Abstract
Acute erythroleukemia is a rare form of acute myeloid leukemia recognized by its distinct phenotypic attribute of erythroblastic proliferation. After a century of its descriptive history, many diagnostic, prognostic, and therapeutic implications relating to this unique leukemia subset remain uncertain. The rarity of the disease and the simultaneous involvement of its associated myeloid compartment have complicated in vitro studies of human erythroleukemia cell lines. Although murine and cell line erythroleukemia models have provided valuable insights into pathophysiology, translation of these concepts into treatment are not forthcoming. Integration of knowledge gained through a careful study of these models with more recent data emerging from molecular characterization will help elucidate key mechanistic pathways and provide a much needed framework that account for erythroid lineage-specific attributes. In this article, we discuss the evolving diagnostic concept of erythroleukemia, translational aspects of its pathophysiology, and promising therapeutic targets through an appraisal of the current literature.
Keywords: Pure erythroid leukemia, Acute Erythroleukemia – M6a subtype, Acute Erythroleukemia – M6b subtype, Erythroblasts, GATA1 protein, PU.1 protein, TP53, bromodomain protein, microRNA
1. Introduction and historical perspective
Erythroleukemia was first described as a leukemic condition by Giovanni Di Guglielmo in the early 1920s, and it was recognized as a distinct morphological and pathological entity from myeloid leukemia based on the predominant erythroid involvement of the involved marrow (1). Di Guglielmo subsequently distinguished between two variants of the disease, including a pure acute (Di Guglielmo disease) and a more chronic (Di Guglielmo syndrome) form. It gradually became apparent through sequential histological evaluations that erythroleukemia can exist in mixed erthyroblastic-myeloblastic forms and can progress variably through stages, ranging from no myeloblastosis to myeloblastic excess in an erythroid-predominant marrow (2). This morphologic process of evolution from erythroid predominance to myeloblastic leukemia was coined ‘Di Guglielmo syndrome’ (3). In 1951, William Dameshek grouped Di Guglielmo syndrome under the broad umbrella of myeloproliferative diseases (MPDs, now known as myeloproliferative neoplasms) based on the hypothesis that a common hematopoietic cell of origin was responsible for these disorders (4). At the time, it was unclear whether Di Guglielmo syndrome should include the pure erythroblastic manifestations of disease (5). Subsequent cytogenetic characterization studies in erythroleukemia revealed various complex cytogenetic abnormalities akin to those observed in acute myeloid leukemias (AML) (6, 7). Erythroleukemia (EL) was ultimately removed from the rubric of chronic MPDs as it was increasingly recognized as a forerunner of an acute leukemic process (8, 9).
The French-American-British (FAB) cooperative group, in their first proposal in 1976, included EL within the AML classification system, defining it based on elevated myeloblasts (≥30%) in the overall marrow. However, the proposed criteria frequently left the diagnosis under established as the non-erythroid fraction, the compartment to which myeloblasts were confined in, usually constituted only a minor fraction of the overall marrow (10). A diagnostic consequence of this was that it would often not be possible to make a diagnosis of erythroleukemia if the non-erythroid marrow were less than 30%. The criteria were revised in the 1985 FAB classification which re-defined erythroleukemia (AML M6) as requiring at least 30% of the non-erythroid compartment to be myeloblasts (specifically, ≥30% of non-erythroid compartment) in a >50% erythroblastic marrow background. However, the diagnostic constraints of AML M6 fell short of recognizing pure erythroblastic forms of disease, which were designated AML M6 variants due to lack of better fit into other morphologic categories (11, 12). Pure erythroid leukemia (PEL) first emerged as a distinct sub-entity in the 2001 WHO classification which categorized acute erythroid leukemia into two subtypes: erythroid/myeloid leukemia (EML, FAB M6a) and PEL (FAB M6b) (13, 14). Under this classification, EML was defined by the following two criteria: (1) erythroid cells comprising ≥50% of total nucleated marrow cells and, (2) myeloblasts comprising ≥20% of non-erythroid cells. PEL was defined by maturation-arrested primitive erythroblasts making up at least 80% of nucleated marrow cells.
Myelodysplastic syndromes (MDS), especially refractory anemia with excessive blasts (RAEB) or MDS with erythroid predominance, show overlapping morphological and immunophenotypic features with erythroleukemia and AML with myelodysplasia-related changes, and thus possibly represent a disease continuum evolving through phases from dyserythropoiesis to myeloid proliferation (15). The presence of increased blasts and multilineage dysplasia in ≥50% of cells in two or more lineages favors the diagnosis of AML with myelodysplasia related changes (AML-MRD) over erythroleukemia. The 2008 WHO criteria refined these diagnoses, and classified AML-MRD as a distinct subcategory (9). A majority of cases that otherwise fulfilled criteria for acute erythroid leukemia (AEL) were thus reclassified as AML-MRD. The WHO 2008 classification, although retaining the subcategories of WHO 2001, transformed erythroleukemia into a diagnosis of exclusion. Therefore, AML patients previously classified under the heterogeneous AML M6 FAB category, would fall into three subgroups under the 2008 WHO classification: (1) AML with multilineage dysplasia; (2) therapy-related AML and myelodysplastic syndromes; (3) acute erythroid leukemia subdivided into AML, NOS (erythro/myeloid leukemia type) and AML, NOS (pure erythroid leukemia type) (9, 16).
The 2008 WHO classification still left areas of diagnostic uncertainty, especially with categorization of cases with less than 20% total marrow blasts as erythroleukemia (diagnosis based on non-erythroid percentage of >20% blasts). Poor inter-observer reproducibility of blast percentages, were cited as concerns that could alternate diagnosis between AML and MDS, with significant implications on therapeutic decision making. In addition, MDS and AEL may represent a disease continuum, the prognosis and clinical course of which is dictated by shared molecular/cytogenetic characteristics than by arbitrary blast percentages (17). This resulted in the WHO revising the blast counting schema in their most recent 2016 update and eliminating the category of the term AML, NOS (erythro/myeloid leukemia type) to merge it under MDS. The category of PEL has been retained under the term AML, NOS and is now the only WHO type of acute erythroid leukemia (8). The current definition of PELrequires 80% erythroid precursor marrow involvement with at least 30% of cells being pro-erythroblasts. This category also excludes cases arising after prior cytotoxic therapy, which are instead classified as ‘therapy related myeloid neoplasms’. The 2016 WHO classification, while resolving many diagnostic challenges, has brought about new complexities, particularly with regard to the elimination of erythroid/myeloid category and the consideration of whether myeloblasts should be calculated as a percentage of non-erythroid marrow (Figure 1) (18). The requirement that 30% proerythroblasts is needed for AML, NOS (PEL type) has also highlighted the need for continued clarification (19). The diagnostic criteria for AEL (erythroid/myeloid) type and PEL type are illustrated in Figure 1. It is important these erythroid maturation disorders from polycythemia vera, a chronic myeloproliferative neoplasm characterized by hypercellular marrow with trilineage proliferation without dysplasia or dysmaturation. Current diagnostic criteria warrant further studies to better define optimal and important criteria for treatment and prognostication. Additionally, to date there is little evidence to suggest these criteria definitively distinguish separate biologically entities. Updated diagnostic criteria will undoubtedly impact future research designs and comparison with historical outcomes in AEL studies. In this manuscript, the terms erythroleukemia, AEL, and PEL are used in various sections to reflect the terminology used in referenced studies.
Figure 1.

Modern diagnostic approach to acute erythroid leukemia. M6 leukemia subtype is highlighted in yellow. Pure erythroid leukemia remains only the true form of erythroleukemia based on the updated WHO 2016 classification. Abbreviations used: MRC-myelodysplasia related changes, NOS- not otherwise specified.
2. Morphologic and clinical features
Acute erythroleukemia (by WHO 2008 Criteria) represents 1% of overall de novo AML and typically occurs in older patients, predominantly in men (male to female ratio = 2:1) (20). Pure erythroid leukemia is even more rare, constituting about 3% of acute erythroid leukemia, and has historically been associated with a dismal prognosis, with a median survival of about 2–3 months (20, 21). This compares much less favorably with the 16 month median survival observed in patients with Di Guglielmo syndrome (21). Importantly, the prognostic relevance of PEL is retained irrespective of whether it occurs de novo, arises from antecedent disease such as MDS, or is therapy-related (20).
Pure erythroid leukemia differs from erythro/myeloid leukemia variant not only in the absence of the myeloblastic component but also by arrests in erythroid maturation. PEL is characterized by proliferations of maturation-arrested primitive erythroid blastic populations (Figure 2). The earliest morphologically recognizable forms are proerythroblasts identified in the bone marrow by their large size, central round nuclei, dispersed chromatin, one to several nucleoli, and deeply basophilic agranular cytoplasm (22). However, leukemic blasts may be arrested at various stages of maturation, with the minimally differentiated erythroleukemias affecting cells in the BFU-E stage and the more mature morphological forms characterized by cells affected in the later stages of maturation such as in CFU-E stage. In this context, immunohistochemistry or flow cytometry may distinguish cells based on markers expressed in early stage precursors including CD71 (transferring receptor-1) and ferritin H. CD-71 is highly and selectively expressed in erythroid precursors, in all stages of maturation (Figure 2). Ferritin H is a soluble iron storage protein expressed in early erythroid precursors, including in acute erythroid leukemia (23). These markers may be crucial in diagnosing acute erythroleukemia versus other morphologic forms of myeloid leukemia (22–24). E-cadherin is another highly sensitive and specific (in this context) marker for immature erythroblasts, and helps in differentiating PEL from erythroid neoplasms. The sequential intact maturation of erythroid proliferations, observed in the MDS-erythroid/myeloid leukemia spectrum, does not occur in PEL (Figure 3). It is necessary to discriminate between the two diagnoses since prognosis and treatments may differ.
Figure 2.

A, Bone marrow core biopsy of pure erythroid leukemia often shows sheets of immature erythroid precursors replacing marrow cellular spaces. Cells are large with round to irregular nuclei, pale chromatin and frequent one to several distinct nucleoli. Background trilineage hematopoiesis is significantly decreased to absent. B, CD71 immunostain highlights leukemic cells with strong membranous staining pattern. C, on bone marrow smears, leukemic cells have dispersed chromatin, deep basophilic cytoplasm and cytoplasmic vacuoles. Some also have cytoplasmic blebs. Background erythroid dysplasia is present in this case. D. Coarsely granular pattern by PAS stain.
Figure 3.
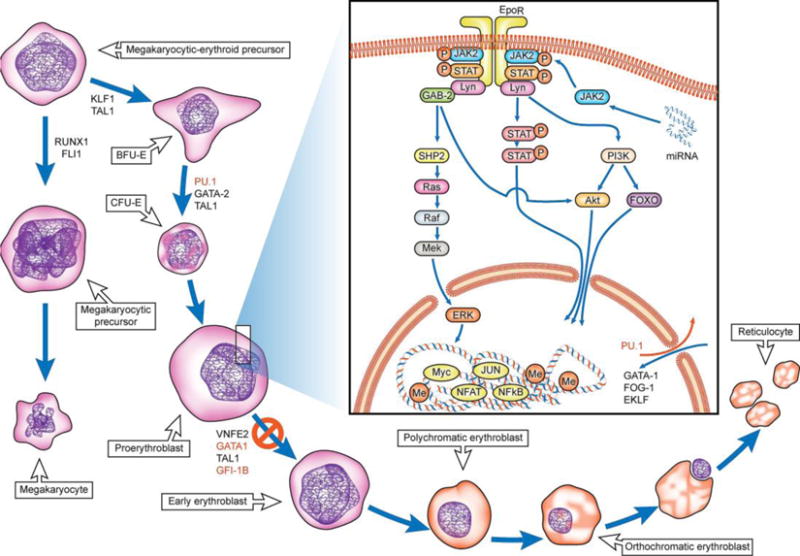
Model for erythroid differentiation and pathways implicated in human/murine erythroleukemia. Scheme outlines the stages of differentiation and maturation from the common megakaryocytic-erythroid precursor stage (70). The transcription factors, signaling proteins and miRNAs aberrantly expressed in human erythroleukemia models are highlighted in red (87). There exists a differentiation blockade at the proerythroblast stage preventing further differentiation to more mature blasts. The underlying mechanisms responsible for the blockade are an active area of investigation.
3. Current Management and prognosis
Santos et al reported on clinical outcomes of AML M6 in 91 patients treated at a single institution (25). The study investigators found no statistically significant difference in survival between M6 and other AML subtypes (p = 0.60). This was confirmed on a multivariate analysis for overall survival where AML-M6 was not an independent risk factor. The median overall survival of the overall M6 cohort was approximately 9 months and significantly lower in M6b cohort (Pure Erythoid) compared with M6a (15 weeks vs 39 weeks, p = 0.007). Interestingly, the subtype of AML-M6 (6a and 6b) was not an independent prognostic factor for disease-free and overall survival. The authors concluded that AML-M6 by itself did not carry additional prognostic import.
Recent emphasis has been the evaluation of efficacy of hypomethylating agents in the treating TP53 mutated leukemia due their ability to function through p53 independent mechanisms to effect responses. A recent clinical study supporting these mechanisms of action reported high response rates with a 10-day regimen of decitabine in TP53 mutated AML and myelodysplastic syndrome (MDS) (26). In a study of 36 AEL patients (81% reclassifiable as MDS, per 2016 WHO), decitabine-10 day regimens showed comparable overall survival and non-significant trend towards improved event free survival when compared with cytarabine-based regimens (27).
PEL is associated with complex and high-risk karyotypes including chromosomes 5q and 7q abnormalities (20). In a study evaluating patients with erythroid-predominant myeloid neoplasms, morphologic features of PEL, adverse risk cytogenetics, and other features such as hypoalbuminemia and high serum lactate dehydrogenase emerged as independent prognostic factors of death (28). Whether prognosis in erythroleukemia links solely to its association with unfavorable karyotype, or relates also to additional disease-specific characteristics, is not well-understood. Median survival among PEL patients is 1–3 months, with no survival differences observed in patients treated with intensive chemotherapy versus hypomethylating agents (HMA) (20, 28, 29). A more recent multinational study evaluating clinical outcomes of 217 patients with acute erythroleukemia demonstrated that intensive chemotherapy was superior to hypomethylating agents in affecting overall response but not associated with superior progression free or overall survival. Importantly, however, patients with high risk cytogenetics treated with HMA lived longer compared with intensive chemotherapy. The excellent therapeutic sensitivity to hypomethylating agents appears selective to the decitabine-10 day regimen and given the dismal outcomes with PEL and its association with high risk cytogenetics, prospective evaluation of 10-day decitabine is needed. It must be noted that while HMAs may represent a good treatment option for patients not ultimately going to transplant, the standard of care for a transplant-eligible patient would be induction chemotherapy based on current body of evidence Allogeneic hematopoietic cell transplantation improves outcomes of AEL and should be considered in all AEL patients with high risk cytogenetic features eligible for transplantation (30). Nevertheless, studies are yet to reassess the role of transplant in PEL as currently defined in the 2016 WHO classification.
Novel therapies based on a more detailed understanding of dysregulated TP53-related molecular pathways may predictably improve current outcomes in PEL patients.
4. Pathophysiology
4.1. Models of erythroleukemia
It is conceivable that the molecular oncogenesis of PEL, characterized by the features of both early differentiation block and proliferation in its blast populations, would share the paradigm of the proposed double-hit model involved in AML evolution (31). While the transformative processes in human erythroid progenitors are incompletely understood, described murine and avian models have proven valuable in this regard. The conceptual model of multistage carcinogenesis is exemplified by the Friend disease, a model system of erythroleukemia first described in 1957 (32). Friend erythroleukemia can be induced in susceptible strains of mice by infection with the ‘Friend’ retrovirus complex, constituted by the replication-defective spleen focus-forming virus (SFFV) and a replication competent Friend murine leukemia virus (33, 34). The disease evolves through two stages; the first stage being preleukemic and involving polyclonal expansion of erythroblasts. Glycoprotein 55 (Gp55), encoded by the envelope (Env) gene of SFFV, activates the erythropoietin receptor (EpoR) to cause erythropoietin-independent erythroblastosis, the effects of which are modulated by the expression of SF-Stk (a truncated form of STK receptor tyrosine kinase) (35, 36). The second stage is transformative and is initiated by pro-viral integration upstream to the promoter of the Sfpi-1 gene that encodes PU.1, a transcription factor of the ETS (E26 transformation-specific) oncogene family. These tumorogenic erythroblasts are clonal and have shown to be consistently altered with respect to two genes, Sfpi-1 and the TP53 gene (37, 38). Deregulated expression of PU.1 is believed to induce blockade in the erythroid differentiation program through upregulation of various potential targets including Fli1, another pro-oncogenic transcription factor, to complete the causal chain of leukemogenesis (38–40). Later developed murine models, such as the Spi-1 transgenic model system, share more similarities with the human erythroleukemic disease than the Friend model. At the onset of disease in the Spi-1 transgenic model, hematopoietic tissues were massively invaded with non-tumorigenic proerythroblasts that expressed a high level of Spi-1 protein causing profound anemia. The transgenic proerythroblasts are dependent on erythropoietin for their proliferation during this stage. In the second stage, proerythroblasts became tumorogenic and achieved growth factor autonomy through acquisition of additional genetic events including TP53 alterations (38). Unlike in the Friend disease, polycythemia does not precede the occurrence of erythroleukemia despite autocrine stimulation by Epo. This clinical feature is shared by human erythroleukemic disease in which EPO autocrine stimulation is similarly observed and may represent a secondary event related to acquisition of malignant state (41) (38, 42). With the currently available evidence, it is not yet known if the two-hit model of mouse leukemogenesis may be an oversimplification of human erythroleukemic disease. Unfortunately, the rarity of this leukemia variant and associated involvement of the myeloid compartment complicates in vitro studies of primary erythroleukemia.
Erythroleukemic cell lines
Other in vitro models for study include an extensive repertoire of human erythroleukemic cell lines, the oldest and most extensively studied of which is the K-562 cell line. Cell line characterization studies, while confirming the shared similarities of karyotypic complexity marking the disease, have identified them to be heterogeneous in their cytogenetic, molecular and cytokine-related profiles (43). A list of leukemic cell lines with erythroid features is outlined in supplemental table 1. These available erythroleukemia cell lines exhibit a wide range of functional characteristics, such as variable cytokine requirements and differing differentiation potential when grown in culture with or without differentiation agents. Still, significant information has been acquired from studying these cell lines, especially related to transcriptional and epigenetic programs in AEL. Some of these cell lines (K562, HEL, OCIMI, OCIM2, LAMA-84) express markers of multiple cell lineages, a characteristic found only infrequently in other primary leukemias (44). Few of the erythroleukemia lines are bipotential precursors, with an ability to differentiate, in vitro, along megakaryocytic or erythroid lineage pathways in the presence of appropriate inducers (45).
4.2. The role of transcription factors, molecular mutations and epigenetic alterations
4.2.1. GATA1 and PU.1
GATA1 and PU.1 function as two opposing lineage specific transcription factors that regulate erythroid and myeloid programs (46). GATA1 plays a critical role in erythroid survival and terminal erythroid differentiation (47, 48). The pivotal functional significance of these interactions in determining lineage fate and lineage-specific differentiation may underlie the leukemogenic mechanisms of maturation arrest consequent to dysregulated expression of these transcription factors (49).
In vitro studies have shown that although upregulation of PU.1 contributed to erythroleukemia by causing differentiation arrest, it induced growth inhibition and apoptosis in the presence of differentiating agents such as dimethyl sulfoxide (DMSO) (50). The growth inhibitory and apoptotic effects of PU.1 on erythroleukemic cells are associated with the downregulation of pro-survival oncogenes such as c-MYC and BCL2, and reduced DNA binding activity of the GATA1 transcription factor (50, 51). PU.1 interacts directly with GATA1 and represses its erythroid differentiating action; introducing exogenous GATA1 was able to induce differentiation in PU.1 blocked murine erythroleukemia (MEL) cells (52). GATA1 represses PU.1 expression by binding to the PU.1 promotor and its other regulatory upstream response elements. Available evidence suggests that the antagonistic influence of these transcription factors’ function in the erythroid maturation-differentiation program is finely modulated by their relative expression. While complete loss is lethal, graded reductions in PU.1 gene expression associate with an increasingly aggressive AML phenotype (53). Unlike in normally differentiating erythroid precursors where GATA1 completely shuts down PU.1 to repress the myeloid program, expression of PU.1 in human AEL is maintained, albeit at reduced levels, from its incomplete repression by GATA1 (54). It appears that a complete transcriptional repression of PU.1 leads to the loss of PU.1-dependent repression of GATA1 targets, facilitating erythroid differentiation. On the other hand, blockade of GATA1 mediated repression on PU.1 would increase PU.1 expression leading to differentiation and growth arrest (55). One may speculate that an attenuated expression of PU.1 may paradoxically enhance leukemogenesis through changes in the nuclear environment brought about by interactions with GATA1.
GATA-1 mediated repression mechanisms exhibit distinct inter-species differences in human and murine AEL cell lines. Unlike in murine AML-EL, PU.1 repression by GATA1 additionally involves DNA binding along with H3K9 and H3K27 trimethylation at regulatory upstream enhancer and promoter regions. Along these lines, the upper response enhancer elements in human AEL cells contain a DNA methylation mark. Reversal of the DNA methylation mark via inhibition of DNMT3A, a co-occupant of the GATA1 repression complex, by hypomethylating agents has been demonstrated to inhibit leukemic growth and induce differentiation (54). This deregulated mechanism of upper response element DNA methylation is also shared by MDS, and may even predict for clinical response to hypomethylation therapy (55).
4.2.2. TP53
Rose et al reported on molecular mutation data in a cohort of 166 AEL (M6) patients and showed that TP53 was the only gene occurring at a higher frequency within M6 as compared with the remaining overall AML cohort (36% vs 11%, respectively). Relatively lower mutational frequencies were observed for other genes including ASXL1, DNMT3A, FLT3-ITD, IDH2, NPM1, NRAS, RUNX1, and TET2 (56). Recent data reported by Montalban-Bravo and Benton et al. revealing an especially high prevalence of at least two TP53 abnormalities (including both mutations and aberrant or deleted chromosome 17p) in >90% of PEL patients (29). The high frequency of TP53 alterations suggest a crucial role of TP53 in the leukemic transformation to AEL. It remains to be determined whether TP53 alterations are a proximate effect of excess cytoplasmic iron sequestered within the dysfunctional pronormoblasts. In this context, there appears to be a link between iron/heme homeostasis and p53 signaling, with p53 downregulated during iron excess, via mechanisms operating at various levels influencing nuclear export of the p53, p53 stabilization and p53-DNA interactions (57). Iron induced DNA damage, via free radical oxygen species formation, may well contribute to the high rate of TP53 mutations and complex karyotype observed in this malignancy.
4.2.3. GATA1 and p53 interactions: a role in erythroleukemia?
GATA1 directly influences p53 by interacting with the p53 transactivation domain and inhibiting its transactivation in erythroid precursor cells (58). This interaction is an erythroid cell-specific event with inhibition of p53 by GATA1, not observed in non-erythroid cells. GATA1 is crucial in mediating erythroid differentiation with GATA1 knockdown shown to induce erythroid leukemia in mice (59). In this context, hyperproliferative GATA1 null erythroid cells which escape cell death may accumulate secondary mutations leading to transformation (60). In the absence of GATA1 mediated p53 inhibition in GATA1 deficient cells, functional p53 pathway activation may be crucial in inducing cell cycle arrest with TP53 mutations may allow for the abnormal expansion of leukemic cells predisposing to leukemic transformation (58, 59).
4.2.4. c-MYC and Bromodomain inhibition
The bromodomain (BRD) family are an epigenetic class of histone modification proteins with an ability to ‘read’ the genome and modulate gene expression through transcriptional regulator recruitment to specific genome locations (61). The protein family comprises four homologous proteins: BRD2, BRD3, BRD4, and BRDT, with widely varying roles on cell cycle growth and regulation. It has been demonstrated that BRDs of these reader proteins promote aberrant gene expression and sustain leukemic maintenance, at least in part to sustained MYC expression, thus paving a rationale for developing inhibitors against this class (62) (Figure 4). In vitro experiments with BRD inhibitors, such as JQ1, have demonstrated these agents to carry anti-leukemic activity (63). Consistent with this, JQ1 treatment of UT7, a human erythroleukemia cell line, was able to rescue erythropoietin differentiation within a matter of two days (64). This also highlights the importance of a cellular erythroid cycle break mediated by c-MYC inhibition before initiation of the erythropoiesis program. The therapeutic potential of BRD inhibition merits further exploration within this subtype of leukemia.
Figure 4.

Proposed potential therapeutic targets in Erythroleukemia. A wide range of signaling pathway mutations (JAK2, NTRK1, ALK), epigenetic alterations (Hypomethylating agents, LSD-1 inhibitors, bromodomain inhibitors), and microRNA dysregulation (microRNA therapies) present with multiple options for therapeutic targeting. Abbreviations used: BRD-bromodomain reading proteins, miR-microRNA, EpoR-Erythropoietin receptor
4.2.5. GFI-1B and LSD-1 interactions
Another important transcription factor that has been implicated in erythroid leukemia is the growth factor-independent 1B protein (GFI-1B). GFI-1B plays a crucial role in erythroid progenitor cell growth and differentiation induction (65). Of interest, its overexpression is restricted to the AML-M6 and AML-M7 subtypes and is associated with increased proliferative capacity of progenitor cell lines (66). Silencing its expression through siRNA was shown to decrease the proliferative capacity of HEL cell lines. GFI-1B interacts with histone demethylase LSD1 thereby repressing GBI-1B target genes and consequent differentiation of lineage specific cells. Treatment with an LSD-1 inhibitor, T-3775440, was able to disrupt the GFI-1B LSD1 interaction, leading to transdifferentiation and cell growth arrest suggesting a novel mechanism of action specifically against AEL.
4.2.6. KIT receptor-ligand system
A focused network of lineage-specific transcription factors play a decisive role in determining lineage fate at the crossroads of erythroid and megakaryocytic differentiation. The erythroid and megakaryocytic lines share early lineage similarities in regulatory transcription factors and cell surface marker expression; evidence suggests that the lineages diversify from a common erythroid–megakaryocytic progenitor (43). Flow cytometric analyses have revealed HEL cell lines to exist in two distinct sub-clones: CKIT-positive, CD41b-negative (erythroid lineage markers) and CKIT-negative, CD41b-positive (megakaryocytic lineage markers). KIT receptor expression relates to the expression of lineage specific antigens and also determines phenotypic fate towards erythroid differentiation (67). MiR-221 and miR-222, miRNAs downregulated during erythroid differentiation, down-modulate CKIT protein production through translational repression. MiR-221 and -222 gene transfer impairs proliferation and accelerates differentiation of the CKIT-positive TF-1 erythroleukemic cell lines (68).
4.2.7. RUNX1 and KLF1
Another key transcription factor in AEL is RUNX1 which, along with multiple micro-RNAs, negatively affects erythroid differentiation by repressing an erythroid master regulator krueppel-like factor 1 (KLF1). Repression of KLF1 turns off the erythroid gene expression program and facilitates megakaryocytic lineage specification (69). Recent studies have reported RUNX1 mutations in erythroleukemia, albeit at significantly lower frequencies compared with overall AML (56). There have not yet been studies directly implicating KLF1 in human erythroleukemia.
4.2.8. Other transcription factors
Other relevant transcription factors involved in the erythroid differentiation program such as GATA2, SCL/TAL, NF-E2, nuclear factor-B, forkhead transcription factors (FOXO), EKLF have also been studied in various in vitro models (70). Although these factors seem to have a pathogenic role in murine tumor models and other malignancies, mutations in genes encoding these transcription factors have not been directly implicated in human erythroleukemia.
4.3. Signaling proteins and pathways
4.3.1. JAK-STAT pathway
The proliferative and differentiating effects of erythropoetin (EPO) commences with ligand binding to its cognate EPO receptor. Subsequent subunit dimerization and JAK2 recruitment results in phosphorylation of several tyrosine residues on the receptor. These phosphorylated residues serve as docking sites for signal transducer and activator of transcription (STAT) transcription factors, most prominently STAT5, and phosphorylate them. Phosphorylated STAT transcription factors dimerize and enter the nucleus to activate transcription of specific genes (71).
In vitro studies on freshly isolated human primary erythroleukemic cells demonstrated the constitutive activation of STAT1 and STAT3 and their role in promoting cell growth through c-MYC activation (72). STAT proteins also influence various aspects of erythroid differentiation. In the Friend disease model, activated STAT3 upregulates PU.1 thus promoting progression of erythroleukemia by inhibiting erythroid differentiation (73). Conversely, activation of transcription factors such as STAT5 correlates with erythropoietin mediated erythroid differentiation and its conditional inactivation in erythroleukemic cell lines have been demonstrated to prevent terminal differentiation (74). Furthermore, erythroleukemic cell lines exhibit activation of multiple signaling pathways apart from JAK-STAT, including mTOR, PI3K/Akt pathways, thus serving multiple potential targets for targeted inhibitors (75). In the Friend disease model, activating mutations in receptor tyrosine kinases including CKIT results in clonal expansion through activation of multiple signaling pathways such ERK & MAP kinases, PI3Kinase, and Src kinases (76).
4.3.2. MicroRNAs
MicroRNAs are non-coding RNAs that play a crucial role in cell growth and differentiation through regulation of gene expression (77). A miRNA profiling study in MEL cell lines found more than a hundred miRNAs to be dynamically expressed, with wide variations in expression levels, during the process of terminal differentiation induction after DMSO treatment (78). MiR-451 and miR-144 are upregulated, whereas miR-221, miR-222, miR-24, and miR-223 are downregulated during erythroid differentiation. Among the many identified miRNAs, miR-451, an erythroid differentiation promoting miRNA, in particular, was found to increase significantly with erythroid differentiation. The investigators were further able to demonstrate, through transfection of synthetic anti-sense miR-451 oligonucleotides into MEL cells thereby inducing miR-451 knockdown, that miR-451 positively regulates erythroid differentiation. Bruchova-Votavova et al demonstrated that enforced expression of miR-451 induced erythroid differentiation in K562 cells, an erythroleukemic cell line (79). Comprehensive miRNA profiling data in human PEL while lacking, perhaps due to the rarity of the neoplasm, could be performed to help provide deeper insights into disease pathogenesis and potentially inform miRNA targeted cancer therapies.
Participating member proteins of the JAK-STAT pathway are tightly regulated by a network of regulatory transcription factors and microRNAs. Su et al demonstrated that the miR-23a, -27a, and -24 miRNA cluster in particular, was dramatically downregulated in AEL patients and that restoration of their expression was able to induce apoptosis through inhibition of JAK-STAT3 pathway cascade (80). The investigators were also able to show that the inhibition of the pathway by the miRNA cluster simultaneously involved an upregulated expression of GATA1, which through PU.1 inhibition, was able to induce erythroid differentiation. This attests to the important role of dysregulated GATA1-JAK-STAT pathway in AEL pathogenesis.
4.4. Molecular characterization of clinical samples
Earlier investigational studies had associated AEL with a high frequency of mutations and identified it to carry mutational profiles significantly different from other AML subtypes. AEL is characterized by far lower NPM1 and FLT3-ITD mutation rates and higher mutational rates in TP53 compared with other AML subtypes (81). Also, AEL samples have been associated with lower frequency of ASXL1 and spliceosome-related mutations compared with AML-MDS and erythroid predominant MDS, respectively (82). However, it must be noted that these studies were limited in their investigation to few candidate genes traditionally implicated in overall AML, and did not consider the pure erythroid leukemia type (81, 82).
Recent preliminary data from a more comprehensive characterization of the genomic landscape of AEL by Iacobucci et al, using next-generation sequencing methods, has identified recurrent mutations in multiple genes involved in cell cycle/tumor suppression, cohesin complex formation, RNA splicing, transcription, signaling, DNA methylation and chromatin modification (75, 83). Additionally, chimeric fusions were detected in nearly half of the cases. Interestingly, in vitro modeling of a few selected fusion transcripts identified only the expression of NUP98-JARID1A fusion protein to result in leukemia, suggesting the need for coexistence of other genetic lesions for leukemogenesis (75). Also, mutation profile patterns varied in pediatric and adult AEL with NUP98-fusions, PTPN11, GATA1, and UBTF mutations more frequent in pediatric AEL, and TP53 and MLL mutations predominant in adult AEL. Expectantly, high risk features of complex karyotype, TP53 mutated and therapy-related AEL were associated with poor outcomes. The investigators were so far reportedly able to classify AEL into 6 subtypes based on exclusivity and co-occurrence of mutations (83). Additionally, 33% of cases harbored signaling pathway gene mutations, 3 classes of which were found to be targetable by in vivo and in vitro studies; ALK mutations to crizotinib, tyrosine kinase domain mutations of NTRK1 to entrectinib, and JAK-STAT, mTOR, PI3K pathway targeting to JAK2 inhibitor ruxolitinib (83). Similarly, isolated case reports of fusion genes such as NFIA-CBFA2T3 (84) and ZMYND8-RELA (85) have been described. Several of these fusion transcripts involve epigenetic regulators, transcription factors and signaling mediators and likely promote leukemogenesis through cellular pathways that have yet to be elucidated (75, 85). Although molecular characterization has facilitated prognostication by distinguishing cytogenetic and molecular subsets, it has thus far failed to explain phenotypic and lineage attributes specific to AEL.
4.5 A proposed leukemia model
In summary, human erythroleukemia may viewed as multi-step model of leukemogenesis. Major inciting events in the initial stages are epigenetic and micro-RNA dysregulation, and transcription factor level alterations leading to differentiation arrest and resistance to apopotosis among pro-erythroblasts. Arrested pro-erythroblasts are functionally defective and hyper-proliferative; increased cellular proliferation leads to accumulation of mutations in signaling pathway genes, including TP53, leading to leukemic transformation of arrested pro-erythroblasts (Figure 5). Subsequent maintenance and proliferation of leukemic blasts is facilitated by continued presence of genetic/epi-genetic alterations in the already transformed leukemic blasts. Although, quantitative defects of GATA1 have not yet been reported in human erythroleukemia, attenuated GATA1 expression levels and TP53 alterations provides a multi-step model of leukemogenesis that warrants further investigation.
Figure 5.

A proposed two-step model of erythroleukemia. Epigenetic and micro-RNA dysregulation, and transcription factor level alterations lead to differentiation arrest at the pro-erythroblast stage. Arrested pro-erythroblasts are hyper-proliferative and accumulate secondary mutations, including TP53, leading to leukemic transformation of arrested pro-erythroblasts. Continued maintenance and proliferation of leukemic blasts is facilitated by pre-existing genetic/epi-genetic alterations in the already transformed blasts.
5. Summary and future directions
Erythroleukemia represents a phenotypically distinct form of AML characterized by unfavorable risk karyotype and disease features. With the emergence of pure erythroid leukemia as a pathophysiologic entity in part disconnected from its historical counterpart, erythro/myeloid leukemia variant, future clinical and translational investigational studies should reflect this distinction, especially in the setting of the newly updated WHO 2016 diagnostic criteria. Further refinements in future definitional criteria are expected, incorporating emerging mutation and chromosomal data garnered from studies inspired by a growing interest and recognition of PEL. Data from pre-clinical studies demonstrate that epigenetic alterations and micro RNA dysregulation are important in the pathogenesis of erythroleukemia and novel therapeutic strategies such as hypomethylating agents, bromodomain inhibitors, histone demethylase inhibitors, and micro-RNA targeting therapies should be explored within this subtype of leukemia (Figure 3). The GATA-JAK-STAT circuit represents another important dysregulated pathway that may be targeted with molecular inhibitors. In addition, the high frequency of TP53 alterations in erythroleukemia suggest that erythroleukemia patients may benefit from treatment with therapies circumventing p53 dependent cytotoxic mechanisms such as BCL-2 inhibitors and antibody drug conjugates (86). Next generation sequencing studies have provided valuable insights into the genetic landscape of AEL, and may pave the way for the transformation from a morphologic/phenotypically based classification to an enhanced molecular classification of prognostic and therapeutic relevance. Efforts at defining the genetic and epigenetic landscape of erythroleukemia are underway, and delineating the pathogenic role of identified molecular aberrations, will guide future therapeutic strategies.
Supplementary Material
Practice points
Pure erythroid leukemia represents a distinct clinicopathological entity characterized by high risk chromosomal abnormalities and dismal outcomes, and must be distinguished from other myeloid neoplasms with erythroid features.
TP53 mutations confer therapeutic sensitivity to hypomethylating agents, and while the role of the 10-day decitabine regimen merits further exploration in pure erythroid leukemia, intensive chemotherapy should be the preferred treatment option based on the current body of evidence.
Allogeneic stem cell transplantation improves outcomes in acute erythroleukemia and should be considered in AEL with high risk chromosomal features.
Given poor responses to standard therapy, patients are best enrolled upfront in clinical trials evaluating investigational therapies such as LSD-1 inhibitors, bromodomain inhibitors, BCL2 inhibitors, and antibody drug conjugates.
Research agenda
Improved understanding of erythroleukemia by developing better translational erythroleukemic models.
Identification of pathognomonic signaling pathways for developing pathway specific inhibitors for AEL.
Comprehensive profiling of genetic, epigenetic and miRNA landscape in erythroleukemia.
Acknowledgments
Our funding was from the following sources – NP is supported in part by the MD Anderson Cancer Center Support Grant CA016672 and Award Number P01 CA049639 and SagerStrong Foundation. JDK is supported in part by a University of Texas MD Anderson Cancer Center Institutional Research Grant Award, a Leukemia Specialized Programs of Research Excellence (SPORE) Research Development Program Award, and the University Cancer Foundation via the Sister Institution Network Fund at the University of Texas MD Anderson Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflicts of interest in the publication of this paper.
References
- 1.Schwartz SO, Critchlow J. Erythremic myelosis (DI Guglielmo’s disease); critical review with report of four cases, and comments on erythroleukemia. Blood. 1952;7(8):765–93. [PubMed] [Google Scholar]
- 2.Dameshek W, Baldini M. The Di Guglielmo syndrome. Blood. 1958;13:192–4. [PubMed] [Google Scholar]
- 3.Bain BJ. Di Guglielmo and his syndromes. Br J Haematol. 2003;120:939–43. doi: 10.1046/j.1365-2141.2003.04181.x. [DOI] [PubMed] [Google Scholar]
- 4.Dameshek W. Some speculations on the myeloproliferative syndromes. Blood. 1951;6(4):372–5. [PubMed] [Google Scholar]
- 5.Dameshek W. The DiGuglielmo syndrome revisited. Blood. 1969;34(5):567–72. [PubMed] [Google Scholar]
- 6.Heath CW, Jr, Bennett JM, Whang-Peng J, Berry EW, Wiernick PH. Cytogenetic findings in erythroleukemia. Blood. 1969;33(3):453–67. [PubMed] [Google Scholar]
- 7.Dyment PG, Melnyk J, Brubaker CA. A cytogenetic study of acute erythroleukemia in children. Blood. 1968;32(6):997–1002. [PubMed] [Google Scholar]
- 8.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 9.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 10.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33(4):451–8. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 11.Villeval JL, Cramer P, Lemoine F, Henri A, Bettaieb A, Bernaudin F, et al. Phenotype of early erythroblastic leukemias. Blood. 1986;68(5):1167–74. [PubMed] [Google Scholar]
- 12.Hasserjian RP, Howard J, Wood A, Henry K, Bain B. Acute erythremic myelosis (true erythroleukaemia): a variant of AML FAB-M6. J Clin Pathol. 2001;54(3):205–9. doi: 10.1136/jcp.54.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arber DA, Brunning RD, Orazi A, Porwit A, Peterson L, Thiele J, et al. Acute myeloid leukemia, not otherwise specified. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al., editors. WHO Classifiction of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC; 2008. pp. 134–6. [Google Scholar]
- 14.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, et al. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol. 1999;17(12):3835–49. doi: 10.1200/JCO.1999.17.12.3835. [DOI] [PubMed] [Google Scholar]
- 15.Park S, Picard F, Azgui Z, Viguie F, Merlat A, Guesnu M, et al. Erythroleukemia: a comparison between the previous FAB approach and the WHO classification. Leuk Res. 2002;26(5):423–9. doi: 10.1016/s0145-2126(01)00146-1. [DOI] [PubMed] [Google Scholar]
- 16.van Krieken JH. The 2008 WHO-classification: small and big changes! J Hematop. 2009;2(2):63. doi: 10.1007/s12308-009-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bacher U, Haferlach C, Alpermann T, Kern W, Schnittger S, Haferlach T. Comparison of genetic and clinical aspects in patients with acute myeloid leukemia and myelodysplastic syndromes all with more than 50% of bone marrow erythropoietic cells. Haematologica. 2011;96(9):1284–92. doi: 10.3324/haematol.2011.043687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arber DA. Revisiting erythroleukemia. Curr Opin Hematol. 2017 Mar;24(2):146–151. doi: 10.1097/MOH.0000000000000314. 2016. [DOI] [PubMed] [Google Scholar]
- 19.Mazzella FM, Kowal-Vern A, Shrit MA, Wibowo AL, Rector JT, Cotelingam JD, et al. Acute erythroleukemia: evaluation of 48 cases with reference to classification, cell proliferation, cytogenetics, and prognosis. Am J Clin Pathol. 1998;110(5):590–8. doi: 10.1093/ajcp/110.5.590. [DOI] [PubMed] [Google Scholar]
- 20.Liu W, Hasserjian RP, Hu Y, Zhang L, Miranda RN, Medeiros LJ, et al. Pure erythroid leukemia: a reassessment of the entity using the 2008 World Health Organization classification. Mod Pathol. 2011;24(3):375–83. doi: 10.1038/modpathol.2010.194. [DOI] [PubMed] [Google Scholar]
- 21.Kowal-Vern A, Cotelingam J, Schumacher HR. The prognostic significance of proerythroblasts in acute erythroleukemia. Am J Clin Pathol. 1992;98(1):34–40. doi: 10.1093/ajcp/98.1.34. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Wang SA, Medeiros LJ, Khoury JD. Pure Erythroid Leukemia. Am J Hematol. 2017 Mar;92(3):292–296. doi: 10.1002/ajh.24626. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Grier DD, Woo J, Ward M, Sui G, Torti SV, et al. Ferritin H is a novel marker of early erythroid precursors and macrophages. Histopathology. 2013;62(6):931–40. doi: 10.1111/his.12101. [DOI] [PubMed] [Google Scholar]
- 24.Dong HY, Wilkes S, Yang H. CD71 is selectively and ubiquitously expressed at high levels in erythroid precursors of all maturation stages: a comparative immunochemical study with glycophorin A and hemoglobin A. Am J Surg Pathol. 2011;35(5):723–32. doi: 10.1097/PAS.0b013e31821247a8. [DOI] [PubMed] [Google Scholar]
- 25.Santos FP, Faderl S, Garcia-Manero G, Koller C, Beran M, O’Brien S, et al. Adult acute erythroleukemia: an analysis of 91 patients treated at a single institution. Leukemia. 2009;23(12):2275–80. doi: 10.1038/leu.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welch JS, Petti AA, Miller CA, Fronick CC, O’Laughlin M, Fulton RS, et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med. 2016;375(21):2023–36. doi: 10.1056/NEJMoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King R, Crouch A, Radojcic V, Marini BL, Perissinotti AJ, Bixby DL. Therapeutic Outcomes of Patients with Acute Erythroid Leukemia Treated with Hypomethylating Agents. Blood. 2016;128(22):5203–-. [Google Scholar]
- 28.Ko PS, Liu YC, Yeh CM, Gau JP, Yu YB, Hsiao LT, et al. The uniqueness of morphological features of pure erythroid leukemia in myeloid neoplasm with erythroid predominance: A reassessment using criteria revised in the 2016 World Health Organization classification. PLoS One. 2017;12(2):e0172029. doi: 10.1371/journal.pone.0172029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montalban-Bravo G, Benton CB, Wang SA, Ravandi F, Kadia T, Cortes J, et al. Greater than one TP53 abnormality is a dominant characteristic of pure erythroid leukemia. Blood. 2017 May 4;129(18):2584–2587. doi: 10.1182/blood-2016-11-749903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasserjian RP, Zuo Z, Garcia C, Tang G, Kasyan A, Luthra R, et al. Acute erythroid leukemia: a reassessment using criteria refined in the 2008 WHO classification. Blood. 2010;115(10):1985–92. doi: 10.1182/blood-2009-09-243964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grove CS, Vassiliou GS. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Dis Model Mech. 2014;7(8):941–51. doi: 10.1242/dmm.015974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ney PA, D’Andrea AD. Friend erythroleukemia revisited. Blood. 2000;96(12):3675–80. [PubMed] [Google Scholar]
- 33.Ben-David Y, Bernstein A. Friend virus-induced erythroleukemia and the multistage nature of cancer. Cell. 1991;66(5):831–4. doi: 10.1016/0092-8674(91)90428-2. [DOI] [PubMed] [Google Scholar]
- 34.Ruscetti SK. Deregulation of erythropoiesis by the Friend spleen focus-forming virus. Int J Biochem Cell Biol. 1999;31(10):1089–109. doi: 10.1016/s1357-2725(99)00074-6. [DOI] [PubMed] [Google Scholar]
- 35.Li JP, D’Andrea AD, Lodish HF, Baltimore D. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature. 1990;343(6260):762–4. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 36.Nishigaki K, Thompson D, Hanson C, Yugawa T, Ruscetti S. The envelope glycoprotein of friend spleen focus-forming virus covalently interacts with and constitutively activates a truncated form of the receptor tyrosine kinase Stk. J Virol. 2001;75(17):7893–903. doi: 10.1128/JVI.75.17.7893-7903.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasher JM, Elenitoba-Johnson KS, Kelley LL. Loss of p53 tumor suppressor function is required for in vivo progression of Friend erythroleukemia. Oncogene. 2001;20(23):2946–55. doi: 10.1038/sj.onc.1204395. [DOI] [PubMed] [Google Scholar]
- 38.Moreau-Gachelin F, Wendling F, Molina T, Denis N, Titeux M, Grimber G, et al. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol. 1996;16(5):2453–63. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreau-Gachelin F. Spi-1/PU.1: an oncogene of the Ets family. Biochim Biophys Acta. 1994;1198(2–3):149–63. doi: 10.1016/0304-419x(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 40.Starck J, Doubeikovski A, Sarrazin S, Gonnet C, Rao G, Skoultchi A, et al. Spi-1/PU.1 is a positive regulator of the Fli-1 gene involved in inhibition of erythroid differentiation in friend erythroleukemic cell lines. Mol Cell Biol. 1999;19(1):121–35. doi: 10.1128/mcb.19.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villeval JL, Mitjavila MT, Dusanter-Fourt I, Wendling F, Mayeux P, Vainchenker W. Autocrine stimulation by erythropoietin (Epo) requires Epo secretion. Blood. 1994;84(8):2649–62. [PubMed] [Google Scholar]
- 42.Mitjavila MT, Le Couedic JP, Casadevall N, Navarro S, Villeval JL, Dubart A, et al. Autocrine stimulation by erythropoietin and autonomous growth of human erythroid leukemic cells in vitro. J Clin Invest. 1991;88(3):789–97. doi: 10.1172/JCI115378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drexler HG, Matsuo Y, MacLeod RA. Malignant hematopoietic cell lines: in vitro models for the study of erythroleukemia. Leuk Res. 2004;28(12):1243–51. doi: 10.1016/j.leukres.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 44.Papayannopoulou T, Nakamoto B, Kurachi S, Tweeddale M, Messner H. Surface antigenic profile and globin phenotype of two new human erythroleukemia lines: characterization and interpretations. Blood. 1988;72(3):1029–38. [PubMed] [Google Scholar]
- 45.Grossman Z. The stem cell concept revisited: self-renewal capacity is a dynamic property of hemopoietic cells. Leuk Res. 1986;10(8):937–50. doi: 10.1016/0145-2126(86)90246-8. [DOI] [PubMed] [Google Scholar]
- 46.Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126(4):755–66. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 47.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D’Agati V, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349(6306):257–60. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 48.Yamada T, Kihara-Negishi F, Yamamoto H, Yamamoto M, Hashimoto Y, Oikawa T. Reduction of DNA binding activity of the GATA-1 transcription factor in the apoptotic process induced by overexpression of PU.1 in murine erythroleukemia cells. Exp Cell Res. 1998;245(1):186–94. doi: 10.1006/excr.1998.4251. [DOI] [PubMed] [Google Scholar]
- 49.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21(21):3368–76. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 50.Yamada T, Kondoh N, Matsumoto M, Yoshida M, Maekawa A, Oikawa T. Overexpression of PU.1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood. 1997;89(4):1383–93. [PubMed] [Google Scholar]
- 51.Kihara-Negishi F, Yamada T, Kubota Y, Kondoh N, Yamamoto H, Abe M, et al. Down-regulation of c-myc and bcl-2 gene expression in PU.1-induced apoptosis in murine erythroleukemia cells. Int J Cancer. 1998;76(4):523–30. doi: 10.1002/(sici)1097-0215(19980518)76:4<523::aid-ijc14>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 52.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13(11):1398–411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36(6):624–30. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 54.Burda P, Vargova J, Curik N, Salek C, Papadopoulos GL, Strouboulis J, et al. GATA-1 Inhibits PU.1 Gene via DNA and Histone H3K9 Methylation of Its Distal Enhancer in Erythroleukemia. PLoS One. 2016;11(3):e0152234. doi: 10.1371/journal.pone.0152234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Curik N, Burda P, Vargova K, Pospisil V, Belickova M, Vlckova P, et al. 5-azacitidine in aggressive myelodysplastic syndromes regulates chromatin structure at PU.1 gene and cell differentiation capacity. Leukemia. 2012;26(8):1804–11. doi: 10.1038/leu.2012.47. [DOI] [PubMed] [Google Scholar]
- 56.Rose D, Haferlach T, Schnittger S, Perglerova K, Kern W, Haferlach C. Subtype-specific patterns of molecular mutations in acute myeloid leukemia. Leukemia. 2017;31(1):11–7. doi: 10.1038/leu.2016.163. [DOI] [PubMed] [Google Scholar]
- 57.Shen J, Sheng X, Chang Z, Wu Q, Wang S, Xuan Z, et al. Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function. Cell Rep. 2014;7(1):180–93. doi: 10.1016/j.celrep.2014.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trainor CD, Mas C, Archambault P, Di Lello P, Omichinski JG. GATA-1 associates with and inhibits p53. Blood. 2009;114(1):165–73. doi: 10.1182/blood-2008-10-180489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimizu R, Engel JD, Yamamoto M. GATA1-related leukaemias. Nat Rev Cancer. 2008;8(4):279–87. doi: 10.1038/nrc2348. [DOI] [PubMed] [Google Scholar]
- 60.Crispino JD. GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol. 2005;16(1):137–47. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 61.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 62.Prinjha RK, Witherington J, Lee K. Place your BETs: the therapeutic potential of bromodomains. Trends Pharmacol Sci. 2012;33(3):146–53. doi: 10.1016/j.tips.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goupille O, Penglong T, Lefevre C, Granger M, Kadri Z, Fucharoen S, et al. BET bromodomain inhibition rescues erythropoietin differentiation of human erythroleukemia cell line UT7. Biochem Biophys Res Commun. 2012;429(1–2):1–5. doi: 10.1016/j.bbrc.2012.10.112. [DOI] [PubMed] [Google Scholar]
- 65.Osawa M, Yamaguchi T, Nakamura Y, Kaneko S, Onodera M, Sawada K, et al. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood. 2002;100(8):2769–77. doi: 10.1182/blood-2002-01-0182. [DOI] [PubMed] [Google Scholar]
- 66.Elmaagacli AH, Koldehoff M, Zakrzewski JL, Steckel NK, Ottinger H, Beelen DW. Growth factor-independent 1B gene (GFI1B) is overexpressed in erythropoietic and megakaryocytic malignancies and increases their proliferation rate. Br J Haematol. 2007;136(2):212–9. doi: 10.1111/j.1365-2141.2006.06407.x. [DOI] [PubMed] [Google Scholar]
- 67.Kubota A, Okamura S, Shimoda K, Ikematsu W, Otsuka T, Niho Y. Analysis of c-kit expression of human erythroleukemia cell line, HEL: clonal variation and relationship with erythroid and megakaryocytic phenotype. Leuk Res. 1995;19(4):283–90. doi: 10.1016/0145-2126(94)00160-c. [DOI] [PubMed] [Google Scholar]
- 68.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102(50):18081–6. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuvardina ON, Herglotz J, Kolodziej S, Kohrs N, Herkt S, Wojcik B, et al. RUNX1 represses the erythroid gene expression program during megakaryocytic differentiation. Blood. 2015;125(23):3570–9. doi: 10.1182/blood-2014-11-610519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wickrema A, Crispino JD. Erythroid and megakaryocytic transformation. Oncogene. 2007;26(47):6803–15. doi: 10.1038/sj.onc.1210763. [DOI] [PubMed] [Google Scholar]
- 71.Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113(3):365–71. doi: 10.1038/bjc.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirito K, Nagashima T, Ozawa K, Komatsu N. Constitutive activation of Stat1 and Stat3 in primary erythroleukemia cells. Int J Hematol. 2002;75(1):51–4. doi: 10.1007/BF02981979. [DOI] [PubMed] [Google Scholar]
- 73.Hegde S, Ni S, He S, Yoon D, Feng GS, Watowich SS, et al. Stat3 promotes the development of erythroleukemia by inducing Pu.1 expression and inhibiting erythroid differentiation. Oncogene. 2009;28(38):3349–59. doi: 10.1038/onc.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwatsuki K, Endo T, Misawa H, Yokouchi M, Matsumoto A, Ohtsubo M, et al. STAT5 activation correlates with erythropoietin receptor-mediated erythroid differentiation of an erythroleukemia cell line. J Biol Chem. 1997;272(13):8149–52. doi: 10.1074/jbc.272.13.8149. [DOI] [PubMed] [Google Scholar]
- 75.Iacobucci I, Wen J, Meggendorfer M, Carmichael C, Choi J, Li Y, et al. Genomic Landscape of Acute Erythroid Leukemia. Clin Lymphoma Myeloma Leuk. 2016;16:S26–27. [Google Scholar]
- 76.Moreau-Gachelin F. Multi-stage Friend murine erythroleukemia: molecular insights into oncogenic cooperation. Retrovirology. 2008;5:99. doi: 10.1186/1742-4690-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 78.Zhan M, Miller CP, Papayannopoulou T, Stamatoyannopoulos G, Song CZ. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp Hematol. 2007;35(7):1015–25. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bruchova-Votavova H, Yoon D, Prchal JT. miR-451 enhances erythroid differentiation in K562 cells. Leuk Lymphoma. 2010;51(4):686–93. doi: 10.3109/10428191003629362. [DOI] [PubMed] [Google Scholar]
- 80.Su R, Dong L, Zou D, Zhao H, Ren Y, Li F, et al. microRNA-23a, -27a and -24 synergistically regulate JAK1/Stat3 cascade and serve as novel therapeutic targets in human acute erythroid le ukemia. Oncogene. 2016;35(46):6001–14. doi: 10.1038/onc.2016.127. [DOI] [PubMed] [Google Scholar]
- 81.Grossmann V, Bacher U, Haferlach C, Schnittger S, Potzinger F, Weissmann S, et al. Acute erythroid leukemia (AEL) can be separated into distinct prognostic subsets based on cytogenetic and molecular genetic characteristics. Leukemia. 2013;27(9):1940–3. doi: 10.1038/leu.2013.144. [DOI] [PubMed] [Google Scholar]
- 82.Cervera N, Carbuccia N, Garnier S, Guille A, Adelaide J, Murati A, et al. Molecular characterization of acute erythroid leukemia (M6-AML) using targeted next-generation sequencing. Leukemia. 2016;30(4):966–70. doi: 10.1038/leu.2015.198. [DOI] [PubMed] [Google Scholar]
- 83.Iacobucci Ilaria, Wen Ji, Meggendorfer Manja, Carmichael Catherine, Choi John K, Masih Katherine, et al. The Genomic Landscape of Childhood and Adult Acute Erythroid Leukemia, ASH 2016 (abstract #39) ( https://ash.confex.com/ash/2016/webprogram/Paper95650.html)
- 84.Micci F, Thorsen J, Panagopoulos I, Nyquist KB, Zeller B, Tierens A, et al. High-throughput sequencing identifies an NFIA/CBFA2T3 fusion gene in acute erythroid leukemia with t(1;16)(p31;q24) Leukemia. 2013;27(4):980–2. doi: 10.1038/leu.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Panagopoulos I, Micci F, Thorsen J, Haugom L, Buechner J, Kerndrup G, et al. Fusion of ZMYND8 and RELA genes in acute erythroid leukemia. PLoS One. 2013;8(5):e63663. doi: 10.1371/journal.pone.0063663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.DeAngelo DJ, Stein EM, Ravandi F. Evolving Therapies in Acute Myeloid Leukemia: Progress at Last? Am Soc Clin Oncol Educ Book. 2016;35:e302–12. doi: 10.1200/EDBK_161258. [DOI] [PubMed] [Google Scholar]
- 87.Vicente C, Conchillo A, Garcia-Sanchez MA, Odero MD. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol. 2012;82(1):1–17. doi: 10.1016/j.critrevonc.2011.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


