Abstract
Vaccine control and prevention of porcine reproductive and respiratory syndrome (PRRS), the most important disease of swine, is difficult to achieve. However, the discovery of broadly neutralizing antibody activity against porcine reproductive and respiratory syndrome virus (PRRSV) under typical field conditions opens the door to new immunologic approaches for robust protection. We show here that passive administration of purified immunoglobulins with neutralizing antibodies reduced PRRSV2 infection by up to 96%, and PRRSV1 infection by up to 87%, whereas immune immunoglobulins lacking neutralizing activity had no effect on viral infection. Hence, immune competence of passive immunoglobulin transfer was associated specifically with antibody neutralizing activity. Current models of PRRSV infection implicate a minor envelope glycoprotein (GP) complex including GP2, GP3, and GP4, as critical to permissive cell infection. However, conserved peptides comprising the putative cell attachment structure did not attenuate neutralization or viral infection. The results show that immunological approaches aimed at induction of broadly neutralizing antibodies may substantially enhance immune protection against PRRSV. The findings further show that naturally occurring viral isolates are able to induce protective humoral immunity against unrelated PRRSV challenge, thus removing a major conceptual barrier to vaccine development.
Keywords: PRRSV, neutralizing antibodies, cross-protection, passive transfer, neutralizing epitope, swine
Introduction
Neutralizing antibodies are a powerful tool for protection of animals and humans against viral infections and the diseases they cause (1–3). Porcine reproductive and respiratory syndrome virus (PRRSV), which has caused the most important disease of swine worldwide since its emergence in the late 1980’s, is not reliably controlled by vaccination and often does not elicit strong neutralizing antibody responses (4–9). In addition to an apparently weak neutralizing antibody response, immune protection is perceived as difficult due to a high degree of genetic variation in the virus (10–14). Two major genotypes differ by around 40% at the nucleotide sequence level, with multiple lineages that vary in pairwise comparison by up to 30% within each genotype (10). Swine farms frequently experience outbreaks with multiple PRRSV strains in the lifetime of individual animals in the herd, further reinforcing the idea that cross-protective immunity against PRRSV is lacking.
It was previously shown that neutralizing antibodies administered to naïve animals prevented viremic infection by the homologous PRRSV strain the neutralizing antibodies were raised against (15–17). Although it was established that neutralizing antibodies could prevent infection, cross-neutralization remained a question, and application of the homologous protection concept in the field by live virulent virus inoculation (in which animals are inoculated with a virulent virus preparation isolated from a resident farm strain) has not resulted in consistent control. Recently, cross-reactive sera with broadly neutralizing activity to PRRSV have been described (18–20). In particular, sera from sows in herds exposed to one or more virulent type 2 viruses were shown to neutralize genetically diverse PRRSV strains, including both type 1 and type 2 PRRSV (18). It is therefore critical to determine if serum cross-neutralizing activity translates to cross-protection against diverse PRRSV strains in vivo to better understand the role of neutralizing antibodies in immunity to genetically diverse PRRSV.
Here, we show that neutralizing antibodies significantly reduce viremic infection to heterologous type 1 and type 2 PRRSV. Candidate linear peptide targets of neutralization in the minor envelope glycoproteins, GP2, GP3, and GP4, failed to interfere with neutralizing activity.
Results
PRRSV neutralizing activity of sow serum
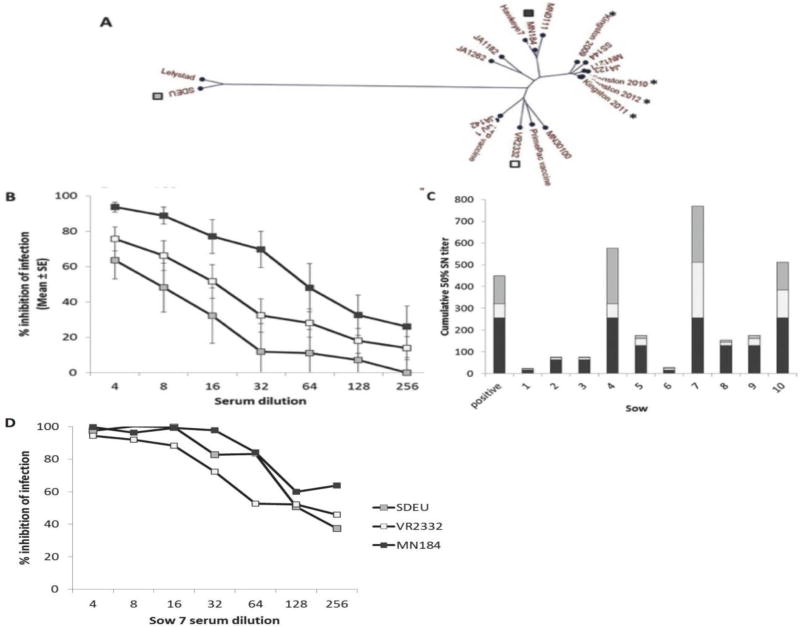
Ten sows at parity ≥6 (equivalent to ≥4 years of age) in a herd that experienced annual PRRSV outbreaks from 2009 to 2012 were tested for PRRSV cross-neutralizing activity. ORF5 sequencing showed that the outbreak strains were distinct from the neutralization test viruses, VR2332, MN184, and SDEU (Fig. 1A). Average 50% neutralizing titers were approximately 1/64 against strain MN184, 1/16 against VR2332, and 1/8 against SDEU, as shown in Figure 1B, and all sera were ELISA-positive for anti-N antibodies with no relationship to neutralizing titers (R2=0.0019). As shown in Figure 1C, 50% neutralizing titers of sows 4, 7, and 10 were consistently high against each of the three virus strains. A further three sows (5, 8, and 9) had less activity that was partially selective for MN184. Sows 2 and 3 had low neutralizing activity, with some cross-reactivity against genotype 2 strains, and sows 1 and 6 had low to negative neutralizing activity (Fig. 1C). The broadly neutralizing activity of sow 7 was high against all three test viruses and was equivalent against SDEU and MN184 (Fig. 1D).
Figure 1.
PRRSV broadly neutralizing activity in sow serum. (A) ORF5 phylogeny of diverse PRRSV isolates. Herd isolates from 2009 –2012 are marked with an asterisk. Viruses representing maximal PRRSV diversity used in neutralizing assays are marked with squares. (B) Comparative neutralizing activity of 10 sows against 3 diverse PRRSV strains. (C) Cumulative 50% neutralizing titers against 3 diverse PRRSV strains for the 10 individual animals. Positive control was a sow from another herd previously tested for neutralizing activity. (D) Sow 7 neutralizing activity in serum. In panels B, C and C the symbols are open squares, VR2332, gray squares, SDEU, and black squares, MN184.
Immunoglobulin isolation from serum
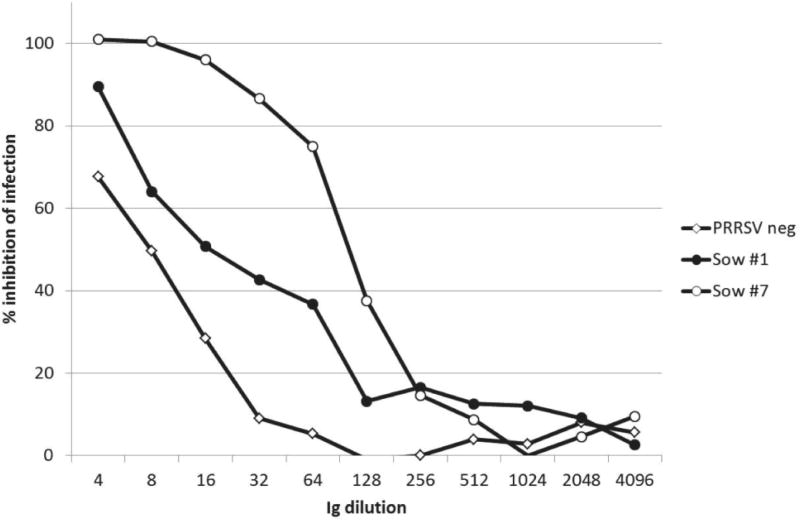
Immunoglobulins were isolated from serum of sow 1 (immune, non-neutralizing), sow 7 (immune, broadly neutralizing) and from pooled serum of PRRSV-negative animals. Ig products were present as anticipated in the caprylic acid supernatant, and absent in the saturated ammonium sulfate (SAS) supernatant. The final dialyzed Ig preparation showed reducing SDS-PAGE bands corresponding to IgG and IgM heavy and light chains. Under non-reducing conditions, bands of the expected size of intact Ig molecules were present. PRRSV-specific antibodies were detected in the purified immunoglobulins of the PRRSV-immune but not the PRRSV-negative animals by ELISA. Total recovery of purified Ig from the original sera was approximately 30–35%. Ig purification and concentration revealed that Sow 1 serum had a low level of neutralizing activity, which was enhanced by Ig enrichment (Fig. 2). Comparison of the three Ig preparations showed that sow 7 maintained high neutralizing activity, and that nonspecific inhibitory activity was present in the PRRSV-negative Ig at low dilutions (Fig. 2).
Figure 2.
Neutralizing activity of purified and concentrated immunoglobulins. ELISA-based neutralization assay against PRRSV VR2332 on MARC 145 cells for PRRSV-negative, PRRSV-immune non-neutralizing sow 1, and PRRSV-immune neutralizing sow 7.
Passive transfer study
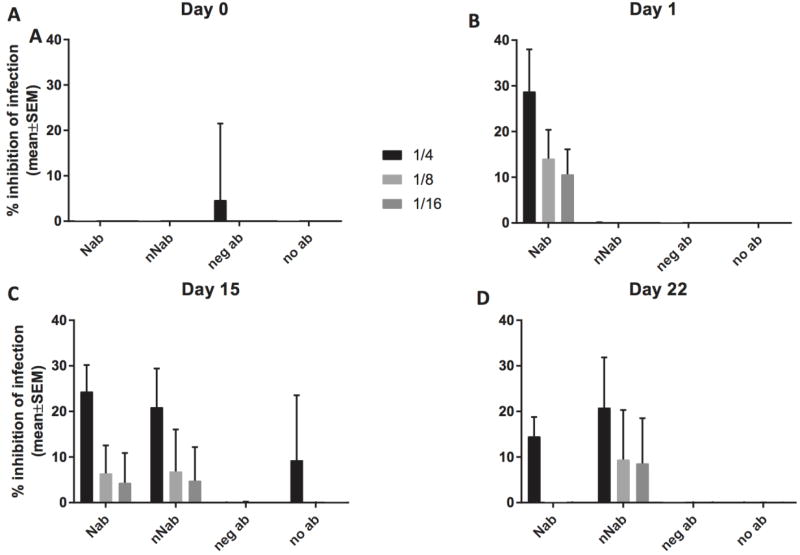
Recipient pigs were infused with 242 mg IgG to achieve an estimated circulating concentration of 0.52 mg/ml of blood, given a pig weight of 6.6±0.6 kg (mean±SD) and a blood volume equal to 7% of body weight. The calculated PRRSV infection inhibition activity, taken from tests on concentrated Ig, was 80% for neutralizing Ig. In vivo viral neutralizing activity 24 hours after Ig administration was present only in pigs that received neutralizing Ig (Fig. 3). Dilution analysis showed that, on day 1, an average 28% inhibitory activity was present in serum diluted 1/4 that decreased with further dilution. There was no detectable neutralizing activity present in pigs receiving immune Ig without neutralizing activity, non-immune Ig, or no Ig (Fig. 3).
Figure 3.
Neutralizing activity after immunoglobulin transfer. ELISA-based serum neutralizing assays against PRRSV VR2332 on MARC 145 cells from 32 animals in passive transfer of immunoglobulin study. Neutralizing activity at (A) time of Ig administration, (B) 24 hours post Ig administration (time of viral challenge), (C) 15 days, and (D) 22 days post Ig administration, for pigs receiving neutralizing (NAb n= 12), non-neutralizing (nNAb n= 12), PRRSV-negative (neg Ab n= 4) or no (no Ab n= 4) immunoglobulins.
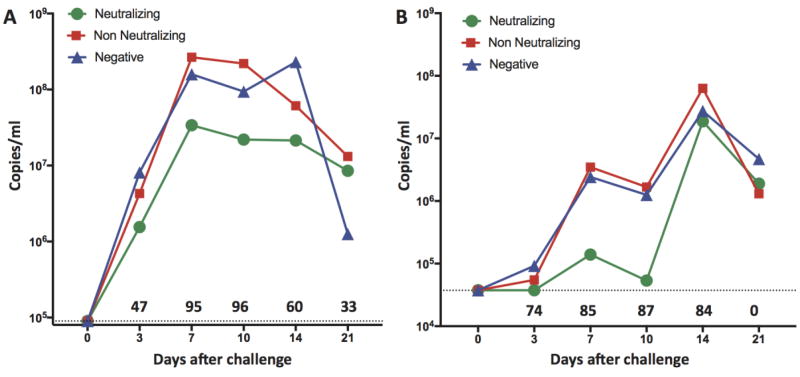
Viral challenge with SDEU and MN184 virus strains resulted in subclinical infection; pigs had no overt signs of respiratory disease. There were no differences in average daily weight gain among groups over the study duration. Administration of MN184 resulted in viremic infection that peaked at day 7, remained high through day 14, and showed equivalent kinetics in the control and non-neutralizing Ig-treated animals. Treatment with neutralizing Ig reduced peak viral loads 95 and 96% at days 7 and 14 compared to pigs not receiving neutralizing Ig (Fig. 4A). Significant reduction was present at the first sampling time and was maintained through day 14. Administration of SDEU resulted in viremic infection with identical kinetics in the control and non-neutralizing Ig-treated animals that peaked at day 14 (Fig. 4B). Pigs treated with neutralizing Ig had viral loads reduced by up to 87% on day 10 (Fig. 4B).
Figure 4.
Passive transfer of neutralizing antibodies reduces viremia. PRRSV RNA quantification from serum by RT-qPCR for (A) genotype 2 MN184, and (B) genotype 1 SDEU, in PRRSV-naïve pigs administered neutralizing immune Ig (n= 12), non-neutralizing immune Ig (n= 12), PRRSV-negative or no Ig (combined control groups, n= 8). Difference in mean viremia between groups receiving neutralizing Ig or not (pooled non-neutralizing and negative Ig) was compared by one-tailed Mann Whitney test of area under the curve. AUC day 0–21 for MN184, p=0.0145. AUC day 0–21 for SDEU, p=0.03. Numbers above the X-axis are percent reduction at the indicated day after challenge. Dotted lines indicate the RT-PCR assay sensitivity limit for MN184 (A) or SDEU (B).
When viral challenge occurred, neutralizing activity was present only in pigs receiving neutralizing Ig, at maximum levels of 20% and 40% (Fig. 3). At 2 and 3 weeks after viral infection, neutralizing activity was observed in pigs receiving PRRSV-positive Ig that lacked neutralizing activity (Fig. 3). The levels were equivalent to or greater than were observed in pigs that received neutralizing Ig, indicating that the higher levels of viral growth were sufficient to elicit a neutralizing immune response.
Targets of broadly cross-neutralizing antibodies
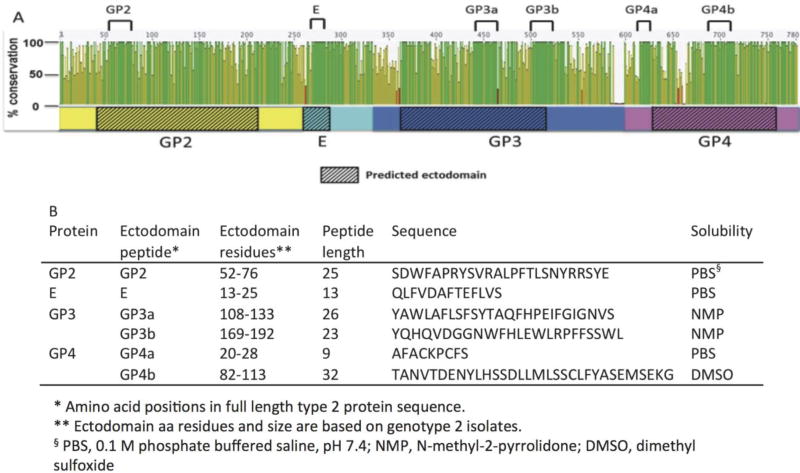
Since passive transfer of neutralizing antibodies provided cross-protective reductions in viremia, we hypothesized that conserved ectodomain regions in the minor envelope glycoproteins were the target. Thus, conserved ectodomain regions were identified by amino acid alignment of the minor envelope proteins. The alignment included 58 PRRSV of maximal diversity, and identified regions of high conservation (Fig. 5). The most conserved linear peptides regions were identified in GP2, E, GP3, and GP4 and synthesized (Fig. 5B).
Figure 5.
Conserved PRRSV minor envelope ectodomain peptides. (A) Alignment of 2 type 1 and 56 type 2 PRRSV sequences. Open reading frames 2, 2b, 3, and 4 sequences were concatenated and translated to yield GP2 (yellow), E (aqua), GP3 (blue), and GP4 (pink) as shown. Sequence conservation is expressed as a percentage, where 100% indicates amino acid sites are completely conserved. Predicted ectodomain regions are marked with hashed boxes. Ectodomain peptide sequences with maximal conservation selected are marked by black lines at top of the figure. (B) Conserved ectodomain peptide position, length and sequences. The solvent used for initial dilution to 1mg/ml is indicated for each peptide.
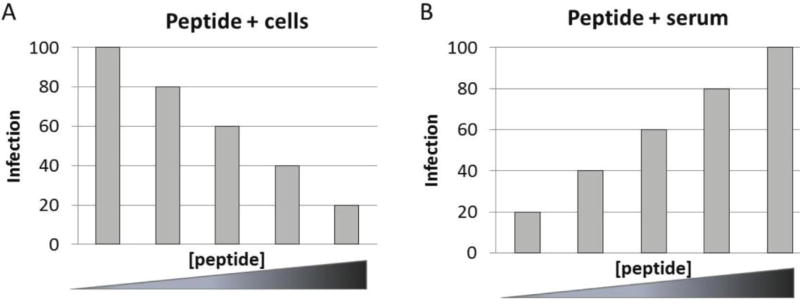
Ectodomain peptides from GP2, E, GP3 and GP4 were evaluated for evidence of neutralizing epitopes by two complementary approaches. First, peptides were added to cells before addition of virus, assuming that peptides which interacted with cellular CD163 would compete with virus in a dose-dependent manner to reduce infection. The second approach was to add peptides to serum, prior to addition of virus and before being placed onto cells. If peptides were the target of neutralizing antibodies, they would compete with virus in a dose-dependent manner for binding, inhibit neutralizing activity, and result in increased infection (Fig. 6).
Figure 6.
Expected outcome from peptide competition with neutralizing targets. (A) Depiction of predicted outcome of peptides added to cells before addition of virus. If peptide(s) bind cellular CD163, mimicking virus binding, they will compete with virus in a dose-dependent manner to result in reduced infection with increasing concentration of peptide. (B) Predicted outcome of peptides added to serum prior to addition of virus and infection of cells. If peptides bind neutralizing antibodies, they will compete with virus in a dose-dependent manner for binding and make more virus available for infection, i.e. they will inhibit neutralizing activity and result in increased infection with increasing concentration of peptide.
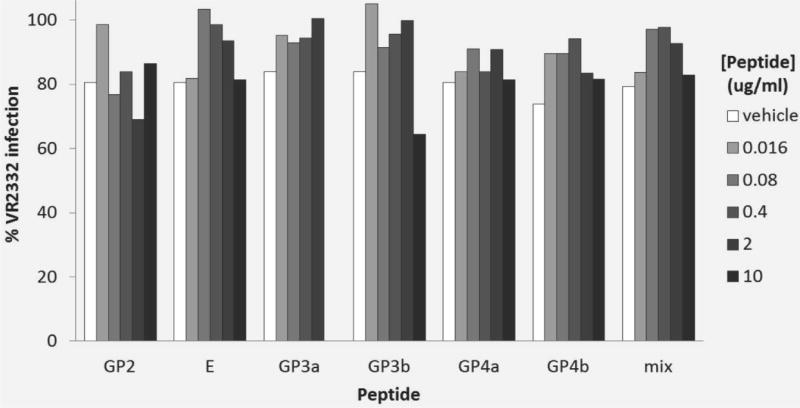
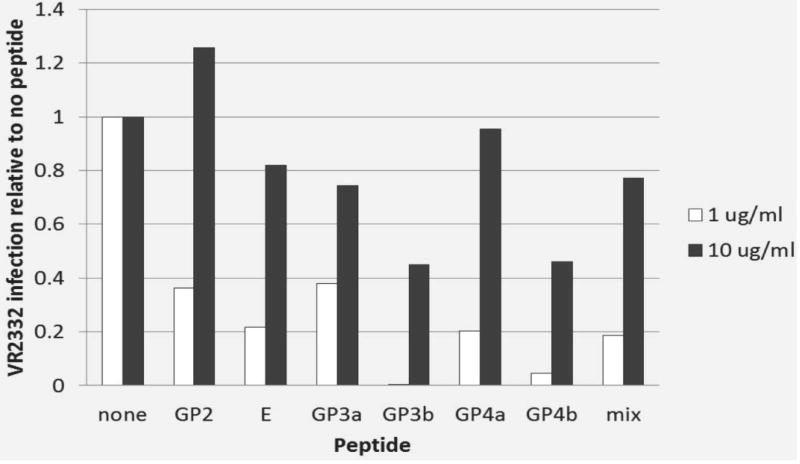
Incubation of cells with increasing concentrations of peptides before viral infection, either alone or as a mixture, did not inhibit infection in a dose-dependent manner beyond that resulting from the vehicle alone (Fig. 7). Incubation of peptides with neutralizing serum did not increase the level of viral infection at a dose of 1 µg/ml. Indeed, the level of infection was reduced substantially in all cases (Fig. 8). Results were more variable at the 10 µg/ml dose, but still without consistent evidence of a significant effect on blockade of neutralizing activity compared to the level of infection with no peptide addition (Fig. 8).
Figure 7.
Peptide competition for cellular receptors. Effect of increasing concentration of six synthetic peptides and mixture of the six peptides on infection of MARC 145 cells by PRRSV VR2332 (MOI of 0.5) relative to infected control wells with no peptides. Vehicle only controls are shown for reference.
Figure 8.
Peptide competition for neutralizing antibodies. Effect on PRRSV infection by two different peptide concentrations (1 and 10 µg/ml) for each of six synthetic peptides, and mixture of the six peptides and incubated with PRRSV-neutralizing serum (1/32 dilution) for 1 h. PRRSV VR2332 (MOI 0.5) was then incubated with the peptide-serum mixture for 1 h prior to inoculation of MARC 145 cells. Data are shown relative to viral growth in the absence of peptide addition.
Peptide ELISA was carried out to determine if anti-peptide antibodies were present in serum from sow 1 (immune, non-neutralizing), sow 7 (immune, broadly neutralizing), and sow 4 (immune, broadly neutralizing). Incubation of sow 1 and sow 7 serum at any serum dilution showed immunoreactivity. with none of the six ectodomain peptides. Interestingly, serum from sow 4, which showed high levels of broadly neutralizing antibodies, was highly reactive under all conditions, including with keyhole limpet hemocyanin and uncoated wells. However, reactivity was abolished by addition of 1 M guanidine HCl, showing that the interactions were nonspecific (21).
Discussion
Cross-genotype in vivo neutralization of PRRSV
The data here show for the first time that immunoglobulins with PRRSV neutralizing activity suppress unrelated, heterologous PRRSV growth in vivo, thereby establishing that cross-neutralizing antibodies may play a significant role in immunological mechanisms controlling PRRSV infection in pigs, and may be relevant to immune memory and vaccine efficacy. Inclusion of controls receiving non-neutralizing, immune Ig argues strongly that the passive inhibition of viral growth in vivo was due solely to the presence of neutralizing antibodies. The suppression of SDEU, a type 1 PRRSV, is particularly notable since type 1 PRRSV are separated by approximately 45% in genetic similarity from type 2 PRRSV, and immunological cross-reactivity has only rarely been demonstrated (22). Indeed, a recent proposal recommends separating them into distinct viral species due to their extensive differences (23). Suppression of viral growth in vivo was robust, especially for MN184. The amount of Ig used was predicted to inhibit viral growth by 80%, and the observed in vivo response was greater than 90%. Reduction in type 1 PRRSV strain SDEU viremia by >80% was significant as well. Since the in vitro SDEU-neutralizing activity in the source sow serum was equivalent to that of MN184, the difference between predicted neutralization in vitro and observed response in vivo for SDEU may have been due to technical differences in amounts of virus used in the neutralization assay, or to a possibly lower affinity of antibodies reacting with the genotype 1 virus in vivo. Anti-PRRSV neutralizing antibody half-life has been estimated at about 6–10 days (15–17, 24). However it is possible that the in vivo neutralization differences were due to individual variation in neutralizing antibody half-life between groups, particularly since SDEU viremia suppression was nearly complete in the first 10 days of infection.
Neutralizing Ig present in serum at time of viral challenge was about 0.5 mg/ml of blood, assuming complete absorption from the peritoneal administration and a 7% blood to body mass ratio. The Ig volume injected was minimized since systemic absorption from the peritoneal cavity is more efficient with lower Ig volumes (25). Immunoglobulins are absorbed from the peritoneal cavity to produce maximal serum titers by 24 hours post-administration (17, 25, 26). At this time, neutralizing activity was only observed in the group receiving neutralizing immune Ig. The level of neutralizing activity was lower than in previous studies investigating passive antibody protection, in which an approximately 20-fold higher dose of Ig (about 11 mg/kg) achieved complete suppression of viremia following homologous viral challenge (15, 16). Differences between the studies, besides analysis of homologous versus heterologous protection, included intensive hyperimmunization with Freund’s adjuvant versus natural exposure by virulent field viruses, and absence versus presence of control immune Ig lacking significant neutralizing activity. The findings show that cross-protection mediated by neutralizing antibodies can occur under field conditions.
It was noted that neutralizing activity was detected in purified, concentrated Igs isolated from immune serum lacking neutralizing activity, as shown in Figure 2. It is possible that neutralizing antibodies were produced during the humoral immune response to PRRSV infection, but were below the limit of detection until after Ig concentration. Alternatively, neutralizing activity may have been a nonspecific result of high Ig concentration. In either case, the passive transfer experiment showed that neutralizing activity was not detected following transfer and that, even if present, it was at a level that did not affect viral growth in vivo.
No antibody-dependent enhancement (ADE) of infection was observed in vivo with sub-neutralizing levels of control immune Ig or during the decline of neutralizing Ig titers to sub-neutralizing levels. Hence, these observations agree with other findings indicating that ADE is not a feature of PRRSV interaction with pigs (15, 27–30).
Broadly neutralizing antibodies in naturally infected pigs
Comparison among animals showed that the presence of broadly neutralizing activity of antibodies at substantial titers was a common feature of adult sows. Three of 10 sows had high neutralizing titers against type 1 SDEU even though there was no evidence of type 1 viral exposure in the herd. The same sows had high titers to type 2 VR2332, which was recovered in 1989 and is the parental virus of the attenuated Ingelvac MLV vaccine, which was never used in the herd. All sows had neutralizing titers to MN184, which appeared in 2000 in the U.S. and is not related to the outbreak viruses identified in the herd from 2009–2012. Broadly neutralizing activity, including neutralization of type 1 PRRSV strains, was recently reported in approximately 90% of sows from herds that had experienced virulent PRRSV outbreaks or were exposed repeatedly to virulent viruses as an immune-enhancing strategy (18). Thus, the presence of cross-neutralizing activity in commercial sows exposed to field viruses appears to be a common event. It remains to be determined whether this activity results from individual, broadly neutralizing immunoglobulin molecules to a conserved neutralizing epitope, or a particular combination of antibodies with specificity against different viral components that provides neutralization of diverse viral strains.
The conditions under which cross-neutralizing antibodies are produced are not known, but may involve multiple exposures to the same or different virus isolates. Consistent with this idea, the source herd experienced annual PRRS outbreaks for 4 consecutive years. Likewise, previously described sows with high titered, broadly neutralizing activity were found in herds with multiple exposures to virulent field viruses (18). In an experimental study, cross-neutralization was observed in animals exposed first to a PRRSV vaccine strain followed by homologous or heterologous virus challenge (31). However, the significance of the findings was not clear since the majority of data analyzed were below the neutralization assay cutoff. In another experimental study using a single PRRSV isolate, a longer duration of viremia, up to 42 days, was associated with increased breadth of neutralizing antibodies (32). Since cross-neutralization and titer data were not presented, it was not possible to further interpret the results. The findings also were unusual since significant neutralizing antibody responses are not commonly observed during viremic infection of young pigs (33–35). The animals were not subsequently challenged, so it is not known if the cross-neutralizing activity in serum was predictive of protection.
Age by itself is not sufficient for production of high neutralizing antibody titers, as increased age does not equate to increased SN titers (36). Duration of antigen exposure might facilitate development of increased breadth of neutralization. In humans, broadly neutralizing anti-HIV antibodies have been shown to appear after a period of years, during which mutations accrue throughout the variable region of the antibody molecule to increase affinity (37, 38). Nevertheless, the occurrence of highly active neutralizing antibodies found in humans is rare even though persistent infection is common (39). By contrast, high-titered, broadly neutralizing activity was frequently observed in our studies, whereas PRRSV infection, though prolonged, is not truly persistent (12).
Minor envelope protein targets of neutralizing antibody
For many years, the major envelope glycoprotein, GP5, was the focus of investigations aimed at elucidating PRRSV neutralization targets. It is the most abundant envelope protein in PRRSV, existing as a dimer with M protein, and homologues in related Arteriviruses (equine arteritis virus and lactate dehydrogenase elevating virus) have been shown to contain a major neutralizing epitope (40–45). It was previously thought that the GP5-M interaction with sialoadhesin (CD169) on macrophages was responsible for PRRSV infectivity (46–48). However, it was subsequently demonstrated that CD163 (scavenger receptor cysteine rich family) was sufficient for PRRSV infection, and explained permissivity of the simian MARC 145 cell line which express CD163, but not CD169 (49). Recent studies have demonstrated that CD169 is not necessary for infection in vivo with CD169 knockout pigs, and removal of N-glycans to expose the epitopes does not reduce viral infectivity (50, 51). GP5-M ectodomains appear to be relatively unimportant for PRRSV neutralization in genotype 1 and 2 PRRSV, although exceptions have been reported (19, 52–54). Consistent with these observations, similar neutralizing curves obtained in porcine alveolar macrophages and MARC 145 cells, which do and do not express sialoadhesin, support a mechanism of PRRSV neutralization that does not involve CD169 (18).
Minor envelope glycoproteins GP2, GP3 and GP4 form a heterotrimer, GP2 and GP4 have been shown to interact with the CD163 receptor, and they are the major determinant of viral tropism in cell culture (55, 56). It was recently demonstrated by gene editing of pigs that absence of CD163 conferred complete resistance to PRRSV infection compared with productive infection in control animals expressing wild type CD163, confirming that CD163 is necessary for infection in vivo (57). Based on broadly-neutralizing epitope targets identified in other RNA viruses such as influenza and HIV (58, 59), PRRSV cross-neutralizing targets would be anticipated to be conserved structural or functional regions critical for essential pathogen/host cell interactions. Currently, there is no structural information regarding PRRSV envelope proteins apart from that predicted from their amino acid sequences (60). Neutralization epitopes have been described in PRRSV structural proteins GP3, GP4, GP5 and M, however there are inconsistencies between the findings and more comprehensive studies are required to understand the nature of neutralizing epitopes and their contribution to a protective immune response (12, 61). Importantly, epitopes that induce or are susceptible to cross-neutralizing antibodies have not been evaluated.
Linear peptides were screened from highly conserved regions of minor envelope proteins for evidence of competition with the PRRSV receptor on MARC 145 cells and for inhibition of neutralizing antibody activity. However, under the conditions evaluated, none of the peptides evaluated competed with PRRSV for CD163 or neutralizing antibody binding either individually, or as a mixture. However, these findings are insufficient to dismiss GP2, GP3 and GP4 ectodomain regions as being important for cross-neutralization. Since the peptides were linear sequences, they may not represent the conformational nature of the peptides in the native protein structure, and conformational epitopes may be required for binding to neutralizing antibody targets. Also, epitopes may be non-contiguous, where antibody molecules bind residues that span different epitopes or even different proteins. Glycoproteins 2, 3 and 4 associate to form trimeric complexes in the viral envelope, which has been shown to interact with E protein, ORF5a protein, and the GP5-M dimer (55, 62). Therefore, the essential features of neutralizing epitopes may bind components of more than one protein in the complex. Hence, negative results from the experiments in this study using linear peptides do not exclude the possibility of minor envelope protein contributions to neutralization.
Physical evidence for structure of PRRSV minor envelope proteins is extremely limited, so the field is currently reliant on bioinformatic predictions (60). Amino acid residues on GP2 and GP4 critical for CD163 interactions are not yet known. This gap in knowledge limits the ability to predict regions that might be likely neutralizing targets based on structural interactions.
A potential confounding factor in this study is the use of polyclonal neutralizing Ig. Serum was treated to inactivate heat-labile fractions such as complement, but serum has many other components along with PRRSV-specific neutralizing antibodies. Even though the Ig fraction was highly purified, we cannot formally exclude that possibility that additional, critical components were co-purified. This possibility is unlikely since neutralizing antibodies are not known to require co-factors. Nevertheless, monoclonal cross-neutralizing antibodies, which have not yet been isolated for PRRSV, would facilitate determination of conserved, broadly neutralizable epitope targets.
Further studies are needed to identify viral targets of cross-neutralization against diverse PRRSV strains and to determine if these are conserved regions with essential roles in infections such as for structure or binding to cellular receptors. Only through elucidation of viral targets of cross-neutralization will it be possible to realize the potential for induction of cross-protective neutralizing antibodies through new immunization approaches, and to develop reliable correlates of neutralizing antibody-mediated cross-protection.
In summary, the findings confirm the existence of high titered, cross-neutralizing antibodies to PRRSV in the field, and experimental passive transfer if immune Ig proves that these antibodies are important for clinical protection. Variation in neutralizing breadth and activity exists between individuals in a relatively uniform population with respect to age, genetics, husbandry and viral exposure conditions. Neutralizing antibodies mediated cross-protection against diverse PRRSV strains across genotypes. The collection of serum and B lymphocytes from sows with cross-neutralizing and non-neutralizing activities described here provides a valuable resource for ongoing studies to identify features and mechanisms of antibody-mediated neutralization of PRRSV.
Materials and methods
Sow blood collection
Serum samples from ten sows of parity ≥6 were obtained from a herd that had experienced virulent virus outbreaks each of the previous 4 years. Blood samples were collected at the farm in serum separator tubes, and shipped to the University of Minnesota. Tubes were centrifuged at 3000 × g for 15 minutes, serum collected and kept at 4°C during testing, and subsequently stored at −20°C. Herd history was obtained as it pertained to PRRSV outbreaks, along with health records for the individual animals tested.
Three sows, selected for further evaluation based on their PRRSV neutralizing profile, were transported to the University of Minnesota and euthanized for collection of serum and lymphoid tissues. The study was approved by and conducted under the guidelines of the University of Minnesota Institutional Animal Care and Use Committee, protocol #1402-31319A.
Sera from pigs in a PRRSV ORF5a protein immunization study were used as negative controls in the virus neutralization assay (63). Pooled sera from three negative control animals that were neither immunized nor challenged with PRRSV were used as a negative control.
Cells and PRRSV isolates
Simian renal epithelial MARC 145 cells were cultured in MEM medium (Gibco, Grand Island NY) supplemented with sodium bicarbonate, non-essential amino acids, HEPES buffer (Sigma, St. Louis MO), gentamycin sulfate (Cellgro Mediatech, Manassas VA) and 10% FBS (Sigma, St. Louis MO).
PRRSV isolates VR2332 (GenBank U87392), MN184 (EF442777), and SD 01–08 (AY395080) (called ‘SDEU’ in this study) were propagated in MARC 145 cells. Infectious titers (TCID50) were determined by evaluation of cytopathic effect on MARC 145 cells in 96 well plates and calculated by the Reed & Muench method (64).
Viral ORF5 sequences were obtained from serum samples collected during outbreaks in the sow herd between 2009 and 2012 and submitted to the Veterinary Diagnostic Laboratory at the University of Minnesota. These sequences, along with reference strains used in the viral neutralization assays and further isolates representing PRRSV diversity, were aligned and subjected to phylogenetic tree construction in Geneious R6 version 6.1.7 (Biomatters Ltd., Auckland, New Zealand) Tree Builder using the Tamura-Nei neighbor-joining method with 100 bootstrap re-samplings and no outgroup to create a consensus tree.
PRRSV ELISA
PRRSV nucleocapsid (N) protein was expressed and purified for indirect ELISA as described (65, 66). Plates were coated with 100 ng N per well, and serum samples were diluted 1:50 in 5% non-fat dry milk (NFDM) in phosphate-buffered saline containing 0.05% Tween 20 (PBST). Detection antibody, horseradish peroxidase-conjugated goat anti-pig IgG (Bethyl Laboratories Inc. Montgomery TX), was used at a 1:100,000 dilution in 5% NFDM in PBST. Immune complexes were revealed by peroxidation of 3,3',5,5'- tetramethylbenzidine (TMB) substrate (KPL, Gaithersburg MD) for 15 min and stopped with 1 M phosphoric acid. Absorbance was read at 450 nm. Controls for the assay included known PRRSV-positive and negative sera.
Peptide Avidity ELISA
One hundred µl of 1 µM peptide solutions (ranging from 100 to 353 ng per well depending on size) in carbonate buffer, pH 9.6, were coated on ELISA plates overnight. Negative controls consisted of keyhole limpet hemocyanin coated at 150 ng/well under the same conditions, and uncoated wells. Wells were blocked and incubations carried out as described above. Test and control serum samples were serially diluted two-fold from 1/50 to 1/400 and incubated in duplicate. Avidity was determined by addition of 1 M guanidine HCl in the final wash step removing unbound serum antibodies as described (21).
ELISA-based PRRSV serum neutralization (SN) assay
SN assays were performed as described by Robinson et al. (18) with the following modifications. Two percent FBS was added to MEM culture media for serum sample and virus dilutions to improve adherence of the MARC 145 cell monolayer. All wells in each plate were scored by light microscopy to evaluate the condition of the monolayer at the conclusion of the SN assay. SN assays were performed across a range of multiplicity of infection (MOI) from 4 (128,000 viral TCID50/well) with 2 fold dilutions down to an MOI of 0.008 (250 TCID50/well). Subsequent assays were performed at an MOI of 0.5, equivalent to 16,000 viral TCID50/well. To detect neutralizing activity against the genotype 1 SD 01–08 (SDEU) strain, cells were incubated for 48 h after infection since this strain replicated more slowly in MARC 145 cells than did genotype 2 viruses (Eric Nelson, personal communication). 50% SN titer was determined by the reciprocal of the serum dilution when the inhibition of infection reached 50%.
Immunoglobulin (Ig) isolation from serum
Antibodies were isolated from PRRSV-immune serum of a sow with high cross-neutralizing activity and a PRRSV-immune sow negative for cross-neutralizing activity. PRRSV-negative control Ig was isolated from serum pooled from three pigs in a PRRSV ORF5a protein immunization study that were neither immunized nor challenged with PRRSV (63).
Total immunoglobulins were isolated by sequential precipitation with caprylic (octanoic) acid and ammonium sulfate (67). Briefly, serum was centrifuged at 10,000 × g for 15 min, filtered and heat-treated at 56°C for 30 min. An equal volume of acetate buffer (60 mM, pH 4.0) was added to 4°C serum, and pH adjusted to 4.5. Caprylic acid (Sigma-Aldrich, St. Louis, MO) was added 1:15 w/w and stirred for 30 min at room temperature. The mixture was then centrifuged at 10,000 × g for 30 min and the supernatant containing the Ig fraction was poured off and passed through a 70 µm mesh filter. Tris-HCl (1 M, pH 8.0) was used to adjust the pH to 7.4. Saturated ammonium sulfate (Sigma-Aldrich, St. Louis, MO), pH 7.4, was slowly added to serum samples with constant mixing to a final concentration of 40%, stirred at 4°C overnight, and centrifuged at 10,000 × g at 4°C for 30 min. Pellets were resuspended in PBS, pH 7.0, to 10 to 25% of the original volume. Purified Ig were dialyzed in PBS, pH 7.0, at 4°C in Spectra/Por 2 membrane dialysis tubing with molecular weight cutoff 12 – 14,000 (Spectrum Laboratories, Rancho Dominquez, CA).
Immunoglobulin concentrations were measured using a porcine IgG ELISA quantitation kit (Bethyl Laboratories, Inc., Montgomery TX). Antibody, standards and sample dilutions were in 5% NFDM in PBST and the stop solution was 1M phosphoric acid.
Protein samples with and without 5% β-mercaptoethanol to reduce disulfide bonds were subjected to denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in 4 to 20% gradient Mini-Protean Precast Gels (Bio-Rad Laboratories, Hercules CA) to evaluate purity. Bands were visualized with Imperial protein stain (Thermo Scientific, Rockford, IL), and destained in deionized water.
SN assays were performed on purified immunoglobulins as described for serum samples with two-fold dilutions.
Passive transfer experimental design
Thirty-two weaned pigs approximately 3 weeks of age were sourced from a PRRSV-negative breeding herd. Pigs were weighed and randomly assigned to seven groups upon arrival at the University of Minnesota College of Veterinary Medicine animal isolation facility (Supplemental Figure S1). Each group of pigs was housed in a separate room and fed a complete ration, with ad lib access to water and acclimated for 3 days.
On the day prior to viral challenge pigs were anesthetized with Telazol in xylazine by intramuscular injection. Blood was collected into serum separator tubes to test for PRRSV by RT-PCR and for SN. Purified neutralizing or non-neutralizing PRRSV immune Ig or PRRSV-negative Ig was injected via a 14 gauge catheter into the peritoneal cavity.
Twenty-four hours later, blood was collected from all pigs to evaluate the PRRSV-neutralizing activity in serum at time of viral challenge (day 0 with respect to challenge). At this time, pigs in groups 2–7 were infected by intramuscular inoculation in the neck of 1 × 105 TCID50 of PRRSV MN184 or SDEU in 1 ml tissue culture media supernatant. The intramuscular route was used due to its high reproducibility and sensitivity compared to intranasal administration (68). Uninfected animals were sham-inoculated with the same volume of tissue culture media intramuscularly. All animals were evaluated daily thereafter for clinical signs of illness, including fever, coughing/sneezing, lethargy, and anorexia. Blood samples were collected from all animals on days 3, 7, 10, and 14 after viral challenge. At 21 days after infection, pigs were weighed and anesthetized by intramuscular injection of Telazol/xylazine as described above. Blood was collected under anesthesia prior to euthanasia by intravenous barbiturate overdose. Necropsies were performed on all animals for evaluation of gross lung morphology. Tissues were kept at 4°C overnight and frozen to −20°C. Blood samples were collected in serum separator tubes, and were centrifuged to collect serum which was frozen at −20°C for further analysis.
Viremia was compared between the groups of interest to determine whether immune Igs were protective against infection, and whether they provided cross-protection against different virus strains compared to animals receiving control Igs. The number of pigs needed per group was estimated by a power analysis based on mean level of viremia and variation between individuals estimated from previous studies (63). Calculations were based on a 2 log reduction in viremia, (from 106 RNA copies/ml of serum to 104 with ±5×103 standard deviation in the control group, and ±5×105 in the Ig groups). To achieve 95% power at 1% alpha level, we calculated that each group would require a minimum of 3 animals (69). To ensure that valid results would be obtained in case of animals requiring early endpoints, or wider variation between individuals, the study was conducted with 4 or 5 animals per group. In addition, the design allowed for combining groups for statistical analysis based on Ig status. Details of treatment groups is shown in Table 1.
Table 1.
Treatment groups for passive transfer of immunoglobulins.
| Group | Category | Viral Challenge |
Treatment (number of pigs) |
|---|---|---|---|
| 1 | Uninfected negative control | None | Neutralizing Ig (n=2) |
| Non-neutralizing Ig (n=2) | |||
| 2 | PRRSV immune neutralizing Ig | SDEU | Neutralizing Ig (n=5) |
| 3 | PRRS immune non-neutralizing Ig | SDEU | Non-neutralizing (n=5) |
| 4 | Infection positive control | SDEU | Negative Ig (n=2) |
| No Ig (n=2) | |||
| 5 | PRRSV immune neutralizing Ig | MN184 | Neutralizing Ig (n=5) |
| 6 | PRRS immune non-neutralizing Ig | MN184 | Non-neutralizing (n=5) |
| 7 | Infection positive control | MN184 | Negative Ig (n=2) |
| No Ig (n=2) |
The study was approved by and conducted under the guidelines of the University of Minnesota Institutional Animal Care and Use Committee and Institutional Biosafety Committee (protocol #1310-31023A).
Viral RNA isolation and reverse transcription quantitative PCR (RT-qPCR)
Viral RNA was isolated from serum using a QIAmp Viral RNA Mini kit (Qiagen, Valencia CA), eluted into 50 µl of RNase-free water and stored at −80 °C. Complementary cDNA was synthesized with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad CA) with random hexamer primers. Primer sequences TCA ACT GTC CCA GTT GCT GG (forward) and AAA TGT GGC TTC TCA GGC TTT T (reverse) amplified a 96 bp ORF7 fragment for genotype 1 PRRSV. Primer sequences AAC CAC GCA TTT GTC GTC (forward) and TGG CAC AGC TGA TTG ACT GG (reverse) amplified a 198 bp ORF6–7 fragment for genotype 2 PRRSV. PCR reactions were performed in a total volume of 20 µl, containing 5 µl of cDNA, 10 µl of SYBR Green PCR Master Mix (PerfeCTa SYBR Green FastMix, Quanta Biosciences, Gaithersburg MD) and 200 ng of each primer. Reactions were run in a Stratagene Mx3000P thermal cycler (Agilent Technologies, Inc. Santa Clara CA), with activation at 95°C for 1 minute, 35 cycles of denaturation at 95°C for 3 seconds and annealing/extension at 60°C for 25 seconds, followed by a dissociation step. All samples were run in duplicate.
PRRSV was quantified using the standard curve method with gel purified PCR products from MN184 and SDEU prepared as described above (70). The cDNA concentration was determined by spectrophotometry and number of copies of the template was calculated as described (71). Serial 10-fold dilutions were used to construct a standard curve, ranging from >106 to <1 copy per reaction. Identification of positive samples was determined by the presence of a quantification cycle value and thermal denaturation analysis. Negative controls without cDNA were used in all PCR plates.
Statistical analysis for differences in mean levels of viremia between groups at each timepoint was conducted by the Wilcoxon Rank Sum Test (Mann-Whitney U Test). Conservative p values of ≤0.05 were considered significant. Area under the curve analysis to determine percent reduction in total viremia was performed in GraphPad Prism (GraphPad Software, La Jolla, CA).
Identification and synthesis of conserved ectodomain peptides
Whole genome sequences from 58 unique PRRSV strains, including genotypes 1 and 2 were obtained from GenBank. Sequences of ORF2a, ORF2b, ORF3 and ORF4, representing GP2, E, GP3, and GP4, respectively, were concatenated for each PRRSV strain, aligned using Geneious R6 version 6.1.7 (Biomatters Ltd., Auckland, New Zealand) and translated to identify regions of amino acid conservation. Predicted ectodomain regions were identified based on (60). The most conserved linear peptides regions were identified within the ectodomain fragments.
Six linear peptides, one each from GP2 and E, and two each from GP3 and GP4, were synthesized by GenScript (Piscataway, NJ). Based on testing at GenScript, peptides were solubilized to 1 mg/ml in either 0.1M PBS, pH 7.4 (GP2, E, GP4a), N-methyl-2-pyrrolidone (NMP) (GP3a, GP3b), or dimethyl sulfoxide (DMSO) (GP4b). Further dilutions from the 1 mg/ml stock were made in MEM containing 2% FBS.
Cellular receptor competition assay
Each of the 6 peptides, a mixture of the 6 peptides, and vehicle only controls were diluted to 10 µg/ml in MEM containing 2% FBS. Five-fold serial dilutions of the peptides were made in MEM 2% FBS to 0.016 µg/ml. Dilutions, and control wells with no peptides (containing MEM with 2% FBS) or vehicle only (at the equivalent dilution for the most concentrated condition of 10 µg/ml) were added to duplicate wells of MARC 145 cells and incubated at 37°C for 1 hour. PRRSV strain VR2332 was added at an MOI of 0.5 in the presence of peptides and incubated at 37°C for an hour. The peptide/virus mixture was decanted, cells were washed once with warmed PBS, and complete MEM with 10% FBS was added for an additional 23 hour incubation. Cells were washed with PBS and processed for the PRRSV neutralization assay.
Neutralizing serum competition assay
Peptides (each alone and a mixture of all 6, at a final concentration of 10 µg/ml and 1 µg/ml) were mixed with serum (1/32 dilution of PRRSV neutralizing serum, the same Ig concentration of negative serum, or no serum control) for 1 hour at 37°C. PRRSV VR2332 was added to an MOI of 0.5 and incubated for one hour, and the mixtures were transferred to cells. Plates, also containing untreated control monolayers, were incubated for 1 hour, and washed with PBS. After 23 hour, cells were washed with PBS, fixed, and permeabilized and processed as described to quantify PRRSV infection in cells.
Supplementary Material
Highlights.
Broadly neutralizing anti-PRRSV antibodies control PRRSV1 and PRRSV2 infection.
Non-neutralizing immune serum does not reduce PRRSV infection.
Conserved putative cell attachment structure peptides did not block infection.
Naturally occurring PRRSV induce protective humoral immunity against unrelated PRRSV.
Induction of broadly neutralizing antibodies may enhance immune protection.
Acknowledgments
Dr. Paul Yeske and Dr. Laura Bruner of Swine Vet Center, MN and Curt Froehle and Holden Farms provided access to sows, information on PRRSV outbreaks, disease and management, individual sow data and blood samples. Technical and bioinformatic assistance was provided by Kyra Martins, Cheryl Dvorak, Lucas dos Santos, Suzanne Stone, Xiong Wang, and Qinye Song. University of Minnesota Veterinary Diagnostic Laboratory staff assisted with animal procedures. Research was supported by National Pork Board Project #14-213, USDA National Needs Fellowship Grant #2008-38420-18726 for support of SRR, Agriculture and Food Research Initiative Competitive Grant no. 2016-67015-24928 from the USDA National Institute of Food and Agriculture, and NIH award T32 RR018719 for support of MCR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
MPM conceptualized the idea, acquired funding, and supervised and administered the project. SRR conducted the research, and was responsible for resources, validation, visualization and preparing the original draft. MCR provided resources, assisted in methodology and validation. KVM and DKG provided key methodology and resources. MPM, SRR, and MCR carried out review and editing of the manuscript.
The authors declare no conflicts of interest.
References
- 1.Burton DR. Antibodies, viruses and vaccines. Nat Rev Immunol. 2002;2(9):706–13. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 2.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337(6091):183–6. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plotkin SA. Correlates of protection induced by vaccination. Clinical and Vaccine Immunology. 2010;17(7):1055–65. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, et al. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q. 1991;13(3):121–30. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 5.Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, et al. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. 1992;4(2):117–26. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- 6.Martínez-Lobo FJ, Díez-Fuertes F, Simarro I, Castro JM, Prieto C. Porcine Reproductive and Respiratory Syndrome Virus isolates differ in their susceptibility to neutralization. Vaccine. 2011;29(40):6928–40. doi: 10.1016/j.vaccine.2011.07.076. [DOI] [PubMed] [Google Scholar]
- 7.Scortti M, Prieto C, Alvarez E, Simarro I, Castro JM. Failure of an inactivated vaccine against porcine reproductive and respiratory syndrome to protect gilts against a heterologous challenge with PRRSV. Vet Rec. 2007;161(24):809–13. [PubMed] [Google Scholar]
- 8.Lopez OJ, Osorio FA. Role of neutralizing antibodies in PRRSV protective immunity. Vet Immunol Immunopathol. 2004;102(3):155–63. doi: 10.1016/j.vetimm.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Geldhof MF, Vanhee M, Van Breedam W, Van Doorsselaere J, Karniychuk UU, Nauwynck HJ. Comparison of the efficacy of autogenous inactivated Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) vaccines with that of commercial vaccines against homologous and heterologous challenges. BMC veterinary research. 2012;8:182. doi: 10.1186/1746-6148-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brar MS, Shi M, Murtaugh MP, Leung FC. Evolutionary diversification of type 2 porcine reproductive and respiratory syndrome virus. J Gen Virol. 2015;96(Pt 7):1570–80. doi: 10.1099/vir.0.000104. [DOI] [PubMed] [Google Scholar]
- 11.Stadejek T, Stankevicius A, Murtaugh MP, Oleksiewicz MB. Molecular evolution of PRRSV in Europe: current state of play. Vet Microbiol. 2013;165(1–2):21–8. doi: 10.1016/j.vetmic.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 12.Murtaugh MP, Genzow M. Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS) Vaccine. 2011;29(46):8192–204. doi: 10.1016/j.vaccine.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Costers S, Lefebvre DJ, Van Doorsselaere J, Vanhee M, Delputte PL, Nauwynck HJ. GP4 of porcine reproductive and respiratory syndrome virus contains a neutralizing epitope that is susceptible to immunoselection in vitro. Arch Virol. 2010;155(3):371–8. doi: 10.1007/s00705-009-0582-7. [DOI] [PubMed] [Google Scholar]
- 14.Vu HL, Kwon B, Yoon KJ, Laegreid WW, Pattnaik AK, Osorio FA. Immune evasion of porcine reproductive and respiratory syndrome virus through glycan shielding involves both glycoprotein 5 as well as glycoprotein 3. J Virol. 2011;85(11):5555–64. doi: 10.1128/JVI.00189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez OJ, Oliveira MF, Garcia EA, Kwon BJ, Doster A, Osorio FA. Protection against porcine reproductive and respiratory syndrome virus (PRRSV) infection through passive transfer of PRRSV-neutralizing antibodies is dose dependent. Clinical and Vaccine Immunology. 2007;14(3):269–75. doi: 10.1128/CVI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osorio FA, Galeota JA, Nelson E, Brodersen B, Doster A, Wills R, et al. Passive transfer of virus-specific antibodies confers protection against reproductive failure induced by a virulent strain of porcine reproductive and respiratory syndrome virus and establishes sterilizing immunity. Virology. 2002;302(1):9–20. doi: 10.1006/viro.2002.1612. [DOI] [PubMed] [Google Scholar]
- 17.Yoon KJ, Wu LL, Zimmerman JJ, Hill HT, Platt KB. Antibody-dependent enhancement (ADE) of porcine reproductive and respiratory syndrome virus (PRRSV) infection in pigs. Viral Immunology. 1996;9(1):51–63. doi: 10.1089/vim.1996.9.51. [DOI] [PubMed] [Google Scholar]
- 18.Robinson SR, Li J, Nelson EA, Murtaugh MP. Broadly neutralizing antibodies against the rapidly evolving porcine reproductive and respiratory syndrome virus. Virus research. 2015;203:56–65. doi: 10.1016/j.virusres.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Trible BR, Popescu LN, Monday N, Calvert JG, Rowland RR. A single amino acid deletion in the matrix protein of porcine reproductive and respiratory syndrome virus confers resistance to a polyclonal swine antibody with broadly neutralizing activity. J Virol. 2015;89(12):6515–20. doi: 10.1128/JVI.03287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galliher-Beckley A, Li X, Bates JT, Madera R, Waters A, Nietfeld J, et al. Pigs immunized with Chinese highly pathogenic PRRS virus modified live vaccine are protected from challenge with North American PRRSV strain NADC-20. Vaccine. 2015;33(30):3518–25. doi: 10.1016/j.vaccine.2015.05.058. [DOI] [PubMed] [Google Scholar]
- 21.Dvorak CM, Akkutay-Yoldar Z, Stone SR, Tousignant SJ, Vannucci FA, Murtaugh MP. An indirect enzyme-linked immunosorbent assay for the identification of antibodies to Senecavirus A in swine. BMC veterinary research. 2017;13(1):50. doi: 10.1186/s12917-017-0967-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson EA, Christopher-Hennings J, Drew T, Wensvoort G, Collins JE, Benfield DA. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. Journal of clinical microbiology. 1993;31(12):3184–9. doi: 10.1128/jcm.31.12.3184-3189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn JH, Lauck M, Bailey AL, Shchetinin AM, Vishnevskaya TV, Bào Y, et al. Reorganization and expansion of the nidoviral family Arteriviridae. Arch Virol. 2016;161(3):755–68. doi: 10.1007/s00705-015-2672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senn MK, Yoon KJ, Zimmerman JJ, Thacker BJ. Decay of colostrum-derived antibodies to porcine reproductive and respiratory syndrome (PRRS) virus in neonatal swine nursing immune dams. Swine Research Report, 1998 Paper 52 [Internet] 1999 Available from: http://lib.dr.iastate.edu/cgi/viewcontent.cgi?article=1051&context=swinereports_1998.
- 25.Barrett JS, Wagner JG, Fisher SJ, Wahl RL. Effect of intraperitoneal injection volume and antibody protein dose on the pharmacokinetics of intraperitoneally administered IgG2a kappa murine monoclonal antibody in the rat. Cancer Res. 1991;51(13):3434–44. [PubMed] [Google Scholar]
- 26.Trautman R, Bennett CE. Relationship between virus neutralization and serum protection bioassays for IgG and IgM antibodies to foot-and-mouth disease virus. Journal of General Virology. 1979;42(3):457–66. doi: 10.1099/0022-1317-42-3-457. [DOI] [PubMed] [Google Scholar]
- 27.Okuda Y, Kuroda M, Ono M, Chikata S, Shibata I. Efficacy of vaccination with porcine reproductive and respiratory syndrome virus following challenges with field isolates in Japan. The Journal of veterinary medical science / the Japanese Society of Veterinary Science. 2008;70(10):1017–25. doi: 10.1292/jvms.70.1017. [DOI] [PubMed] [Google Scholar]
- 28.Martelli P, Gozio S, Ferrari L, Rosina S, De Angelis E, Quintavalla C, et al. Efficacy of a modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine in pigs naturally exposed to a heterologous European (Italian cluster) field strain: Clinical protection and cell-mediated immunity. Vaccine. 2009;27(28):3788–99. doi: 10.1016/j.vaccine.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Cano JP, Dee SA, Murtaugh MP, Pijoan C. Impact of a modified-live porcine reproductive and respiratory syndrome virus vaccine intervention on a population of pigs infected with a heterologous isolate. Vaccine. 2007;25(22):4382–91. doi: 10.1016/j.vaccine.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 30.Delputte PL, Meerts P, Costers S, Nauwynck HJ. Effect of virus-specific antibodies on attachment, internalization and infection of porcine reproductive and respiratory syndrome virus in primary macrophages. Vet Immunol Immunopathol. 2004;102(3):179–88. doi: 10.1016/j.vetimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Galliher-Beckley A, Pappan L, Trible B, Kerrigan M, Beck A, et al. Comparison of host immune responses to homologous and heterologous type II porcine reproductive and respiratory syndrome virus (PRRSV) challenge in vaccinated and unvaccinated pigs. Biomed Res Int. 2014;2014:416727. doi: 10.1155/2014/416727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Islam ZU, Bishop SC, Savill NJ, Rowland RRR, Lunney JK, Trible B, et al. Quantitative analysis of porcine reproductive and respiratory syndrome (PRRS) viremia profiles from experimental infection: a statistical modelling approach. PloS one. 2013;8(12) doi: 10.1371/journal.pone.0083567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molina RM, Cha SH, Chittick W, Lawson S, Murtaugh MP, Nelson EA, et al. Immune response against porcine reproductive and respiratory syndrome virus during acute and chronic infection. Vet Immunol Immunopathol. 2008;126(3–4):283–92. doi: 10.1016/j.vetimm.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Mateu E, Diaz I. The challenge of PRRS immunology. Vet J. 2008;177(3):345–51. doi: 10.1016/j.tvjl.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson EA, Christopher-Hennings J, Benfield DA. Serum immune responses to the proteins of porcine reproductive and respiratory syndrome (PRRS) virus. J Vet Diagn Invest. 1994;6(4):410–5. doi: 10.1177/104063879400600402. [DOI] [PubMed] [Google Scholar]
- 36.Klinge KL, Vaughn EM, Roof MB, Bautista EM, Murtaugh MP. Age-dependent resistance to Porcine reproductive and respiratory syndrome virus replication in swine. Virology journal. 2009;6:177. doi: 10.1186/1743-422X-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–76. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jardine JG, Sok D, Julien JP, Briney B, Sarkar A, Liang CH, et al. Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design. PLoS pathogens. 2016;12(8):e1005815. doi: 10.1371/journal.ppat.1005815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol. 2000;145(4):659–88. doi: 10.1007/s007050050662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mardassi H, Massie B, Dea S. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology. 1996;221(1):98–112. doi: 10.1006/viro.1996.0356. [DOI] [PubMed] [Google Scholar]
- 42.Meulenberg JJ, Petersen-den Besten A, De Kluyver EP, Moormann RJ, Schaaper WM, Wensvoort G. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology. 1995;206(1):155–63. doi: 10.1016/S0042-6822(95)80030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wissink EH, Kroese MV, van Wijk HA, Rijsewijk FA, Meulenberg JJ, Rottier PJ. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. Journal of Virology. 2005;79:12495–506. doi: 10.1128/JVI.79.19.12495-12506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faaberg KS, Balasuriya UB, Brinton MA, Gorbalenya AE, Leung FC, Nauwynck HJ, et al. Virus Taxonomy, Classification and Nomenclature of Viruses Ninth report of the International Committee on Taxonomy of Viruses [Internet] Oxford: Elsevier; 2011. Family Arteriviridae; pp. 796–805. [Google Scholar]
- 45.Wissink EH, van Wijk HA, Pol JM, Godeke GJ, van Rijn PA, Rottier PJ, et al. Identification of porcine alveolar macrophage glycoproteins involved in infection of porcine respiratory and reproductive syndrome virus. Arch Virol. 2003;148(1):177–87. doi: 10.1007/s00705-002-0897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delputte PL, Nauwynck HJ. Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J Virol. 2004;78(15):8094–101. doi: 10.1128/JVI.78.15.8094-8101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Breedam W, Delputte PL, Van Gorp H, Misinzo G, Vanderheijden N, Duan X, et al. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. Journal of General Virology. 2010;91(7):1659–67. doi: 10.1099/vir.0.020503-0. [DOI] [PubMed] [Google Scholar]
- 48.Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J Gen Virol. 2008;89(Pt 12):2943–53. doi: 10.1099/vir.0.2008/005009-0. [DOI] [PubMed] [Google Scholar]
- 49.Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, et al. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J Virol. 2007;81(14):7371–9. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Murtaugh MP. Functional analysis of porcine reproductive and respiratory syndrome virus N-glycans in infection of permissive cells. Virology. 2015;477:82–8. doi: 10.1016/j.virol.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 51.Prather RS, Rowland RR, Ewen C, Trible B, Kerrigan M, Bawa B, et al. An intact sialoadhesin (Sn/SIGLEC1/CD169) is not required for attachment/internalization of the porcine reproductive and respiratory syndrome virus. J Virol. 2013;87(17):9538–46. doi: 10.1128/JVI.00177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Murtaugh MP. Dissociation of porcine reproductive and respiratory syndrome virus neutralization from antibodies specific to major envelope protein surface epitopes. Virology. 2012;433(2):367–76. doi: 10.1016/j.virol.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 53.Vanhee M, Van Breedam W, Costers S, Geldhof M, Noppe Y, Nauwynck H. Characterization of antigenic regions in the porcine reproductive and respiratory syndrome virus by the use of peptide-specific serum antibodies. Vaccine. 2011;29(29–30):4794–804. doi: 10.1016/j.vaccine.2011.04.071. [DOI] [PubMed] [Google Scholar]
- 54.Popescu LN, Trible BR, Chen N, Rowland RRR. GP5 of porcine reproductive and respiratory syndrome virus (PRRSV) as a target for homologous and broadly neutralizing antibodies. Vet Microbiol. 2017 doi: 10.1016/j.vetmic.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 55.Das PB, Dinh PX, Ansari IH, de Lima M, Osorio FA, Pattnaik AK. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J Virol. 2010;84(4):1731–40. doi: 10.1128/JVI.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian D, Wei Z, Zevenhoven-Dobbe JC, Liu R, Tong G, Snijder EJ, et al. Arterivirus minor envelope proteins are a major determinant of viral tropism in cell culture. J Virol. 2012;86(7):3701–12. doi: 10.1128/JVI.06836-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nature biotechnology. 2016;34(1):20–2. doi: 10.1038/nbt.3434. [DOI] [PubMed] [Google Scholar]
- 58.Laursen NS, Wilson IA. Broadly neutralizing antibodies against influenza viruses. Antiviral Res. 2013;98(3):476–83. doi: 10.1016/j.antiviral.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.West AP, Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156(4):633–48. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dokland T. The structural biology of PRRSV. Virus Res. 2010;154(1–2):86–97. doi: 10.1016/j.virusres.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darwich L, Díaz I, Mateu E. Certainties, doubts and hypotheses in porcine reproductive and respiratory syndrome virus immunobiology. Virus research. 2010;154(1–2):123–32. doi: 10.1016/j.virusres.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 62.Sun L, Zhou Y, Liu R, Li Y, Gao F, Wang X, et al. Cysteine residues of the porcine reproductive and respiratory syndrome virus ORF5a protein are not essential for virus viability. Virus research. 2015;197:17–25. doi: 10.1016/j.virusres.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 63.Robinson SR, Figueiredo MC, Abrahante JE, Murtaugh MP. Immune response to ORF5a protein immunization is not protective against Porcine reproductive and respiratory syndrome virus infection. Veterinary Microbiology. 2013;164:281–5. doi: 10.1016/j.vetmic.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. American Journal of Epidemiology. 1938;27(3):493–7. [Google Scholar]
- 65.Brown E, Lawson S, Welbon C, Gnanandarajah J, Li J, Murtaugh MP, et al. Antibody response to porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural proteins and implications for diagnostic detection and differentiation of PRRSV types I and II. Clinical and Vaccine Immunology. 2009;16(5):628–35. doi: 10.1128/CVI.00483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson CR, Yu W, Murtaugh MP. Cross-reactive antibody responses to nsp1 and nsp2 of porcine reproductive and respiratory syndrome virus. Journal of General Virology. 2007;88:1184–95. doi: 10.1099/vir.0.82587-0. [DOI] [PubMed] [Google Scholar]
- 67.McKinney MM, Parkinson A. A simple, non-chromatographic procedure to purify immunoglobulins from serum and ascites fluid. J Immunol Methods. 1987;96(2):271–8. doi: 10.1016/0022-1759(87)90324-3. [DOI] [PubMed] [Google Scholar]
- 68.Hermann JR, Muñoz-Zanzi CA, Roof MB, Burkhart K, Zimmerman JJ. Probability of porcine reproductive and respiratory syndrome (PRRS) virus infection as a function of exposure route and dose. Vet Microbiol. 2005;110(1–2):7–16. doi: 10.1016/j.vetmic.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 69.Uitenbroek DG. Sample size calculation Southampton. 1997 [ http://www.quantitativeskills.com/sisa/calculations/samsize.htm]. Available from: http://www.quantitativeskills.com/sisa/calculations/samsize.htm.
- 70.Wasilk A, Callahan JD, Christopher-Hennings J, Gay TA, Fang Y, Dammen M, et al. Detection of U.S., Lelystad, and European-Like porcine reproductive and respiratory syndrome viruses and relative quantitation in boar semen and serum samples by real-time PCR. Journal of clinical microbiology. 2004;42(10):4453–61. doi: 10.1128/JCM.42.10.4453-4461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Staroscik A. Calculator for determining the number of copies of a template: URI Genomics & Sequencing Center. 2004 Available from: http://www.uri.edu/research/gsc/resources/cndna.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.