Abstract
This work presents a suprathreshold fiber cluster (STFC) method that leverages the whole brain fiber geometry to enhance statistical group difference analyses. The proposed method consists of 1) a well-established study-specific data-driven tractography parcellation to obtain white matter tract parcels and 2) a newly proposed nonparametric, permutation-test-based STFC method to identify significant differences between study populations. The basic idea of our method is that a white matter parcel’s neighborhood (nearby parcels with similar white matter anatomy) can support the parcel’s statistical significance when correcting for multiple comparisons. We propose an adaptive parcel neighborhood strategy to allow suprathreshold fiber cluster formation that is robust to anatomically varying inter-parcel distances. The method is demonstrated by application to a multi-shell diffusion MRI dataset from 59 individuals, including 30 attention deficit hyperactivity disorder patients and 29 healthy controls. Evaluations are conducted using both synthetic and in-vivo data. The results indicate that the STFC method gives greater sensitivity in finding group differences in white matter tract parcels compared to several traditional multiple comparison correction methods.
1. Introduction
Diffusion magnetic resonance imaging (dMRI), which can probe the underlying microstructure of brain white matter (WM) in vivo, provides the only existing technique to study brain WM structures in a non-invasive way. With the growing popularity of dMRI, computational neuroimaging methods have been proposed to identify group WM differences for understanding brain neurological function in health and disease. Applications have included attention deficit hyperactivity disorder (ADHD) [93], autism [2], human aging [27], brain connectome modeling [47], and many more studies, as described in several reviews [97, 82, 3].
Traditional methods to study WM group differences rely on voxel-based image analysis techniques such as voxel-based morphometry (VBM) [4] and tract-based spatial statistics (TBSS) [87]. While these methods have been highly successful, several studies suggest that VBM and TBSS could be affected by highly-localized group differences [25, 46] and partial volume effects [7, 16]. An alternative can be using a region-wise WM atlas where regions are defined as WM tracts, which has been successfully applied in multiple studies [55, 45, 66, 26, 104, 90, 13]. However, using these voxel-based methods, accurate localization of between-population differences to specific WM tracts can be difficult to assess, especially since more than one tract may cross within a single voxel. Fixel-based analysis (FBA) that takes account of multiple fibers within a voxel has been proposed recently to enable enhanced whole-brain statistics [75, 76], showing the potential of incorporating information from multiple fiber populations into WM analyses.
In order to allow simultaneous analysis across multiple brain regions in neuroimaging, a hypothesis test (e.g. Student’s t-test) is normally used to identify local group differences, followed by multiple comparison correction (e.g. false discovery rate (FDR) [11] and Bonferroni [43] methods) for corrected statistical significance. Since these commonly used correction methods can be less sensitive in finding significance, voxel-based multiple comparison correction has been conducted in a cluster-thresholding manner that utilizes spatial neighborhoods to boost belief in extended voxel cluster areas [58]. Multiple studies have applied voxel-cluster-thresholding methods to find WM group differences [81, 72, 94, 75, 15].
An alternative approach for investigating differences in the brain’s WM uses tractography [9], which allows estimation of the trajectories of fiber tracts [102, 74] and thus measurement of microstructural WM properties of fiber pathways [40, 21]. A traditional tractography-based group difference analysis includes manual selection of anatomical tract(s) of interest (such as the corpus callosum [2] or multiple fiber tracts [71]) and then comparison of groups to find statistical differences in WM diffusion features (e.g. diffusion anisotropy), either using feature mean values [2, 23, 71] or along-tract measures [24, 36, 22, 71]. However, these studies are generally limited to a small number of selected tracts due to the requirement for extensive manual interpretation by trained experts.
Compared to the aforementioned studies that apply manual fiber tract selection methods, another strategy can efficiently identify multiple tracts in an automated fashion based on connected brain gray matter (GM) regions [101], anatomical priors [104], and/or WM anatomy (i.e. fiber geometric trajectory) [65]. Several groups have applied this strategy to study multiple tracts for identification of group WM differences [105, 103, 18, 84]. Nonetheless, in general each tract has been studied individually without any correction for multiple comparisons, preventing simultaneous analysis across multiple tracts. Recently, two research studies did perform simultaneous statistical analysis of multiple tracts using FDR for multiple comparison correction [100, 114]. Both works estimated direct fiber correspondences across subjects using fiber geometry. One work analyzed 30 major fiber tracts [100], while the other work performed a whole brain white matter statistical analysis [114]. These approaches demonstrated the potential of simultaneous multi-tract analysis. However, neither group investigated strategies for improving sensitivity during multiple comparison correction.
Another important approach for white matter analysis leverages a cortical parcellation to subdivide the white matter according to its cortical tract terminations and generates a structural connectome matrix that stores data such as fiber count. Statistical analyses of structural connectome matrices have employed FDR, Bonferroni, and permutation testing methods for correcting multiple comparisons [34, 48, 79, 47]. Cluster-formation methods that leverage cortical graph geometry have also been proposed for connectome matrix analysis [106]. However, there are challenges in the application of connectome-style analyses to dMRI [91] including anatomical variability of fiber tract terminations and cortical anatomy [8, 42, 96], spatial distortions [50], and reduced reproducibility due to the non-continuous nature of the matrix representation [56]. In contrast, white-matter-centric fiber analysis approaches can leverage information about the full course of a fiber tract, which is the anatomical basis for fiber tract definition [101], and empirical results indicate that white-matter-centric approaches benefit from low variability across subjects in terms of the WM parcellations that can be defined [109, 64, 19, 37].
In light of the above, we propose a suprathreshold fiber cluster (STFC) method to identify WM group differences using whole-brain tractography. For the first time, the proposed method leverages permutation testing and whole brain fiber geometry to enhance the statistical analysis of tractography. The method design is inspired by the idea that fiber tract parcels with similar WM anatomy, e.g. neighboring parcels, can support each other to obtain statistical significance. Important considerations in proposing such a method are the definition of the parcel neighborhood and the method for STFC formation using the parcel neighborhoods. This investigation extends our initial conference publication that demonstrated feasibility of STFC analysis [112] to 1) improve suprathreshold fiber cluster formation with anatomically varying inter-parcel distances and 2) improve robustness to parameter selection, an important goal in statistical neuroimaging analyses [89].
Our proposed method is based on a study-specific data-driven whole brain WM parcellation that finely divides all input tractography into many regions (a total of 1416 tract parcels in our study) and hence allows identification of potential local WM group differences from the whole brain. Recent research has shown the potential benefit of fine scale WM/GM parcellations to provide better group difference information compared to coarse-grained parcellations [39, 95, 111, 53]. In addition, we propose a fiber-cluster-thresholding-based multiple comparison correction to enable simultaneous analysis across the multiple tract parcels. Different from the voxel-cluster-thresholding approach [58, 89], our proposed method leverages fiber spatial neighborhoods that are defined according to the whole brain WM anatomy. To our knowledge, this work represents the first fiber-cluster-thresholding method to identify tractography-based group differences for whole brain analysis.
In the rest of this paper, we first describe our proposed method and then demonstrate its application to a multi-shell dMRI dataset from attention deficit hyperactivity disorder (ADHD) patients and healthy controls (HCs). A tract-specific study and a whole-brain WM study are conducted to illustrate the proposed method’s ability in both tract-of-interest and whole-brain tractography analyses.
2. Methods
2.1. Overview
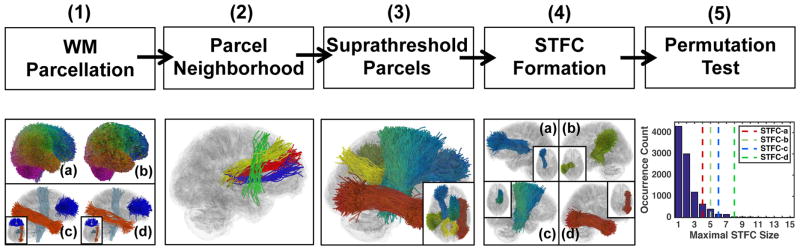
The proposed STFC method is designed to enhance statistical group difference analyses using whole-brain tractography. Figure 1 gives an overview of the proposed STFC method, including several steps: study-specific data-driven whole brain WM parcellation (Section 2.2), locally adaptive WM parcel neighborhood construction (Section 2.3), and computation of STFC statistics (Section 2.4) composed of suprathreshold parcel identification (Section 2.4.1), STFC formation (Section 2.4.2) and nonparametric, permutation-test-based group difference computation (Section 2.4.3). Overall, these steps enable identification of corresponding WM parcels in the whole brain of all subjects under study, followed by a statistical group difference analysis while correcting for multiple comparisons leveraging fiber geometry.
Figure 1.
Method overview: (1) WM Parcellation: (a) and (b) show the study-specific data-driven groupwise WM parcellation (atlas) and a subject-specific WM parcellation; (c) and (d) show several example atlas WM tract parcels and the corresponding subject-specific WM tract parcels (d); (2) Parcel Neighborhood: example neighbors of the red tract parcel include the yellow and the blue parcels but not the more distant green one with dissimilar WM anatomy to the others; (3) Suprathreshold Parcels: example fiber parcels that passed a parcel-level statistic test from the whole brain according to a quantitative parameter extracted from each parcel (the diffusion feature of return-to-the-origin probability (RTOP) was used in this study), computed using a Student’s t-test given a user-given threshold (uncorrected p-value = 0.05 is used in this study); (4) STFC Formation: example STFCs obtained using clique percolation (CP) to join neighboring parcels with uncorrected significance; (5) Permutation Test: example null distribution obtained from a non-parametric permutation test with summary statistic of maximal STFC size. For each STFC, its corrected significance is computed by comparing its STFC size (dashed lines) to the null distribution.
2.2. Data-driven WM Parcellation
A study-specific WM parcellation was first conducted using a well established data-driven pipeline to divide the whole brain tractography into multiple WM parcels according to the common WM anatomy from the whole population [63]. An open-source implementation of the WM parcellation pipeline is available online as the whitematteranalysis1 software. Our recent research studies have shown high performance of the pipeline in WM parcellation [111, 108, 64, 109, 110]. Here, we provide a brief introduction of this WM parcellation pipeline.
First, we computed an unbiased groupwise whole brain tractography registration to align all subjects’ tractography into a common space (i.e. atlas space) [62]. The method performed an entropy-based registration in a multiscale manner based on the pairwise fiber trajectory distances (the popular mean closest point distance was used [54, 63]) across all subjects. An affine transform was first performed, followed by a b-spline transform, in a coarse-to-fine fashion. This enabled nonrigid deformations of the fibers for improvements in parcellation consistency across subjects. Registration was conducted in a symmetric manner by registering all subjects, including midsagittal plane reflected copies of the subjects. In this way, the registration could effectively perform tractography alignment across the midsagittal plane. This benefited the bilateral clustering for atlas generation, which is introduced in the following paragraph.
Next, we created a study-specific data-driven groupwise WM parcellation atlas using spectral embedding [63], as follows. Each fiber was first represented as a point in a space useful for clustering. This was achieved by performing spectral embedding based on pairwise fiber affinities across all subjects. Then fiber affinities were obtained by converting the pairwise fiber distances using a Gaussian-like kernel with sigma of 60 mm. Nystrom sampling [33] was performed to reduce the computations considering the large number of fiber pairs across subjects. Bilateral clustering that simultaneously segments fibers in both hemispheres was applied to improve parcellation robustness [63]. To achieve bilateral clustering, the fiber distance measurement used all fiber connections as well as their reflections through the midsagittal plane of the brain. In this study, a study-specific WM parcellation atlas consisting of a total of K=800 WM parcels, including both inter- and intra-hemispheric parcels, was generated from all subjects under study. K=800 was chosen to provide a good parcellation of the whole WM based on our recent study [64]. (Please see the video S1 included in the supplementary material for a visualization of each individual WM parcel within this atlas.)
Then, we applied the parcellation atlas to each individual subject to obtain a subject-specific WM parcellation [63]. The spectral embedding for each new fiber (already affine and non-rigid transformed) was calculated by comparison to the fibers stored in the atlas (the stored Nystrom sample) using the same fiber affinity as in the atlas generation. The subject-specific WM parcels were then obtained by assigning the fibers to their closest atlas parcel. Given the bilateral clustering of the atlas, the subject WM parcels were detected bilaterally. A hemisphere-based parcel separation was conducted, where bilateral (intra-hemispheric) parcels were separated into left and right hemisphere parcels.
After obtaining the hemisphere-based-separated parcels in all subjects, valid parcels were defined as the ones that were commonly detected in the population (shared structural connections). We identified the valid parcels using a nonparametric one-tailed sign test. For each parcel, the sign test was performed with the null hypothesis that there were no fibers in the parcel across all subjects (Bonferroni corrected at a significance level of 0.05). This method has been applied in multiple studies to identify valid white matter connections [34, 79, 111, 109]. In this study, we obtained a total of 1416 valid hemispheric and commissural parcels from the dataset under study. Parameters involved in the WM parcellation pipeline were set to the default values in the whitematteranalysis package.
2.3. WM Parcel Neighborhood Construction
The goal of constructing a parcel neighborhood is to identify nearby WM connections with similar WM anatomy. This will be further used to find neighboring WM parcels to form the STFCs (Section 2.4.2). The parcel neighborhood construction includes two steps of an inter-parcel distance computation and a locally adaptive neighborhood construction to adapt to local WM geometry.
The distance between parcels (p1 and p2) was computed as the mean of the pairwise fiber distances between parcels in the WM parcellation atlas (Section 2.2), i.e.
| (1) |
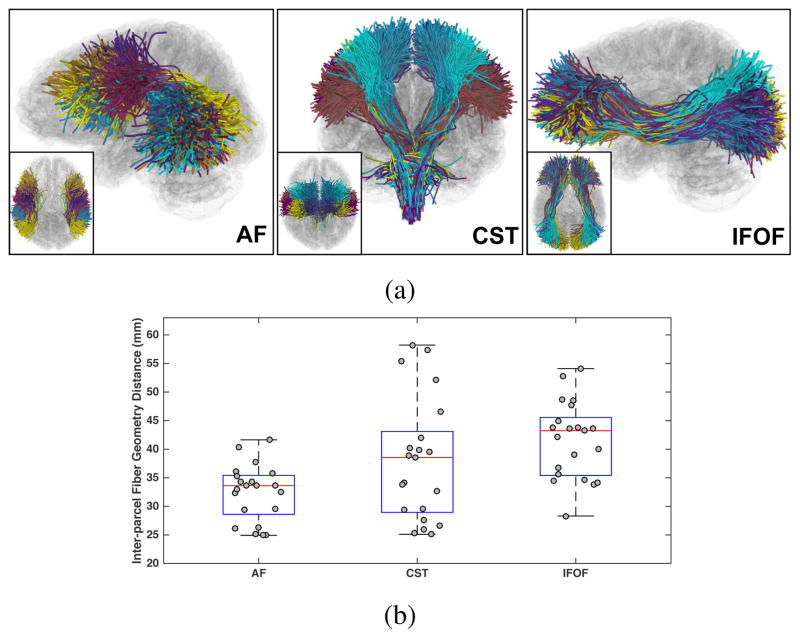
where di j is the fiber pair distance and I and J are the total numbers of fibers in the two parcels. We applied the mean closest point fiber distance [63], the same distance used in the spectral clustering WM parcellation, to measure di j. In this way, the distances D could capture the anatomical similarity between the parcels, with a low distance representing a high WM anatomy similarity and vice versa. Figure 2 shows the inter-parcel distances from several example fiber tracts.
Figure 2.
Inter-parcel distances across all parcel pairs in several example fiber tracts, including arcuate fasciculus (AF), corticospinal tracts (CST) and inferior fronto-occipital fasciculus (IFOF). (a) shows the WM parcels (colored differently) within each tract. These example fiber tracts were selected for visualization using expert knowledge following the anatomical hierarchy creation process using 3D Slicer as described in [64]. (b) shows that the inter-parcel distances in these tracts have different scales, e.g. the inter-parcel distances within the AF tract are in general lower than those within the IFOF tract, motivating our design of a locally adaptive neighborhood formation.
The strategy of constructing neighborhoods using locally surrounding information has been widely used in multiple fields, such as data mining [107, 28] and computer vision [73, 1] for its good performance in finding similar structures from data with heterogeneous distance scales. Here we propose to use this strategy to define WM parcel neighborhoods, considering that the inter-parcel distances in different anatomical WM structures are expected to have different scales (Figure 2b). We propose that the neighborhood of a WM parcel should be locally adaptive, where each parcel’s neighbors are decided considering its nearest parcels.
In machine learning, local neighborhood construction is often modeled as a graph-based problem. A robust method for “good” neighborhood selection is to use consensus information: first select the top T nearest neighbors for each node in the graph and then decide which neighbors should be retained to form a tight neighborhood where the nodes are highly similar to each other [107, 113, 73]. To apply this method to our data, we considered the WM parcels as graph nodes, where the internode distance was the inter-parcel distance (Eq. 1). For each individual parcel p, we first selected the top T nearest parcels, which we referred to as candidate neighbors. Then, among the T candidates, we considered the ones that formed the largest weakly connected sub-graph to be the neighborhood of p, as illustrated in Figure 3a. This ensured that all parcel neighbors had similar WM anatomy to each other while adapting to the different anatomical size scales. As a result, we could obtain a parcel neighborhood graph, where each node represented a WM parcel and an edge represented that two parcels were neighbors. Here, we note that compared to neighborhood construction using a strongly connected sub-graph that could be too strict in forming neighborhoods, a weakly connected sub-graph could reduce the possibility of missing potential neighbors. For example, in the CST shown in Figure 2a, in a weakly-connected-sub-graph-based way, the lateral brown cluster belonged to the neighborhood of the more superior blue cluster due to the cyan cluster between them, while using a strongly connected subgraph, such a neighborhood could be missed.
Figure 3.
Schema of (a) locally adaptive WM parcel neighborhood construction (Section 2.3) and (b) clique-percolation-based STFC formation (Section 2.4.2). (a) For an example parcel p1 (blue circle), the top four nearest parcels p2 to p5 (under T = 4) are selected as candidates (orange circles). The candidates weakly (directly or indirectly) connect to each other (red edges) and thus are considered as the neighbors of p1. (b) Here, we assume all parcels (colored circles) passed a parcel-level statistic test as the suprathreshold parcels (Section 2.4.1). Following the example in (a) with more parcels p6 to p9, there are one 4-clique ([p1, p3, p4, p5]) and three 3-cliques ([ p1, p2, p3], [ p4, p5, p9] and [ p4, p8, p9]), which are 2-connected by sharing two common parcels (red edges). Therefore, they form an STFC of size 7 (under a parameter setting h = 2 that two parcel cliques are considered adjacent if they share at least two parcels). The parcels p6 and p7 do not form any cliques with the aforementioned parcels and thus are not considered as part of the formed STFC.
One benefit of the locally adaptive neighborhood construction was its robustness to parameter selection. The single needed parameter, T, could be chosen according to the WM anatomy of the parcels obtained from the whole brain parcellation, prior to any statistical analyses (see Section 3.2 for details). In all our statistical experiments (both on synthetic and real datasets) we used a single value of T = 4.
2.4. STFC Statistics Computation
Computation of the suprathreshold fiber cluster (STFC) statistics included the following steps: 1) identifying suprathreshold parcels that passed an uncorrected, parcel-level statistic test, 2) forming the suprathreshold parcels as STFCs given the obtained parcel neighborhoods, and 3) calculating the STFC statistics in a nonparametric permutation test while correcting for multiple comparisons.
2.4.1. Suprathreshold Parcel Identification
A suprathreshold parcel was defined as a WM parcel that passed a parcel-level statistic test at a predetermined primary threshold (analogous to suprathreshold voxel in the voxel-cluster-thresholding approach [58]). Specifically, to derive the parcel-level statistic, we performed a null hypothesis test that computed an uncorrected WM group significance (an uncorrected p-value) for each individual parcel. We set the primary threshold to p-value =0.05, i.e. the parcels with uncorrected p-values ≤ 0.05 were suprathreshold parcels. The suprathreshold parcel identification would be conducted in all runs in the permutation test (both correctly labeled and permuted data) (Section 2.4.3).
In the interest of the application to study ADHD, we applied a one-tailed Student’s t-test to compute the uncorrected p-values (see Section 3.1 for details). However, we note that the parcel-level statistics can be computed with many other statistical computation methods and/or other tract quantitative measures under the hypothesis of interest.
2.4.2. Clique-Percolation-Based STFC Formation
An STFC was defined as a fiber cluster of multiple neighboring suprathreshold parcels. Given the defined parcel neighborhood graph (Section 2.3) and the suprathreshold parcels that passed the parcel-level statistic test (Section 2.4.1), we applied clique percolation (CP) [68] to identify the STFCs. We chose CP due to its known good performance in identifying densely connected node clusters on a graph. Applying CP to our parcel neighborhood graph therefore enabled formation of STFCs consisting of WM parcels with highly similar WM anatomy.
In graph theory, CP is a popular method for detecting densely connected node clusters given known edges between nodes. It has been successfully used in a variety of applications, such as analyzing structural and functional brain connectivity [92], community detection in protein networks [51] and studying social networks [35]. In brief, given the neighborhood relationships among all nodes on a graph, CP first detects all cliques, i.e., fully connected subgraphs. A clique with k nodes is called a k-clique and two cliques are adjacent if they share at least h nodes. Then, a node cluster is calculated as the maximal union of cliques that could be reached from each other through a series of adjacent cliques.
Given the parcel neighborhood graph from all WM parcels (this was computed before statistical analysis (Section 2.3)), we first extracted a subgraph consisting of only the suprathreshold parcel nodes and edges. After that, CP was applied to identify all parcel cliques and then the adjacent parcel cliques as the STFCs, as illustrated in Figure 3b. The CP-based STFC formation had an advantage that the only parameter h to define clique adjacency could be determined according to the number of neighbor candidates T. In our experiments, given T = 4, we set h = 2 that made two parcel cliques adjacent if there were at least two shared parcels. Detailed explanations of parameter selections will be provided in Section 3.2. We note that the parameter selections of h and T were determined together before any statistical analyses.
2.4.3. STFC Statistics Using Nonparametric Permutation Test
STFC statistics were computed in a nonparametric permutation test to obtain a final significance of each STFC with correction for multiple comparisons. A nonparametric permutation test is used because it is conceptually simple, relies only on minimal assumptions, and allows multiple comparison correction for simultaneous analysis of multiple brain regions [44, 58]. In related work, we and other research groups have successfully applied a permutation test for identifying group WM statistic differences along single fiber tracts within certain WM regions [65, 94, 100]. In the proposed method, we uses a permutation test for a whole brain WM analysis. Our proposed STFC method determines a corrected significance for each STFC based on its STFC size, i.e. the number of WM parcels within each STFC, under the null hypothesis of no group difference in any parcel in this STFC. Similar to the suprathreshold voxel cluster test in [58], we used the maximal statistic, i.e. the maximal STFC size, as the summary statistic in the permutation test, where the maximal STFC size was computed in each permutation run to build the null distribution to provide family-wise error control of false positives.
Algorithm 1 shows the pseudocode of the overall STFC method. After performing the groupwise WM parcellation (line 1) and the locally adaptive neighborhood construction (line 2), we first produced a null distribution of the summary statistic across the permutation runs (N=10000 was used in all experiments) (lines 3.1 to 3.3). Then, for each STFC from the correctly labeled groups (i.e. the real data) (lines 4 to 6), we compared its STFC size to the null distribution to obtain a corrected significance of the STFC, as well as the corrected p-value of each parcel in the STFC (lines 7.1 to 7.3). The STFCs that had corrected significance values lower than or equal to a user-given significance level were considered to have statically significant group differences. We set the significance level to 0.05 in all statistical experiments in this study.
3. Evaluation Dataset, Experiments and Results
3.1. Dataset
We demonstrate the proposed method using a dataset from 59 individuals (30 ADHD, 7 females and 23 males; 29 HC, 10 females and 19 males) that were locally recruited from multiple sources including the Departments of Psychiatry and Neurology at a tertiary care university hospital, a community based health center, and through online advertisements. The two groups were matched for age (ADHD: 10.6±1.7 years; HC: 10.7±1.7 years; two-tailed t-test, p = 0.9) and socioeconomic status (SES) via household annual income (ADHD: 64.2 ±3.68 thousand dollars; HC: 78.6 ± 34.2 thousand dollars; two-tailed t-test, p-value = 0.2).
Algorithm 1.
Suprathreshold Fiber Cluster (STFC) Method
| 1: | Perform study-specific groupwise WM parcellation (Section 2.2) |
| 2: | Build parcel neighborhood graph from all WM parcels (Section 2.3) |
| 3: | for each permutation run in [1, N] do |
| 3.1: | Randomly permute group labels of all subjects |
| 3.2: | Identify suprathreshold parcels from the permuted data (uncorrected p-values ≤ 0.05) (Section 2.4.1) |
| 3.3: | Extract all STFCs and record the maximal STFC size (Section 2.4.2) |
| 4: | Calculate the histogram of the maximal STFC size to produce the null distribution |
| 5: | Identify suprathreshold parcels in the correctly labeled data (uncorrected p-values ≤ 0.05) (Section 2.4.1) |
| 6: | Extract all STFCs and compute their STFC sizes (Section 2.4.2) |
| 7: | for each STFC s do |
| 7.1: | Locate its STFC size in the histogram of the maximal STFC size |
| 7.2: | Obtain number of permutations with the maximal STFC size larger than or equal to the size of STFC s as N(s) |
| 7.3: | Compute the corrected significance value as (N(s) +1)/(N +1) |
3.1.1. Data Preprocessing
High-resolution MR images were obtained on a Siemens 3T scanner at Boston Childrens Hospital, Boston, USA, with approval of the local ethics board. Multi-shell diffusion-weighted imaging (DWI) data were acquired using simultaneous multi-slice acquisition factor of 2 [83] at a spatial resolution of 2×2×2 mm3 with 70 gradient directions spread over the three b-value shells of 1000/2000/3000 s/mm2. Additionally, T1- and T2-weighted images were acquired at a spatial resolution of 1 mm3 and 8 mm3 respectively.
A semi-automated quality control (using in-house developed Matlab scripts) was conducted on all datasets, and all gradient directions with a signal drop were removed. Brain masks for dMRI images were first obtained using BET [86], followed by manual inspection and correction in 3D Slicer2 via SlicerDMRI3 [60]. Eddy current and head motion correction was performed using FSL FLIRT software [88]. Further, dMRI images were corrected for EPI distortions by registration (restricted to the phase-encode direction) to the T2-weighted images using ANTS [5]. All processed images were manually inspected to ensure high quality of the data sets.
3.1.2. Tractography, Diffusion Feature and Suprathreshold Parcel
Whole brain tractography was conducted using the unscented Kalman filter tractography method [77, 80], as implemented in the ukftractography4 package. Tractography was seeded 5 times per voxel, in all voxels within the brain mask. We employed a multi-fiber model, where each fiber is represented using a two-tensor bi-exponential model [78, 77]. This framework enables robust analytical estimation of fiber-specific diffusion propagator and microstructure measures, while simultaneously tracing the white matter tracts. Fibers that were longer than 40 mm were retained to avoid any bias towards implausible short fibers [38, 49, 52].
Return-to-the-origin probability (RTOP) was estimated using the bi-exponential model to provide a fiber-specific microstructure measure [77, 59]. RTOP estimates the probability of water molecules with zero net displacements during the diffusion time of the diffusion MRI scan. RTOP is known to be sensitive to the anisotropy of WM tissue and may increase pathophysiological specificity compared to traditional diffusion anisotropy measures, e.g. fractional anisotropy (FA) [67, 6]. The median of the RTOP values of all points in each WM parcel was measured, i.e. MRTOP. The median value has been previously shown to provide a robust measure of the central tendency of diffusion properties in WM regions for group difference analysis [10, 99].
Studies in ADHD have widely suggested decreased diffusion anisotropy [41, 70, 57]. Therefore, we performed an one-tailed Student’s t-test under the null hypothesis H0: μHC (MRTOP) ≤ μADHD(MRTOP) to compute the parcel-level statistic for each individual WM parcel. The primary threshold to define suprathreshold parcels was set to p-value = 0.05, i.e., the parcels that had the uncorrected p-values ≤ 0.05.
3.1.3. Synthetic Dataset
A synthetic dataset with simulated true group difference was created to validate the proposed method. To simplify the assessment and tractography visualization, we created a realistic synthetic dataset in the corpus callosum (CC). The synthetic data was generated as follows. We identified a total of 34 CC parcels from the whole brain parcellation, as shown in Figure 4a. For each CC parcel, we added white Gaussian noise (signal-to-noise ratio at 1 [89]) to the actual measured features (i.e. RTOP) of all HC subjects. Repeating this process twice generated two synthetic groups of G1 and G2, each with 29 subjects. We then modified 15 CC parcels of interest to have true group difference by adding synthetic feature changes to the G2 subjects. For each of the 15 parcels, we decreased its group mean MRTOP values in G2 (as a percent of its original feature mean) for a null hypothesis test: H0: μG1(MRTOP) ≤ μG2(MRTOP). These 15 parcels were selected to form 3 different synthetic ground truth fiber clusters (with sizes of 4, 5 and 6, as shown in Figure 4b). This synthetic dataset will be used in two experiments to validate our method (Section 3.3) after we introduce the parameter decisions in the next subsection (Section 3.2)
Figure 4.
(a) All CC parcels (size of 34) obtained in the whole brain parcellation. (b) 15 CC parcels-of-interest with synthetic ground truth group differences. These parcels formed three fiber clusters from the genu (blue), body (green) and splenium (purple) sections of the CC, with sizes of 4, 5 and 6 respectively.
3.2. Parameter Selection According to Population Whole Brain WM Anatomy
We first performed an experiment for parameter selection. Our method has two parameters: 1) T that is the number of candidate neighbors per parcel (Section 2.3) for local neighborhood construction, and 2) h that is the number of shared parcels between cliques and defines clique adjacency (Section 2.4.3) to form STFCs. The two parameters together control if parcels with similar WM anatomy can be grouped into an STFC. We propose that reasonable settings of T and h are the ones that do not overly limit the size of potential STFCs that may be formed, while avoiding grouping parcels with dissimilar WM anatomy together.
To determine reasonable settings, we investigated how the two parameters affected fiber cluster (FC) formation according to the whole brain parcellation in the population. We note the experiment was conducted based on the whole brain fiber geometry and WM anatomy only, without using any measured diffusion data. Thus, the parameter selection was independent of any statistical analysis. Specifically, given certain settings of T and h, CP (under the given h) was applied directly to the full parcel neighborhood graph (under the given T ) of all WM parcels. This allowed us to detect all possible FCs that could be achieved from the whole brain. Here, we note that FCs were different from STFCs, since the STFCs were calculated based on a sub-neighborhood-graph consisting of only suprathreshold parcel nodes and edges (thus, an STFC was a subset of one of the FCs).
After obtaining all possible FCs under the given T and h, we investigated the largest FC (LFC), i.e. the FC that had the maximum number of parcels. Corresponding to the proposed criteria (introduced at the beginning of this section) for parameter selection, the most reasonable T and h were the settings resulting in the maximal LFC but without dissimilar WM anatomy. For quantitative comparisons, we computed the cluster size (number of parcels) and the maximum inter-parcel distance (Dmax) for the LFC per setting, as shown in Table 1. Here, the experiments were conducted on a reasonable range of T between 3 to 6. While T ≤ 2 was too small to construct informative WM parcel neighborhoods, T ≥ 7 needed long computational time5 as extracting parcel cliques was a time-consuming process [68].
Table 1.
Quantitative assessments for selecting a reasonable h value given a certain T value include the cluster size and the maximum inter-parcel distance (Dmax mm) for the largest fiber cluster (LFC) that formed with each setting. Note that given a certain T value (per column), the maximum possible parcel clique was a (T +1)-clique (see [68] for details) thus the maximum possible h was T (no two parcel cliques could share T +1 or more parcels). Therefore, no FCs formed when h > T. While a large h value (e.g. h = T ) tended to generate the most white-matter-constant FCs (a low Dmax value), it was too strict when combining parcel cliques as FCs. Thus, it could overly limit the size of potential FCs that may be formed. For example, under h = T = 4, the LFC size was 6 (see Figure 5 for tract visualizations and comparisons across different h settings under T = 4), which meant that any FCs with sizes larger than 6 could not be identified. In this study, we set h = T – 2 (bold font per column) as a reasonable setting for a balance between a high WM anatomy consistency (a small Dmax) and a high coverage of a local WM structure of fiber clusters (a large LFC size).
| T = 3 | T = 4 | T = 5 | T = 6 | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| size | Dmax | size | Dmax | size | Dmax | size | Dmax | |
|
| ||||||||
| h = 1 | 18 | 66.2 | 64 | 90.4 | 433 | 142.3 | 604 | 144.7 |
| h = 2 | 13 | 69.4 | 23 | 68.9 | 112 | 114.8 | 555 | 143.9 |
| h = 3 | 5 | 48.7 | 8 | 59.0 | 15 | 51.5 | 56 | 94.0 |
| h = 4 | - | - | 6 | 57.4 | 8 | 59.8 | 13 | 51.1 |
| h = 5 | - | - | - | - | 6 | 58.9 | 7 | 59.8 |
| h = 6 | - | - | - | - | - | - | 6 | 59.0 |
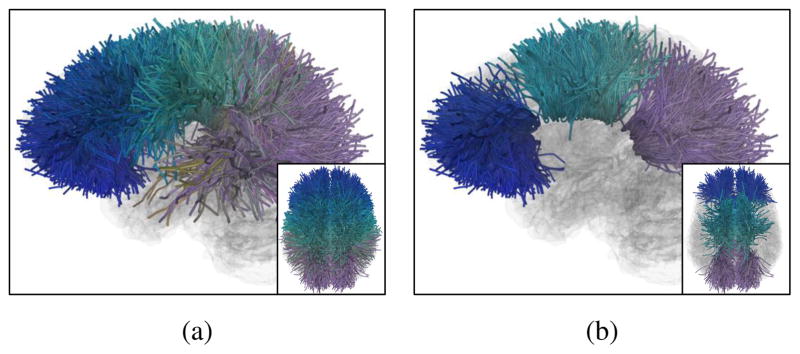
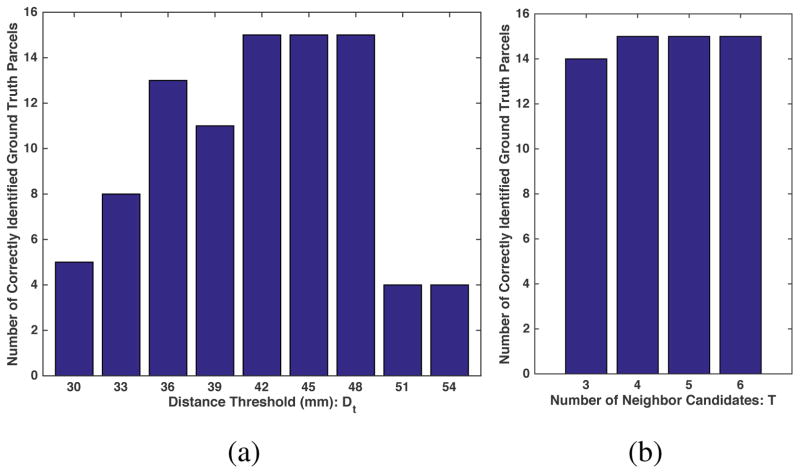
Experimental results indicate that given a certain T value, choosing h = T – 2 can balance between a high WM anatomy consistency (a small Dmax) and a high coverage of a local WM structure of fiber clusters (a large LFC size), as illustrated in Table 1. For visual comparisons, Figure 5 displays the LFC obtained per h value, with T = 4 for an illustration. h = 2 (i.e. T – 2) generated the best result.
Figure 5.
Visual tract comparisons of the largest fiber cluster (LFC) obtained from the whole brain parcellation given different h values under T = 4. (a) to (d) were obtained from h =1 to 4, consisting of 64, 23, 8 and 6 WM parcels, respectively. h = 1 obtained a LFC with the most parcels; however the parcels were from different local WM regions including the temporal lobe and the cerebellum. h = 3 and 4 generated a LFC with only a small number of parcels, which provided a small coverage of the fiber connections from a local WM region. h = 2 generated a LFC with 23 parcels that were all from the corona radiata region, providing a visual confirmation of our choice of setting of h = T – 2.
Under h = T – 2, we considered that T = 4 (across T ∈ [3,6]) was the most reasonable setting, which was used in all following experiments. This setting derived a maximal LFC (Figure 5b) for a high coverage of a local WM structure (cluster size 23) while the parcels within the LFC were highly white-matter-consistent (Dmax = 68.9 mm). Given this setting, in Figure 6, we show several example FCs obtained in the whole brain parcellation and the inter-parcel distances per FC. The proposed STFC formation could identify FCs from different local WM structures that have different distance scales.
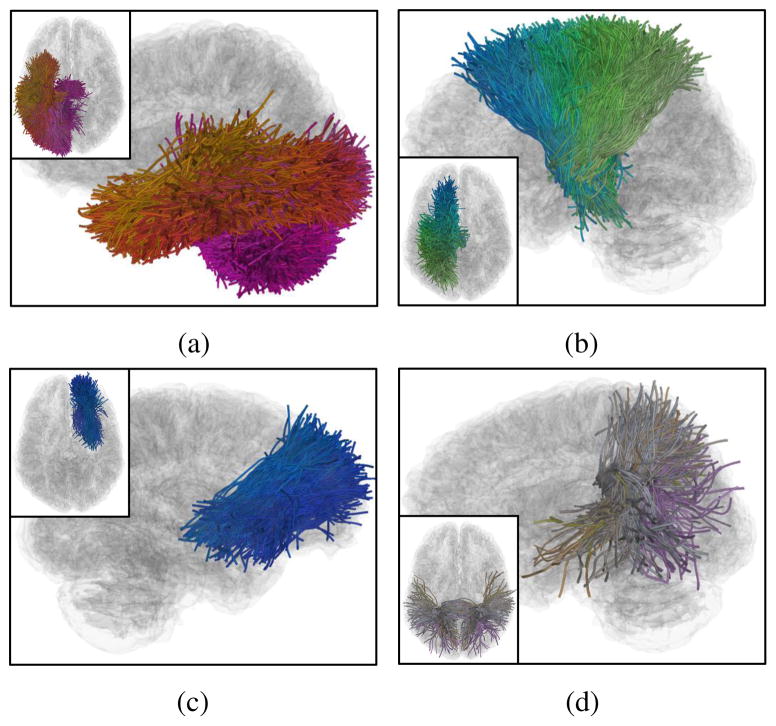
Figure 6.
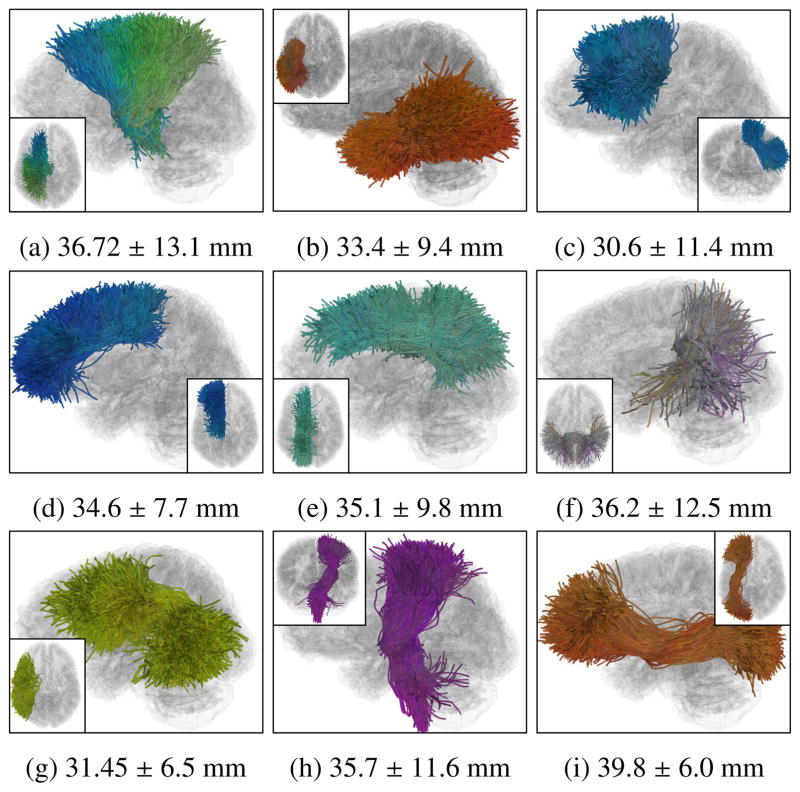
Visualizations of several example fiber clusters (FCs) obtained from the whole brain WM parcellation under the chosen parameter settings (T = 4 and h = 2), including the fiber connections belonging the local WM structures of (a) corona radiata (CR) (23 parcels), (b) inferior longitudinal fasciculus (ILF) (18 parcels), (c) frontal aslant tract (FAT) (14 parcels), (d) anterior cingulum (ACG) (11 parcels), (e) middle cingulum (MCG) (11 parcels), (f) posterior corpus callosum (CC) following the lateral paths to temporal lobes (6 parcels), (g) arcuate fasciculus (AF) (5 parcels), (h) corticalspinal tracts (CST) (5 parcels) and (i) inferior fronto-occipital fasciculus (IFOF) (5 parcels). Inter-parcel distances (mean ± standard deviation) within each of the FCs are displayed in the sub-captions. The proposed locally adaptive STFC formation was able to identify FCs from the different local WM structures that are expected to have different distance scales.
3.3. Synthetic Data Analysis
Given the chosen parameter settings, we first illustrate our STFC method on the synthetic data (Section 3.1.3) in the following two experiments.
3.3.1. Comparison of Multiple Comparison Correction Methods
We first compared the proposed STFC method with several multiple comparison correction methods using the synthetic datasets. The evaluation goal was to test if a method could correctly identify the parcels with true significance, even when the added change was small. To do this, we added synthetic feature changes to the ground truth parcels at different scales.
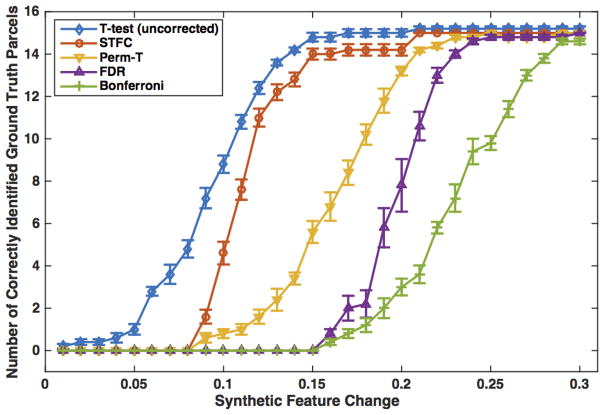
Comparisons were conducted among the uncorrected t-test, the proposed STFC method, a standard permutation test (Perm-T, N=10000) that used the minimal t-test-based p-value of all parcels for the summary statistic, and two traditional FDR (as applied in [100, 114]) and Bonferroni multiple comparison correction methods. The same significance level of α = 0.05 was used for all compared methods. Figure 7 displays the number of significantly different parcels that were correctly identified in each method. Here, we note that the four multiple comparison correction methods corrected for significance of the parcels that passed the initial uncorrected t-test. These were a subset of the ground truth parcels, i.e. the parcels with feature changes. (We note that in some experimental settings, the feature changes in the selected ground truth parcels were of small magnitude and therefore a subset of these parcels survived the initial t-test.) No false positives were detected by any of the compared methods.
Figure 7.
Comparison among different multiple comparison correction methods on the synthetic dataset: Number of correctly identified CC parcels per method, versus the level of synthetic group difference, is displayed. For each feature change scale (from 0.01 to 0.3), we reported the mean result from 10 randomly generated feature changes, with a quarter of the standard deviation displayed for better visualization.
3.3.2. Comparison of Locally Adaptive Neighborhood vs Global Distance Threshold
We then compared the proposed method to a simpler global-distance-threshold-based (GDTB) STFC formation approach as reported in our preliminary work [112]. Briefly, in GDTB, two parcels were considered as neighbors if their distance D (as in Eq. 1) was smaller than a user-given parcel distance threshold Dt, and neighboring suprathreshold parcels were joined to form STFCs.
The initially proposed GDTB method was relatively sensitive to the choice of distance threshold [112]; therefore we designed the current method to be highly robust to parameter selection. Here, we performed an experiment by varying the parameters of both methods to assess robustness of statistical findings in the synthetic data. Specifically, the GDTB approach has one parameter of the distance threshold Dt and the proposed approach has one parameter of the number of neighbor candidates T (given h = T – 2 as discussed in Section 3.2). The two methods were compared using a synthetic dataset with feature change at the level of 0.21, the minimum feature change to allow all ground truth parcels to be successfully detected by the initial t-test (Figure 7). Figure 8 shows the numbers of correctly identified parcels among those with feature changes given different parameter settings of the two methods. The proposed method was less sensitive to the parameter changes compared to the GDTB approach. The two STFC-based methods corrected for significance of the parcels that passed the initial uncorrected t-test. Therefore, no false positives were detected by the GDTB or the proposed method.
Figure 8.
Comparison between the proposed and a simpler global-distance-threshold-based (GDTB) STFC formations using the synthetic dataset: in GDTB (a), the numbers of correctly identified parcels were sensitive to the different distance threshold Dt values, while in the proposed method (b), the numbers of ground truth parcels that were correctly identified were similar across different T ∈ [3,6].
3.3.3. Receiver operating characteristic evaluations
We then performed a receiver operating characteristic (ROC) experiment to investigate potential false positives arising during the statistical analyses for the two STFC approaches: the GDTB method and the proposed method that uses a locally adaptive parcel neighborhood construction. We applied the alternative free-response ROC (AFROC) [17] method that has been used to measure the family-wise errors that could happen anywhere in the brain in the presence of multiple comparisons for neuroimage analyses [89, 75]. Specifically, in these studies the false positive rate (FPR) was computed using “noise-only” images as the probability of any false positives detected anywhere in the image, while the true positive rate (TPR) was computed using “signal+noise” images as the average fraction of true positives correctly detected.
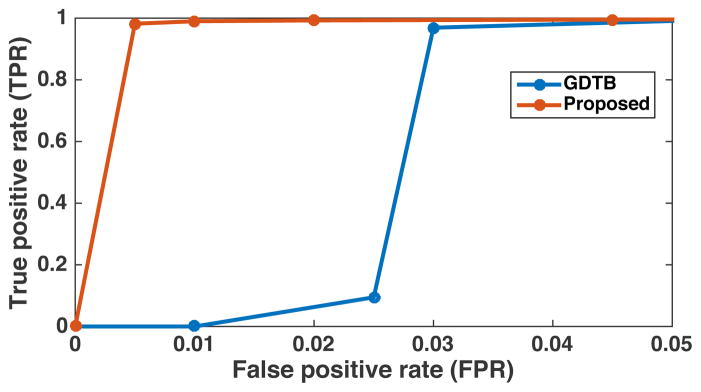
To perform an AFROC experiment using our tractography data, we extended the dataset from the previous experiment. We generated 1000 synthetic datasets, where each dataset was created by adding random Gaussian noise to the actual measured feature to create two synthetic groups (as described in Section 3.1.3). These were used as the “noise-only images” in our experiment. Then, for each synthetic dataset, we changed features of the ground truth parcels in the second synthetic group (as described in Section 3.1.3) at the level of 0.21 (as used in the above experiment). These were used as the “signal+noise images.” For the two compared STFC methods (i.e. GDTB and proposed), we varied the primary threshold (the uncorrected p-values from 0.01 to 0.1) to compute the ROC curves, as follows. At each threshold level, the FPR was computed as the probability of any false positive parcels detected anywhere across the 1000 synthetic datasets without feature changes (i.e. the “noise-only images”), and the TPR was computed as the average fraction of true positive parcels across the 1000 synthetic datasets after adding feature changes to the ground truth parcels (i.e. the “signal+noise images”). Similar to the studies from Smith and Nichols [89] and Raffelt et al. [75], we limited the ROC analysis to the FPR values smaller than 0.05 (FPR in excess of 0.05 is not of interest). We set the method parameters to Dt = 42 mm for the GDTB method and T = 4 for the proposed method, which were sufficient to detect all ground truth parcels in our previous experiment (Figure 8). Figure 9 shows the obtained ROC curves for the two compared STFC methods. The area under the curve (AUC) of the proposed method was 0.936 compared to that of 0.508 from the GDTB method, suggesting high sensitivity and specificity performances of the proposed method on these synthetic CC datasets.
Figure 9.
Receiver operating characteristic (ROC) curves obtained from the synthetic corpus callosum (CC) datasets using the alternative free-response ROC (AFROC) method for the two STFC methods: the global-distance-threshold-based (GDTB) method and the proposed method that uses a locally adaptive parcel neighborhood construction. The false positive rate (FPR) represents the probability of any false positive parcels detected anywhere across 1000 randomly generated synthetic datasets, and the true positive rate (TPR) is the average fraction of true positive parcels correctly detected across the 1000 datasets. The areas under the curve (AUCs) for the two methods are 0.936 and 0.508, respectively.
3.4. Real Data Analysis
Next, we show experimental results on the real data for a tract-specific study of the CC and a whole brain WM study.
3.4.1. Tract-specific Study of the Corpus Callosum
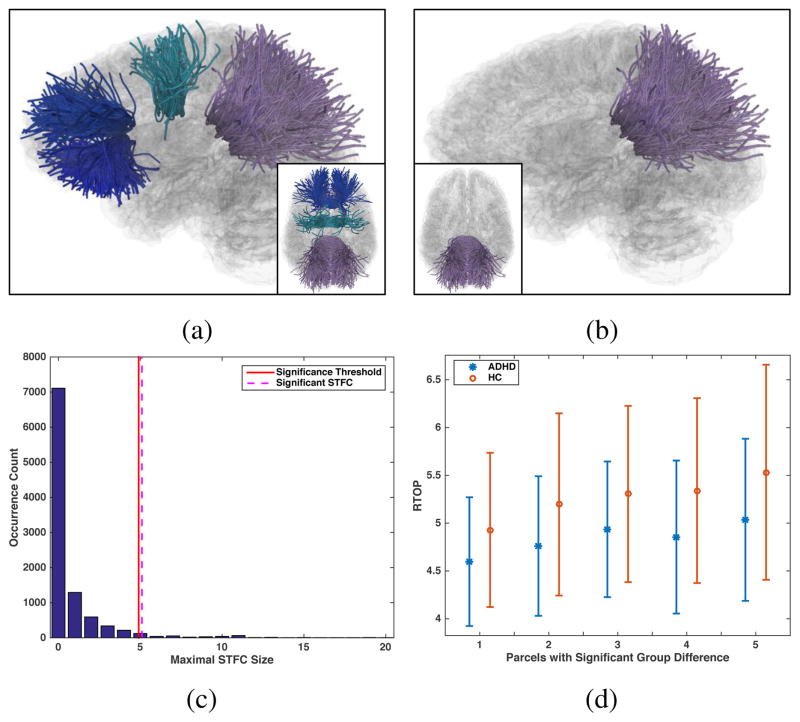
We first performed a tract-specific study to identify group differences specific to a local WM region. The CC tract was selected for a demonstration because it is one of the most commonly studied tracts in ADHD [14, 30, 69, 29]. Given the CC tract that consisted of a total of 34 parcels (Figure 4a), we first performed an one-tailed t-test with the null hypothesis H0: μHC (MRTOP) ≤ μADHD(MRTOP) to compute parcel-level statistics. 8 parcels passed the initial t-test with uncorrected p-values ≤ 0.05, as displayed in Figure 10a. Multiple comparison correction was then conducted using the STFC, the Perm-T, the FDR and the Bonferroni methods respectively, at a significance level of α=0.05. While no parcels survived the Perm-T, the FDR or the Bonferroni methods, a significant STFC of 5 parcels was identified using the proposed STFC method, as displayed in Figure 10b. (Please see the video S2 included in the supplementary material for a visualization of each individual WM parcel within the significant STFC.) The corrected significance value of the identified STFC was 0.046, as illustrated in Figure 10c. Comparisons of the group mean feature values (MRTOP) of the 5 parcels are shown in Figure 10d.
Figure 10.
Real data experiment for tract-specific study of the corpus callosum: (a) 8 WM parcels passed the parcel-level statistic test, with uncorrected p-value ≤ 0.05; (b) The significant STFC consisting of 5 WM parcels from the splenium section of the CC; (c) Null distribution of the maximal STFC size from the permutation runs. Significance threshold of STFC size was 5, i.e. an STFC in the correctly labeled data with the size ≤ 5 would pass the significance level 0.05 as significant. The identified significant STFC had the size of 5, resulting in a corrected significance value of 0.046; (d) Comparison of MRTOP values per identified parcel in the significant STFC, plotted in sorted order.
3.4.2. Whole Brain WM Study
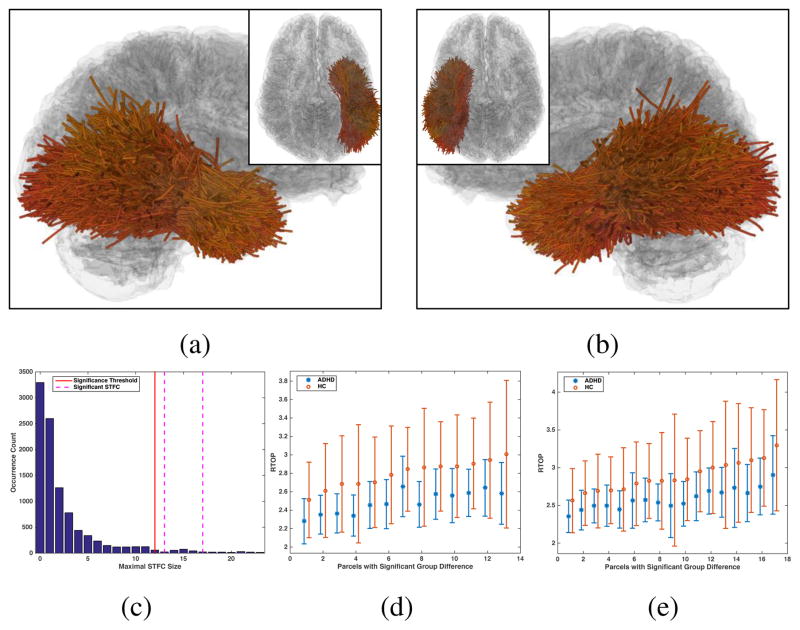
This experiment investigated potential WM alterations in the whole brain, without relying on a region-specific hypothesis. For the whole brain parcellation of a total of 1416 parcels (Section 2.2), 654 parcels had p-values smaller than 0.05 in the initial parcel-level t-test under the null hypothesis H0: μHC (MRTOP) ≤ μADHD(MRTOP). The same multiple comparison correction methods were applied as aforementioned. The FDR and the Bonferroni methods did not find any significantly different parcels between the groups. The Perm-T method identified one individual significant parcel (corrected p-value 0.0272) connecting the middle precentral gyrus to the supra-marginal and the superior-parietal gyri in the right hemisphere. Our method identified two significant STFCs of 13 and 17 WM parcels, which were located in the occipital and temporal lobes in both hemispheres as shown in Figures 11a and 11b. (Please see the video S3 included in the supplementary material for a visualization of each individual WM parcel within the significant STFCs.) The corrected significance values of the identified left and right hemisphere STFCs were p =0.035 and 0.015 respectively, as illustrated in Figure 11c. Comparisons of the group mean feature values (MRTOP) of the parcels are given in Figures 11d and 11e.
Figure 11.
Real data experiment for the whole brain WM study: (a, b) The two significant STFCs consisting of 13 and 17 WM parcels respectively in the left and right occipital and temporal lobes; (c) Null distribution of the maximal STFC size obtained from the permutation runs. Significance threshold of STFC size was 12, i.e. an STFC in the correctly labeled data with the size ≥ 12 was considered as significant. The two significant STFCs had the sizes of 13 and 17, resulting in corrected significance values of 0.035 and 0.015 respectively. (d, e) Comparisons of MRTOP values per identified parcel in the significance STFCs, plotted in sorted order.
4. Discussion
In this paper, we presented an STFC method that leverages the whole brain fiber geometry to enhance statistical analyses in identifying between-group white matter differences. We demonstrated the method’s performance with an application to a dataset of ADHD patients and healthy controls. We have several overall observations about the results, which are discussed below.
We demonstrated a high sensitivity and specificity of the proposed STFC method in identifying WM group differences while correcting for multiple comparisons. In the synthetic dataset with known group differences, results showed that the STFC method was more sensitive to detect the ground truth group differences when compared to three other multiple comparison correction methods (Figure 7). While the FDR and the Bonferroni methods could only find the significance when there were large differences (over 15% change of the group mean feature value), the two permutation-based approaches of the STFC and Perm-T could begin to detect the ground truth parcels after 9% feature changes (Figure 7). However, the STFC method could achieve a higher number of correctly identified parcels than the Perm-T method. The high sensitivity of the STFC method was also demonstrated by the experimental results on the real data analysis. In the tract-specific study, we identified a significant STFC of 5 WM parcels (Figure 10b), while no significance was found by the other three compared methods. In the whole brain WM study, two significant STFCs of a total of 30 WM parcels were identified using the STFC method (Figures 11a and 11b). Only one significant parcel was identified from the whole brain using the Perm-T method and no significance was detected using the FDR or Bonferroni methods. In addition, results from our AFROC experiment in the synthetic data analysis showed the proposed method’s high specificity, demonstrated by a high AUC value (0.936) obtained from the synthetic CC datasets (Figure 9).
The proposed method enabled a robust parameter selection, which was demonstrated from the following perspectives. First, reasonable parameter settings could be determined according to the whole brain fiber geometry in the population, prior to any statistical analyses (Section 3.2). Second, the proposed method was adaptive to the heterogeneous inter-parcel fiber distance scales in different WM structures (Figure 6). Third, the proposed method’s results demonstrated a low sensitivity to the changes of parameter settings (Figure 8).
Given the chosen parameters (T = 4 and h = 2), in the tract-specific study we found that the splenium could be most affected in the whole CC in ADHD. We used the median RTOP feature to explore potential changes in WM anisotropy in ADHD (Section 3.1). The feature comparison between the two groups showed significantly reduced median RTOP values in the ADHD individuals (Figure 10d), suggesting potentially reduced WM anisotropy in the splenium of the CC. This was consistent with the finding of altered splenium of the CC in ADHD in a review paper by Dougherty et al. [29], and with that of reduced anisotropy in this local CC region in dMRI-based studies [30, 69].
Regarding the experiments for the whole brain WM study, we identified WM regions with significant group differences in bilateral temporal and occipital lobes (Figures 11a and 11b). While previous studies have suggested these regions in either left [85, 20] or right [98, 61] hemispheres affected in ADHD, the alterations in both hemispheres have also been reported by recent studies [93, 31] similar to our analysis result. Our identified WM parcels had lower median RTOP values in the ADHD group when compared to the HC group, showing potentially reduced WM anisotropy in ADHD. Decreased WM anisotropy in the temporal and occipital lobes in ADHD has also been reported in multiple dMRI-based studies [20, 57]. The results related to the occipital and temporal lobes in our analyses are consistent with the findings in the existing literature in ADHD.
Potential future directions and limitations of the current work are as follows. First, similar to voxel-cluster-thresholding analyses, the proposed method aims to find large clusters of WM parcels with significance; thus it could potentially miss some significantly different parcels located in small neighborhoods, e.g. the one individual significant parcel identified in the standard permutation test (Perm-T). Second, it would be of interest to incorporate additional brain function and anatomy information, such as brain cortical parcellation from Freesurfer [32] or functional MRI data, to enable WM parcel neighborhood construction adaptively to not only local white matter anatomy but also brain functions. For example, only parcels connecting to the same Freesurfer cortical region could be considered as neighbors. Third, in our proposed method, we applied a parcel neighborhood construction to generate a binary neighborhood graph, followed by a CP-based fiber cluster formation, to identify STFCs adaptively to different inter-parcel distances. A further research direction could include applying a weighted-graph-based cluster formation method (e.g. the Lou-vain method [12]) on the pairwise parcel distance affinity D to reduce the number of parameters (e.g. the number of candidate neighbors T). Fourth, our previous work that applied the data-driven tractography parcellation method (Section 2.2) for a machine-learning-based classification task suggested that parcellation of the whole brain tractography into a large number of parcels tended to increase the classification performance [110]. Therefore, it would be of interest to investigate if a finer parcellation (in this study, we used a parcellation of 1416 parcels) could improve the statistical analysis by allowing the measurement of more locally specific WM changes.
5. Conclusion
In this paper, we have presented a novel suprathreshold fiber cluster (STFC) analysis to identify WM group differences using whole brain tractography. The proposed method leverages the whole brain fiber geometry to enhance statistical analyses of tractography while correcting for multiple comparisons to allow simultaneous analysis across the entire white matter. Experimental results suggest that our method in general is more sensitive for identifying WM group differences when compared to several traditional multiple comparison correction methods. The two real data demonstrations show that the proposed method can be generally applied in both hypothesis-driven tract-specific analysis and whole brain tractography analysis for between-population studies.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding provided by the following grants: the Australian Research Council (ARC) grants, National Science Foundation (NSF) of China No.61502117, NSF of Heilongjiang Province QC2016084, and the following National Institutes of Health (NIH) grants: P41 EB015902, P41 EB015898, R01 MH074794, R01 MH097979 and U01 CA199459.
Footnotes
On a standard computer, it could take over an hour or more with T ≥ 7, while with a smaller T value it took several minutes.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S. Slic superpixels compared to state-of-the-art superpixel methods. IEEE transactions on pattern analysis and machine intelligence. 2012;34(11):2274–2282. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- 2.Alexander AL, Lee JE, Lazar M, Boudos R, DuBray MB, Oakes TR, Miller JN, Lu J, Jeong EK, McMahon WM, et al. Diffusion tensor imaging of the corpus callosum in Autism. NeuroImage. 2007;34(1):61–73. doi: 10.1016/j.neuroimage.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 3.Ameis S, Catani M. Altered white matter connectivity as a neural substrate for social impairment in autism spectrum disorder. Cortex. 2015;62:158–181. doi: 10.1016/j.cortex.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner J, Friston KJ. Voxel-based morphometry–the methods. NeuroImage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 5.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avram AV, Sarlls JE, Barnett AS, Özarslan E, Thomas C, Irfanoğlu MO, Hutchinson E, Pierpaoli C, Basser PJ. Clinical feasibility of using mean apparent propagator (MAP) MRI to characterize brain tissue microstructure. NeuroImage. 2016;127:422–434. doi: 10.1016/j.neuroimage.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bach M, Laun FB, Leemans A, Tax CM, Biessels GJ, Stieltjes B, Maier-Hein KH. Methodological considerations on tract-based spatial statistics (TBSS) NeuroImage. 2014;100:358–369. doi: 10.1016/j.neuroimage.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Bajada CJ, Haroon HA, Azadbakht H, Parker GJ, Ralph MAL, Cloutman LL. The tract terminations in the temporal lobe: Their location and associated functions. Cortex. 2016 doi: 10.1016/j.cortex.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magnetic resonance in medicine. 2000;44(4):625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 10.Bazarian JJ, Zhong J, Blyth B, Zhu T, Kavcic V, Peterson D. Diffusion tensor imaging detects clinically important axonal damage after mild traumatic brain injury: a pilot study. Journal of neurotrauma. 2007;24(9):1447–1459. doi: 10.1089/neu.2007.0241. [DOI] [PubMed] [Google Scholar]
- 11.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. Series B (Methodological) 1995:289–300. [Google Scholar]
- 12.Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. Journal of statistical mechanics: theory and experiment. 2008;2008(10):P10008. [Google Scholar]
- 13.Brown CA, Johnson NF, Anderson-Mooney AJ, Jicha GA, Shaw LM, Trojanowski JQ, Van Eldik LJ, Schmitt FA, Smith CD, Gold BT. Development, validation and application of a new fornix template for studies of aging and preclinical Alzheimer’s disease. NeuroImage: Clinical. 2017;13:106–115. doi: 10.1016/j.nicl.2016.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Q, Sun L, Gong G, Lv Y, Cao X, Shuai L, Zhu C, Zang Y, Wang Y. The macrostructural and microstructural abnormalities of corpus callosum in children with attention deficit/hyperactivity disorder: a combined morphometric and diffusion tensor MRI study. Brain research. 2010;1310:172–180. doi: 10.1016/j.brainres.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 15.Carper RA, Treiber JM, DeJesus SY, Müller RA. Reduced hemispheric asymmetry of white matter microstructure in autism spectrum disorder. Journal of the American Academy of Child & Adolescent Psychiatry. 2016;55(12):1073–1080. doi: 10.1016/j.jaac.2016.09.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catani M, DellAcqua F, Budisavljevic S, Howells H, de Schotten MT, Froudist-Walsh S, DAnna L, Thompson A, Sandrone S, Bullmore ET, et al. Frontal networks in adults with autism spectrum disorder. Brain. 2016;139(2):616–630. doi: 10.1093/brain/awv351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty DP, Winter L. Free-response methodology: alternate analysis and a new observer-performance experiment. Radiology. 1990;174(3):873–881. doi: 10.1148/radiology.174.3.2305073. [DOI] [PubMed] [Google Scholar]
- 18.Chamard E, Lefebvre G, Lassonde M, Theoret H. Long-term abnormalities in the corpus callosum of female concussed athletes. Journal of neurotrauma. 2016;33(13):1220–1226. doi: 10.1089/neu.2015.3948. [DOI] [PubMed] [Google Scholar]
- 19.Chekir A, Hassas S, Descoteaux M, Côté M, Garyfallidis E, Oulebsir-Boumghar F. 3d-ssf: A bio-inspired approach for dynamic multi-subject clustering of white matter tracts. Computers in Biology and Medicine. 2017 doi: 10.1016/j.compbiomed.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Chuang TC, Wu MT, Huang SP, Weng MJ, Yang P. Diffusion tensor imaging study of white matter fiber tracts in adolescent attention-deficit/hyperactivity disorder. Psychiatry Research: Neuroimaging. 2013;211(2):186–187. doi: 10.1016/j.pscychresns.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Ciccarelli O, Catani M, Johansen-Berg H, Clark C, Thompson A. Diffusion-based tractography in neurological disorders: concepts, applications, and future developments. The Lancet Neurology. 2008;7(8):715–727. doi: 10.1016/S1474-4422(08)70163-7. [DOI] [PubMed] [Google Scholar]
- 22.Colby JB, Soderberg L, Lebel C, Dinov ID, Thompson PM, Sowell ER. Along-tract statistics allow for enhanced tractography analysis. NeuroImage. 2012;59(4):3227–3242. doi: 10.1016/j.neuroimage.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper M, Thapar A, Jones DK. ADHD severity is associated with white matter microstructure in the subgenual cingulum. NeuroImage: Clinical. 2015;7:653–660. doi: 10.1016/j.nicl.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corouge I, Fletcher PT, Joshi S, Gouttard S, Gerig G. Fiber tract-oriented statistics for quantitative diffusion tensor MRI analysis. Medical image analysis. 2006;10(5):786–798. doi: 10.1016/j.media.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Davatzikos C. Why voxel-based morphometric analysis should be used with great caution when characterizing group differences. NeuroImage. 2004;23(1):17–20. doi: 10.1016/j.neuroimage.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 26.de Schotten MT, Bizzi A, Dell’Acqua F, Allin M, Walshe M, Murray R, Williams SC, Murphy DG, Catani M, et al. Atlasing location, asymmetry and inter-subject variability of white matter tracts in the human brain with MR diffusion tractography. Neuroimage. 2011;54(1):49–59. doi: 10.1016/j.neuroimage.2010.07.055. [DOI] [PubMed] [Google Scholar]
- 27.Dennis EL, Jahanshad N, McMahon KL, de Zubicaray GI, Martin NG, Hickie IB, Toga AW, Wright MJ, Thompson PM. Development of brain structural connectivity between ages 12 and 30: a 4-Tesla diffusion imaging study in 439 adolescents and adults. NeuroImage. 2013;64:671–684. doi: 10.1016/j.neuroimage.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domeniconi C, Peng J, Gunopulos D. Locally adaptive metric nearest-neighbor classification. IEEE transactions on pattern analysis and machine intelligence. 2002;24(9):1281–1285. [Google Scholar]
- 29.Dougherty CC, Evans DW, Myers SM, Moore GJ, Michael AM. A comparison of structural brain imaging findings in autism spectrum disorder and attention-deficit hyperactivity disorder. Neuropsychology review. 2016;26(1):25–43. doi: 10.1007/s11065-015-9300-2. [DOI] [PubMed] [Google Scholar]
- 30.Dramsdahl M, Westerhausen R, Haavik J, Hugdahl K, Plessen KJ. Adults with attention-deficit/hyperactivity disordera diffusion-tensor imaging study of the corpus callosum. Psychiatry Research: Neuroimaging. 2012;201(2):168–173. doi: 10.1016/j.pscychresns.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Duffy FH, Shankardass A, McAnulty GB, Als H. A unique pattern of cortical connectivity characterizes patients with attention deficit disorders: a large electroencephalographic coherence study. BMC medicine. 2017;15(1):51. doi: 10.1186/s12916-017-0805-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischl B. FreeSurfer. NeuroImage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fowlkes C, Belongie S, Chung F, Malik J. Spectral grouping using the nystrom method. IEEE transactions on pattern analysis and machine intelligence. 2004;26(2):214–225. doi: 10.1109/TPAMI.2004.1262185. [DOI] [PubMed] [Google Scholar]
- 34.Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, Beaulieu C. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cerebral cortex. 2009;19(3):524–536. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.González MC, Herrmann HJ, Kertész J, Vicsek T. Community structure and ethnic preferences in school friendship networks. Physica A: Statistical mechanics and its applications. 2007;379(1):307–316. [Google Scholar]
- 36.Goodlett CB, Fletcher PT, Gilmore JH, Gerig G. Group analysis of DTI fiber tract statistics with application to neurodevelopment. NeuroImage. 2009;45(1):S133–S142. doi: 10.1016/j.neuroimage.2008.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guevara M, Román C, Houenou J, Duclap D, Poupon C, Mangin JF, Guevara P. Reproducibility of superficial white matter tracts using diffusion-weighted imaging tractography. NeuroImage. 2017;147:703–725. doi: 10.1016/j.neuroimage.2016.11.066. [DOI] [PubMed] [Google Scholar]
- 38.Guevara P, Duclap D, Poupon C, Marrakchi-Kacem L, Fillard P, Le Bihan D, Leboyer M, Houenou J, Mangin JF. Automatic fiber bundle segmentation in massive tractography datasets using a multi-subject bundle atlas. NeuroImage. 2012;61(4):1083–1099. doi: 10.1016/j.neuroimage.2012.02.071. [DOI] [PubMed] [Google Scholar]
- 39.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS biology. 2008;6(7):e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagmann P, Kurant M, Gigandet X, Thiran P, Wedeen VJ, Meuli R, Thiran JP. Mapping human whole-brain structural networks with diffusion MRI. PloS one. 2007;2(7):e597. doi: 10.1371/journal.pone.0000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamilton LS, Levitt JG, ONeill J, Alger JR, Luders E, Phillips OR, Caplan R, Toga AW, McCracken J, Narr KL. Reduced white matter integrity in attention-deficit hyperactivity disorder. Neuroreport. 2008;19(17):1705. doi: 10.1097/WNR.0b013e3283174415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hau J, Sarubbo S, Perchey G, Crivello F, Zago L, Mellet E, Jobard G, Joliot M, Mazoyer BM, Tzourio-Mazoyer N, et al. Cortical terminations of the inferior fronto-occipital and uncinate fasciculi: anatomical stem-based virtual dissection. Frontiers in neuroanatomy. 2016:10. doi: 10.3389/fnana.2016.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm S. A simple sequentially rejective multiple test procedure. Scandinavian journal of statistics. 1979:65–70. [Google Scholar]
- 44.Holmes AP, Blair R, Watson J, Ford I. Nonparametric analysis of statistic images from functional mapping experiments. Journal of Cerebral Blood Flow & Metabolism. 1996;16(1):7–22. doi: 10.1097/00004647-199601000-00002. [DOI] [PubMed] [Google Scholar]
- 45.Hua K, Zhang J, Wakana S, Jiang H, Li X, Reich DS, Calabresi PA, Pekar JJ, van Zijl PC, Mori S. Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. Neuroimage. 2008;39(1):336–347. doi: 10.1016/j.neuroimage.2007.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingalhalikar M, Parker D, Bloy L, Roberts T, Verma R. Diffusion based abnormality markers of pathology: toward learned diagnostic prediction of ASD. NeuroImage. 2011;57(3):918–927. doi: 10.1016/j.neuroimage.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ingalhalikar M, Smith A, Parker D, Satterthwaite TD, Elliott MA, Ruparel K, Hakonarson H, Gur RE, Gur RC, Verma R. Sex differences in the structural connectome of the human brain. Proceedings of the National Academy of Sciences. 2014;111(2):823–828. doi: 10.1073/pnas.1316909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jahanshad N, Aganj I, Lenglet C, Joshi A, Jin Y, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, et al. Sex differences in the human connectome: 4-tesla high angular resolution diffusion imaging (hardi) tractography in 234 young adult twins. Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on. IEEE; 2011. pp. 939–943. [Google Scholar]
- 49.Jin Y, Shi Y, Zhan L, Gutman BA, de Zubicaray GI, McMahon KL, Wright MJ, Toga AW, Thompson PM. Automatic clustering of white matter fibers in brain diffusion mri with an application to genetics. NeuroImage. 2014;100:75–90. doi: 10.1016/j.neuroimage.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones DK, Cercignani M. Twenty-five pitfalls in the analysis of diffusion mri data. NMR in Biomedicine. 2010;23(7):803–820. doi: 10.1002/nbm.1543. [DOI] [PubMed] [Google Scholar]
- 51.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22(18):2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lefranc S, Roca P, Perrot M, Poupon C, Le Bihan D, Mangin JF, Rivière D. Groupwise connectivity-based parcellation of the whole human cortical surface using watershed-driven dimension reduction. Medical image analysis. 2016;30:11–29. doi: 10.1016/j.media.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Lauer KK, Ward D, Roberts C, Liu S, Suneeta G, Rohloff R, Gross W, Xu Z, Chen G, Binder J, Li S-J, Hudetz AG. Fine-grained parcellation of brain connectivity improves differentiation of states of consciousness during graded propofol sedation. Brain Connectivity. 2017 doi: 10.1089/brain.2016.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moberts B, Vilanova A, Van Wijk JJ. VIS 05. IEEE Visualization. IEEE; 2005. Evaluation of fiber clustering methods for diffusion tensor imaging; pp. 65–72. [Google Scholar]
- 55.Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziotta J. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40(2):570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moyer D, Gutman BA, Faskowitz J, Jahanshad N, Thompson PM. Continuous representations of brain connectivity using spatial point processes. Medical Image Analysis. 2017 doi: 10.1016/j.media.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagel BJ, Bathula D, Herting M, Schmitt C, Kroenke CD, Fair D, Nigg JT. Altered white matter microstructure in children with attention-deficit/hyperactivity disorder. Journal of the American Academy of Child & Adolescent Psychiatry. 2011;50(3):283–292. doi: 10.1016/j.jaac.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Human brain mapping. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ning L, Westin CF, Rathi Y. Estimating diffusion propagator and its moments using directional radial basis functions. IEEE Transactions on Medical Imaging. 2015;34(10):2058–2078. doi: 10.1109/TMI.2015.2418674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Norton I, Essayed WI, Zhang F, Pujol S, Yarmarkovich A, Golby AJ, Kindlmann G, Wasserman D, Estepar RSJ, Rathi Y, et al. SlicerDMRI: Open source diffusion MRI software for brain cancer research. Cancer Research. 2017;77(21):e101–e103. doi: 10.1158/0008-5472.CAN-17-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Conaill CR, Malisza KL, Buss JL, Bolster RB, Clancy C, de Gervai PD, Chudley AE, Longstaffe S. Visual search for feature conjunctions: an fMRI study comparing alcohol-related neurodevelopmental disorder (ARND) to ADHD. Journal of neurodevelopmental disorders. 2015;7(1):10. doi: 10.1186/s11689-015-9106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Donnell L, Wells W, Golby A, Westin C-F. Unbiased groupwise registration of white matter tractography. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI); 2012. pp. 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Donnell L, Westin C. Automatic tractography segmentation using a high-dimensional white matter atlas. IEEE Transactions on Medical Imaging. 2007;26(11):1562–1575. doi: 10.1109/TMI.2007.906785. [DOI] [PubMed] [Google Scholar]
- 64.O’Donnell LJ, Suter Y, Rigolo L, Kahali P, Zhang F, Norton I, Albi A, Olubiyi O, Meola A, Essayed WI, et al. Automated white matter fiber tract identification in patients with brain tumors. NeuroImage: Clinical. 2017;13:138–153. doi: 10.1016/j.nicl.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Donnell LJ, Westin CF, Golby AJ. Tract-based morphometry for white matter group analysis. NeuroImage. 2009;45(3):832–844. doi: 10.1016/j.neuroimage.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oishi K, Faria A, Jiang H, Li X, Akhter K, Zhang J, Hsu JT, Miller MI, van Zijl PC, Albert M, Lyketsos CG, Woods R, Toga AW, Pike GB, Rosa-Neto P, Evans A, Mazziotta J, Mori S. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer’s disease participants. Neuroimage. 2009;46(2):486–499. doi: 10.1016/j.neuroimage.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Özarslan E, Koay CG, Shepherd TM, Komlosh ME, Irfanoğlu MO, Pierpaoli C, Basser PJ. Mean apparent propagator (MAP) MRI: a novel diffusion imaging method for mapping tissue microstructure. NeuroImage. 2013;78:16–32. doi: 10.1016/j.neuroimage.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palla G, Derényi I, Farkas I, Vicsek T. Uncovering the over-lapping community structure of complex networks in nature and society. Nature. 2005;435(7043):814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 69.Pastura G, Doering T, Gasparetto EL, Mattos P, Arauújo AP. Exploratory analysis of diffusion tensor imaging in children with attention deficit hyperactivity disorder: evidence of abnormal white matter structure. ADHD Attention Deficit and Hyperactivity Disorders. 2016;8(2):65–71. doi: 10.1007/s12402-015-0185-y. [DOI] [PubMed] [Google Scholar]
- 70.Pavuluri MN, Yang S, Kamineni K, Passarotti AM, Srinivasan G, Harral EM, Sweeney JA, Zhou XJ. Diffusion tensor imaging study of white matter fiber tracts in pediatric bipolar disorder and attention-deficit/hyperactivity disorder. Biological psychiatry. 2009;65(7):586–593. doi: 10.1016/j.biopsych.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pecheva D, Yushkevich P, Batalle D, Hughes E, Aljabar P, Wurie J, Hajnal JV, Edwards AD, Alexander DC, Counsell SJ, Zhang H. A tract-specific approach to assessing white matter in preterm infants. NeuroImage. 2017 doi: 10.1016/j.neuroimage.2017.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pérez-Iglesias R, Tordesillas-Gutiérrez D, Barker GJ, McGuire PK, Roiz-Santiañez R, Mata I, de Lucas EM, Quintana F, Vazquez-Barquero JL, Crespo-Facorro B. White matter defects in first episode psychosis patients: a voxelwise analysis of diffusion tensor imaging. NeuroImage. 2010;49(1):199–204. doi: 10.1016/j.neuroimage.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 73.Premachandran V, Kakarala R. Consensus of k-nns for robust neighborhood selection on graph-based manifolds. IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2013. pp. 1594–1601. [Google Scholar]
- 74.Pujol S, Wells W, Pierpaoli C, Brun C, Gee J, Cheng G, Vemuri B, Commowick O, Prima S, Stamm A, Goubran M, Khan A, Peters T, Neher P, Maier-Hein KH, Shi Y, Tristan-Vega A, Veni G, Whitaker R, Styner M, Westin CF, Gouttard S, Norton I, Chauvin L, Mamata H, Gerig G, Nabavi A, Golby A, Kikinis R. The DTI challenge: toward standardized evaluation of diffusion tensor imaging tractography for neurosurgery. Journal of Neuroimaging. 2015;25(6):875–882. doi: 10.1111/jon.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raffelt DA, Smith RE, Ridgway GR, Tournier JD, Vaughan DN, Rose S, Henderson R, Connelly A. Connectivity-based fixel enhancement: Whole-brain statistical analysis of diffusion MRI measures in the presence of crossing fibres. Neuroimage. 2015;117:40–55. doi: 10.1016/j.neuroimage.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raffelt DA, Tournier JD, Smith RE, Vaughan DN, Jackson G, Ridgway GR, Connelly A. Investigating white matter fibre density and morphology using fixel-based analysis. Neuroimage. 2017;144:58–73. doi: 10.1016/j.neuroimage.2016.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rathi Y, Gagoski B, Setsompop K, Michailovich O, Grant PE, Westin C-F. Diffusion propagator estimation from sparse measurements in a tractography framework. International Conference on Medical Image Computing and Computer-Assisted Intervention (MIC-CAI); 2013. pp. 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rathi Y, Michailovich O, Setsompop K, Bouix S, Shenton M, Westin C-F. Sparse multi-shell diffusion imaging. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI); Springer; 2011. pp. 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ratnarajah N, Rifkin-Graboi A, Fortier MV, Chong YS, Kwek K, Saw SM, Godfrey KM, Gluckman PD, Meaney MJ, Qiu A. Structural connectivity asymmetry in the neonatal brain. NeuroImage. 2013;75:187–194. doi: 10.1016/j.neuroimage.2013.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reddy CP, Rathi Y. Joint multi-fiber NODDI parameter estimation and tractography using the unscented information filter. Frontiers in neuroscience. 2016;10:166. doi: 10.3389/fnins.2016.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rotarska-Jagiela A, Oertel-Knoechel V, DeMartino F, van de Ven V, Formisano E, Roebroeck A, Rami A, Schoenmeyer R, Haenschel C, Hendler T, Maurer K, Vogeley K, Linden DE. Anatomical brain connectivity and positive symptoms of schizophrenia: a diffusion tensor imaging study. Psychiatry Research: Neuroimaging. 2009;174(1):9–16. doi: 10.1016/j.pscychresns.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 82.Samartzis L, Dima D, Fusar-Poli P, Kyriakopoulos M. White matter alterations in early stages of schizophrenia: a systematic review of diffusion tensor imaging studies. Journal of Neuroimaging. 2014;24(2):101–110. doi: 10.1111/j.1552-6569.2012.00779.x. [DOI] [PubMed] [Google Scholar]
- 83.Setsompop K, Cohen-Adad J, Gagoski B, Raij T, Yendiki A, Keil B, Wedeen VJ, Wald LL. Improving diffusion mri using simultaneous multi-slice echo planar imaging. NeuroImage. 2012;63(1):569–580. doi: 10.1016/j.neuroimage.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shaffer JJ, Ghayoor A, Long JD, Kim REY, Lourens S, O’donnell LJ, Westin CF, Rathi Y, Magnotta V, Paulsen JS, et al. Longitudinal diffusion changes in prodromal and early HD: Evidence of white-matter tract deterioration. Human brain mapping. 2017;38(3):1460–1477. doi: 10.1002/hbm.23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Silk TJ, Vance A, Rinehart N, Bradshaw JL, Cunnington R. White-matter abnormalities in attention deficit hyperactivity disorder: a diffusion tensor imaging study. Human brain mapping. 2009;30(9):2757–2765. doi: 10.1002/hbm.20703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith SM. Fast robust automated brain extraction. Human brain mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31(4):1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 88.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 89.Smith SM, Nichols TE. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. NeuroImage. 2009;44(1):83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- 90.Sølsnes AE, Sripada K, Yendiki A, Bjuland KJ, Østgård HF, Aanes S, Grunewaldt KH, Løhaugen GC, Eikenes L, Håberg AK, Rimol LM, Skranes J. Limited microstructural and connectivity deficits despite subcortical volume reductions in school-aged children born preterm with very low birth weight. NeuroImage. 2016;130:24–34. doi: 10.1016/j.neuroimage.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 91.Sotiropoulos SN, Zalesky A. Building connectomes using diffusion mri: Why, how and but. NMR in Biomedicine. 2017 doi: 10.1002/nbm.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sporns O, Betzel RF. Modular brain networks. Annual review of psychology. 2016;67:613–640. doi: 10.1146/annurev-psych-122414-033634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Svatkova A, Nestrasil I, Rudser K, Goldenring Fine J, Bledsoe J, Semrud-Clikeman M. Unique white matter microstructural patterns in ADHD presentationsa diffusion tensor imaging study. Human brain mapping. 2016;37(9):3323–3336. doi: 10.1002/hbm.23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taquet M, Scherrer B, Commowick O, Peters J, Sahin M, Macq B, Warfield SK. Registration and analysis of white matter group differences with a multi-fiber model. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI); Springer; 2012. pp. 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van den Heuvel MP, Stam CJ, Boersma M, Pol HH. Small-world and scale-free organization of voxel-based resting-state functional connectivity in the human brain. NeuroImage. 2008;43(3):528–539. doi: 10.1016/j.neuroimage.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 96.Van Essen DC. A population-average, landmark-and surface-based (pals) atlas of human cerebral cortex. Neuroimage. 2005;28(3):635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- 97.van Ewijk H, Heslenfeld DJ, Zwiers MP, Buitelaar JK, Oosterlaan J. Diffusion tensor imaging in attention deficit/hyperactivity disorder: a systematic review and meta-analysis. Neuroscience & Biobehavioral Reviews. 2012;36(4):1093–1106. doi: 10.1016/j.neubiorev.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 98.Vance A, Silk T, et al. Right parietal dysfunction in children with attention deficit hyperactivity disorder, combined type: a functional MRI study. Molecular psychiatry. 2007;12(9):826–832. doi: 10.1038/sj.mp.4001999. [DOI] [PubMed] [Google Scholar]
- 99.Veeramuthu V, Hariri F, Narayanan V, Tan LK, Ramli N, Ganesan D. Microstructural change and cognitive alteration in maxillofacial trauma and mild traumatic brain injury: A diffusion tensor imaging study. Journal of Oral and Maxillofacial Surgery. 2016;74(6):1197–e1. doi: 10.1016/j.joms.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 100.Wang D, Luo Y, Mok VC, Chu WC, Shi L. Tractography atlas-based spatial statistics: Statistical analysis of diffusion tensor image along fiber pathways. NeuroImage. 2016;125:301–310. doi: 10.1016/j.neuroimage.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 101.Wassermann D, Makris N, Rathi Y, Shenton M, Kikinis R, Kubicki M, Westin CF. The white matter query language: a novel approach for describing human white matter anatomy. Brain Structure and Function. 2016;221(9):4705–4721. doi: 10.1007/s00429-015-1179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamada K, Sakai K, Akazawa K, Sachiko Y, Nishimura T. MR tractography: a review of its clinical applications. Magnetic resonance in medical sciences. 2009;8(4):165–174. doi: 10.2463/mrms.8.165. [DOI] [PubMed] [Google Scholar]
- 103.Yeatman JD, Dougherty RF, Myall NJ, Wandell BA, Feldman HM. Tract profiles of white matter properties: automating fibertract quantification. PloS one. 2012;7(11):e49790. doi: 10.1371/journal.pone.0049790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yendiki A, Panneck P, Srinivasan P, Stevens A, Zöllei L, Augustinack J, Wang R, Salat D, Ehrlich S, Behrens T, et al. Automated probabilistic reconstruction of white-matter pathways in health and disease using an atlas of the underlying anatomy. Front Neuroinform. 2011;5(23):12–23. doi: 10.3389/fninf.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yushkevich PA, Zhang H, Simon TJ, Gee JC. Structure-specific statistical mapping of white matter tracts. NeuroImage. 2008;41(2):448–461. doi: 10.1016/j.neuroimage.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zalesky A, Cocchi L, Fornito A, Murray MM, Bullmore E. Connectivity differences in brain networks. Neuroimage. 2012;60(2):1055–1062. doi: 10.1016/j.neuroimage.2012.01.068. [DOI] [PubMed] [Google Scholar]
- 107.Zelnik-Manor L, Perona P. Advances in Neural Information Processing Systems. 2004. Self-tuning spectral clustering; pp. 1601–1608. [Google Scholar]
- 108.Zhang F, Kahali P, Suter Y, Norton I, Rigolo L, Savadjiev P, Song Y, Rathi Y, Cai W, Wells WM, III, Golby AJ, O’Donnell LJ. Automated connectivity-based groupwise cortical atlas generation: Application to data of neurosurgical patients with brain tumors for cortical parcellation prediction. IEEE International Symposium on Biomedical Imaging (ISBI); IEEE; 2017. pp. 774–777. [Google Scholar]
- 109.Zhang F, Norton I, Cai W, Song Y, Wells WM, III, O’Donnell LJ. Comparison between two white matter segmentation strategies: an investigation into white matter segmentation consistency. IEEE International Symposium on Biomedical Imaging (ISBI); IEEE; 2017. pp. 796–799. [Google Scholar]
- 110.Zhang F, Savadjiev P, Cai W, Song Y, Rathi Y, Tunç B, Parker D, Kapur T, Schultz RT, Makris N, et al. Whole brain white matter connectivity analysis using machine learning: An application to autism. NeuroImage. 2017 doi: 10.1016/j.neuroimage.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang F, Savadjiev P, Cai W, Song Y, Verma R, Westin C-F, O’Donnell LJ. Fiber clustering based white matter connectivity analysis for prediction of autism spectrum disorder using diffusion tensor imaging. IEEE International Symposium on Biomedical Imaging (ISBI); IEEE; 2016. pp. 564–567. [Google Scholar]
- 112.Zhang F, Wu W, Ning L, McAnulty G, Waber D, Gagoski B, Sarill K, Hamoda H, Song Y, Cai W, Rathi Y, O’Donnell LJ. Supra-threshold fiber cluster statistics for data-driven whole brain tractography analysis. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI); 2017. pp. 556–565. [Google Scholar]
- 113.Zhang W, Wang X, Zhao D, Tang X. Graph degree linkage: Agglomerative clustering on a directed graph. European Conference on Computer Vision (ECCV); 2012. pp. 428–441. [Google Scholar]
- 114.Zimmerman-Moreno G, Ben Bashat D, Artzi M, Nefussy B, Drory V, Aizenstein O, Greenspan H. Whole brain fiber-based comparison (FBC)–A tool for diffusion tensor imaging-based cohort studies. Human brain mapping. 2016;37(2):477–490. doi: 10.1002/hbm.23043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.