Abstract
Although nonpathogenic bacterial endosymbionts have been shown to contribute to their arthropod host’s fitness by supplying them with essential vitamins and amino acids, little is known about the nutritional basis for the symbiotic relationship of endosymbionts in ticks. Our lab has previously reported that Rickettsia species phylotype G021 in Ixodes pacificus carries all five genes for de novo folate synthesis, and that these genes are monophyletic with homologs from other Rickettsia species. In this study, the rickettsial folate synthesis folA gene, coding for dihydrofolate reductase, was PCR amplified, cloned into an expression vector, and overexpressed in E. coli. Bioinformatic analysis identified that the FolA protein of phylotype G021 has the conserved DHFR domain, NADP binding sites, and substrate binding sites of bacterial dihydrofolate reductase. SDS-PAGE results showed that recombinant rickettsial FolA protein was overexpressed in BL21(DE3) E. coli in its soluble form. Affinity chromatography was used to purify the protein, and in vitro enzyme assays were performed to assess the biochemical activity of dihydrofolate reductase. The specific activity of recombinant FolA from phylotype G021 was determined to be 16.1 U/mg. This study has revealed that Rickettsia species phylotype G021 of I. pacificus is capable of producing a functional enzyme of the folate biosynthesis pathway, addressing the nutritional interactions behind the symbiosis between Rickettsia species phylotype G021 and its host.
Keywords: Rickettsia species phylotype G021, Ixodes pacificus, dihydrofolate reductase, recombinant protein, in vitro enzymatic assay
Introduction
Among invertebrate animals, arthropods are especially prone to establishing symbiotic relationships with intracellular bacteria (Rio, et al., 2016; Zug and Hammerstein, 2015). These relationships contribute to the ecological success of arthropods by providing protection from natural predators (Tsuchida et al., 2010) and parasite induced mortality (Brownlie and Johnson, 2009), producing detoxifying enzymes for degradation of insecticides (Indiragandhi et al., 2007), increasing heat tolerance (Moran and Yun, 2015) and reproductive success (Himler et al., 2011), and conferring direct fitness benefits under conditions of nutritional stress (Brownlie and Johnson, 2009). There are many examples of nutrient provisioning by intracellular bacteria to arthropods with low nutrient diets (Baumann, 2005; Dale and Moran, 2006; Moran, 2006). In fact, the reduced genomes of arthropod symbionts have retained only the genes required for maintaining a symbiotic lifestyle: those involved in host fitness and indispensable molecular processes, such as DNA and protein synthesis (Dale and Moran, 2006; McCutcheon and Moran, 2011; Moran, 2006).
The western black-legged tick, Ixodes pacificus, is a vector of Borrelia burgdorferi and Anaplasma phagocytophilum, the causative agents of Lyme borreliosis and anaplasmosis, respectively, in the western United States and western Canada (Lane et al., 1994; Reubel et al., 1998; Richter et al., 1996). I. pacificus is an ectoparasite and relies on a strict host blood diet that contains very low concentrations of nutrients and essential vitamins (Rio et al., 2016). I. pacificus feeds on vertebrate hosts three times in their life, each occurring before moving on to the next stage of the life cycle (Padgett and Lane, 2001). As revealed by the I. scapularis (a close relative of I. pacificus) genome project (NCBI Bioproject Accession number PRJNA34667), some of the genes required for synthesizing vitamins such as folate (vitamin B9) are not possessed by ixodid ticks. Remarkably, juveniles and adults of I. pacificus are capable of surviving for up to a year between blood meals (Padgett and Lane, 2001).
Physiological changes occur in response to a blood meal including engorgement and molting. During the early phase of the blood meal, a period of intermolt growth is undertaken in which a new cuticle and visceral tissues are grown (Sonenshine, 1993). Based on this knowledge, research has been aimed at investigating the mechanisms by which the tick is capable of surviving on the limited nutrients available in blood, with such long periods between feedings, and growing new tissues before molting to the next life stage without an abundant vitamin source. A major obstacle in understanding tick biology is the paucity of information concerning the functions of endosymbiotic bacteria. For example, little light has been shed on the nutritive exchanges choreographing the symbiotic architecture of endosymbionts in ticks.
A novel species of endosymbiotic Rickettsia classified as Rickettsia species phylotype G021 was discovered in I. pacificus ticks by PCR and sequencing. Phylotype G021 is grouped with the Spotted fever group rickettsiae with close relation to R. buchneri, R. akari, and R. australis (Phan et al., 2011). Our lab reported that each I. pacificus tick carries phylotype G021 (Cheng et al., 2013b), and that the bacterium is passed through inheritance and maintained through all four stages of tick development (Cheng et al., 2013a). Inferences have been made regarding the virulence of Rickettsia species phylotype G021 based on the ubiquitous prevalence and the 100% efficiency of transovarial transmission and transstadial passage of this species in I. pacificus compared to its fellow Rickettsia species phylotype G022 in I. pacificus (Cheng et al., 2013a; Cheng et al., 2013b). The authors suppose that phylotype G021 is nonpathogenic, but further characterization is required. Thus far, no data has been shown that would suggest that Rickettsia species phylotype G021 induces pathogenic effects in vertebrates. Furthermore, the closely related R. buchneri in I. scapularis, and R. peacockii show no indications of pathogenicity in vertebrates (Felsheim et al., 2009; Kurtti et al., 2015; Weller et al., 1998).
Although the function of the Rickettsia species phylotype G021 in I. pacificus is unknown, recent metabolic reconstructions carried out in our lab showed that all five genes of the folate (vitamin B9) biosynthetic pathway exist in the genome of Rickettsia species phylotype G021, including folA, folC, folE, folkP, and ptpS (Hunter et al., 2015). These are the genes necessary to complete folate synthesis (Hanson and Gregory, 2002; Pribat et al., 2009). These data, along with the nutrient poor diet, nature of life cycle, and lack of folate biosynthesis capabilities of I. pacificus, are convincing evidence of nutrient provisioning by phylotype G021.
In this report, we used rickettsial FolA as an indicator into the nature of the symbiosis between phylotype G021 and I. pacificus ticks, specifically the rickettsial folate biosynthesis in I. pacificus. The ability of Rickettsia species phylotype G021 to synthesize the essential vitamin B9 has been determined through bioinformatic gene annotation followed by isolation and biochemical characterization of the recombinant FolA protein. If it can be demonstrated that Rickettsia species phylotype G021 does indeed generate functional folate biosynthesis enzymes, further research may be aimed at in vivo evaluation of the specific benefits received by the I. pacificus ticks harboring them.
Materials and Methods
Materials
N-[Tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid (TES) buffer was purchased from Acros Organics (Geel, Belgium). Dihydrofolic acid (DHF) and nicotinamide adenine dinucleotide phosphate (NADPH) were purchased from Sigma Aldrich (St. Louis, MO). BSA was purchased from Promega (Madison, WI).
Tick collection and DNA Extraction
Flat ticks collected from Humboldt county California (GPS coordinate: N 40 55.200 W 123 50.400) in March 2012 were identified by light microscopy using key features of the Ixodes pacificus species according to the standard morphological key (Furman and Loomis, 1984). The ticks were ground with a pestle (Fisher Scientific, Tampa, FL) in liquid nitrogen and boiled in sealed Eppendorf tubes with a heating block at 100°C for 15 minutes in 100 μl of 0.7 M ammonium hydroxide, followed by heating at 100°C for 10 minutes to remove the ammonia (Guy and Stanek, 1991). The extracted DNA was quantified using a Nanodrop ND-1000 spectrophotometer at OD260 (Thermo Fisher Scientific, Waltham, MA) and stored at −20°C.
Amplification of the rickettsial folA gene by Polymerase Chain Reaction (PCR)
The complete coding sequence for the folA gene of phylotype G021 was amplified using PCR. Primers were designed using sequence data of the folA gene previously obtained in our lab (Hunter et al., 2015). The open reading frame (ORF) was amplified using the following primers: forward primer 5’-GAATGACAAATGTCAAATGAAAAATAGAAAAATCATCGGTATAATGG -3’; reverse primer 5’-GATTGACACGAGTCTTACCTCCTTTTAGTAAATTTATAAATCTGATAATTA -3’ (Elim Biopharmaceuticals, Hayward, CA), both containing the PshAI restriction enzyme recognition site (shown underlined in bold). The amplification reaction contained 50 ng genomic DNA from I. pacificus, 1X EconoTaq Plus Green Master Mix (0.1 units/μl EconoTaq DNA polymerase, 50 mM Tris-HCL, pH 9.0, 50 mM NaCl, 0.1 mg/ml BSA, 200 μM dNTPs, 1.5 mM MgCl2) (Lucigen, Middleton, WI), and 1 μM forward and reverse primers in 25 μl of total volume. The PCR cycling conditions consisted of a single cycle at 94°C for 5 minutes, followed by 40 cycles of 94°C for 30 seconds, primer annealing at 50.3°C for 1 minute and extension at 72°C for 1 minute 20 seconds. The PCR included a final extension cycle at 72°C for 7 minutes. The amplified product was subjected to electrophoresis in 1X TAE buffer (40 mM Tris, pH 8.6, 20 mM acetate, 1 mM EDTA) using a 1% agarose gel and visualized by ethidium bromide (0.5 μg/ml) staining. The gel image was documented using an AlphaImager HP (Alpha Innotech, San Leandro, CA).
Construction of expression plasmid and DNA sequencing
The pET-41a(+) expression vector (EMD Millipore, Billerica, MA) was used for cloning and subsequent protein expression. This plasmid expresses recombinant proteins under control of a T7 lac promoter for high levels of expression, attaches an N-terminal GST fusion tag for affinity purification, and has a kanamycin resistance gene for transformant selection.
The amplified rickettsial folA gene PCR product was gel purified and ligated into the linearized, dephosphorylated pET-41a(+) expression vector using T4 DNA ligase (New England Biolabs, Ipswich, MA). The ligation reaction was transformed into NovaBlue competent cells (EMD Millipore, Billerica, MA) according to the manufacturer’s protocol. Transformed cells were plated on LB–kanamycin (50 mg/L) and incubated overnight at 37°C. Individual colonies were inoculated in LB broth plus 50 mg/L kanamycin. Plasmid DNA was then purified using the Wizard Plus SV Minipreps DNA Purification System (Promega, Madison, WI), according to the manufacturer’s centrifugation protocol. The purified clone DNA was quantified by Nanodrop at OD260 and stored at −20°C.
folA clone DNA was sequenced at Elim Biopharmaceuticals (Elim, Hayward, CA) with vector specific forward and reverse primers (Forward primer: 5’-AAGAAACCGCTGCTGCTAAA - 3’; Reverse primer: 5’-AAGCTTGTCGACGGAGCT-3’), whose sequences are located upstream and downstream of the target gene, respectively. Sequences were uploaded into CodonCode Aligner (CodonCode Corporation, Centerville, MA) and the sequences with the highest quality were chosen for analysis. The GenBank accession number of the folA gene of phylotype G021 is KT225568.
To serve as a control for the subsequent in vitro enzymatic assay, the gltA gene of phylotype G021 was also PCR amplified and cloned in the pET-41a(+) expression vector. The construct was confirmed by DNA sequencing at Elim Biopharmaceuticals (Elim, Hayward, CA). The GenBank accession number of the gltA gene of phylotype G021 is ALS88439.
Bioinformatics
The correct ORF of the folA gene was identified by NCBI’s ORF Finder. Because the folA gene of phylotype G021 is located on an operon which begins with the adjacent folKP gene, the GenBank submissions previously generated in our lab for these genes were used to annotate key features of the genes (Hunter et al., 2015). A putative promoter for the folKP gene was predicted using Softberry’s BPROM (www.softberry.com) selecting oligonucleotides from the 5’ untranslated region (UTR) of the folKP gene. Prediction of ribosome binding sites was performed by “RBS Calculator” on https://salislab.net/software/. Expasy’s Translate Tool was used to covert the nucleotide sequence to its corresponding amino acids. The protein domains were identified using PROSITE (http://prosite.expasy.org/) and NCBI CD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Induction and expression of recombinant GST-FolA fusion protein
The folA clone DNA was transformed into lon(-), ompT(-) BL21(DE3) strain E. coli competent cells (EMD Millipore, Billerica, MA) for protein expression according to the manufacturer’s protocol. A 200 ml starter culture was added to 800 ml of LB broth supplemented with 100 mg/L kanamycin and 0.05% glucose. The culture was grown at 37°C while shaking at 230 rpm until an optical density of 0.5 at 590 nm was reached. Isopropyl-beta-D-thiogalactopyranoside (IPTG) (Gold Biotechnology, St. Louis, MO) was then added to a final concentration of 0.4 mM, and the culture was incubated at 30°C overnight on a shaker. To maintain strong selection for the clone within the culture, administration of excess kanamycin was delivered as follows: 1 volume immediately following IPTG addition, and 1 volume at about 8 hours after induction. The cells were harvested by centrifugation for 5 min at 10,000 X g, resuspended in phosphate buffered saline with 1 mM phenylmethylsulfonyl fluoride (PMSF) (Thermo Scientific, Waltham, MA), and chilled on ice. Cell resuspensions were lysed by sonication. Lysates were transferred to Eppendorf tubes and centrifuged at 16,100 X g for 30 minutes at 4°C. Supernatants were transferred to sterile conical tubes (Thermo Scientific, Waltham, MA) and purified immediately.
SDS-PAGE and Western blot analysis
Total protein expression for the recombinant GST-FolA fusion protein was assessed by SDS-PAGE followed by Coomassie blue staining. 2% of the total of each protein sample, in SDS sample buffer (0.75 M Tris-HCl, pH 6.8, 40% glycerol, 0.002% Bromophenol blue, 8% SDS) with 5% β-mercaptoethanol (β-ME) (Fisher Scientific, Tampa, FL), was loaded onto a 12% Criterion TGX precast polyacrylamide protein electrophoresis gel (Bio-Rad, Hercules, CA) and electrophoresed in SDS running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.3) for 55 min at 200 V with a current of 55 mA. The gel was stained with Coomassie blue and visualized over transilluminating white light with an AlphaImager HP (Alpha Innotech, San Leandro, CA).
Verification of protein identity was achieved by Western blotting. In performing this, the above procedure for SDS-PAGE was followed, but immediately following electrophoresis, the proteins were transferred to a PVDF (polyvinyl difluoride) membrane (Bio-Rad, Hercules, CA), probed for 2 hours at room temperature with a 1:500 dilution of mouse monoclonal anti-GST antibody (Thermo Scientific, Waltham, MA), followed by incubation with Immun-Star Goat Anti-Mouse-Horse Radish Peroxidase (HRP) conjugated secondary antibody (Bio-Rad, Hercules, CA) at a 1:20,000 dilution for 1 hour at room temperature. Detection was performed using the chemiluminescence detection reagents Clarity Western ECL Substrates (Bio-Rad, Hercules, CA). The image was documented using the C-Digit digital imager (Li-Cor, Lincoln, NE).
Solubility Analysis
For the protein solubility analysis, induction conditions for recombinant FolA expression were held as described above. Cells were harvested by centrifugation, resuspended in PBS, pH 7.2, and lysed by sonication. The lysate fractions were separated by centrifugation at 16,100 X g for 30 min at 4°C. The soluble fraction was transferred to a new tube. 5% of the total protein samples (soluble fraction and pellet) were loaded into a 12% Criterion TGX precast polyacrylamide protein electrophoresis gel (Bio-Rad, Hercules, CA) and electrophoresed by SDS-PAGE as described above. The gel was stained with Coomassie blue and visualized over transilluminating white light using an AlphaImager HP (Alpha Innotech, San Leandro, CA).
Purification of the recombinant rickettsial GST-FolA fusion protein
Soluble GST-FolA was purified by affinity column chromatography using the Amicon Pro Affinity Concentration Kit-GST (EMD Millipore, Billerica, MA) according to the manufacturer’s instructions. Briefly, the entire FolA-containing lysate supernatant was filtered with a 0.45 μm nitrocellulose filter (Corning, Corning, NY) and incubated with GST Bind Resin (EMD Millipore, Billerica, MA) on a column at room temperature for 2 hours with gentle agitation. The sample was centrifuged at 1,000 X g for 5 min to allow the sample to flow through the column. The resin was washed with 15 column volumes GST Bind/Wash Buffer (EMD Millipore, Billerica, MA) by centrifugation at 1,000 X g for 2 min. Finally, GST-FolA protein was eluted with 10 column volumes 1X elution buffer (50 mM Tris, pH 8.0, 10.2 mM reduced glutathione) (EMD Millipore, Billerica, MA) supplemented with 1 mM Triton X-100 (Thermo Fisher Scientific, Waltham, MA) by centrifugation at 4,000 X g for 20 min in a swinging bucket rotor. The concentration of eluted protein was quantified by Nanodrop at OD280 and stored at −80°C in 20% glycerol.
In vitro enzyme assay
Dihydrofolate reductase (DHFR) activity for purified recombinant rickettsial GST-FolA fusion protein was monitored for 10 min at 25°C by the decrease in absorbance at 340 nm associated with NADP+ formation (Hillcoat et al., 1967). The reaction consisted of 50 mM TES buffer, pH 7.0, 1 mg/ml BSA, 75 mM β-ME, 0.1 mM DHF, 0.1 mM NADPH, and varying concentrations of the GST-FolA protein in a 96 well Costar clear UV microplate (Corning, Corning, NY) in a final volume of 200 μl. One unit of enzyme is defined as the amount that reduces 1 μmol of dihydrofolate per minute using a molar extinction coefficient of 12,300 M−1 (Hillcoat et al., 1967). The absorbance change was observed using a SpectraMax i3x (Molecular Devices, Sunnyvale, CA) plate reader. Affinity column-purified GST-GltA fusion protein and the reaction buffer of the assay served as two negative controls. The DHFR activity was measured three times for both purified GST-FolA or GST-GltA.
Results and Discussion
Our previous study reported that Rickettsia species phylotype G021 has the genetic capacity for de novo folate synthesis (Hunter et al., 2015). In this study, we have demonstrated the vitamin B9 synthesizing dihydrofolate reductase function of the FolA protein of phylotype G021 through performed in silico assessment of the protein structure and overexpression of the recombinant protein as well as characterization through in vitro enzyme assay.
To overexpress recombinant FolA protein in E. coli, full-length folA gene (642 base pairs, bp) of Rickettsia species phylotype G021 was amplified by PCR and cloned into the pET-41a(+) plasmid. Clones verified by PshA1 digestion were sequenced. Based on the sequencing results, the correct orientation of the folA gene in the vector was verified and the nucleotide sequence of the folA gene of phylotype G021 was confirmed (Hunter et al., 2015).
folA is part of an operon downstream of folKP (Hunter et al., 2015). Therefore, the nucleotide sequences including the 5’ and 3’ untranslated regions (UTRs) of rickettsial folA and folKP were used to identify features for regulatory elements of the folA gene from Rickettsia species phylotype G021. The ORF and putative E. coli sigma-70-like promoter region for the operon were identified by bioinformatics approaches. The G + C content of the folA gene of phylotype G021 amounts to 30.22%. The putative ribosome binding site for the folate genes were AGCAGA and AGGAGA, for folA and folKP, respectively. The putative -35 and -10 regions of the E. coli sigma-70-like promoter, which is upstream of the folKP gene, are TTCAGG and TTTCTTTT, respectively, with 19 bp between the two regions. These sequences are positioned correctly when compared with the consensus E. coli -35 (TTGACA) and -10 (TATAAT) regions of the sigma-70-like promoters. These sequences also show homology to previously characterized rickettsial promoters (Cai et al., 1995; Cai and Winkler, 1993; Shaw et al., 1997). The ribosome binding site (or Shine-Delgarno sequence, SD) of the folA gene has two mismatches to the consensus sequence. Previous studies reported that 5'UTR of prokaryotic genes are highly diverse (Chang et al., 2006; Sakai et al., 2001). This is actually a common phenomenon, as many non-SD-led genes and leadless genes have been detected in the genomes of Archaea and Bacteria (Slupska et al., 2001). In fact, less than 20% of genes from some microbial genomes were found to be SD-led (Chang et al., 2006). It seems that the base-pairing between mRNA and 16S rRNA of the folA gene of phylotype G021 takes place in regions other than sites of SD – anti-SD base paring, indicating that Rickettsia species phylotype G021, similar to other prokaryotic species, has great elasticity and variation to translation initiation (Jenner et al., 2005; Wu and Janssen, 1996).
We compared the FolA amino acid sequences of phylotype G021 with orthologs from other microbial organisms. By PROSITE and NCBI CD-Search, a dihydrofolate reductase (DHFR) domain was identified in the central region of the FolA protein from Rickettsia species phylotype G021. The DHFR domain is responsible for reducing dihydrofolate to tetrahydrofolate with NADPH as a cofactor. The NCBI CD-Search also identified ten NADPH and substrate binding sites of the FolA protein of phylotype G021, including alanine at residue 11, isoleucine at residue 18, arginine-lysine-tyrosine at residues 48–50, arginine at residue 69, and glycine-glycine-threonine-glutamic acid at residues 153–156. Alignments of the predicted FolA protein of phylotype G021 with DHFR proteins of several bacterial species showed that the ten NADPH and substrate binding sites are conserved among bacterial dihydrofolate reductases (Fig. 1). Among the ten NADPH and substrate binding sties, the glycine-glycine dipeptide at residues 153–154 is conserved among all known DHFR (Bolin et al., 1982; Roos, 1993). The glycine-glycine residues are connected with a cis-peptide bond in all DHFR proteins (Blakley, 1984). In addition, seven folate binding sites are also identified in the FolA protein of phylotype G021 by the NCBI CD-Search, including isoleucine at residue 9, tryptophan at residue 26, glutamic acid at residue 31, cysteine at residue 62, valine at residue 152, alanine at residue 158, and threonine at residue 173. Alignments of the predicted FolA protein of phylotype G021 with DHFR proteins of several bacterial species showed that the seven folate binding sites are conserved among bacterial dihydrofolate reductase (Fig. 1). Among the seven folate binding sties, tryptophan at residue 26 of the FolA protein of phylotype G021 belongs to the proline-tryptophan dipeptide that is conserved in all dihydrofolate reductases (Bolin et al., 1982).
Figure 1.
Amino acid sequence alignment for dihydrofolate reductase (DHFR) from Rickettsia species phylotype G021 and other bacteria as well as phage species, including Campylobacter jejuni (GenBank accession number CAC19929), Chlamydophila caviae GPIC (AAP05737), Ruminiclostridium thermocellum ATCC 27405 (ABN52458), Enterococcus faecalis (WP_049097722), Escherichia coli (WP_063844334), Lactobacillus johnsonii NCC 533 (WP_011162224), Lactococcus lactis subsp. Lactis (EHE92570), Mycoplasma penetrans (WP_011077508), Pseudomonas phage phiKZ (NP_803570), Rickettsia conorii str Malish 7 (WP_010976722), Salmonella enterica subsp. Enterica (WP_032490189), Salmonella phage FelixO1 (NP_944929), and Streptococcus agalactiae (ODG94437). The first 205 (of 213) amino acids of Rickettsia species phylotype G021 FolA protein, and the corresponding amino acids from the other species listed, are shown. Residues for NADPH and substrate binding sites in DHFR enzymes are labeled with asterisks. The folate binding sites are noted with hashtags.
The amino acid sequence alignment indicates that the DHFR domain is widely distributed among bacteria. Surprisingly, both Salmonella phage Felix01 and Pseudomonas phage phiKZ, in addition to bacterial species, possess the conserved DHFR domain of dihydrofolate reductase. This result indicates that the folA gene of phylotype G021 could be obtained by phage-mediated horizontal transfer events (Fig. 1). Being an obligate endosymbiont undergoing reductive genomic evolution, phylotype G021 would tend to lose identifiable functional domains over time for proteins that are not necessary for survival or maintenance of its endosymbiosis. The fact that the FolA protein of phylotype G021 contains the conserved DHFR domain and NADPH and substrate binding sites indicates a strong selective pressure for maintaining the function of this protein. Also, the loss of this gene altogether in several closely related bacterial species indicates that there may be a general trend toward losing this gene. Taken together, the obligate endosymbiotic nature of phylotype G021, the lack of folates in mammalian blood, and the maintained DHFR domain and NADPH and substrate binding sites of the FolA protein of phylotype G021, provide preliminary evidence to support our hypothesis that this symbiosis between phylotype G021 and I. pacificus is sustained through indispensable nutritional interactions.
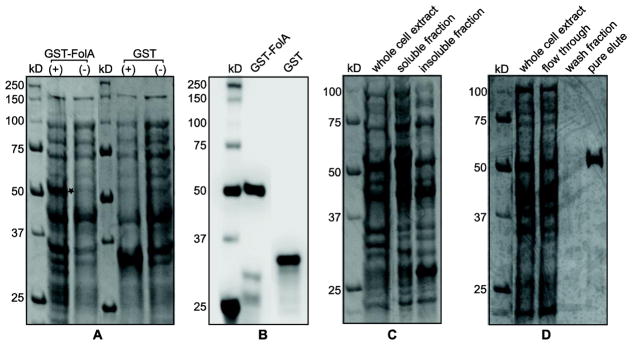
The recombinant GST-FolA fusion protein of Rickettsia species phylotype G021 was expressed using BL21(DE3) E. coli in LB broth. Whole-cell protein expression was determined by SDS-PAGE and Coomassie blue staining. Early log-phase addition of 0.4 mM IPTG with overnight induction at 30°C proved to be effective conditions for producing recombinant GST-FolA protein. Also, three administrations of kanamycin provided sufficient selection during overexpression. SDS-PAGE analysis of the expression of GST-FolA showed that the recombinant protein was observed in the correct theoretical locations on the gel: 56 kilodaltons (kDa) for the GST-FolA fusion protein compared to 35 kDa for the GST protein alone. The uninduced control lanes gave no corresponding bands (Fig. 2a). Using the Western blot assay, the specific recombinant GST and GST-FolA fusion proteins were identified. The sizes of the GST proteins were also confirmed. No bands corresponding to the recombinant proteins were observed in any of the uninduced negative controls (Fig. 2b).
Figure 2.
Overexpression, soluble expression, and purification of recombinant GST-FolA fusion protein of Rickettsia species phylotype G021 in E. coli (DE3). 2A) Overexpression of recombinant GST-FolA fusion protein of Rickettsia species phylotype G021 in E. coli (DE3). The expression of GST proteins (GST-FolA or GST) was induced by adding 0.4 mM IPTG into E. coli cultures, which were incubated at 30°C overnight. Coomassie blue staining of total protein expression, resolved on a 12% polyacrylamide gel. The location of the overexpressed GST-FolA is labeled as a star. (+) = induced by 0.4 mM IPTG at 30°C; (−) = uninduced negative control; kDa = kilodaltons. 2B) Western blot of GST-FolA fusion protein using mouse monoclonal anti-GST antibody, visualized on a PVDF membrane with chemiluminescent detection. 2C) Soluble expression of recombinant GST-FolA of Rickettsia species phylotype G021. Recombinant GST-FolA is expressed at its correct theoretical location (53 kDa). After the induction period, cells were harvested by centrifugation and lysed by sonication. The lysate fractions were separated by centrifugation. The soluble fraction and pellet were subjected to SDS-PAGE on a 12% polyacrylamide gel, stained with Coomassie blue, and visualized using transilluminating white light. 2D) The purity analysis of the GST-FolA protein by SDS-PAGE. GST-FolA fusion protein was purified using on-column affinity chromatography. Whole cell extract after induction, flow through, wash fraction, and pure eluate were analyzed for purity by SDS-PAGE followed by Coomassie blue staining and transilluminating white light.
Protein solubility during expression of the GST-FolA fusion protein of Rickettsia species phylotype G021 was determined following induction with IPTG. E. coli cells were first lysed by sonication, insoluble and soluble fractions of E. coli cells were separated by centrifugation and analyzed for the presence of the overexpressed GST-FolA protein by SDS-PAGE. Using the above parameters of overexpression, the majority (~80%) of the GST-FolA protein was present in the soluble fraction, though some GST-FolA protein was detected in the insoluble pellet (Fig. 2c). For overproduction and purification of the recombinant FolA protein, 200 ml culture volumes, grown overnight in 250 ml flasks while shaking at 165 rpm, were harvested by centrifugation, lysed, and fractionated. We can only postulate that the atypically high culture-to-flask volume ratio leading to the best FolA expression involves decreased toxicity of the protein under the conditions used in the E. coli expression system. Also, the possibility exists that more anaerobic conditions are necessary to produce this protein using this system. Soluble recombinant FolA was purified by affinity chromatography. SDS-PAGE results showed one predominant band with a molecular mass of 56 kDa in the pure eluate (Fig. 2d). It was estimated that the affinity chromatography yield about 2.5 mg of over 90% pure GST-FolA per liter of culture.
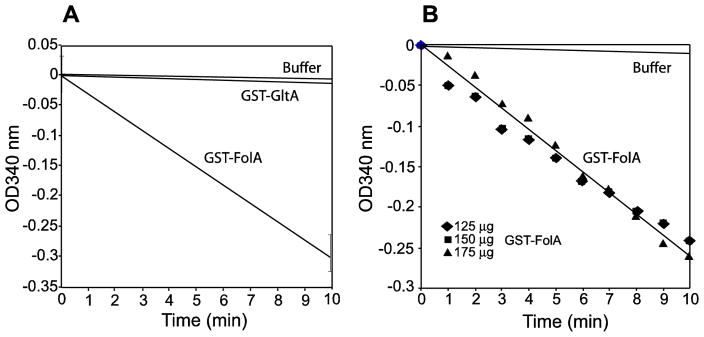
To verify the functionality of the Rickettsia species phylotype G021, the GST-FolA protein was assayed for enzyme activity. Purified GST-FolA was assayed at 25ºC as previously reported (Hillcoat et al., 1967; Sirawaraporn et al., 1993; Zywno-Van Ginkel et al., 1997). The FolA enzyme assay used for detecting its dihydrofolate reductase activity is based on the conversion of dihydrofolate to tetrahydrofolate, resulting in the oxidation of NADPH to NADP+, which was can be monitored on a kinetic plate reader by measurement of the absorbance at 340 nm. At 25ºC, a linear decrease in absorption over a 10 minute time period was observed (Fig. 3a). Using a molar extinction coefficient of 12,300 M−1 cm−1, the conversion of DHF and NADPH to THF and NADP+ was determined. The specific activity of the GST-FolA enzyme was found to be 16.1 U/mg, with a unit being defined as the amount of enzyme that reduces 1 μmol of DHF/min (Fig. 3b). The specific activity of 16.1 U/mg for the FolA protein of phylotype G021 is within the same range of activity as other folate producing bacteria such as Mycobacterium avium (1.2 U/mg), and Bacillus cereus (11.1 U/mg) (Joska and Anderson, 2006; Zywno-Van Ginkel et al., 1997). To rule out the possibility that the pure elute contained a small amount of dihydrofolate reductase from E. coli, we expressed recombinant GST-GltA fusion protein of phylotype G021 in E. coli and purified the GST-GltA protein under the same conditions as the GST-FolA fusion protein. We found that there was relatively no detectable decrease in absorbance at 340 nm for both negative controls (150 μg GltA, and no enzyme) in the in vitro enzyme assay (Fig. 3a).
Figure 3.
Enzyme activity assays of GST-FolA of Rickettsia species phylotype G021. The dihydrofolate reductase activity of the GST-FolA protein was monitored at 25ºC for 10 min. The absorbance change at 340 nm was observed using a SpectraMax i3x plate reader. 3A) The graph represents the FolA specific activity assay. 150 μg GST-FolA or GST-GltA protein was used in the enzymatic assay. GST-GltA and buffer only serve as two controls for the assay. 3B) This graph represents the determination of the optimum enzyme concentration for the specific activity assay. 125 μg, 150 μg, and 175 μg GST-FolA were used in the enzymatic assay.
This study continues an earlier project (Hunter et al., 2015) to determine the role of Rickettsia species phylotype G021 as a potential mutualist in the Lyme borreliosis vector, I. pacificus, evaluating the putative nutritional benefit received by ticks harboring phylotype G021. It is also noteworthy that the bacterial microbiome of I. pacificus (Swei and Kwan, 2017), other than the genus of Rickettsia, also have the capacity of producing folate and other nutrients. Surprisingly, not all vitamins are indispensable for the growth of bacteria in ticks. B. burgdorferi does not have the genes for de novo biosynthesis of thiamin, which is a cofactor required for the oxydative decarboxylation of pyruvate. However, the growth of B. burgdorferi is not affected by thiamin deprivation in vitro and in vivo (Zhang et al., 2016), indicating the existence of evolutionary constraints in the adaptive evolution dynamics between bacteria and tick hosts (Howick and Lazzaro, 2014.). The genetic capacity of bacteria is known to differ among bacterial species in ticks; such differences may play important roles in the evolution of the tick hosts (Zhang et al., 2016; Hunter et al., 2015).
The described work uses molecular biology and biochemistry to narrow the gap of knowledge in interactions involving bacteria-tick relationships, particularly folate production by phylotype G021. With the genetic capacity of de novo biosynthesis of folate (Hunter et al., 2015), the production of a functional dihydrofolate reductase enzyme, the predominant infection in the midgut and ovary tissues of I. pacificus (Phan et al., 2011; Bagheri et al., 2017), and the increased bacterial burden in I. pacificus following a blood meal (Cheng, et al., 2013a), Rickettsia species phylotype G021 may significantly contribute to its host’s physiological homeostasis.
Conclusions
This research has helped increase our understanding of tick physiology, the biosynthesis of folate from Rickettsia species phylotype G021 in ticks, and the nutritional interactions of the symbiosis between phylotype G021 and I. pacificus. Our description of a functional folate biosynthesis protein from Rickettsia species phylotype G021 allows us to make inferences about the role of phylotype G021 in I. pacificus ticks. Although our ability to detect enzyme activity in a recombinant rickettsial folate biosynthesis protein awards phylotype G021 the capacity for folate biosynthesis, we need to bear in mind the in vitro context of the tests reported here. Future studies will be needed to further characterize this endosymbiont. Indeed, in vitro and in vivo analyses, such as infecting pure tick-cell culture lines with the isolate of Rickettsia species phylotype G021 and monitoring developmental and transcriptional effects, and analyzing folate levels between flat and engorged ticks, should prove quite rewarding in our understanding of this symbiosis.
Acknowledgments
The authors thank all of the staff from the departments of Chemistry and Biological Sciences at Humboldt State University who provided critical advice for optimization of protein purification, in vitro enzyme assays, proper usage of equipment, and troubleshooting of experiments in general. Dr. Brian Kyte and Dr. Jenny Cappuccio from the Department of Chemistry at Humboldt State University provided critical advice for optimization of protein purification. Numerous undergraduate researchers and Dr. David Baston from the CNRS Core Research Facility at Humboldt State University also provided great assistance. All of the funding for this project came from the National Institute of Health, grant number 1 R15 AI099902-01.
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest in this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bagheri G, Lehner JD, Zhong J. Enhanced detection of Rickettsia species in Ixodes pacificus using highly sensitive fluorescence in situ hybridization coupled with Tyramide Signal Amplification. Ticks Tick Borne Dis. 2017;8:915–921. doi: 10.1016/j.ttbdis.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. Biology bacteriocyte-associated endosymbionts of plant sapsucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- Blakley RL. Dihydrofolate reductase. In: Blakley RL, Benkovic SJ, editors. Folates and pteridines. Wiley; New York: 1984. pp. 191–253. [Google Scholar]
- Bolin JT, Filman DJ, Matthews DA, Hamlin RC, Kraut J. Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I General features and binding of methotrexate. J Biol Chem. 1982;257:13650–13662. [PubMed] [Google Scholar]
- Brownlie JC, Johnson KN. Symbiont-mediated protection in insect hosts. Trends Microbiol. 2009;17:348–354. doi: 10.1016/j.tim.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Cai J, Pang H, Wood DO, Winkler HH. The citrate synthase-encoding gene of Rickettsia prowazekii is controlled by two promoters. Gene. 1995;163:115–119. doi: 10.1016/0378-1119(95)00365-d. [DOI] [PubMed] [Google Scholar]
- Cai J, Winkler HH. Identification of tlc and gltA mRNAs and determination of in situ RNA half-life in Rickettsia prowazekii. J Bacteriol. 1993;175:5725–5727. doi: 10.1128/jb.175.17.5725-5727.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Halgamuge S, Tang SL. Analysis of SD sequences in completed microbial genomes: Non-SD-led genes are as common as SD-led genes. Gene. 2006;373:90–99. doi: 10.1016/j.gene.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Cheng D, Lane RS, Moore BD, Zhong J. Host blood meal-dependent growth ensures transovarial transmission and transstadial passage of Rickettsia sp phylotype G021 in the western black-legged tick (Ixodes pacificus) Ticks Tick Borne Dis. 2013a;4:421–426. doi: 10.1016/j.ttbdis.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Vigil K, Schanes P, Brown RN, Zhong J. Prevalence and burden of two rickettsial phylotypes (G021 and G022) in Ixodes pacificus from California by real-time quantitative PCR. Ticks Tick Borne Dis. 2013b;4:280–287. doi: 10.1016/j.ttbdis.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Felsheim RF, Kurtti TJ, Munderloh UG. Genomic sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. Plos One. 2009;4:e8361. doi: 10.1371/journal.pone.0008361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman DP, Loomis EC. The ticks of California (Acari:Ixodida) Berkeley: University of California Press; 1984. [Google Scholar]
- Guy EC, Stanek G. Detection of Borrelia burgdorferi in patients with Lyme disease by the polymerase chain reaction. J Clin Pathol. 1991;44:610–611. doi: 10.1136/jcp.44.7.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Gregory JF., 3rd Synthesis and turnover of folates in plants. Curr Opin Plant Biol. 2002;5:244–249. doi: 10.1016/s1369-5266(02)00249-2. [DOI] [PubMed] [Google Scholar]
- Hillcoat BL, Nixon PF, Blakley RL. Effect of substrate decomposition on the spectrophotometric assay of dihydrofolate reductase. Anal Biochem. 1967;21:178–189. doi: 10.1016/0003-2697(67)90179-0. [DOI] [PubMed] [Google Scholar]
- Himler AG, Adachi-Hagimori T, Bergen JE, Kozuch A, Kelly SE, Tabashnik BE, Chiel E, Duckworth VE, Dennehy TJ, Zchori-Fein E, Hunter MS. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science. 2011;332:254–256. doi: 10.1126/science.1199410. [DOI] [PubMed] [Google Scholar]
- Howick VM, Lazzaro BP. Genotype and diet shape resistance and tolerance across distinct phases of bacterial infection. BMC Evol Biol. 2014;14:56. doi: 10.1186/1471-2148-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Torkelson JL, Bodnar J, Mortazavi B, Laurent T, Deason J, Thephavongsa K, Zhong J. The Rickettsia endosymbiont of Ixodes pacificus contains all the genes of de novo folate biosynthesis. Plos One. 2015;10:e0144552. doi: 10.1371/journal.pone.0144552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiragandhi P, Anandham R, Madhaiyan M, Poonguzhali S, Kim GH, Saravanan VS, Sa T. Cultivable bacteria associated with larval gut of prothiofos-resistant, prothiofos-susceptible and field-caught populations of diamondback moth, Plutella xylostella and their potential for, antagonism towards entomopathogenic fungi and host insect nutrition. J Appl Microbiol. 2007;103:2664–2675. doi: 10.1111/j.1365-2672.2007.03506.x. [DOI] [PubMed] [Google Scholar]
- Jenner L, Romby P, Rees B, Schulze-Briese C, Springer M, Ehresmann C, Ehresmann B, Moras D, Yusupova G, Yusupov M. Translational operator of mRNA on the ribosome: How repressor proteins exclude ribosome binding. Science. 2005;308:120–123. doi: 10.1126/science.1105639. [DOI] [PubMed] [Google Scholar]
- Kurtti TJ, Felsheim RF, Burkhardt NY, Oliver JD, Heu CC, Munderloh UG. Rickettsia buchneri sp nov., a rickettsial endosymbiont of the blacklegged tick Ixodes scapularis. Int J Syst Evol Microbiol. 2015;65:965–970. doi: 10.1099/ijs.0.000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joska TM, Anderson AC. Structure-activity relationships of Bacillus cereus and Bacillus anthracis dihydrofolate reductase: toward the identification of new potent drug leads. Antimicrob Agents Chemother. 2006;50:3435–3443. doi: 10.1128/AAC.00386-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane RS, Brown RN, Piesman J, Peavey CA. Vector competence of Ixodes pacificus and Dermacentor occidentalis (Acari: Ixodidae) for various isolates of Lyme disease spirochetes. J Med Entomol. 1994;31:417–424. doi: 10.1093/jmedent/31.3.417. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- Moran NA. Symbiosis. Curr Biol. 2006;16:R866–871. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Moran NA, Yun Y. Experimental replacement of an obligate insect symbiont. Proc Natl Acad Sci USA. 2015;112:2093–2096. doi: 10.1073/pnas.1420037112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett KA, Lane RS. Life cycle of Ixodes pacificus (Acari: Ixodidae): timing of developmental processes under field and laboratory conditions. J Med Entomol. 2001;38:684–693. doi: 10.1603/0022-2585-38.5.684. [DOI] [PubMed] [Google Scholar]
- Phan JN, Lu CR, Bender WG, Smoak RM, Zhong J. Molecular Detection and identification of Rickettsia species in Ixodes pacificus in California. Vector Borne Zoonotic Dis. 2011;11:957–961. doi: 10.1089/vbz.2010.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribat A, Jeanguenin L, Lara-Nunez A, Ziemak MJ, Hyde JE, de Crecy-Lagard V, Hanson AD. 6-pyruvoyltetrahydropterin synthase paralogs replace the folate synthesis enzyme dihydroneopterin aldolase in diverse bacteria. J Bacteriol. 2009;191:4158–4165. doi: 10.1128/JB.00416-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubel GH, Kimsey RB, Barlough JE, Madigan JE. Experimental transmission of Ehrlichia equi to horses through naturally infected ticks (Ixodes pacificus) from northern California. J Clin Microbiol. 1998;36:2131–2134. doi: 10.1128/jcm.36.7.2131-2134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter PJ, Jr, Kimsey RB, Madigan JE, Barlough JE, Dumler JS, Brooks DL. Ixodes pacificus (Acari: Ixodidae) as a vector of Ehrlichia equi (Rickettsiales: Ehrlichieae) J Med Entomol. 1996;33:1–5. doi: 10.1093/jmedent/33.1.1. [DOI] [PubMed] [Google Scholar]
- Rio RV, Attardo GM, Weiss BL. Grandeur alliances: symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol. 2016;32:739–749. doi: 10.1016/j.pt.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos DS. Primary structure of the dihydrofolate reductase-thymidylate synthase gene from Toxoplasma gondii. J Biol Chem. 1993;268:6269–6280. [PubMed] [Google Scholar]
- Sakai H, Imamura C, Osada Y, Saito R, Washio T, Tomita M. Correlation between Shine-Dalgarno sequence conservation and codon usage of bacterial genes. J Mol Evol. 2001;52:164–170. doi: 10.1007/s002390010145. [DOI] [PubMed] [Google Scholar]
- Shaw EI, Marks GL, Winkler HH, Wood DO. Transcriptional characterization of the Rickettsia prowazekii major macromolecular synthesis operon. J Bacteriol. 1997;179:6448–6452. doi: 10.1128/jb.179.20.6448-6452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirawaraporn W, Prapunwattana P, Sirawaraporn R, Yuthavong Y, Santi DV. The dihydrofolate reductase domain of Plasmodium falciparum thymidylate synthase-dihydrofolate reductase. Gene synthesis, expression, and anti-folate-resistant mutants. J Biol Chem. 1993;268:21637–21644. [PubMed] [Google Scholar]
- Slupska MM, King AG, Fitz-Gibbon S, Besemer J, Borodovsky M, Miller JH. Leaderless transcripts of the crenarchaeal hyperthermophile Pyrobaculum aerophilum. J Mol Biol. 2001;309:347–360. doi: 10.1006/jmbi.2001.4669. [DOI] [PubMed] [Google Scholar]
- Sonenshine DE. Biology of ticks. 2. Oxford University Press; Oxford: 1993. [Google Scholar]
- Swei A, Kwan JY. Tick microbiome and pathogen acquisition altered by host blood meal. ISME J. 2017;11:813–816. doi: 10.1038/ismej.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Koga R, Horikawa M, Tsunoda T, Maoka T, Matsumoto S, Simon JC, Fukatsu T. Symbiotic bacterium modifies aphid body color. Science. 2010;330:1102–1104. doi: 10.1126/science.1195463. [DOI] [PubMed] [Google Scholar]
- Weller SJ, Baldridge GD, Munderloh UG, Noda H, Simser J, Kurtti TJ. Phylogenetic placement of rickettsiae from the ticks Amblyomma americanum, and Ixodes scapularis. J Clin Microbiol. 1998;36:1305–1317. doi: 10.1128/jcm.36.5.1305-1317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CJ, Janssen GR. Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of the 5' untranslated leader. Mol Microbiol. 1996;22:339–355. doi: 10.1046/j.1365-2958.1996.00119.x. [DOI] [PubMed] [Google Scholar]
- Zhang K, Bian J, Deng Y, Smith A, Nunez RE, Li MB, Pal U, Yu A, Qiu W, Ealick SE, Li C. Lyme disease spirochaete Borrelia burgdorferi does not require thiamin. Nat Microbiol. 2016;2:16213. doi: 10.1038/nmicrobiol.2016.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug R, Hammerstein P. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol Rev Camb Philos Soc. 2015;90:89–111. doi: 10.1111/brv.12098. [DOI] [PubMed] [Google Scholar]
- Zywno-Van Ginkel S, Dooley TP, Suling WJ, Barrow WW. Identification and cloning of the Mycobacterium avium folA gene, required for dihydrofolate reductase activity. FEMS Microbiol Lett. 1997;156:69–78. doi: 10.1111/j.1574-6968.1997.tb12707.x. [DOI] [PubMed] [Google Scholar]