 7 in the main manuscript and Figures S2 and S3 in the SI should be seen as depicting variations in energy contributions not absolute values.
Supplementary Material
Acknowledgments
This work was supported by The Netherlands Organisation for Scientific Research (NWO) (X.P and S.J.M., ECHO.08.BM.041). P.C.A.v.d.W. acknowledges grant support from the National Institutes of Health (R01 GM112678). Computer time was allocated by The Netherlands National Computing Facilities Foundation (NCF grant SH-113-V-08). We thank the Electron Microscopy Resource Facility at Rockefeller University.
References
- 1.Sipe JD, Benson MD, Buxbaum JN, Ikeda SI, Merlini G, Saraiva MJM, Westermark P. Nomenclature 2014: Amyloid Fibril Proteins and Clinical Classification of the Amyloidosis. Amyloid. 2014;21(4):221–224. doi: 10.3109/13506129.2014.964858. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Annual Review of Biochemistry. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg D, Jucker M. The Amyloid State of Proteins in Human Diseases. Cell. 2012;148(6):1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maury CPJ. The Emerging Concept of Functional Amyloid. Journal of Internal Medicine. 2009;265(3):329–334. doi: 10.1111/j.1365-2796.2008.02068.x. [DOI] [PubMed] [Google Scholar]
- 5.Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KPR, Simon R, Schubert D, et al. Functional Amyloids as Natural Storage of Peptide Hormones in Pituitary Secretory Granules. Science. 2009;325(5938):328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenwald J, Riek R. Biology of Amyloid: Structure, Function, and Regulation. Structure. 2010;18(10):1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Watt B, Tenza D, Lemmon MA, Kerje S, Raposo G, Andersson L, Marks MS. Mutations in or Near the Transmembrane Domain Alter PMEL Amyloid Formation From Functional to Pathogenic. PLoS Genet. 2011;7(9):e1002286. doi: 10.1371/journal.pgen.1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S. Fabrication of Novel Biomaterials Through Molecular Self-Assembly. Nature Biotechnology. 2003;21(10):1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 9.Riek R, Eisenberg DS. The Activities of Amyloids From a Structural Perspective. Nature. 2016;539(7628):227–235. doi: 10.1038/nature20416. [DOI] [PubMed] [Google Scholar]
- 10.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson CM. Designing Conditions for in Vitro Formation of Amyloid Protofilaments and Fibrils. Proc Natl Acad Sci US A. 1999;96(7):3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Structure of the Cross-B Spine of Amyloid-Like Fibrils. Nature. 2005;435(7043):773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh P, Simonetti K, Sharpe S. Core Structure of Amyloid Fibrils Formed by Residues 106-126 of the Human Prion Protein. Structure. 2009;17(3):417–426. doi: 10.1016/j.str.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen JT, Bjerring M, Jeppesen MD, Pedersen RO, Pedersen JM, Hein KL, Vosegaard T, Skrydstrup T, Otzen DE, Nielsen NC. Unique Identification of Supramolecular Structures in Amyloid Fibrils by Solid-State NMR Spectroscopy. Angew Chem Int Ed. 2009;48(12):2118–2121. doi: 10.1002/anie.200804198. [DOI] [PubMed] [Google Scholar]
- 14.Cheng PN, Pham JD, Nowick JS. The Supramolecular Chemistry of B-Sheets. J Am Chem Soc. 2013;135(15):5477–5492. doi: 10.1021/ja3088407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoop CL, Lin HK, Kar K, Magyarfalvi G, Lamley JM, Boatz JC, Mandal A, Lewandowski JR, Wetzel R, van der Wel PCA. Huntingtin Exon 1 Fibrils Feature an Interdigitated β-Hairpin-Based Polyglutamine Core. Proc Natl Acad Sci US A. 2016;113(6):1546–1551. doi: 10.1073/pnas.1521933113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, et al. Solid-State NMR Structure of a Pathogenic Fibril of Full-Length Human [Alpha]-Synuclein. Nat Struct Mol Biol. 2016;23(5):409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular Structure of B-Amyloid Fibrils in Alzheimer’s Disease Brain Tissue. Cell. 2013;154(6):1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, et al. Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J Am Chem Soc. 2016;138(30):9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid Fibrils of the HET-S(218-289) Prion Form a Beta Solenoid with a Triangular Hydrophobic Core. Science. 2008;319(5869):1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 20.Makin OS, Serpell LC. Structures for Amyloid Fibrils. The FEBS Journal. 2005;272(23):5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- 21.Astbury WT, Dickinson S, Bailey K. The X-Ray Interpretation of Denaturation and the Structure of the Seed Globulins. Biochem J. 1935;29(10):2351–2360.2351. doi: 10.1042/bj0292351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eanes ED, Glenner GG. X-Ray Diffraction Studies on Amyloid Filaments. Journal of Histochemistry & Cytochemistry. 2016;16(11):673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 23.Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, Serpell LC. The Common Architecture of Cross-B Amyloid. J Mol Biol. 2010;395(4):717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 24.Volpatti LR, Vendruscolo M, Dobson CM, Knowles TPJ. A Clear View of Polymorphism, Twist, and Chirality in Amyloid Fibril Formation. ACS Nano. 2013;7(12):10443–10448. doi: 10.1021/nn406121w. [DOI] [PubMed] [Google Scholar]
- 25.Kodali R, Wetzel R. Polymorphism in the Intermediates and Products of Amyloid Assembly. Curr Opin Struct Biol. 2007;17(1):48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Aguzzi A. Unraveling Prion Strains with Cell Biology and Organic Chemistry. Proc Natl Acad Sci US A. 2008;105(1):11–12. doi: 10.1073/pnas.0710824105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones EM, Surewicz WK. Fibril Conformation as the Basis of Species- and Strain-Dependent Seeding Specificity of Mammalian Prion Amyloids. Cell. 2005;121(1):63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 28.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-Propagating, Molecular-Level Polymorphism in Alzheimer’s β-Amyloid Fibrils. Science. 2005;307(5707):262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 29.Schleeger M, vandenAkker CC, Deckert-Gaudig T, Deckert V, Velikov KP, Koenderink G, Bonn M. Amyloids: From Molecular Structure to Mechanical Properties. Polymer. 2013;54(10):2473–2488. [Google Scholar]
- 30.Knowles TPJ, Buehler MJ. Nanomechanics of Functional and Pathological Amyloid Materials. Nat Nanotechnol. 2011;6(8):469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 31.Qiang W, Yau WM, Lu JX, Collinge J, Tycko R. Structural Variation in Amyloid-β Fibrils From Alzheimer’s Disease Clinical Subtypes. Nature. 2017;541(7636):217–221. doi: 10.1038/nature20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandersteen A, Masman MF, De Baets G, Jonckheere W, van der Werf K, Marrink SJ, Rozenski J, Benilova I, De Strooper B, Subramaniam V, et al. Molecular Plasticity Regulates Oligomerization and Cytotoxicity of the Multipeptide-Length Amyloid-B Peptide Pool. J Biol Chem. 2012;287(44):36732–36743. doi: 10.1074/jbc.M112.394635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller Y, Ma B, Nussinov R. Polymorphism in Alzheimer Abeta Amyloid Organization Reflects Conformational Selection in a Rugged Energy Landscape. Chem Rev. 2010;110(8):4820–4838. doi: 10.1021/cr900377t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chothia C. Conformation of Twisted B-Pleated Sheets in Proteins. J Mol Biol. 1973;75(2):295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- 35.Weatherford DW, Salemme FR. Conformations of Twisted Parallel Beta-Sheets and the Origin of Chirality in Protein Structures. Proc Natl Acad Sci US A. 1979;76(1):19–23. doi: 10.1073/pnas.76.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salemme FR. Structural Properties of Protein B-Sheets. Progress in Biophysics and Molecular Biology. 1983;42:95–133. doi: 10.1016/0079-6107(83)90005-6. [DOI] [PubMed] [Google Scholar]
- 37.Fishwick CWG, Beevers AJ, Carrick LM, Whitehouse CD, Aggeli A, Boden N. Structures of Helical B-Tapes and Twisted Ribbons: the Role of Side-Chain Interactions on Twist and Bend Behavior. Nano Lett. 2003;3(11):1475–1479. [Google Scholar]
- 38.Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fändrich M. Aβ(1-40) Fibril Polymorphism Implies Diverse Interaction Patterns in Amyloid Fibrils. J Mol Biol. 2009;386(3):869–877. doi: 10.1016/j.jmb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoyer W, Antony T, Cherny D, Heim G, Jovin TM, Subramaniam V. Dependence of A-Synuclein Aggregate Morphology on Solution Conditions. J Mol Biol. 2002;322(2):383–393. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- 40.Seuring C, Verasdonck J, Ringler P, Cadalbert R, Stahlberg H, Böckmann A, Meier BH, Riek R. Amyloid Fibril Polymorphism: Almost Identical on the Atomic Level, Mesoscopically Very Different. J Phys Chem B. 2017;121(8):1783–1792. doi: 10.1021/acs.jpcb.6b10624. [DOI] [PubMed] [Google Scholar]
- 41.Klement K, Wieligmann K, Meinhardt J, Hortschansky P, Richter W, Fändrich M. Effect of Different Salt Ions on the Propensity of Aggregation and on the Structure of Alzheimer’s Aβ(1-40) Amyloid Fibrils. J Mol Biol. 2007;373(5):1321–1333. doi: 10.1016/j.jmb.2007.08.068. [DOI] [PubMed] [Google Scholar]
- 42.Berryman JT, Radford SE, Harris SA. Thermodynamic Description of Polymorphism in Q- and N-Rich Peptide Aggregates Revealed by Atomistic Simulation. Biophys J. 2009;97(1):1–11. doi: 10.1016/j.bpj.2009.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berryman JT, Radford SE, Harris SA. Systematic Examination of Polymorphism in Amyloid Fibrils by Molecular-Dynamics Simulation. Biophys J. 2011;100(9):2234–2242. doi: 10.1016/j.bpj.2011.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiménez JL, Tennent G, Pepys M, Saibil HR. Structural Diversity of Ex Vivo Amyloid Fibrils Studied by Cryo-Electron Microscopy. J Mol Biol. 2001;311(2):241–247. doi: 10.1006/jmbi.2001.4863. [DOI] [PubMed] [Google Scholar]
- 45.Fitzpatrick AWP, Debelouchina GT, Bayro MJ, Clare DK, Caporini MA, Bajaj VS, Jaroniec CP, Wang L, Ladizhansky V, Müller SA, et al. Atomic Structure and Hierarchical Assembly of a Cross-B Amyloid Fibril. Proc Natl Acad Sci US A. 2013;110(14):5468–5473. doi: 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, Scheres SHW. Cryo-EM Structures of Tau Filaments From Alzheimer’s Disease. Nature. 2017;547(7662):185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin HK, Boatz JC, Krabbendam IE, Kodali R, Hou Z, Wetzel R, Dolga AM, Poirier MA, van der Wel PCA. Fibril Polymorphism Affects Immobilized Non-Amyloid Flanking Domains of Huntingtin Exon1 Rather Than Its Polyglutamine Core. Nat Comms. 2017;8:15462. doi: 10.1038/ncomms15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith JF, Knowles TPJ, Dobson CM, MacPhee CE, Welland ME. Characterization of the Nanoscale Properties of Individual Amyloid Fibrils. Proc Natl Acad Sci US A. 2006;103(43):15806–15811. doi: 10.1073/pnas.0604035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adamcik J, Mezzenga R. Adjustable Twisting Periodic Pitch of Amyloid Fibrils. Soft Matter. 2011;7(11):5437–5437. [Google Scholar]
- 50.Knowles TPJ, De Simone A, Fitzpatrick AW, Baldwin A, Meehan S, Rajah L, Vendruscolo M, Welland ME, Dobson CM, Terentjev EM. Twisting Transition Between Crystalline and Fibrillar Phases of Aggregated Peptides. Phys Rev Lett. 2012;109(15):158101. doi: 10.1103/PhysRevLett.109.158101. [DOI] [PubMed] [Google Scholar]
- 51.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI. Atomic Structures of Amyloid Cross-β Spines Reveal Varied Steric Zippers. Nature. 2007 doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 52.van der Wel Patrick CA, Lewandowski Józef R, Griffin RG. Solid-State NMR Study of Amyloid Nanocrystals and Fibrils Formed by the Peptide GNNQQNY From Yeast Prion Protein Sup35p. J Am Chem Soc. 2007;129(16):5117–5130. doi: 10.1021/ja068633m. [DOI] [PubMed] [Google Scholar]
- 53.van der Wel PCA, Lewandowski JR, Griffin RG. Structural Characterization of GNNQQNY Amyloid Fibrils by Magic Angle Spinning NMR. Biochemistry. 2010;49(44):9457–9469. doi: 10.1021/bi100077x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marshall KE, Hicks MR, Williams TL, Hoffmann SV, Rodger A, Dafforn TR, Serpell LC. Characterizing the Assembly of the Sup35 Yeast Prion Fragment, GNNQQNY: Structural Changes Accompany a Fiber-to-Crystal Switch. Biophys J. 2010;98(2):330–338. doi: 10.1016/j.bpj.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kodali R, Williams AD, Chemuru S, Wetzel R. Abeta(1-40) Forms Five Distinct Amyloid Structures Whose Beta-Sheet Contents and Fibril Stabilities Are Correlated. J Mol Biol. 2010;401(3):503–517. doi: 10.1016/j.jmb.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Periole X, Rampioni A, Vendruscolo M, Mark AE. Factors That Affect the Degree of Twist in Beta-Sheet Structures: a Molecular Dynamics Simulation Study of a Cross-Beta Filament of the GNNQQNY Peptide. J Phys Chem B. 2009;113(6):1728–1737. doi: 10.1021/jp8078259. [DOI] [PubMed] [Google Scholar]
- 57.Esposito L, Pedone C, Vitagliano L. Molecular Dynamics Analyses of Cross-Beta-Spine Steric Zipper Models: Beta-Sheet Twisting and Aggregation. Proc Natl Acad Sci US A. 2006;103(31):11533–11538. doi: 10.1073/pnas.0602345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koh E, Kim T. Minimal Surface as a Model of β-Sheets. Proteins: Structure. 2005;61(3):559–569. doi: 10.1002/prot.20636. [DOI] [PubMed] [Google Scholar]
- 59.Fändrich M, Meinhardt J, Grigorieff N. Structural Polymorphism of Alzheimer Abeta and Other Amyloid Fibrils. Prion. 2009;3(2):89–93. doi: 10.4161/pri.3.2.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wälti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Böckmann A, Güntert P, Meier BH, Riek R. Atomic-Resolution Structure of a Disease-Relevant Aβ(1-42) Amyloid Fibril. Proc Natl Acad Sci US A. 2016;113(34):E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masman MF, Eisel ULM, Csizmadia IG, Penke B, Enriz RD, Marrink SJ, Luiten PGM. In Silico Study of Full-Length Amyloid Beta 1-42 Tri- and Penta-Oligomers in Solution. J Phys Chem B. 2009;113(34):11710–11719. doi: 10.1021/jp901057w. [DOI] [PubMed] [Google Scholar]
- 62.Lemkul JA, Bevan DR. Assessing the Stability of Alzheimer’s Amyloid Protofibrils Using Molecular Dynamics. J Phys Chem B. 2010;114(4):1652–1660. doi: 10.1021/jp9110794. [DOI] [PubMed] [Google Scholar]
- 63.Lemkul JA, Bevan DR. Lipid Composition Influences the Release of Alzheimer&Apos;S Amyloid B-Peptide From Membranes. Protein Sci. 2011;20(9):1530–1545. doi: 10.1002/pro.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han W, Schulten K. Fibril Elongation by Aβ17–42: Kinetic Network Analysis of Hybrid-Resolution Molecular Dynamics Simulations. J Am Chem Soc. 2014;136(35):12450–12460. doi: 10.1021/ja507002p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buchete NV, Tycko R, Hummer G. Molecular Dynamics Simulations of Alzheimer’s B-Amyloid Protofilaments. J Mol Biol. 2005;353(4):804–821. doi: 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 66.Buchete NV, Hummer G. Structure and Dynamics of Parallel B-Sheets, Hydrophobic Core, and Loops in Alzheimer’s Aβ Fibrils. Biophys J. 2007;92(9):3032–3039. doi: 10.1529/biophysj.106.100404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tofoleanu F, Brooks BR, Buchete NV. Modulation of Alzheimer’s Aβ Protofilament-Membrane Interactions by Lipid Headgroups. ACS Chem Neurosci. 2015;6(3):446–455. doi: 10.1021/cn500277f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chou KC, Némethy G, Scheraga HA. Role of Interchain Interactions in the Stabilization of the Right-Handed Twist of B-Sheets. J Mol Biol. 1983 doi: 10.1016/s0022-2836(83)80025-4. [DOI] [PubMed] [Google Scholar]
- 69.Yang AS, Honig B. Free Energy Determinants of Secondary Structure Formation: II. Antiparallel B-Sheets. J Mol Biol. 1995 doi: 10.1006/jmbi.1995.0503. [DOI] [PubMed] [Google Scholar]
- 70.Wang L, O’Connell T, Tropsha A, Hermans J. Molecular Simulations of B-Sheet Twisting. J Mol Biol. 1996 doi: 10.1006/jmbi.1996.0513. [DOI] [PubMed] [Google Scholar]
- 71.Lewandowski JR, van der Wel PCA, Rigney M, Grigorieff N, Griffin RG. Structural Complexity of a Composite Amyloid Fibril. J Am Chem Soc. 2011;133(37):14686–14698. doi: 10.1021/ja203736z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu JX, Bayro MJ, Tycko R. Major Variations in HIV-1 Capsid Assembly Morphologies Involve Minor Variations in Molecular Structures of Structurally Ordered Protein Segments. J Biol Chem. 2016;291(25):13098–13112. doi: 10.1074/jbc.M116.720557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merg AD, Boatz JC, Mandal A, Zhao G, Mokashi-Punekar S, Liu C, Wang X, Zhang P, van der Wel PCA, Rosi NL. Peptide-Directed Assembly of Single-Helical Gold Nanoparticle Superstructures Exhibiting Intense Chiroptical Activity. J Am Chem Soc. 2016;138(41):13655–13663. doi: 10.1021/jacs.6b07322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grimsley GR, Scholtz JM, Pace CN. A Summary of the Measured pK Values of the Ionizable Groups in Folded Proteins. Protein Sci. 2009;18(1):247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richarz R, Wüthrich K. High-Field 13C Nuclear Magnetic Resonance Studies at 90. 5 MHz of the Basic Pancreatic Trypsin Inhibitor. Biochemistry. 1978;17(12):2263–2269. doi: 10.1021/bi00605a002. [DOI] [PubMed] [Google Scholar]
- 76.Platzer G, Okon M, McIntosh LP. pH-Dependent Random Coil (1)H, (13)C, and (15)N Chemical Shifts of the Ionizable Amino Acids: a Guide for Protein pK a Measurements. J Biomol NMR. 2014;60(2–3):109–129. doi: 10.1007/s10858-014-9862-y. [DOI] [PubMed] [Google Scholar]
- 77.van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, Flexible, and Free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 78.van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG. Biomolecular Simulation: the GROMOS96 Manual and User Guide. vdf Hochschulverag 33 AG an der ETH, Zurich/Biomos BV; Groningen: 1996. [Google Scholar]
- 79.Berendsen HJ, Postma JPM, Van Gunsteren WF, Hermans J. Intermolecular Forces. 1981. [Google Scholar]
- 80.Berendsen HJC, Postma JPM, Van Gunsteren WF, DiNola A, Haak JR. Molecular Dynamics with Coupling to an External Bath. J Chem Phys. 1984;81(8):3684–3690. [Google Scholar]
- 81.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a Linear Constraint Solver for Molecular Simulations. J Comput Chem. 1997;18(12):1463–1472. [Google Scholar]
- 82.Miyamoto S, Kollman PA. Settle: an Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J Comput Chem. 1992;13(8):952–962. [Google Scholar]
- 83.Tironi IG, Sperb R, Smith PE, van Gunsteren WF. A Generalized Reaction Field Method for Molecular Dynamics Simulations. J Chem Phys. 1995;102(13):5451–5459. [Google Scholar]
- 84.Torrie GM, Valleau JP. Monte Carlo Study of a Phase-Separating Liquid Mixture by Umbrella Sampling. J Chem Phys. 1977 [Google Scholar]
- 85.Periole X, Knepp AM, Sakmar TP, Marrink SJ, Huber T. Structural Determinants of the Supramolecular Organization of G Protein-Coupled Receptors in Bilayers. J Am Chem Soc. 2012;134(26):10959–10965. doi: 10.1021/ja303286e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. THE Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. I. the Method. J Comput Chem. 1992;13(8):1011–1021. [Google Scholar]
- 87.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. Multidimensional Free-Energy Calculations Using the Weighted Histogram Analysis Method. J Comput Chem. 1995;16(11):1339–1350. [Google Scholar]
- 88.Souaille M, Roux B. Extension to the Weighted Histogram Analysis Method: Combining Umbrella Sampling with Free Energy Calculations. Computer Physics Communications. 2001;135(1):40–57. [Google Scholar]
- 89.MacCallum JL, Moghaddam MS, Chan HS, Tieleman DP. Hydrophobic Association of -Helices, Steric Dewetting, and Enthalpic Barriers to Protein Folding. Proc Natl Acad Sci US A. 2007;104(15):6206–6210. doi: 10.1073/pnas.0605859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.