Abstract
The inflammasome is a complex of proteins that through the activity of caspase-1 and the downstream substrates gasdermin D, IL-1β, and IL-18 execute an inflammatory form of cell death termed pyroptosis. Activation of this complex often involves the adaptor protein ASC and upstream sensors including NLRP1, NLRP3, NLRC4, AIM2, and pyrin, which are activated by different stimuli including infectious agents and changes in cell homeostasis. Here we discuss new regulatory mechanisms that have been identified for the canonical inflammasomes, the most recently identified NLRP9b inflammasome, and the new gasdermin family of proteins that mediate pyroptosis and other forms of regulated cell death.
INTRODUCTION
The innate immune system recognizes infection and changes in cellular homeostasis to initiate responses to clear pathogens and repair tissue damage. One of the major complexes involved in these processes is the inflammasome, a multimeric protein complex that activates pro-caspase-1, which then proceeds to cleave multiple substrates including the pro-inflammatory cytokines IL-1β and IL-18 [1]. The inflammasome further initiates an inflammatory form of cell death termed pyroptosis via an activating cleavage of gasdermin D, which forms pores in the plasma membrane and acts as the executioner molecule for pyroptosis [2–4]. As an inflammatory form of cell death, dysregulation of the inflammasome’s activity is associated with many different autoimmune, metabolic, and infectious diseases.
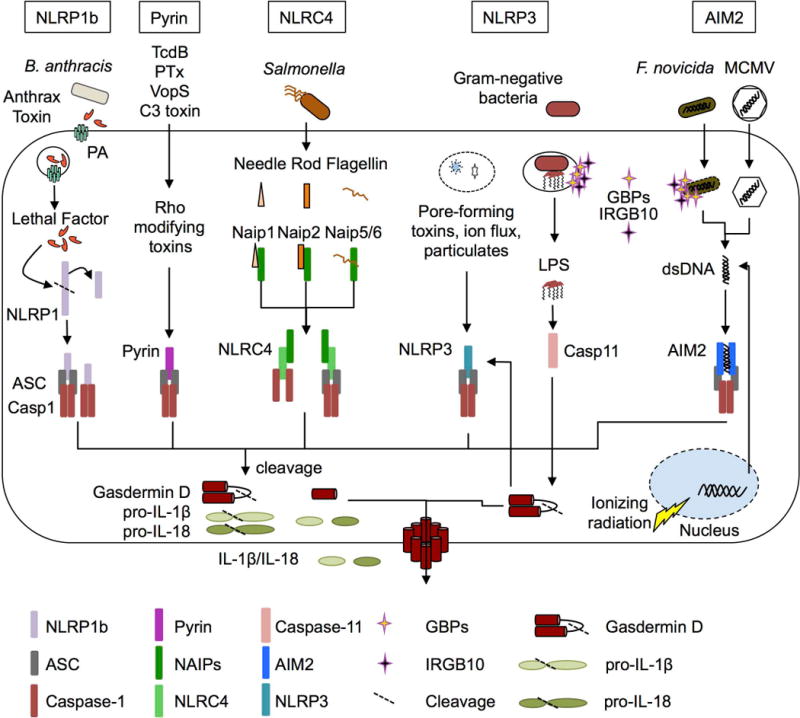
Upstream of caspase-1 oligomerization and activation are adaptor and sensor proteins that recognize specific stimuli, which are named after their structural domains: the NLRs (nucleotide-binding domain and leucine-rich repeat containing), ALRs (absent in melanoma 2-like receptors), the pyrin receptor, and the PYD (pyrin domain) and CARD (Caspase activation and recruitment domain) containing adaptor molecule ASC (apoptosis-associated speck—like protein containing a CARD, also known as Pycard). Of these, NLRP1, NLRP3, NLRC4, AIM2, and pyrin are well established to assemble a canonical inflammasome complex, while other sensors have also been proposed (Figure 1). These include the human NLRP2, NLRP7, and IFI16 and murine NLRP6 and NLRP9b inflammasomes [5–10]. One hallmark of activation is the recruitment of the sensor protein, ASC, and caspase-1 into a single large macromolecular complex called the ASC speck, which can be visualized by microscopy. Within this complex, caspase-1 is proposed to undergo autocatalytic cleavage to produce enzymatically active p20 and p10 subunits [11]. In this review we will describe the recent advances in our understanding of the canonical inflammasomes as well as the most recently identified NLRP9b inflammasome and the gasdermin family.
Figure 1.
NLRP1 Inflammasome
The first NLR described to form an inflammasome complex was NLRP1, which in humans is encoded by a single NLRP1 gene containing a PYD (pyrin) domain, FIIND (function-to-find domain), and CARD domain or as NLRP1(a-c) in mice, which lack the PYD domain [12,13]. Bacillus anthracis anthrax lethal toxin, which is composed of the two proteins protective antigen and lethal factor, activates mouse NLRP1b through the toxin’s ability to form pores in the host cell membrane (via protective antigen) and subsequent cleavage of NLRP1b within the N-terminal NR100 domain (via lethal factor) [14]. Cleavage of murine NLRP1b by lethal toxin within the FIIND domain is sufficient to cause assembly of an inflammasome complex, but the ligand for human NLRP1 remains unknown [15]. Murine NLRP1b does not contain a PYD domain, unlike human NLRP1, suggesting that NLRP1b can assemble with caspase-1 directly via CARD-CARD interactions rather than through ASC, similar to NLRC4 [13]. Interestingly, the NLRP1b inflammasome does not lead to autoproteolytic cleavage of caspase-1 in all cases, but it still able to cleave IL-1β and undergo pyroptosis, unlike the NLRP3 inflammasome [16]. In human NLRP1, the PYD and LRR domains exert an autoinhibitory function, with gain-of-function mutations leading to spontaneous activation. Mutations in these domains lead to skin diseases including multiple self-healing palmoplantar carcinomas and familial keratosis lichenoides chronica [17]. NLRP1a has also been implicated in driving pyroptosis of hematopoietic cells following a gain-of-function mutation in the region between the NACHT and LRR domain (Q593P), which leads to overactive inflammasome activation and IL-1β release [18]. The physiological triggers of human NLRP1 and murine NLRP1a/c remain to be discovered, but current biochemical studies suggest all are capable of forming inflammasome complexes.
NLRP3 Inflammasome
One of the most studied inflammasome sensors is NLRP3, which was first associated with hereditary autoimmune diseases called cryopyrin-associated periodic syndromes, which present with skin rashes and fever and has since been associated with over 90 disease-associated mutations [19–21]. NLRP3 forms an inflammasome complex containing ASC and caspase-1 and responds to a wide range of infections and stress stimuli with no single trigger. The model for NLRP3 activation is a two-step process, with signal one acting as a priming and licensing step including upregulation of NLPR3 and post-translational modifications to license NLRP3 to be activated by signal two, which through a still-debated mechanism leads to NLRP3 oligomerization with inflammasome components. Many studies have recently identified regulators of both signal one and signal 2 but the complete mechanism by which NLRP3 is activated is still under investigation.
Signal one, which primes/licenses NLRP3, occurs downstream of TLRs and MyD88 signaling and leads to NF-κB activation and the upregulation of NLRP3, IL-1β, and IL-18 [22]. Recent work, however, suggests that NLRP3 licensing occurs independently from upregulation, with rapid NLRP3 inflammasome activation observed downstream of TLR/IL-1R/MyD88 signaling through IRAK4/IRAK1 kinase activity and TRAF6 E3 ubiquitin ligase activity [23–25].
Signal two results in the activation of the NLRP3 inflammasome, provided by extracellular ATP, uric acid crystals, bacterial pore-forming toxins, Gram-positive and Gram-negative bacteria, viruses, fungal, or protozoan pathogens [2,26,27]. The mechanism by which signal two activates NLRP3 is disputed, but these diverse stimuli likely act on a common cellular mechanism, with many candidates proposed, including: changes in ionic homeostasis via potassium efflux, calcium influx, or toxin-mediated membrane disruption; lysosomal rupture and release of cathepsin B; changes in cell volume; cardiolipin translocation to the outer membrane of mitochondria; or, release of oxidized mitochondrial DNA [2,27]. Interestingly, human monocytes undergo NLRP3 inflammasome activation independently from an observed signal two [28].
Further, NLRP3 is also regulated by its subcellular localization and by cytoskeleton components. Endoplasmic reticulum-resident NLRP3 is spatially isolated from ASC, which under basal conditions resides in mitochondria, cytosol, and the nucleus, and is recruited to NLRP3 via microtubules [29]. Another key regulator of NLRP3 activation is NEK7 (NIMA-related kinase 7), a protein involved in regulating the cell cycle, mitotic spindle formation, and cytokinesis [30–32]. Interaction between the NLRP3 LRR domain and NEK7, independent of its kinase activity, leads to NLRP3 activation in interphase cells downstream of potassium efflux. Another novel regulator of NLRP3 activation, PKD (protein kinase D), was recently found to recruit to the Golgi, where its phosphorylation of NLRP3 was shown to be required and sufficient for NLRP3 inflammasome activation [33]. In another study, dephosphorylation of NLRP3 at Tyr861 by PTPN22 (protein tyrosine phosphatase 22) was involved in activation of the NLRP3 inflammasome [34]. Many other proteins have been implicated in regulation of NLRP3 and are described in detail in other reviews [2,26,27].
NLRP3 inflammasome activation also occurs downstream of caspase-11 and gasdermin D cleavage and pore-formation in a process called non-canonical NLRP3 inflammasome activation [3,35,36]. More recently, NLRP3 inflammasome activation downstream of necroptosis and MLKL pore-formation have also been reported [37,38]. Together, these data suggest that pore formation, similar to toxin-mediated pore formation, activates the NLRP3 inflammasome through disruption of the cellular ion homeostasis.
NAIP-NLRC4 Inflammasomes
The NAIP-NLRC4 inflammasomes responds to the evolutionarily related bacterial components of the T3SS (type three secretion system) and flagellin proteins of Gram-negative bacteria [2,39]. In humans, a single NAIP protein senses the bacterial T3SS needle and flagellin proteins, while 7 murine Naip proteins recognize distinct ligands: Naip1, T3SS needle protein; Naip2, T3SS rod protein; Naip5/6, flagellin; while the ligands for the remaining Naip proteins remains unknown [40–42]. Cryo-electron microscopy has revealed that a single NAIP protein is sufficient to recruit multiple NLRC4 proteins into an inflammasome complex [43]. Recent studies have identified that the NLRC4 inflammasome is also regulated by an activating phosphorylation event on S533 during Salmonella infection, which was mediated by PKCδ or by LRRK2 kinase activity [44–46]. Other studies showed that the phosphorylation of S533 occurs independently from Naip5 binding to flagellin and that Salmonella and Shigella infections could activate the NAIP-NLRC4 inflammasome independent from PKCδ, suggesting that both LRRK2 and PKCδ regulate S533 phosphorylation. Another study showed that a S533A mutant of NLRC4 was minimally defective in inflammasome activation by Salmonella and flagellin due to enhanced activation of the NLRP3 inflammasome, suggesting this modification may regulate inflammasome scaffolding functions [47]. These data and previous studies have shown that NLRC4 is able to form inflammasome complexes incorporating NLRP3, ASC, caspase-1 and caspase-8 during Salmonella infection, suggesting other modifications may be able to regulate how specific NLRC4 inflammasomes and oligomeric complexes form [48].
AIM2 Inflammasome
The AIM2 inflammasome detects dsDNA in the cytosol of cells during viral and intracellular bacterial infections, as well as self-DNA. The HIN-200 domain of AIM2 mediates the binding of dsDNA in a sequence-independent manner [49]. Many of the advances in understanding the AIM2 inflammasome have recently focused on the mechanisms by which bacterial DNA, which is sequestered within the bacterial cell walls, is exposed for sensing by cytosolic sensors. The interferon-inducible proteins GBP2, GBP5, and IRGB10 have been identified as required for activation of AIM2 during Francisella novicida infections in a series of recent studies, downstream of the type I IFN/IRF1 signaling axis [50–52]. GBP2 and GBP5 are both required for activation of the AIM2 inflammasome, as absence of either restricts the activation of the AIM2 inflammasome. Similar to the GBPs, a newly characterized protein, IRGB10, has also been identified as a factor required for AIM2 inflammasome activation during F. novicida infection [52]. These proteins act by attacking either the vacuolar membrane or bacterial membrane and causing lysis, thereby exposing DNA or other bacterial components to cytosolic sensors. Furthermore, GBPs appear to act upstream of IRGB10, and other studies have suggested that these proteins are recruited to pathogens in a hierarchical manner [53]. Future work with other infection models may reveal novel functions for these interferon-inducible proteins in inflammasome activation.
Other recent work has examined the role of AIM2 in sensing of self-DNA. Following ionizing radiation exposure, double-stranded DNA breaks occur and lead to extensive cell death and tissue damage via AIM2 [54]. An interesting feature of this recognition is that AIM2 localizes to the nucleus of DNA-damaged cells, rather than sensing release of dsDNA into the cytosol, as is observed in most other situations [54]. This study also showed that AIM2 contributes to the pathology associated with DNA damaging chemotherapy drugs but did not limit the anti-cancer properties, suggesting that AIM2 activity could be modulated to limit side effects associated with these drugs. In addition to radiation, drugs that stress the nuclear envelope integrity have been linked to AIM2 inflammasome activation [55]. Together, these findings suggest that AIM2 inflammasome activation contributes to the negative side effects of chemotherapy.
Pyrin Inflammasome
Gain-of-function mutations in the pyrin protein were first associated with familiar Mediterranean fever (FMF), an autoinflammatory disease of humans and later pyrin was identified as a component of an inflammasome complex[56]. In normal conditions, Rho-inactivating toxins from bacterial pathogens including Clostridium difficile (TcdB), Vibrio parahemolyticus (VopS), and others were discovered to activate the pyrin inflammasome [57]. Each toxin targets Rho activity by different covalent modifications including glycosylation, adenylation, ADP ribosylation, or deamination, but these modifications all lead to pyrin inflammasome activation, suggesting other common factors help to induce pyrin activation [2]. More recent work has identified that Rho activates PKN1 and PKN2 (protein kinase N1/2), which phosphorylate and maintain pyrin in an inactive [58]. Phosphorylation of human pyrin at S242 leads to binding of 14-3-3 proteins, which limit the activation of pyrin as S242 mutations which disrupt the phosphorylation site lead to pyrin-associated autoimmunity and neutrophilic dermatoses [59]. Another recent discovery showed that pyrin inflammasomes are regulated by the mevalonate pathway, as cells deficient in protein geranylgeranylation have defective PI(3)K activity, which limits excessive TLR-mediated signaling and constitutive pyrin inflammasome activation[60]. Upon stimulation with pyrin-activating stimuli, the repressive phosphorylation of pyrin is removed and inflammasome activation proceeds. Further work has shown that microtubule dynamics also regulate pyrin inflammasome assembly and activation independently from the dephosphorylation events, but microtubules are not required for FMF-associated inflammasome activation, suggesting the cytoskeleton also has distinct regulatory functions in normal pyrin inflammasome activation and in FMF-associated pyrin inflammasomes [61,62].
NLRP9b Inflammasome
The murine NLRP9b inflammasome was very recently described to be activated by rotavirus infection of intestinal epithelial cells [63]. Rotaviruses specifically infect the small intestine epithelial cells, where the inflammasome components NLRP6, NLRC4, NAIPs, and NLRP9b were expressed. The authors proposed that Dhx9, an RNA helicase, recognizes viral double-stranded RNA and leads to NLRP9b association with the inflammasome adaptor ASC and caspase-1, maturation of IL-18, and gasdermin D-mediated pyroptosis (Figure 2).
Figure 2.
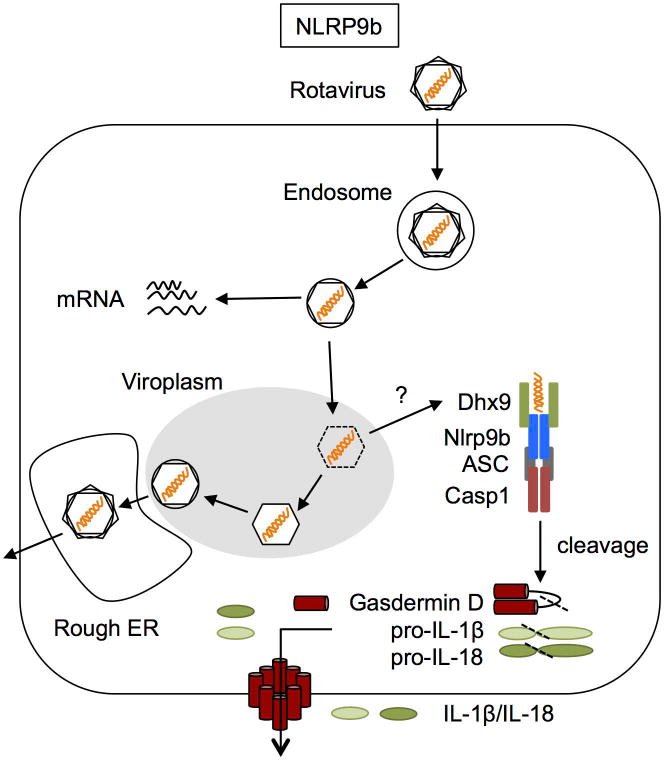
The proximal sensor of rotavirus dsRNA, Dhx9, co-immunoprecipitated preferentially with short dsRNA (low molecular weight poly(I:C)) and slightly with tri-phosphate RNA (which preferentially is recognized by the RNA sensor RIG-I). Previous studies have identified that Dhx9 is able to unwind dsDNA, dsRNA, and DNA/RNA hybrids, so future studies may reveal other nucleic acid species capable of activating the NLRP9b inflammasome via Dhx9 [64]. The authors did not show which domains in Dhx9 are required for NLRP9b inflammasome activation or rotavirus RNA recognition, but the protein contains a dsRNA binding domain, helicase domain, and RGG box domain (involved in ssRNA binding). Further work confirming the assembly of a true inflammasome and downstream cleavage of gasdermin D, IL-1β, and IL-18 must still be done to firmly establish Nlrp9b as a genuine inflammasome complex. Whether Dhx9 or other helicases help to activate other inflammasomes in different cell types remains another interesting area of future research.
Gasdermins as Executioner Molecules
One of the most significant discoveries in the last few years was that caspase-1 and caspase-11 cleavage of the substrate gasdermin D, first identified in a proteomic screen of caspase-1 substrates, leads to cell death following activation of the inflammasome [3,4,65]. Upon cleavage by caspase-1 or caspase-11, the N-terminal domain of gasdermin D is released from the autoinhibitory C-terminal domain and translocates to the plasma membrane where it forms large, 10–15 nanometer pores that lead to cell death. A related molecule, DFNA5 (or GSDME), which is cleaved by caspase-3, also has similar N-terminal domain that is capable of forming pores in the cell membrane, leading to cell death similar to secondary necrosis, suggesting that in specific cell types, specific gasdermin molecules function to execute cell death [66]. Of the remaining gasdermin family members (gasdermin A (in mice A1-3), B (absent in mice), C (in mice C1-4), D, E, and DFNB59), all contain an N-terminal pore- forming domain with all but DFNB59 shown to have pore-forming activity [67]. It will be interesting to see where other gasdermin molecules are involved in regulated cell death and whether new “inflammasome” complexes may contribute to these processes.
CONCLUSIONS
The most well studied inflammasomes including NLRP1, NLRP3, NLRC4, AIM2, and pyrin each have been solidly established as activators of caspase-1 and gasdermin D, two hallmarks of pyroptosis. While these have been established for years, current work has begun to unravel many of the unique post-translational regulatory mechanisms that modulate their activation and repression and how gain-of-function mutations in these molecules leads to autoinflammatory diseases and susceptibility to various infections. Other less well studied inflammasomes including the human NLRP2 inflammasome (activated in astrocytes), human NLRP7 (activated by pathogen-derived acylated lipopeptides), human IFI16 (activated within the nucleus by KHSV and HIV), and murine NLRP6 (activated by the taurine, histamine, and spermine in the intestine), and NLRP9b (activated by dsRNA in intestinal epithelial cells) are still poorly characterized but with careful investigation may be established as genuine inflammasome complexes or regulate other aspects of immunity [5–10,63]. How each of these inflammasomes is regulated presents new targets for drugs that could limit their pathological effects in many diseases.
HIGHLIGHTS.
The NLRP3, NLRC4, and pyrin inflammasomes are regulated by post-translational modifications
The Dhx9-NLRP9b inflammasome recognizing viral dsRNA was recently identified in intestinal cells
Gasdermin family proteins are cleaved by caspases to form pores in cells and execute cell death
Acknowledgments
We would like to thank members of the Kanneganti lab for editing of this manuscript. This work was supported by the National Institutes of Health [grant numbers AI101935, AI124346, AR056296 and CA163507] and the American Lebanese Syrian Associated Charities (ALSAC). Funders had o involvement in the writing of this review. We apologize to all investigators whose research could not be appropriately cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Man SM, Kanneganti T-D. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat Rev Immunol. 2016;16:7–21. doi: 10.1038/nri.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kesavardhana S, Kanneganti T-D. Mechanisms governing inflammasome activation, assembly and pyroptosis induction. Int Immunol. 2017 doi: 10.1093/intimm/dxx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 4.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 5.Janowski AM, Sutterwala FS. NLR Proteins. Humana Press; New York, NY: 2016. Atypical Inflammasomes; pp. 45–62. [DOI] [PubMed] [Google Scholar]
- 6.Minkiewicz J, de Rivero Vaccari JP, Keane RW. Human astrocytes express a novel NLRP2 inflammasome. Glia. 2013;61:1113–1121. doi: 10.1002/glia.22499. [DOI] [PubMed] [Google Scholar]
- 7.Khare S, Dorfleutner A, Bryan NB, Yun C, Radian AD, de Almeida L, Rojanasakul Y, Stehlik C. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity. 2012;36:464–476. doi: 10.1016/j.immuni.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. IFI16 DNA Sensor Is Required for Death of Lymphoid CD4 T-cells Abortively Infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 2015;163:1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 2010;8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinon F, Burns K, Tschopp J. The Inflammasome. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 13.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 14.Chavarría-Smith J, Vance RE. Direct Proteolytic Cleavage of NLRP1B Is Necessary and Sufficient for Inflammasome Activation by Anthrax Lethal Factor. PLOS Pathog. 2013;9:e1003452. doi: 10.1371/journal.ppat.1003452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu C-H, Moecking J, Geyer M, Masters SL. Mechanisms of NLRP1-Mediated Autoinflammatory Disease in Humans and Mice. J Mol Biol. 2017 doi: 10.1016/j.jmb.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Guey B, Bodnar M, Manié SN, Tardivel A, Petrilli V. Caspase-1 autoproteolysis is differentially required for NLRP1b and NLRP3 inflammasome function. Proc Natl Acad Sci U S A. 2014;111:17254–17259. doi: 10.1073/pnas.1415756111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong FL, Mamaï O, Sborgi L, Boussofara L, Hopkins R, Robinson K, Szeverényi I, Takeichi T, Balaji R, Lau A, et al. Germline NLRP1 Mutations Cause Skin Inflammatory and Cancer Susceptibility Syndromes via Inflammasome Activation. Cell. 2016;167:187–202 e17. doi: 10.1016/j.cell.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Masters SL, Gerlic M, Metcalf D, Preston S, Pellegrini M, O’Donnell JA, McArthur K, Baldwin TM, Chevrier S, Nowell CJ, et al. NLRP1 Inflammasome Activation Induces Pyroptosis of Hematopoietic Progenitor Cells. Immunity. 2012;37:1009–1023. doi: 10.1016/j.immuni.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*) Annu Rev Immunol. 2009;27:621–668. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanneganti T-D, Ozören N, Body-Malapel M, Amer A, Park J-H, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 22.Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu J, Monks BG, Fitzgerald KA, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol Baltim Md 1950. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandes-Alnemri T, Kang S, Anderson C, Sagara J, Fitzgerald KA, Alnemri ES. Cutting edge: TLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. J Immunol Baltim Md 1950. 2013;191:3995–3999. doi: 10.4049/jimmunol.1301681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin K-M, Hu W, Troutman TD, Jennings M, Brewer T, Li X, Nanda S, Cohen P, Thomas JA, Pasare C. IRAK-1 bypasses priming and directly links TLRs to rapid NLRP3 inflammasome activation. Proc Natl Acad Sci U S A. 2014;111:775–780. doi: 10.1073/pnas.1320294111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing Y, Yao X, Li H, Xue G, Guo Q, Yang G, An L, Zhang Y, Meng G. Cutting Edge: TRAF6 Mediates TLR/IL-1R Signaling–Induced Nontranscriptional Priming of the NLRP3 Inflammasome. J Immunol. 2017 doi: 10.4049/jimmunol.1700175. [DOI] [PubMed] [Google Scholar]
- 26.Sharma D, Kanneganti T-D. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J Cell Biol. 2016;213:617–629. doi: 10.1083/jcb.201602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y, Hara H, Núñez G. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AAB, Cooper MA, Graf T, Hornung V. Human Monocytes Engage an Alternative Inflammasome Pathway. Immunity. 2016;0 doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 30.He Y, Zeng MY, Yang D, Motro B, Núñez G. Nek7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530:354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid-Burgk JL, Chauhan D, Schmidt T, Ebert TS, Reinhardt J, Endl E, Hornung V. A Genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J Biol Chem. 2016;291:103–109. doi: 10.1074/jbc.C115.700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, Wang Y, Li X, Zhan X, Tang M, Fina M, Su L, Pratt D, Bu CH, Hildebrand S, et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol. 2016;17:250–258. doi: 10.1038/ni.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Meszaros G, He W, Xu Y, Magliarelli H de F, Mailly L, Mihlan M, Liu Y, Gámez MP, Goginashvili A, et al. Protein kinase D at the Golgi controls NLRP3 inflammasome activation. J Exp Med. 2017 doi: 10.1084/jem.20162040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spalinger MR, Kasper S, Gottier C, Lang S, Atrott K, Vavricka SR, Scharl S, Gutte PM, Grütter MG, Beer H-D, et al. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J Clin Invest. 2016;126:1783–1800. doi: 10.1172/JCI83669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 36.Rühl S, Broz P. Caspase-11 activates a canonical NLRP3 inflammasome by promoting K(+) efflux. Eur J Immunol. 2015;45:2927–2936. doi: 10.1002/eji.201545772. [DOI] [PubMed] [Google Scholar]
- 37.Gutierrez KD, Davis MA, Daniels BP, Olsen TM, Ralli-Jain P, Tait SWG, Gale M, Oberst A. MLKL Activation Triggers NLRP3-Mediated Processing and Release of IL-1β Independently of Gasdermin-D. J Immunol. 2017 doi: 10.4049/jimmunol.1601757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conos SA, Chen KW, Nardo DD, Hara H, Whitehead L, Núñez G, Masters SL, Murphy JM, Schroder K, Vaux DL, et al. Active MLKL triggers the NLRP3 inflammasome in a cell-intrinsic manner. Proc Natl Acad Sci. 2017;114:E961–E969. doi: 10.1073/pnas.1613305114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Shao F. The NAIP–NLRC4 inflammasome in innate immune detection of bacterial flagellin and type III secretion apparatus. Immunol Rev. 2015;265:85–102. doi: 10.1111/imr.12293. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Zhao Y, Shi J, Shao F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci U S A. 2013;110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franchi L, Amer A, Body-Malapel M, Kanneganti T-D, Ozören N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 42.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs dictates inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Chen S, Ruan J, Wu J, Tong AB, Yin Q, Li Y, David L, Lu A, Wang WL, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015 doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W, Liu X, Li Y, Zhao J, Liu Z, Hu Z, Wang Y, Yao Y, Miller AW, Su B, et al. LRRK2 promotes the activation of NLRC4 inflammasome during Salmonella Typhimurium infection. J Exp Med. 2017 doi: 10.1084/jem.20170014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu Y, Misaghi S, Izrael-Tomasevic A, Newton K, Gilmour LL, Lamkanfi M, Louie S, Kayagaki N, Liu J, Kömüves L, et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature. 2012;490:539–542. doi: 10.1038/nature11429. [DOI] [PubMed] [Google Scholar]
- 46.Matusiak M, Van Opdenbosch N, Vande Walle L, Sirard J-C, Kanneganti T-D, Lamkanfi M. Flagellin-induced NLRC4 phosphorylation primes the inflammasome for activation by NAIP5. Proc Natl Acad Sci U S A. 2015;112:1541–1546. doi: 10.1073/pnas.1417945112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu Y, Misaghi S, Newton K, Maltzman A, Izrael-Tomasevic A, Arnott D, Dixit VM. NLRP3 recruitment by NLRC4 during Salmonella infection. J Exp Med. 2016;213:877–885. doi: 10.1084/jem.20132234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Man SM, Hopkins LJ, Nugent E, Cox S, Glück IM, Tourlomousis P, Wright JA, Cicuta P, Monie TP, Bryant CE. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057– 1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 50.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Rühl S, Dussurgey S, Dick MS, Kistner A, Rigard M, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Man SM, Karki R, Malireddi RKS, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti T-D. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Man SM, Karki R, Sasai M, Place DE, Kesavardhana S, Temirov J, Frase S, Zhu Q, Malireddi RKS, Kuriakose T, et al. IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell. 2016;167:382–396.e17. doi: 10.1016/j.cell.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Britzen-Laurent N, Bauer M, Berton V, Fischer N, Syguda A, Reipschläger S, Naschberger E, Herrmann C, Stürzl M. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PloS One. 2010;5:e14246. doi: 10.1371/journal.pone.0014246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu B, Jin C, Li H-B, Tong J, Ouyang X, Cetinbas NM, Zhu S, Strowig T, Lam FC, Zhao C, et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science. 2016;354:765–768. doi: 10.1126/science.aaf7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Micco AD, Frera G, Lugrin J, Jamilloux Y, Hsu E-T, Tardivel A, Gassart AD, Zaffalon L, Bujisic B, Siegert S, et al. AIM2 inflammasome is activated by pharmacological disruption of nuclear envelope integrity. Proc Natl Acad Sci. 2016;113:E4671–E4680. doi: 10.1073/pnas.1602419113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chae JJ, Cho Y-H, Lee G-S, Cheng J, Liu PP, Feigenbaum L, Katz SI, Kastner DL. Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity. 2011;34:755–768. doi: 10.1016/j.immuni.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong Y-N, Peng X, Xi JJ, Chen S, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 58.Park YH, Wood G, Kastner DL, Chae JJ. Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat Immunol. 2016;17:914–921. doi: 10.1038/ni.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masters SL, Lagou V, Jéru I, Baker PJ, Eyck LV, Parry DA, Lawless D, Nardo DD, Garcia-Perez JE, Dagley LF, et al. Familial autoinflammation with neutrophilic dermatosis reveals a regulatory mechanism of pyrin activation. Sci Transl Med. 2016;8:332ra45–332ra45. doi: 10.1126/scitranslmed.aaf1471. [DOI] [PubMed] [Google Scholar]
- 60.Akula MK, Shi M, Jiang Z, Foster CE, Miao D, Li AS, Zhang X, Gavin RM, Forde SD, Germain G, et al. Control of the innate immune response by the mevalonate pathway. Nat Immunol. 2016;17:922–929. doi: 10.1038/ni.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao W, Yang J, Liu W, Wang Y, Shao F. Site-specific phosphorylation and microtubule dynamics control Pyrin inflammasome activation. Proc Natl Acad Sci U S A. 2016;113:E4857–4866. doi: 10.1073/pnas.1601700113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Gorp H, Saavedra PHV, de Vasconcelos NM, Van Opdenbosch N, Vande Walle L, Matusiak M, Prencipe G, Insalaco A, Van Hauwermeiren F, Demon D, et al. Familial Mediterranean fever mutations lift the obligatory requirement for microtubules in Pyrin inflammasome activation. Proc Natl Acad Sci U S A. 2016;113:14384–14389. doi: 10.1073/pnas.1613156113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu S, Ding S, Wang P, Wei Z, Pan W, Palm NW, Yang Y, Yu H, Li H-B, Wang G, et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 2017;546:667–670. doi: 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee T, Pelletier J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget. 2016;7:42716–42739. doi: 10.18632/oncotarget.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agard NJ, Maltby D, Wells JA. Inflammatory Stimuli Regulate Caspase Substrate Profiles. Mol Cell Proteomics MCP. 2010;9:880–893. doi: 10.1074/mcp.M900528-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8 doi: 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kovacs SB, Miao EA. Gasdermins: Effectors of Pyroptosis. Trends Cell Biol. 2017;27:673–684. doi: 10.1016/j.tcb.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
ANNOTATED REFERENCES
- 1*.Xing Y, Yao X, Li H, Xue G, Guo Q, Yang G, An L, Zhang Y, Meng G. Cutting Edge: TRAF6 Mediates TLR/IL-1R Signaling–Induced Nontranscriptional Priming of the NLRP3 Inflammasome. J Immunol. 2017 doi: 10.4049/jimmunol.1700175. a. This study shows how TRAF6 E3 ubiquitin ligase activity downstream of TLR/IL-1R-MyD88-IRAK signaling is required for the oligomerization of NLRP3 inflammasomes, extending our understanding of how priming occurs. [DOI] [PubMed] [Google Scholar]
- 2**.He Y, Zeng MY, Yang D, Motro B, Núñez G. Nek7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530:354–357. doi: 10.1038/nature16959. a. One of three contemporaneous publications (Schmid-Burgk JL et al. and Shi H et al.) showing NEK7 is required for NLRP3 inflammasome activation, independent of its kinase activity. This study showed that NEK7 was required for mediating NLRP3 activation in macrophages with the CAPS-associated activating mutation NLRP3R258W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Schmid-Burgk JL, Chauhan D, Schmidt T, Ebert TS, Reinhardt J, Endl E, Hornung V. A Genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J Biol Chem. 2016;291:103–109. doi: 10.1074/jbc.C115.700492. a. One of three contemporaneous publications (He Y et al. and Shi H et al.) showing NEK7 is required for NLRP3 inflammasome activation, independent of its kinase activity. This work utilized CRISPR to identify cells resistant to NLRP3 inflammacome activation by flow cytometry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4**.Shi H, Wang Y, Li X, Zhan X, Tang M, Fina M, Su L, Pratt D, Bu CH, Hildebrand S, et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol. 2016;17:250–258. doi: 10.1038/ni.3333. a. One of three contemporaneous publications (He Y et al. and Schmid-Burgk JL et al.) showing NEK7 is required for NLRP3 inflammasome activation, independent of its kinase activity. This work shows that the cell cycle can restrict when inflammasome activation occurs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5*.Zhang Z, Meszaros G, He W, Xu Y, Magliarelli H de F, Mailly L, Mihlan M, Liu Y, Gámez MP, Goginashvili A, et al. Protein kinase D at the Golgi controls NLRP3 inflammasome activation. J Exp Med. 2017 doi: 10.1084/jem.20162040. a. This study showed that PKD regulates NLRP3 inflammasome activation by phosphorylation and subsequent release from mitochondria-associated endoplasmic reticulum membranes adjacent to Golgi membranes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Gutierrez KD, Davis MA, Daniels BP, Olsen TM, Ralli-Jain P, Tait SWG, Gale M, Oberst A. MLKL Activation Triggers NLRP3-Mediated Processing and Release of IL-1β Independently of Gasdermin-D. J Immunol. 2017 doi: 10.4049/jimmunol.1601757. a. Published back to back with Conos SA et al this paper showed that MLKL oligomerization alone is sufficient to activate the NLRP3 inflammasome via potassium efflux and independently from gasdermin D pore formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7*.Conos SA, Chen KW, Nardo DD, Hara H, Whitehead L, Núñez G, Masters SL, Murphy JM, Schroder K, Vaux DL, et al. Active MLKL triggers the NLRP3 inflammasome in a cell-intrinsic manner. Proc Natl Acad Sci. 2017;114:E961–E969. doi: 10.1073/pnas.1613305114. a. Published back-to-back with Gutierrez KD et al this paper showed that MLKL oligomerization alone is sufficient to activate the NLRP3 inflammasome via potassium efflux and independently from gasdermin D pore formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Man SM, Karki R, Sasai M, Place DE, Kesavardhana S, Temirov J, Frase S, Zhu Q, Malireddi RKS, Kuriakose T, et al. IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell. 2016;167:382–396.e17. doi: 10.1016/j.cell.2016.09.012. a. This paper is the first to describe a function for the protein IRGB10 and its role in AIM2 inflammasome activation downstream of GBP proteins during Francisella infection. GBPs and IRGB10 are required to lyse intracellular bacteria and release their DNA for sensing in the cytosol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9*.Gao W, Yang J, Liu W, Wang Y, Shao F. Site-specific phosphorylation and microtubule dynamics control Pyrin inflammasome activation. Proc Natl Acad Sci U S A. 2016;113:E4857–4866. doi: 10.1073/pnas.1601700113. a. This paper shows that the microtubule cytoskeleton regulates pyrin inflammasome activation, helping to explain how colchicine functions in treating familial Mediterranean fever (FMF). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Zhu S, Ding S, Wang P, Wei Z, Pan W, Palm NW, Yang Y, Yu H, Li H-B, Wang G, et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 2017;546:667–670. doi: 10.1038/nature22967. a. In this study the authors identify Nlrp9b as a novel inflammasome sensor in intestinal epithelial cells that recognizes rotavirus dsRNA via Dhx9-Nlrp9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. a. This back-to-back paper with Kayagaki N et al. showed that gasdermin D cleavage by caspases and release of the pore-forming N-terminal domain leads to the cell death associated with pyroptosis. [DOI] [PubMed] [Google Scholar]
- 12**.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. a. This back-to-back paper with Shi J et al. showed that gasdermin D cleavage by caspases and release of the pore-forming N-terminal domain leads to the cell death associated with pyroptosis. [DOI] [PubMed] [Google Scholar]


