Abstract
Recently, many studies have demonstrated the significant advantages of loop-mediated isothermal amplification (LAMP) based methods over serological tests and PCR for rapid detection of microbial pathogens. Here, a rapid LAMP assay was developed to detect the hepatitis B virus (HBV) from DNA, and particularly, blood samples from infected patients using a commercially available master mix and portable real-time fluorometer. The final optimized fluorescence-based LAMP assay provided significant amplification time of less than 15 minutes compared with over 1 hour for PCR and an opened tube LAMP system described previously. Results indicated that fluorescence-based LAMP assay was more sensitive than PCR as a rapid, sensitive, efficient, and highly reliable approach for rapid detection of HBV.
Keywords: Medicine, Evidence-based medicine, Infectious disease, Pathology, Biochemistry, Bioengineering, Microbiology, Virology
1. Introduction
Hepatitis B virus (HBV) is considered to be one of the major causes of acute hepatitis, chronic hepatitis, and hepatocellular carcinoma (HCC). According to the World Health Organization, 2 billion people have been infected globally and 686,000 die as a result of hepatitis B every year (WHO, 2016). Based on the similarity of genomic sequences, HBV can be divided into 8 different genotypes denoted from A to H following the order of discovery, with a difference of 8% over the entire genome (Okamoto et al., 1988; Norder et al., 1994; Kramvis et al., 2005). Genotype distribution also depends on geography (Okamoto et al., 1988; Norder et al., 1993; Sunbul, 2014). Limited development of HBV therapy worldwide has contrasted with considerable advances in the epidemiology of the virus. Thus, finding and accurately detecting HBV genotypes is vital to reduce global HBV infection rates and boost both prevention and treatment.
Various techniques including enzyme-linked immunosorbent assay (ELISA) and real-time PCR have been considered for detecting HBV surface antigens, antibodies and DNA-HBV (Abe et al., 1999; Usuda et al., 1999; Yeh et al., 2004; Wu et al., 2008). However, such methods are normally both labor and time intensive and can only be undertaken in well-equipped labs (Kao, 2008; Caliendo et al., 2011; Wang et al., 2012). Therefore, an alternative approach known as loop-mediated isothermal amplification (LAMP) has been intensively studied and developed worldwide as a rapid, simple and low-cost diagnostic tool. The LAMP assay can detect clinical microbial pathogens less than half an hour including sample treatment and is appropriate for poor and developing countries. Development of LAMP based detection of all the major HBV genotypes in peripheral blood has been previously reported, with amplification results of plasma and HBV DNA samples evaluated based on multiple inverted repeats of target amplicons (ladder-like banding pattern) and colorimetric assays (Nyan et al., 2014; Moslemi et al., 2009). This LAMP technique is considered as an opened tube system with the possibility of causing false positive results. It is also time-consuming due to cross DNA contamination (multiple cap opening) and running agarose gel for electrophoresis, indicating some disadvantages for portable detection of HBV at the point of care. Using commercially available master mix and a real-time fluorometer such as Genie II (OptiGene, UK), various studies using portable devices to avoid the obstacles of an opened LAMP system reported rapid detection of human viral pathogens, measuring the amplification and melting curves of target amplicons in real-time (Mahony et al., 2013; Randhawa et al., 2013). Here, a modified sensitive LAMP assay was developed for the rapid detection of hepatitis B virus (HBV) using the Genie III system (OptiGene, UK) and compared with LAMP testing for HBV DNA and heat-treated donor blood samples to provide diagnosis results in less than 30 minutes.
2. Material and methods
2.1. Specimens and DNA preparation
A total of 30 healthy and HBV-infected blood samples were obtained from the medical clinic/hospital in Ho Chi Minh City and clinically validated by real-time PCR. Written informed consent was attained from all patients and the Human Research Ethics Committee, Oncology Hospital (#08/BVUB-HDDD) approved the research protocol. DNA extraction was performed using a DNA extraction kit (ZR Viral DNA Kit™, USA) according to the manufacturer's protocol. DNA was extracted from 200 μL of plasma standards and eluted in 800 μL of ZR Viral DNA buffer. DNA was measured using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, USA), aliquoted and stored at −20 °C until needed for testing. The DNA concentration of HBV positive sample for PCR assays and serial dilutions was 22.18 ng/μL.
2.2. Heat treatment of donor blood and plasma samples as substrate
Substrate for HBV-LAMP was prepared by heat treatment of donor blood and plasma samples without DNA extraction (Nyan et al., 2014). For plasma specimens, 25 μL of samples were diluted 2-fold with nuclease-free water. The mixture was then centrifuged at 12,000×g for 3 minutes. The supernatant was reserved and 3–10 μL were used in the isothermal amplification reactions. Heat treatment for the plasma samples was applied in a similar manner to the blood samples.
2.3. PCR reaction and electrophoresis
PCR reaction was carried out in a total of 25 μl reaction mixture containing 12.5 μL Go Taq Master Mix (Promega, USA), 1 μmol of each forward and reverse primers, and 2 μL of template DNA. PCR cycling parameter incubation cycles consisted of denaturation at 95 °C for 30 s, annealing at 53 °C for 60 s, and extension at 74 °C for 30 s, repeated for 35 cycles. The PCR amplified product was electrophoresed on 1% agarose gel (0.5 × TBE).
2.4. LAMP reaction and amplification condition
The sequence of primers in Table 1 was used as described previously (Nyan et al., 2014). LAMP reaction was carried out using a total of 25 μL of the reaction mixture containing 0.5 μL each of HBU-F3 and HBU-B3 primers (25 pmol/μL) equivalent to 5 μM final concentration, 2.0 μL each of HBU-FIP and HBU-BIP primers (25 pmol/μL) equivalent to 20 μM final concentration, and 1.0 μL each of HBU-LF and HBU-LB primers (25 pmol/μL) equivalent to 10 μM final concentration. Fifteen microliters of Isothermal Master Mix (OptiGene, UK) and 3 μL DNA was added as a template. The LAMP assay was run at 63 °C for 30 minutes with a melting curve analysis step (annealing curve 98°–80 °C ramping at 0.1 per min) in a portable real-time fluorometer like the Genie III (OptiGene, UK). A no template (water) control and DNA extracted from HBV genotypes were used as negative and positive controls, respectively.
Table 1.
Sequences of LAMP and PCR primers (Nyan et al., 2014; Gunter et al., 1998).
| Primer | Primer sequence | Reference |
|---|---|---|
| HBU-F3 | 5′ TCCTCACAATACCGCAGAGT 3′ | Nyan et al., 2014 |
| HBU-B3 | 5′ GCAGCAGGATGAAGAGGAAT 3′ | |
| HBU-FIP | 5′ GTTGGGGACTGCGAATTTTGGCTTTTTAGACTCGTGGTGGACTTCT 3′ | |
| HBU-BIP | 5′ TCACTCACCAACCTCCTGTCCTTTTTAAAACGCCGCAGACACAT 3′ | |
| HBU-LF | 5′ GGTGATCCCCCTAGAAAATTGAG 5′ | |
| HBU-LB | 5′ AATTTGTCCTGGTTATCGCTGG 3′ | |
| ATL-F | 5′ CCCTGATGAAGAACTTGTATCTC 3′ | Gunter et al., 1998 |
| ATL-R | 5′ GAAATTACACACATAGGTGGCACT 3′ |
2.5. Detection of HBV from DNA, plasma and blood samples using Genie III
Detection of hepatitis B virus (HBV) was performed by LAMP reactions as described above with HBV-DNA templates extracted from plasma, heat-treated plasma and blood samples. Positive and negative results of samples were evaluated based on amplification plots, time to positivity (less than 15 minutes) and melting curves of target amplicons analyzed automatically by the Genie III system (OptiGene, UK). To check the reliability of the Genie III system, LAMP reaction products were run on 1% agarose gel electrophoresis in 1 × TBE buffer at 100 V for 50–55 minutes, followed by staining with GelRed DNA dye (TBR, Vietnam) and visualized with a UV transilluminator at 302 nm.
2.6. Analytical sensitivity of HBV assays with DNA, heat-treated plasma and blood samples
Analytical sensitivity was evaluated by testing 10-fold serial dilutions of HBV DNA in LAMP reactions. Assay detection limit was determined by analyzing 4–7 replicates of serially diluted HBV DNA. All LAMP reactions were run using the Genie III system (OptiGene, UK). PCR reactions were also conducted with HBU-F3 and HBU-B3 (Table 1) to compare with the LAMP technique, followed by sequencing for evaluation. A pair of ATL1 primers was also used as internal amplification control in PCR assays to guard against poor quality DNA and PCR inhibition, nucleic acid extraction failure or template degradation (Table 1).
3. Results
3.1. Detection of HBV using different templates for PCR and LAMP assays
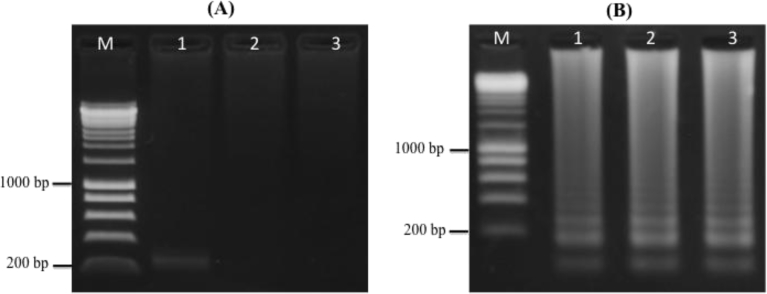
Templates used for PCR reaction in this assay included HBV DNA extracted from plasma, heat-treated plasma, and blood samples of HBV-infected patients. Electrophoresis analysis of amplified amplicons showed successful detection of HBV DNA extracted from plasma (263 bp) with a pair of universal HBV primers (F3 and B3) (Table 1). No amplified PCR products were observed for heat-treated plasma and blood templates (Fig. 1A). Results of BLAST running indicated that the sequence of PCR-amplified product from DNA template was HBV-DNA (data not shown).
Fig. 1.
Gel electrophoresis of PCR and LAMP assays with different templates. (A) PCR reactions with HBV-DNA (lane 1), heat-treated plasma specimen (lane 2), heat-treated blood sample (lane 3). (B) LAMP assays with HBV-DNA (lane 1), heat-treated plasma specimen (lane 2), heat-treated blood sample (lane 3).
To compare detectable capacity with PCR assay, LAMP products were amplified with a universal set of LAMP primers at 63 °C for 30 mins using the Genie III system, run on 1% agarose gel with 1 × TBE and UV visualized with GelRed dye. The appearance of a unique laddering pattern of amplicons observed for all three different templates revealed that they were positive for HBV DNA, resulting in a detectable capacity of LAMP assays with templates that could not be amplified by conventional approaches due to the inhibition of chemicals in blood or plasma specimens (Fig. 1B).
3.2. Limits of HBV-DNA detection between PCR and LAMP assays
To compare the limits of HBV detection by PCR and LAMP assays, at least 7 replicates of serially diluted HBV DNA (10−1–10−7) extracted from HBV-infected plasma sample were assayed and analyzed. Concentrations of HBV-DNA corresponding to above serial dilutions were from 2218 pg/μL to 0.002218 pg/μL, respectively. Results of LAMP assays indicated a 100% detection rate from 10−1 to 10−7 of HBV DNA molecules per reaction (Table 2). For PCR assays, 100% rates of HBV detection were observed from 10−1 to 10−4 HBV DNA, whereas 10−5 and 10−6 – 10−7 diluted HBV DNA templates were detected at 57% and 0%, respectively (Table 2).
Table 2.
Environment of HBV DNA detection by PCR and LAMP assays.
| Serial diluted HBV-DNA | DNA concentration (pg/μL) | PCR |
LAMP |
||||
|---|---|---|---|---|---|---|---|
| No. of replicates tested | No. of detections | Detection rate (%) | No. of replicates tested | No. of detections | Detection rate (%) | ||
| 10−1 | 2218 | 7 | 7 | 100 | 7 | 7 | 100 |
| 10−2 | 221.8 | 7 | 7 | 100 | 7 | 7 | 100 |
| 10−3 | 22.18 | 7 | 7 | 100 | 7 | 7 | 100 |
| 10−4 | 2.218 | 7 | 7 | 100 | 7 | 7 | 100 |
| 10−5 | 0.2218 | 7 | 4 | 57 | 7 | 7 | 100 |
| 10−6 | 0.02218 | 7 | 0 | 0 | 7 | 7 | 100 |
| 10−7 | 0.002218 | 7 | 0 | 0 | 7 | 7 | 100 |
3.3. Evaluation of HBV LAMP assays with blood samples
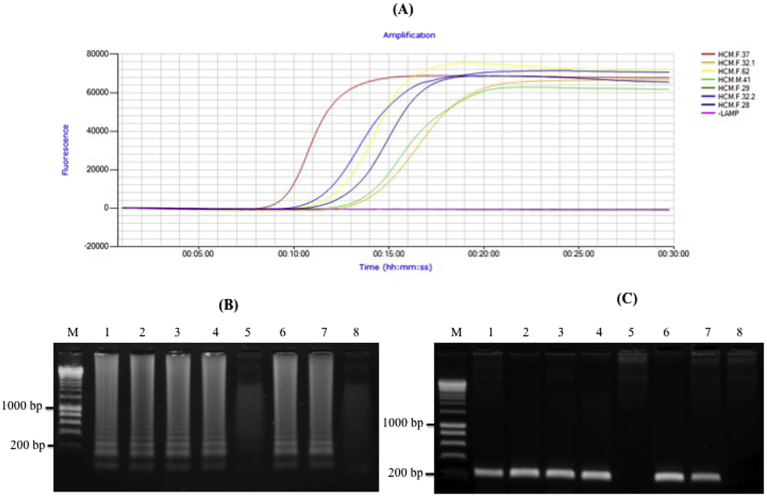
The use of both extracted DNA and heat-treated plasma samples for the detection of HBV DNA was described previously (Nyan et al., 2014). Here, experiments were conducted to determine a detectable capacity of HBV with donor blood samples by LAMP assay in the shortest possible time. Aliquots of identical blood samples were also heat-treated (without DNA and plasma extraction) within 3 minutes and directly tested in the LAMP reactions. HBV DNA extraction of these blood samples was conducted to confirm the results of LAMP based assay using polymerase chain reaction (PCR) with universal HBV primers (F3 and B3). Amplicons, amplified with ATL1 primers known as internal amplification control, indicated good quality DNA for PCR and LAMP assays (Fig. S1) (Gunter et al., 1998). Six of the seven blood samples showed amplification plots within 9–13 minutes, revealing positive results for amplified HBV DNA (Fig. 2A). LAMP products were also confirmed by electrophoresis analysis. Results were consistent with amplification plots displayed in the Genie III system (Fig. 2B.) and PCR amplification (Fig. 2C).
Fig. 2.
Detection of Hepatitis B Virus (HBV) from heat-treated blood samples using the Genie III system (OptiGene, UK). (A) Electrophoresis result of LAMP to blood heat-treatment. (B) Amplification plots of the LAMP assays from blood heat-treatment. (C) Electrophoresis results of HBV-DNA from blood samples using PCR method with F3 and B3 primers. Lane M = 1 kp marker; Lanes 1–7 = HCM.F.37, HCM.F.32.1, HCM.F.62, HCM.M.41, HCM.F.29, HCM.F.32.2, HCM.F.28 DNA-HBV samples; Lane 8 = Negative LAMP assay.
3.4. Time of LAMP based detection with heat-treated HBV-infected blood samples
Ten-fold serial dilutions of heat-treated HBV-infected blood samples were tested using LAMP assays with three replicates. Results indicated that assay detection of HBV DNA appeared after approximately 10 minutes using blood samples directly, compared with less than 14 minutes at 10−5 dilution for heat-treated HBV-infected blood samples (Table 3). To evaluate the productivity and diagnostic sensitivity of LAMP assay, donor blood samples (n = 30) were tested. Results revealed that LAMP assays detected 19 out of 30 donor blood samples (63%) as HBV positive (Fig. S2) and demonstrated the closed tube HBV-LAMP assay as a rapid and sensitive diagnostic tool for screening blood donors and on-site testing of patient specimens (Table S1).
Table 3.
Times for LAMP based detection with heat-treated HBV-infected blood samples.
| Serial dilution of heat-treated HBV-infected blood samples | Peak values |
Mean of peak values | Standard deviation | ||
|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | |||
| Positive sample | 09:45 | 10:15 | 09:45 | 09:55 | 0.01 |
| 10−1 | 10:15 | 11:00 | 09:15 | 10:10 | 0.04 |
| 10−2 | 11:15 | 11:45 | 08:45 | 10:35 | 0.07 |
| 10−3 | 13:00 | 13:00 | 09:00 | 11:40 | 0.1 |
| 10−4 | 12:15 | 13:00 | 10:15 | 11:50 | 0.06 |
| 10−5 | 12:00 | 18:15 | 10:45 | 13:40 | 0.17 |
4. Discussion
Advantages of LAMP based HBV detection were described previously as a simple and rapid diagnostic and screening tool that is useful for undeveloped and developing countries with limited resources, and also in other global regions with high HBV prevalence (Nyan et al., 2014; Jain et al., 2012; Stramer et al., 2011). However, although LAMP universal primers for the detection of HBV genotypes (A-F) were successfully designed and showed high detection specificity and accuracy in clinical diagnostics, many reports highlighted major drawbacks of the opened tube LAMP technique, as cross contamination when the lids of the reaction tubes were opened at the end of the reaction for gel electrophoresis, or when adding dye for result visualization (Nyan et al., 2014; Parida et al., 2008; Njiru et al., 2008; Lau et al., 2010; Angamuthu et al., 2011). In addition, running gel electrophoresis at the end of the LAMP reaction requires this step to be conducted in the lab which is time-consuming and, therefore, not suitable for on-site rapid detection.
To surmount such obstacles of an opened tube LAMP system, closed tube LAMP assays using a fluorometer (Genie II/III systems) and commercially available master mix (OptiGene, UK) were developed and validated for rapid detection of HBV in DNA and blood samples. This approach has been demonstrated by few previous studies; the visualization of LAMP reaction via closed tube system decreased cross contamination (Tao et al., 2011; Karthik et al., 2014; Prusty et al., 2016). Furthermore, the LAMP master mix can be used for Genie II/III systems (OptiGene, UK) and also for other mobile/molecular devices such as Smart-DART (Diagenetix, USA), PCR, real-time PCR, and even colorimetric assays (Reddy et al., 2011; Davoodian and Sadeghifard, 2011). The sensitivity and clinical applicability of closed tube LAMP assays for HBV detection were validated and compared with different templates including HBV-DNA, plasma specimens, and blood samples. The HBV LAMP assay detected all these templates, whereas the PCR method detected HBV-DNA but not HBV-infected plasma specimens and blood samples that could be interfered by PCR reaction inhibitors. Sensitivity of the HBV LAMP assay over the PCR method gave the following advantages: (i) the assay did not require well-equipped laboratories, (ii) it reduced time to within 15 minutes for the detection of HBV, without the necessity of HBV DNA or plasma extraction from blood samples, (iii) it was performed on a closed tube LAMP system operated simply, accurately and quickly to reduce cross contamination, and (iv) the amplification plots and melting curves could be easily and directly observed via the Genie III system, with reduced time for generating results and the potential for differentiating genotypes of HBV. These advantages indicated that closed tube HBV LAMP assay is applicable for on-site detection and useful for developing or developed economies with limited resources to conduct affordable, quick and accurate diagnoses for HBV infection. Costs of LAMP and PCR assays were calculated by determining reagent component cost, consumables and primer cost, depreciation of equipment and a technologist's salary of $1.25/hour in Vietnam. Cost of HBV-DNA extraction was calculated using the purchase price of the HBV-DNA extraction kit included in the PCR cost determination. Costs were based on a run size of 8 samples in each assay format. There were no significant differences between costs of PCR and LAMP assay with plasma based HBV-DNA extraction; however, the advantage of LAMP was observed in hands-on-time at 30 minutes compared with 3 hours using PCR assay (Table 4). Specificity and accuracy of primers in LAMP assays were reported by Nyan et al. (2014), and closed tube LAMP assays were developed to detect heat-treated blood samples directly. This approach does not require plasma extraction which reduces the time for sample preparation, running cost at $5.1/test, and requires a smaller blood sample volume (approximately 20–25 μl) (Table 4). Healthy donor blood samples (n = 11) showed no amplification of HBV DNA, whereas HBV-infected blood samples gave amplification of HBV DNA within 10–15 minutes. LAMP based assays were consistent with clinical validation of these samples using real-time PCR (Table S1).
Table 4.
Relative costs of LAMP and PCR assays.a
| Assay type | No. of tests/run | HBV-DNA extraction kit cost ($) | Reagent cost/run ($) | Cost of consumables and primers ($) | Hands-on-time (h) | Labor cost/runb ($) | Depreciation of equipment ($) | Total cost/test ($) |
|---|---|---|---|---|---|---|---|---|
| LAMP | 8 | 17.6 | 16 | 16 | 0.5 | 0.63 | 8 | 7.3 |
| Modified LAMP | 8 | 0 | 16 | 16 | 0.5 | 0.63 | 8 | 5.1 |
| PCR | 8 | 17.6 | 8 | 16 | 3 | 3.75 | 10 | 7.0 |
All costs are in US dollars.
Based on the basic salary of technicians in Vietnam.
This approach can, therefore, be used to investigate HBV prevalence rapidly, accurately and economically. Furthermore, the master mix (OptiGene, UK) is adaptable for LAMP assays using various mobile devices including Genie II, Genie III, Smart-DART, PCR, and real-time PCR. Master mix is stable and robust under thermally stressed conditions and applicable for in-field environments and on-site detection of HBV-DNA.
5. Conclusion
The closed tube HBV-LAMP assay reported here could be used directly with blood samples; it is specific with high reliability, low cost, and easy implementation for rapid detection of the hepatitis B virus (HBV). This method is particularly applicable for on-site detection of HBV infection for accurate treatment and prognosis of HBV-infected patients.
Declarations
Author contribution statement
Nguyen Bao Quoc: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Nguyen Doan Nguyen Phuong: Performed the experiments.
Nguyen Ngoc Bao Chau: Analyzed and interpreted the data.
Do Thi Phuong Linh: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported in part by the grant of Ministry of Education and Training, Vietnam (B2015-12-11) and Nong Lam University grant (CS-SV16-CNSH-04).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Abe A., Inoue K., Tanaka T., Kato J., Kajiyama N., Kawaguchi R., Kohara M. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J. Clin. Microbiol. 1999;37:2899–2903. doi: 10.1128/jcm.37.9.2899-2903.1999. http://jcm.asm.org/content/37/9/2899.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angamuthu R., Baskaran S., Gopal D.R., Deverajan J., Kathaperumal K. Rapid detection of the Marek's disease viral genome in chicken feathers by loop-mediated isothermal amplification. J. Clin. Microbiol. 2011;50:961–965. doi: 10.1128/JCM.05408-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliendo A.M., Valsamakis A., Bremer J.W., Ferreira-Gonzalez A., Granger S., Sabatini L. Multilaboratory evaluation of real-time PCR tests for hepatitis B virus DNA quantification. J. Clin. Microbiol. 2011;49:2854–2858. doi: 10.1128/JCM.00471-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodian A., Sadeghifard N. Detection of hepatitis B virus DNA by real-time PCR in chronic hepatitis B patients, Ilam, Iran. Middle-East J. Sci. Res. 2011;9:478–480. https://www.idosi.org/mejsr/mejsr9(4)11/9.pdf [Google Scholar]
- Gunter C., Paradee W., Crawford D.C., Meadows K.A., Newman J., Kunst C.B. Re-examination of factors associated with expansion of CGG repeats using a single nucleotide polymorphism in FMR1. Hum. Mol. Genet. 1998;7(12):1935–1946. doi: 10.1093/hmg/7.12.1935. [DOI] [PubMed] [Google Scholar]
- Jain R., Aggarwal P., Gupta G.N. Need for nucleic acid testing in countries with high prevalence of transfusion-transmitted infections. ISRN Hematol. 2012;2012 doi: 10.5402/2012/718671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao J.H. Diagnosis of hepatitis B virus infection through serological and virological markers. Expert Rev. Gastroenterol. Hepatol. 2008;2:553–562. doi: 10.1586/17474124.2.4.553. [DOI] [PubMed] [Google Scholar]
- Karthik K., Rathore R., Thomas P., Arun T.R., Viswas K.N., Agarwal R.K. Loop-mediated isothermal amplification (LAMP) test for specific and rapid detection of Brucella abortus in cattle. Vet. Q. 2014;34(4):147–179. doi: 10.1080/01652176.2014.966172. [DOI] [PubMed] [Google Scholar]
- Kramvis A., Kew M., Francois G. Hepatitis B virus genotypes. Vaccine. 2005;23(19):2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- Lau Y.L., Meganathan P., Sonaimuthu P., Thiruvengadam G., Nissapatorn V., Chen Y. Specific, sensitive and rapid diagnosis of active toxoplasmosis by a loop-mediated isothermal amplification method using blood samples from patients. J. Clin. Microbiol. 2010;48(10):3698–3702. doi: 10.1128/JCM.00462-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J., Chong S., Bulir D., Ruyter A., Mwawasi K., Waltho D. Multiplex loop-mediated isothermal amplification (M-LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen-to-result diagnosis in 40 min with single genome copy sensitivity. J. Clin. Virol. 2013;58:127–131. doi: 10.1016/j.jcv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Moslemi E., Shahhosseiny M.H., Javadi G., Praivar K., Sattari T.N., Amini H.K. Loop mediated isothermal amplification (LAMP) for rapid detection of HBV in Iran. Afr. J. Microbiol. Res. 2009;3:439–445. http://www.academicjournals.org/journal/AJMR/article-abstract/E70925B13699 [Google Scholar]
- Njiru Z.K., Mikosza A.S.J., Armstrong T., Enyaru J.C., Ndung’u J.M. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Neglected Trop. Dis. 2008;2(2):147. doi: 10.1371/journal.pntd.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norder H., Hammas B., Lee S.D., Bile K., Courouce A.M., Mushahwar I.K., Magnius L.O. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J. Gen. Virol. 1993;74:1341–1348. doi: 10.1099/0022-1317-74-7-1341. [DOI] [PubMed] [Google Scholar]
- Norder H., Courouce A.M., Magnius L.O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the Hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- Nyan D.C., Ulitzky L.E., Cehan N., Williamson P., Winkelman V., Rios M., Taylor D.R. Rapid detection of hepatitis B virus in blood plasma by a specific and sensitive loop-mediated isothermal amplification assay. Clin. Infect. Dis. 2014;59:16–23. doi: 10.1093/cid/ciu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Tsuda F., Sakugawa H., Sastrosoewinjo R.I., Imai M., Miyakawa Y., Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- Parida M., Sannarangaiah S., Dash P.K., Rao P.V.L., Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008;18:407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusty B.R., Chaudhuri P., Chaturvedi V.K., Saini M., Mishra B.P., Gupta P.K. Visual detection of Brucella spp. in spiked bovine semen using loop-mediated isothermal amplification (LAMP) assay. Indian J. Microbiol. 2016;56(2):142–147. doi: 10.1007/s12088-015-0563-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa G.J., Singh M., Morisset D., Sood P., Žel J. Loop-mediated isothermal amplification: rapid visual and real-time methods for detection of genetically modified crops. J. Agric. Food Chem. 2013;61:11338–11346. doi: 10.1021/jf4030085. [DOI] [PubMed] [Google Scholar]
- Reddy P.B., Mukherjee R.M., Aparna J., Mitnala S., Rao P.N., Gupta R., Reddy D.N. Detection of diagnosis escape variants of hepatitis B virus by in house polymerase chain reaction assay. J. Gen. Mol. Virol. 2011;3:43–48. http://www.academicjournals.org/journal/JGMV/article-full-text-pdf/4B0CC9510928 [Google Scholar]
- Stramer S.L., Wend U., Candotti D., Foster G.A., Hollinger F.B., Dodd R.Y. Nucleic acid testing to detect HBV infection in blood donors. N. Engl. J. Med. 2011;364:236–247. doi: 10.1056/NEJMoa1007644. [DOI] [PubMed] [Google Scholar]
- Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J. Gastroenterol. 2014;20(18):5427–5434. doi: 10.3748/wjg.v20.i18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z.Y., Zhou H.Y., Xia H., Xu S., Zhu H.W., Culleton R.L. Adaption of visualized loop-mediated isothermal amplification technique for field detection of Plasmodium vivax infection. Parasites Vectors. 2011;4:115. doi: 10.1186/1756-3305-4-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuda S., Okamoto H., Iwanari H., Baba K., Tsuda F., Miyakawa Y., Mayumi M. Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J. Virol. Meth. 1999;80:97–112. doi: 10.1016/s0166-0934(99)00039-7. [DOI] [PubMed] [Google Scholar]
- Wang F., Ma C., Zeng X., Li C., Deng Y., He N. Chemiluminescence molecular detection of sequence-specific HBV-DNA using magnetic nanoparticles. J. Biomed. Nanotechnol. 2012;8:786–790. doi: 10.1166/jbn.2012.1446. [DOI] [PubMed] [Google Scholar]
- WHO fact sheet . 2016. Hepatitis B.http://www.who.int/mediacentre/factsheets/fs204/en/ [Google Scholar]
- Wu X., Zhou C., Huang W.J., Qi Z.B., Liang Z.L., Li H.M., Zhuang H. Sensitivity and specificity of 4 domestic ELISA kits for detection of hepatitis B virus markers. Zhonghua Liuxingbingxue Zazhi. 2008;29(9):915–918. http://europepmc.org/abstract/med/19173858 [PubMed] [Google Scholar]
- Yeh S.H., Tsai C.Y., Kao J.H., Liu C.J., Kuo T.J., Lin M.W., Chen P.J. Quantification and genotyping of hepatitis B virus in a single reaction by real-time PCR and melting curve analysis. J. Hepatol. 2004;41:659–666. doi: 10.1016/j.jhep.2004.06.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.