Abstract
Protein expression is regulated by the production and degradation of mRNAs and proteins but the specifics of their relationship are controversial. Although technological advances have enabled genome-wide and time-series surveys of mRNA and protein abundance, recent studies have shown paradoxical results, with most statistical analyses being limited to linear correlation, or analysis of variance applied separately to mRNA and protein datasets. Here, using recently analyzed genome-wide time-series data, we have developed a statistical analysis framework for identifying which types of genes or biological gene groups have significant correlation between mRNA and protein abundance after accounting for potential time delays. Our framework stratifies all genes in terms of the extent of time delay, conducts gene clustering in each stratum, and performs a non-parametric statistical test of the correlation between mRNA and protein abundance in a gene cluster. Consequently, we revealed stronger correlations than previously reported between mRNA and protein abundance in two metabolic pathways. Moreover, we identified a pair of stress responsive genes (ADC17 and KIN1) that showed a highly similar time series of mRNA and protein abundance. Furthermore, we confirmed robustness of the analysis framework by applying it to another genome-wide time-series data and identifying a cytoskeleton-related gene cluster (keratin 18, keratin 17, and mitotic spindle positioning) that shows similar correlation. The significant correlation and highly similar changes of mRNA and protein abundance suggests a concerted role of these genes in cellular stress response, which we consider provides an answer to the question of the specific relationships between mRNA and protein in a cell. In addition, our framework for studying the relationship between mRNAs and proteins in a cell will provide a basis for studying specific relationships between mRNA and protein abundance after accounting for potential time delays.
Keywords: Bioinformatics, Computational biology, Mathematical biosciences, Cell biology
1. Introduction
Protein expression is known to be regulated by the production and degradation of mRNAs and proteins, but details about their specific relationships are controversial [1]. Although technological advances have enabled genome-wide and time-series surveys of mRNA and protein abundance that should deepen our understanding of the relationships, recent studies using cells under non-steady state (perturbed by biological stress) have instead shown paradoxical results [2, 3, 4].
For example, in a study of time-dependent changes of the transcriptome and proteome in Saccharomyces cerevisiae (yeast) subjected to osmolarity stress, the authors found that the maximum mRNA and protein levels were well correlated for the upregulated genes, but not for the downregulated ones [5]. Another study examined the correlation between mRNA and protein abundance changes in yeast in response to rapamycin, an anticancer and immunosuppressive drug, where it was found that most of the proteins that had decreased in abundance were correlated with a decrease in mRNA expression, although 26 of 56 proteins increasing in abundance were not correlated with an mRNA increase [6]. These studies indicate that the relationships between mRNA and protein abundance vary depending on gene categories.
In addition, the rapamycin treatment study [6] reported a temporal delay in the correlation of mRNA and protein expression among 328 genes, where mRNA expression levels at 1 and 2 h were the most highly correlated with protein expression changes after 6 h of the treatment. The study also conducted a clustering analysis of genes based on distance in terms of mRNA and protein time-series profiles, and defined 12 patterns of correlation between mRNA and protein expression changes, indicating that such expression relationships between mRNA and protein are not linear [6]. Another study focused on the time-delayed correlation and nonlinearity, and took an approach based on Spearman's rank correlation to investigate the global coordination between mRNAs and proteins in the cell using transcriptome and proteome data measured across the life cycle of Plasmodium falciparum (a malaria parasite). They detected statistically significant correlations in 1840 genes, 1408 of which showed time-delayed correlations [7].
A more recent study introduced the SWATH-MS method and demonstrated its ability to efficiently generate reproducible, consistent, and quantitatively accurate measurements of a large fraction of a proteome (over 2500 proteins) across multiple samples, by investigating cell cultures in biological triplicates at six time points following osmolarity stress [8]. The study examined the correlation between the proteome data obtained by SWATH-MS and their corresponding transcript profiles by using a transcriptome dataset that had been previously generated for yeast treated under similar experimental conditions [5]. As a result, 50% of the protein profiles measured for two of the four most regulated pathways showed no clear correlation between the protein abundance and their corresponding RNA profiles (i.e., –0.5 < Pearson's correlation coefficient <0.5), which may be mainly due to a slight delay observed for the protein response compared with the mRNA response [8].
Although these previous studies revealed the nonlinearity and complexity of the relationship between mRNA and protein abundance, most of their statistical analyses were limited to linear correlation, or analysis of variance applied separately to mRNA and protein datasets [9]. Another study introduced above [7] was based on non-parametric rank correlation rather than linear correlation, but only reported the overall correlation or the proportion of genes showing statistically significant correlations. However, all of these studies have not specifically identified the kind of genes or biological gene groups that have significant correlation between mRNA and protein abundance after accounting for the potential time delay. In the present study, we have used recently analyzed genome-wide time-series data in yeast cell responding to the osmolarity stress [8] and developed a statistical analysis framework for the identification of such genes. First, we stratified all genes in terms of the extent of time delay of the correlation between mRNA and protein abundance changes by using a method originally developed for the relationships of gene expression levels [10]. Second, we conducted gene clustering in each stratum in terms of concordance of the time course of mRNA and protein abundance changes. Third, for each gene group found by the clustering, we performed a non-parametric statistical test of the correlation between mRNA and protein abundance after accounting for natural correlations among repeated measures in a time series, similar to what was done in the previous study [7]. Furthermore, we confirmed robustness of the analysis framework by applying it to another genome-wide time-series data in mammalian cells responding to stress of the endoplasmic reticulum (ER) [4].
Our study revealed stronger correlations between mRNA and protein abundance in two metabolic pathways than that found without the stratification in terms of the extent of time delay. In addition, we identified a pair of stress responsive genes (ADC17 and KIN1) that showed a highly similar time series of mRNA and protein abundance, particularly their evident increase within 30 min after the osmolarity stress. Furthermore, analysis of the other dataset identified a cluster of three genes related to cytoskeleton that similarly increased mRNA and protein abundance until 8 h after the ER stress. The significant correlations and highly similar changes of the mRNA and protein abundance of these genes provide an answer to the question of the specific relationships between mRNA and protein in a cell.
2. Materials and methods
2.1. Datasets and software
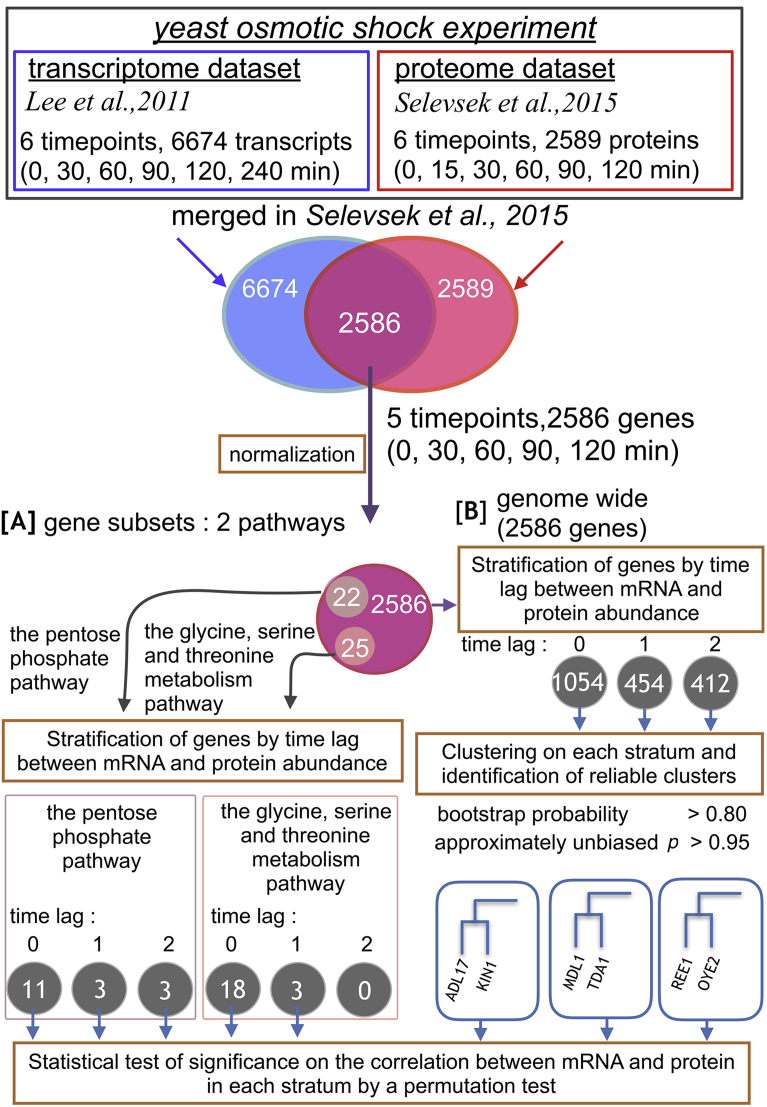
We used the proteome and transcriptome datasets that were combined and analyzed in a previous study [8]. They consist of data on the genome-wide mRNA and protein abundance in yeast cells measured at six time points following osmolarity stress (NaCl added to the culture). The proteome dataset, consisting of 2589 proteins, was obtained by the SWATH-MS method at 0, 15, 30, 60, 90, and 120 min following the osmolarity stress (shown in the red box in Fig. 1). The SWATH-MS technique for proteome measurements is a MS strategy that mines the complete fragment ion maps (spectra) generated using a data-independent acquisition method, and vastly extends the number of peptides/proteins quantified per sample [8, 11]. The transcriptome dataset, consisting of 6674 transcripts, was originally obtained at 0, 30, 60, 90, 120, and 240 min in another study that used a microarray experiment to treat yeast cells under a similar condition [5] (shown in the blue box in Fig. 1). The samples for transcriptome measurements were hybridized to custom Nimblegen tiled arrays after RNA extraction, RNA purification and cDNA synthesis. Arrays were scanned and analyzed with a GenePix4000 scanner (Molecular Devices, Sunnyvale, CA), and signal was extracted with the program NimbleScan [5]. These two datasets combined, consisting of a total of 2586 genes observed at 0, 30, 60, 90, and 120 min following the osmolarity stress (in magenta in Fig. 1), were provided to us by the author [8]. These datasets are available in the compressed supplementary data file “Dataset1.zip”
Fig. 1.
Overview of the analysis framework. Each circle indicates a set of genes, and the numbers in each circle indicate the number of genes in the set. [A] and [B] indicate flows of “Analysis of genes of two metabolic pathways” and “Genome-wide analysis” in Results, respectively.
The data provided to us are the log2 ratios of abundance changes relative to the first time point (e.g., 0 at the first time point). In this study, to make the values more intuitive, we converted them into relative expression values that were specified to be 1 at the first time point. In the following analyses, the relative expression values of the mRNA and protein of gene g at the t-th time point (t = 1, 2, 3, 4, and 5 corresponding to 0, 30, 60, 90, and 120 min, indicated by τt) are denoted as and , respectively. (For simplicity, the subscript g is omitted in the equations below.)
All analyses described below were performed using R ver. 3.2.2, and the source code and data are available at https://github.com/Shimpeim/time_delayed_2017.
2.2. Normalization
The values of and are normalized as follows so that they can be readily compared between genes:
| (1) |
where
| (2) |
| (3) |
We call and the “normalized relative expression” of the mRNA and protein, respectively.
2.3. Inference of the extent of time lag between the time series of mRNA and protein abundance changes
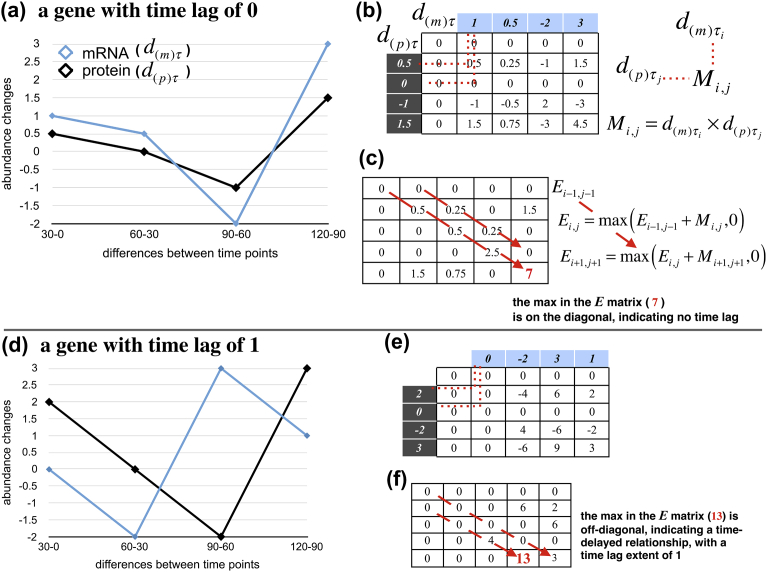
For each gene, we detected the extent of time delay of the correlation between mRNA and protein abundance changes on the basis of a “local clustering” algorithm originally developed for the relationships of gene expression levels in a previous study [10]. We applied the algorithm to the mRNA and protein abundance changes between the time points, denoted as and , respectively:
| (4) |
| (5) |
We defined a matrix M for each gene as the direct product of d(m)τi and d(p)τj (Fig. 2b and e). The element (i, j) in a matrix M is denoted as Mi,j. M1˙ and M˙1 are fixed to be 0, as in the previous study [10]. Next, a matrix E is defined as follows (Fig. 2c and f):
| (6) |
Fig. 2.
Algorithm to infer the extent of the time lag between mRNA and protein abundance changes. The algorithm explained in the section “Inference of the extent of time lag between the time series of mRNA and protein abundance changes” is illustrated. (a) and (d) are time courses of mRNA and protein abundance changes with time lags of 0 and 1, respectively. The red-dotted lines in (b) and (e) represent multiplication, and all elements in the matrix are products calculated in the same way. The red arrows in (c) and (f) show the direction of summation. The matrices were filled according to the summation.
If the maximum Ei′,j′ in the matrix E is off-diagonal (for example, Fig. 2f), then the time series of mRNA and protein abundance changes have a time-delayed relationship, with a time lag extent of i′ – j′ (an example of a gene with time lag of 1 and corresponding matrices M and E are shown in the lower in Fig. 2). Otherwise, there is no time lag between the two time series.
The rationale is as follows: Mi,j becomes positive when d(m)τi and d(p)τj have the same sign. In other words, Mi,j is interpreted as a score of concordance of mRNA and protein abundance changes at τi and τj from one time point before. The maximum Ei′,j′ is obtained by taking the summation of the score along the main or shifted diagonal that maximizes the concordance (red dashed arrows in Fig. 2). The extent of the shift corresponds to that of the time lag between the time series of mRNA and protein abundance changes.
2.4. Stratification of genes
The genes were stratified by time-lag extent, as defined by the method in the previous section. For genome-wide analyses, they were further stratified by clustering on the basis of the time-course distance of mRNA and protein abundance changes between genes. For that purpose, we used the E matrix (explained in the previous section) to define the distance between genes (g and g′):
| (7) |
Using this distance, we conducted hierarchical gene clustering using the Ward's method.
We evaluated the confidence of each gene cluster using bootstrap probability (BP) [12]. In addition, we calculated the approximately unbiased (AU) p-value [13] developed for reducing the known bias of the BP test. If a cluster has an AU p-value of >0.95, then the hypothesis that “the cluster does not exist” is rejected with a significance level of 0.05. In this study, we focused on clusters of genes with BP >0.80 and AU >0.95 as being reliable clusters. We used BP in addition to AU because we sometimes saw considerable differences between these two values in a cluster, which seemed to be unreliable.
We calculated BP and AU using the R ver. 3.2.2 (2015-08-14) pvclust package (ver. 2.0–0).
2.5. Statistical test
We conducted a statistical test of significance on the correlation between the time series of the normalized relative expression of mRNA and protein in each stratum found in the previous section. The stratum-based test increases the statistical power relative to that of the gene-based test because of the increased sample size. Taking the natural correlation of repeated measures (so-called serial correlation [14]) in a time series into account, we conducted a similar permutation test as done in the previous study [7] by using Spearman's rank correlation coefficient as a test statistic. If a stratum showed a time lag in which the mRNA preceded the protein, then the correlation coefficient was calculated after shifting the time series of the normalized relative expression of the protein to that of the mRNA according to its extent: where u is the extent of the time lag.
The significance of the rank correlation coefficient was tested by calculating the empirical p-value from a null distribution generated by permuting the observed time series of mRNA and protein abundance of each gene 10000 times, respectively. If the p-value after false discovery rate (FDR) correction (PFDR) was <0.05, then we rejected the null hypothesis (no correlation between the two time series).
We assessed the type-I error rate of the permutation test using simulated data generated by random sampling from multivariable normal distribution. We used a variance-covariance matrix calculated from a time series of normalized relative expression (2586 genes across the genome, 5 time points) of mRNA and protein (denoted as Σ(m) and Σ(p), respectively). We randomly sampled 1000 sets of mRNA and protein time courses (denoted as X and Y) from a five-dimensional normal distribution with mean 0s and covariance of Σ(m) and Σ(p), respectively.
where
Corresponding to the gene cluster consisting of the two genes we analyzed (detailed in Results), we sampled two xk from X and two yk from Y, calculated Spearman's rank correlation coefficient between them, and conducted the permutation test. We conducted the test 500 times and counted the number of tests out of the 500 that showed p < 0.05, resulting in estimation of the type I error rate of the permutation test to be 5.2%.
2.6. Another dataset
For confirmation of robustness of the analysis framework, we also applied it to genome-wide time-series data of mRNA and protein abundance in mammalian cells responding to stress of the endoplasmic reticulum (ER) [4]. The dataset consisted of two biological replicates of a total of 1237 genes measured at eight time points (0, 0.5, 1, 2, 8, 16, 24 and 30 h following the ER stress), and was available in Supporting information (“Dataset EV1”) [4]. The dataset is available in the compressed supplementary data file “Dataset2.zip”. We selected genes with Pearson's correlation coefficient >0.7 between the biological replicates, and used average values of the abundance. We then calculated the relative expression values of the mRNA and protein of gene g at the t-th time point, and followed the procedures above.
3. Results
3.1. Analysis of genes of two metabolic pathways
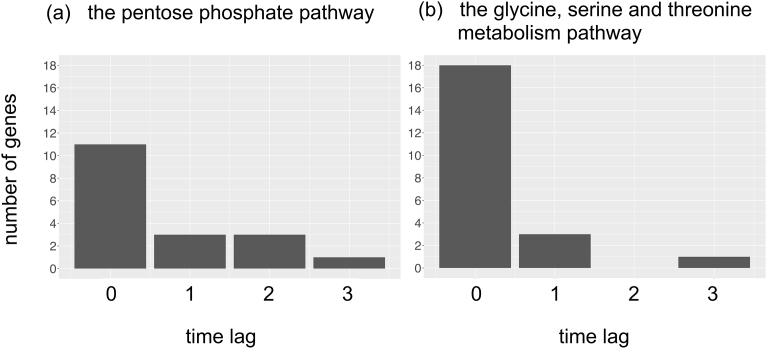
First, we analyzed genes involved in the pentose phosphate pathway and the glycine, serine and threonine metabolism pathway, for which a previous study had suggested a potential delay of the protein response compared with the mRNA response following osmolarity stress. The flowchart of the analyses is shown in [A] in Fig. 1. For each gene, we inferred the extent of the time delay of the correlation between mRNA and protein abundance changes as explained in Materials and Methods. As a result, we found 11, 3, and 3 out of 22 genes with time lags of 0, 1, and 2, respectively, in the pentose phosphate pathway (Fig. 3a, and grey circles at the bottom left panel of Fig. 1). Similarly, we found 18, 3, and 0 out of 25 genes with time lags of 0, 1, and 2, respectively, in the glycine, serine and threonine metabolism pathway (Fig. 3b, and grey circles at the bottom middle panel of Fig. 1). We also found that 18.2% of the genes involved in the pentose phosphate pathway and 12.0% of genes involved in the glycine, serine and threonine metabolism pathway showed negative time-lag values (i.e., the change of the protein abundance proceeded that of mRNA); these genes were excluded from Fig. 3 and the subsequent analyses because of difficulty of biological interpretation. Overall, only 31.8% and 16.0% of genes in the pentose phosphate pathway and the glycine, serine and threonine metabolism pathway, respectively, had positive time-lag values, whereas the genes without a time lag were in the majority in the two metabolic pathways.
Fig. 3.
Distribution of inferred time lags (0, 1, 2, and 3) between time series of mRNA and protein abundance changes. (a) Genes involved in the pentose phosphate pathway. (b) Genes involved in the glycine, serine and threonine metabolism pathway.
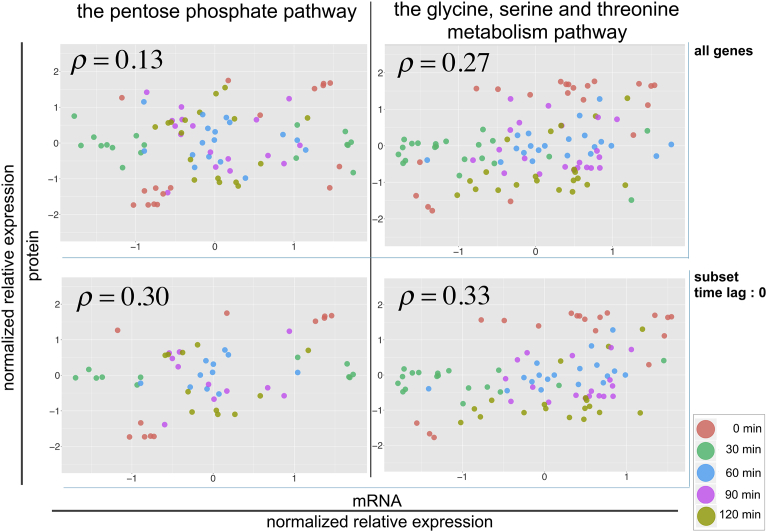
When we did not take the time lag into account and instead calculated the global correlation between mRNA and protein abundance among all genes in each pathway, Spearman's rank correlation coefficient was 0.13 and 0.27, respectively (upper panels in Fig. 4). However, when we stratified the genes by the inferred time lag and performed the correlation analysis in each stratum, the genes with a time lag of 0 showed an increased correlation (0.30 in the pentose phosphate pathway, and 0.33 in the glycine, serine and threonine metabolism pathway) (lower panels in Fig. 4).
Fig. 4.
Correlation between mRNA and protein expression before and after data stratification. Left: genes involved in the pentose phosphate pathway. Right: genes involved in the glycine, serine and threonine metabolism pathway. Upper: all genes. Lower: a subset of genes that showed the inferred time lag of 0. Each dot corresponds to the normalized relative expression level of mRNA and protein of a gene at each time point (0, 30, 60, 90, and 120 min).
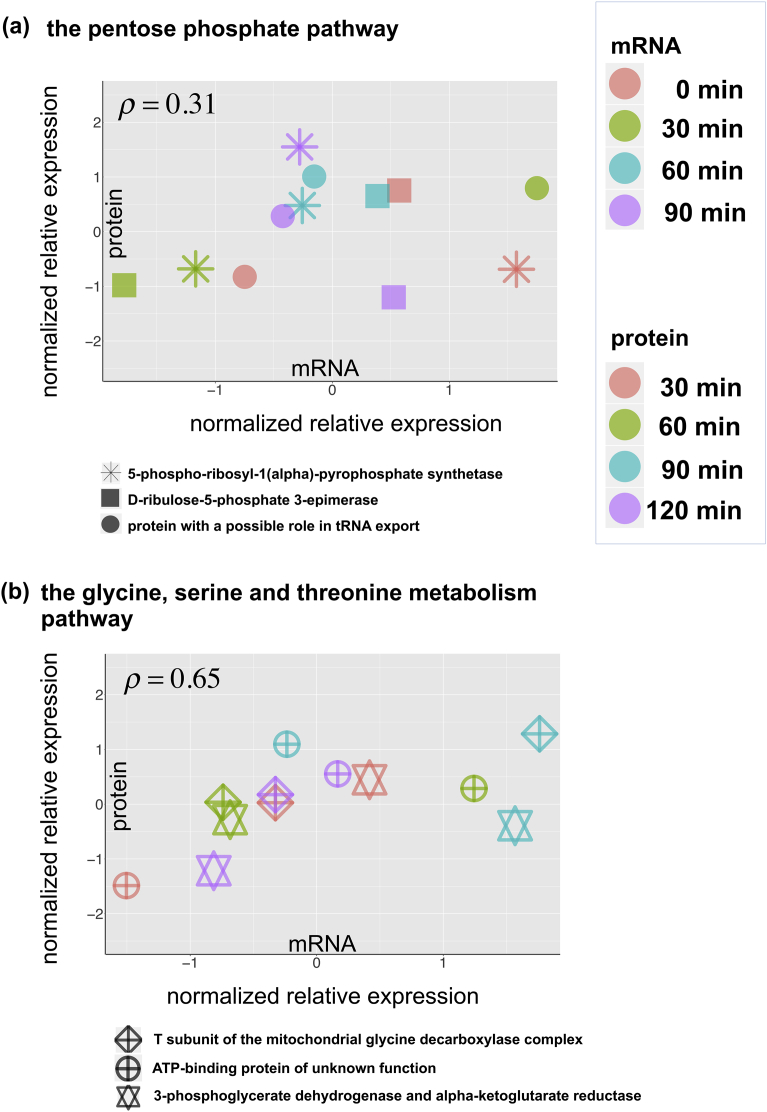
For the strata of genes with a time lag of 1, we conducted the correlation analysis after shifting one time point of the protein abundance values to adjust for the time lag. As a result, the stratified genes showed a correlation coefficient of 0.31 and 0.65 in the respective pathways (Fig. 5a and b), which were also larger than those of the global correlation analysis above without the stratification (0.13 and 0.27, respectively).
Fig. 5.
Correlation between mRNA and protein expression among subsets of genes that showed a time lag of 1. (a) Genes involved in the pentose phosphate pathway. (b) Genes involved in the glycine, serine and threonine metabolism pathway. Different shapes and colors of the dots indicate different genes and time points. The scatter plots were created after shifting the time series of the normalized relative expression of the protein to that of the mRNA.
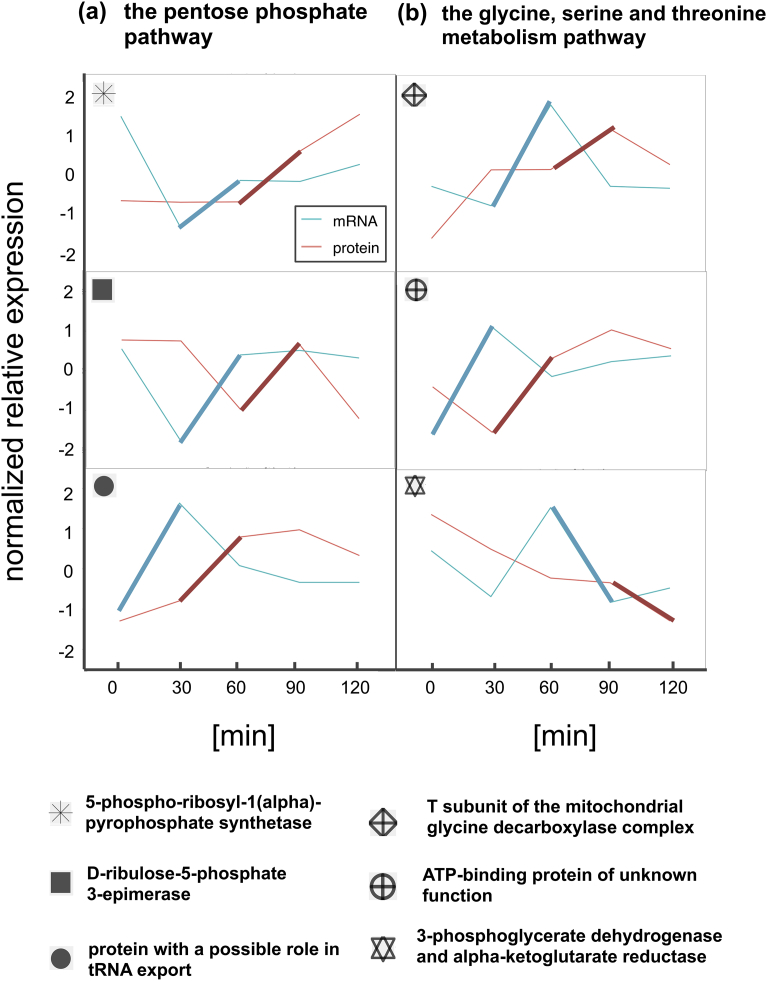
The time courses of mRNA and protein abundance of the three genes with a time lag of 1 in each pathway are shown in Fig. 6a and b. These genes were PRS4, RPE1, and SOL1, encoding 5-phospho-ribosyl-1(α)-pyrophosphate synthetase, d-ribulose-5-phosphate 3-epimerase, and a protein with a possible role in tRNA export, respectively, in the pentose phosphate pathway; and GCV1, TDA10, and SER33, encoding the T subunit of the mitochondrial glycine decarboxylase complex, an ATP-binding protein of unknown function, and 3-phosphoglycerate dehydrogenase, respectively, in the glycine, serine and threonine metabolism pathway. Indeed, we can see the time-delayed correlation with a time lag of 1, which is clarified as the thick lines in the plots corresponding to the largest value in {Mi, i + 1; i = 2, 3, 4} that measures the concordance of abundance changes between mRNA and protein from one time point before as explained in Materials and Methods. The stratified correlation analyses were not conducted for strata of genes with a time lag of ≥2 because of their small sample size.
Fig. 6.
Time courses of the normalized relative expression of genes that showed a time lag of 1. (a) Genes involved in the pentose phosphate pathway. (b) Genes involved in the glycine, serine and threonine metabolism pathway. The bold lines correspond to the largest value in {Mi, i + 1; i = 2, 3, 4} (as explained in Materials and methods) to clarify the time lag.
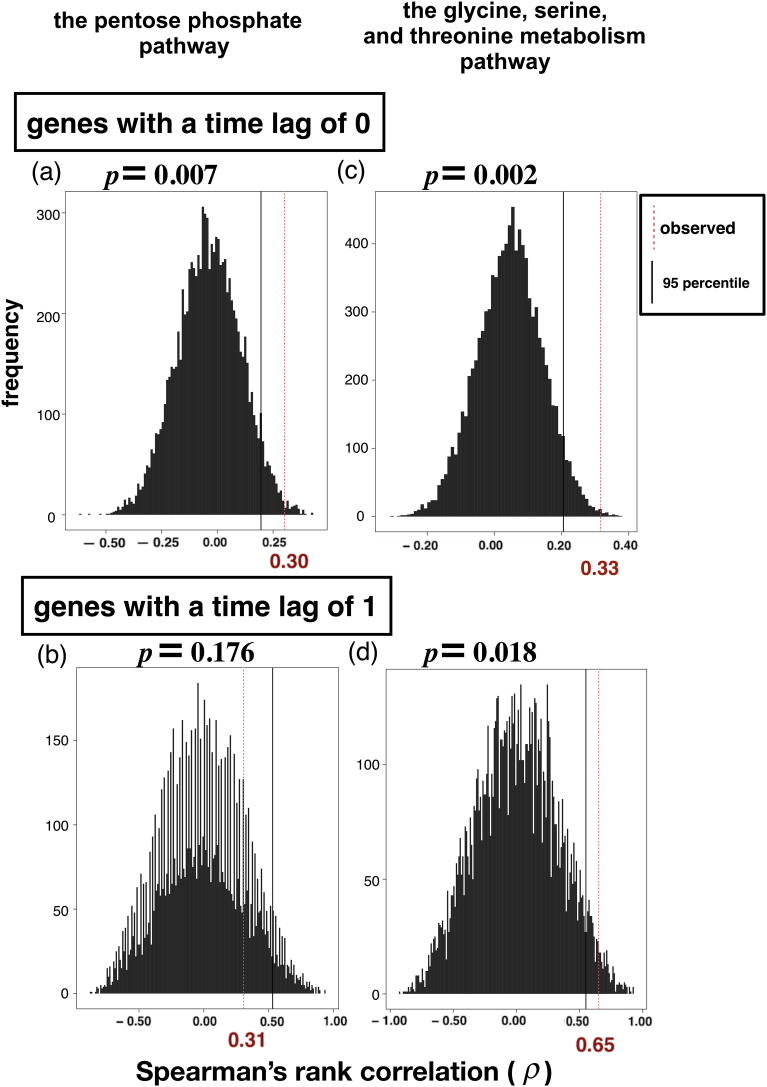
In order to test the significance of the increased correlation between mRNA and protein abundance after the stratification accounting for natural correlation among repeated measures in a time series, we conducted a permutation test for each stratum. We found that the observed increased correlations in the following strata of genes were significant: time lag of 0 (p = 0.007) in the pentose phosphate pathway, and time lags of 0 (p = 0.002) and 1 (p = 0.018) in the glycine, serine and threonine metabolism pathway (Fig. 7). Only the stratum with a time lag of 1 in the pentose phosphate pathway was tested to be not significant (p = 0.176, Fig. 7, bottom left panel).
Fig. 7.
Null distributions and observed values of the rank correlation statistics. The red values on the x-axes are observed rank correlation coefficients. The black vertical lines are the 95 percentiles in the null distributions. (a) Genes with a time lag of 0 involved in the pentose phosphate pathway, (b) genes with a time lag of 1 involved in the pentose phosphate pathway, (c) genes with a time lag of 0 involved in the glycine, serine and threonine metabolism pathway, and (d) genes with a time lag of 1 involved in the glycine, serine and threonine metabolism pathway.
In summary, we first identified the time-delayed correlation between mRNA and protein abundance in the two metabolic pathways that was suggested in a previous study. In addition, we were able to reveal clearer and significant correlations between the time series of mRNA and protein abundance by the stratification and statistical tests that accounted for the inferred time lag.
3.2. Genome-wide analysis
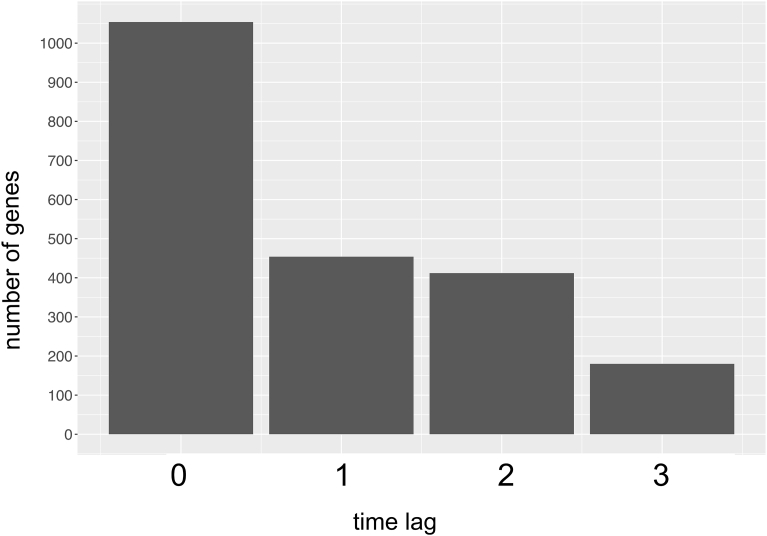
Next, we conducted similar analyses on the 2586 genes across the genome that had time-course data of both mRNA and protein abundance ([B] in Fig. 1). In 40.8% of the genes, the time lags between mRNA and protein abundance changes were inferred to be 0, whereas 40.5% of the genes showed positive inferred time-lag values (Fig. 8). The remaining genes showed negative inferred time-lag values, and were not included in Fig. 8 and the subsequent analyses because of difficulty of biological interpretation.
Fig. 8.
Distribution of inferred time lags (0, 1, 2, and 3) between the time series of mRNA and protein abundance changes in the genome-wide data.
After stratification of the genes by the inferred time lags, we conducted hierarchical clustering on each stratum with time lags of 0, 1, and 2 (gray circles in [B] in Fig. 1), respectively. Among a total of 1920 gene clusters across the strata, we identified 34 clusters that satisfied BP > 0.80 and AU p-value > 0.95. Among them, we excluded 3 clusters that showed negative correlation between mRNA and protein abundance because of difficulty of biological interpretation. The genes in these 31 clusters are listed in Table S1.
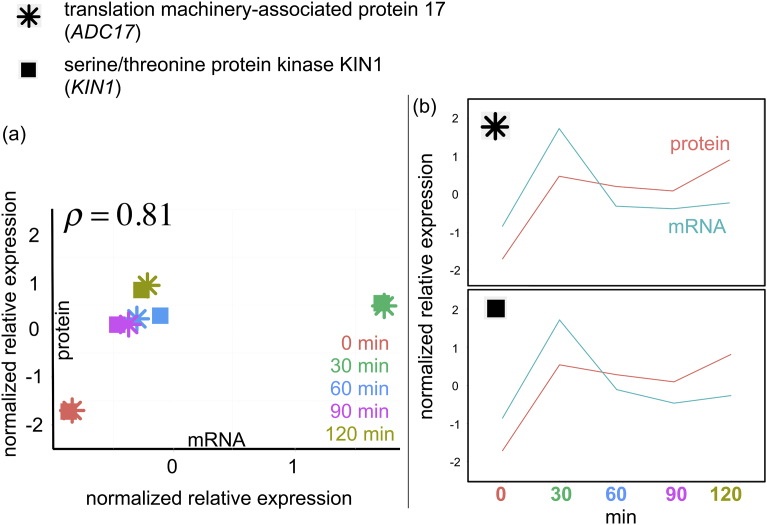
Among them, we found only one cluster that showed a statistically significant correlation between mRNA and protein abundance (Spearman's rank correlation coefficient = 0.81 in Fig. 9a; PFDR = 0.022 by the permutation test). Two genes were included in this cluster: translation machinery-associated protein (TMA17) also known as ADC17, and serine/threonine protein kinase (KIN1).
Fig. 9.
Examination of the gene cluster that showed statistically significant correlation between the mRNA and protein abundance. (a) Scatter plot expression. (b) Time courses.
ADC17 encodes a chaperone for proteasome assembly during stress response that is vital for cells to survive conditions such as an accumulation of misfolded proteins, and has recently gained attention as a key protein in maintaining proteasome homeostasis in yeast cells [15, 16]. Its absence aggravates proteasome defects [15] that are associated with numerous diseases in humans [17]. Cells generally increase proteasome abundance when demand increases upon environmental stress, and the abundance of Adc17 also increases upon the stress condition [15]. KIN1 plays a central role in regulating cell polarity and exocytosis [18], and in the unfolded protein response in the endoplasmic reticulum (ER), a process that resolves the unfolded and misfolded proteins during ER stress [19]. KIN1 is related to cellular sensitivity to stress in fission yeast, and its deletion makes cells hypersensitive to several stress conditions, including upward shifts in osmotic pressure [20]. Recently, Kin1 and its homolog Kin2 were reported to play a role in the unfolded protein response in the ER, a process that resolves unfolded and misfolded proteins during ER stress [19, 21].
The time courses of mRNA and protein abundance changes for these two genes are shown in Fig. 9b. Clearly, the abundance changes after the osmotic stress are highly similar without time lag across the two genes, suggesting a concerted role of these genes in cellular stress response (see Discussion).
3.3. Application to another dataset
Furthermore, we confirmed robustness of the analysis framework by applying it to another genome-wide time-series data in mammalian cells responding to stress of the endoplasmic reticulum (ER). As a result, we identified two gene clusters, consisting of genes with time-lag of 0 and 2, respectively, which showed a statistically significant correlation between mRNA and protein abundance. Spearman's rank correlation coefficient was 0.75 (PFDR = 0.003, the permutation test) and 0.66 (PFDR = 0.022, the permutation test), respectively.
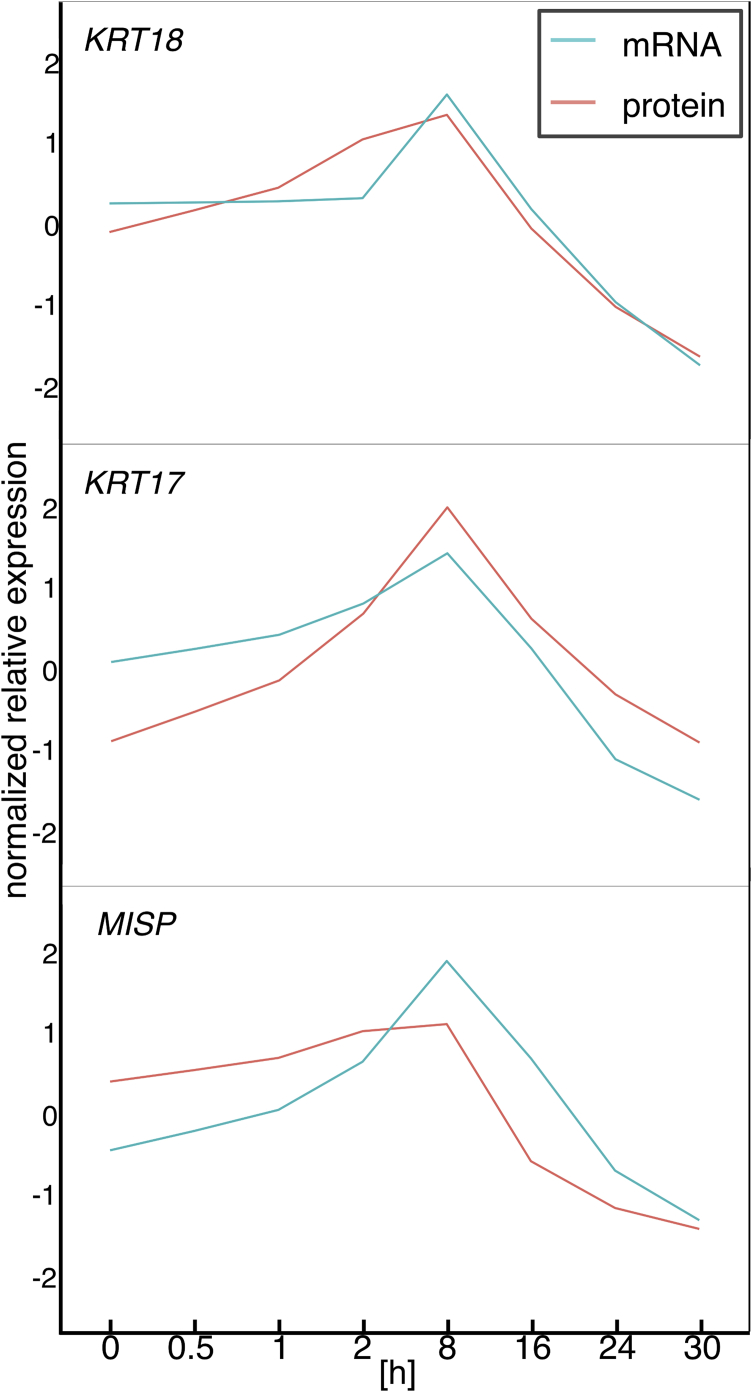
The cluster of genes with time lag of 0 was interpretable, and consisted of three cytoskeleton-related genes: keratin 18 (KRT18), keratin 17 (KRT17), and mitotic spindle positioning (MISP). KRT18 and KRT17 encodes the type I intermediate filament chain keratin 18 and 17, respectively. The protein encoded by MISP, mitotic spindle positioning, is an actin-bundling protein involved in determining cell morphology and mitotic progression.
Time courses of mRNA and protein abundance changes for these three genes are shown in Fig. 10, indicating highly similar increase until 8 h after the stress. That is a period in which the previous study [4] reported enrichment of mRNA expression changes of genes for apoptosis. Indeed, at least keratin 18 and keratin 17 are known to be related to apoptosis (see Discussion).
Fig. 10.
Time courses of mRNA and protein abundance in the gene cluster that showed statistically significant correlation in the data of mammalian cells. The gene cluster consists of the three cytoskeleton-related genes: keratin 18 (KRT18), keratin 17 (KRT17), and mitotic spindle positioning (MISP).
4. Discussion
In the present study, we first showed the existence of a time-delayed correlation between mRNA and protein abundance changes among genes of two metabolic pathways. Although such correlation was suggested in a previous study [8], we verified it here by inferring the time-lag extent for each gene. Stratification of the genes in terms of the inferred time lag enabled us to find a higher correlation between the mRNA and protein abundance. Second, we extended our analysis to the genome-wide data, and performed the stratification in terms of the inferred time lag, followed by gene clustering in terms of time-course concordance of the mRNA and protein abundance changes. As a result, we identified a cluster consisting of a pair of genes that showed a statistically significant correlation between mRNA and protein abundance (PFDR = 0.022). This is the first report that has revealed specifically which genes increased their mRNA and protein abundance in a concerted manner after osmolarity stress. We consider that it provides an answer to the question of the specific relationships between mRNA and protein in a cell [1].
The pair of genes was ADC17 and KIN1. The Adc17 protein, which is crucial for maintaining homeostatic proteasome levels, is known as a stress-induced regulatory particle assembly chaperone protein (RAC) that increases upon proteasome stress. Cells have mechanisms to adjust proteasome assembly when demands increase, with the Adc17 protein being a critical effector of this process [15]. An increase in Adc17 leads to upregulation of the proteasome, which would increase amino acid pools and permit the translation of proteins important for survival [22]. With regard to its regulation, it was recently reported that increases in the abundance of Adc17 and of the proteasome in yeast were caused by inhibition of the central stress and growth controller, target of rapamycin complex 1 (TORC1) kinase [16].
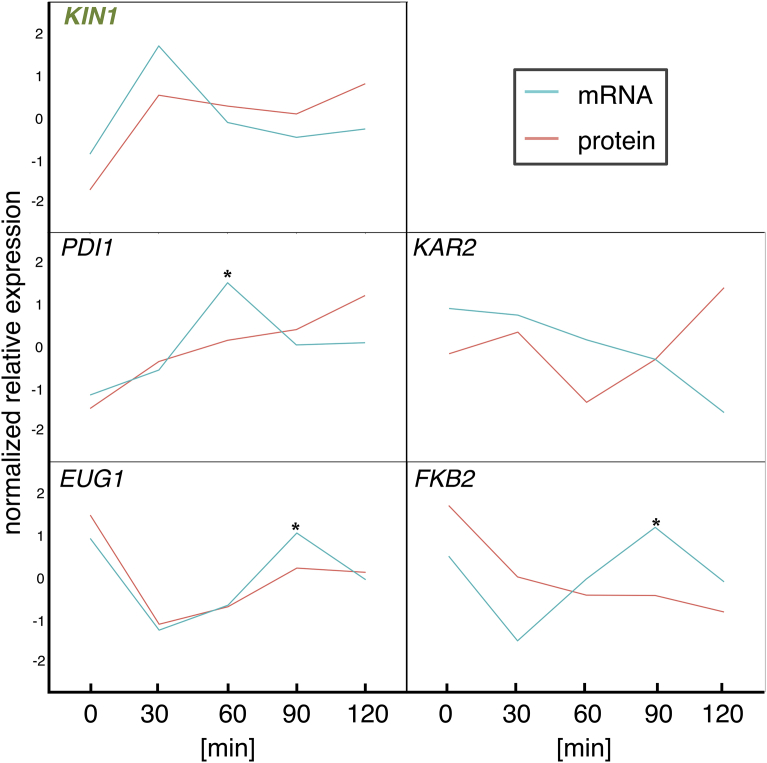
The Kin1 protein was recently reported to play a role in the unfolded protein response in the ER [19, 21]. The unfolded protein response is a signal transduction cascade that allows eukaryotic cells to respond to changing conditions, and resolves unfolded and misfolded proteins during ER stress by regulating the targeting, splicing, and translation of HAC1 mRNA [19, 23]. The Hac1 protein is a key transcription activator that binds to the promoter of unfolded protein response-regulated genes [24], such as KAR2, PDI1, EUG1, and FKB2. These genes encode enzymes that help to catalyze the correct folding of proteins [23, 25, 26]. In the absence of ER stress, ribosomes are stalled on unspliced HAC1 mRNA. ER stress is sensed by Ire1, which initiates the nonconventional splicing of HAC1 mRNA, thereby allowing synthesis of Hac1 protein from the spliced mRNA [23, 27, 28]. Although the Hac1 protein was not included in the dataset that we analyzed, we confirmed that the mRNA abundance of three out of the four unfolded protein response-regulated genes (PDI1, EUG1, and FKB2) had increased after the Kin1 protein's abundance peak (30 min after the osmotic shock, Fig. 11), which is consistent with the known function of this protein [19].The ADC17 and KIN1 genes are located in the same chromosome IV, but are approximately 430 kb distant from each other. Further studies are warranted to investigate what kind of mechanism enables the genes to be expressed in a quite similar manner upon exposure to osmotic stress both at mRNA and protein levels. It could be a novel common transcriptional regulation in these genes that are distantly located in the same chromosome.
Fig. 11.
Time series of mRNA and protein abundance of unfolded protein response (UPR)-regulated genes. The asterisks indicate peaks of mRNA abundance of the UPR-regulated genes (PDI1, KAR2, EUG1, and FKB2) at 30 or 60 min after the increase in Kin1 protein expression.
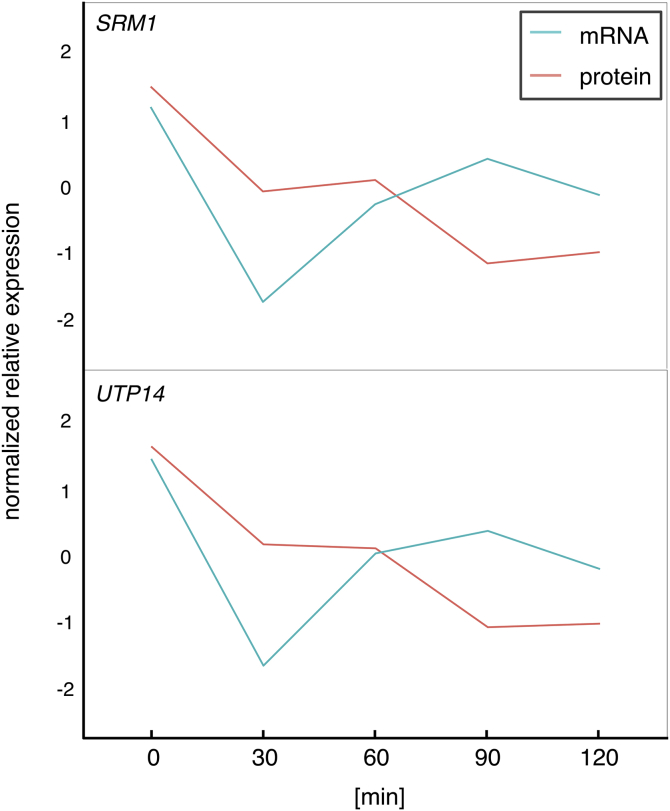
The present study spotlighted the pair of genes that showed a statistically significant correlation between mRNA and protein abundance. On the other hand, clear majority of genes did not show the statistically significant correlation, suggesting that protein abundance often could not be simply explained by the changes in mRNA but rather might be regulated by unknown mechanisms. An example is shown in Fig. 12 for SRM1 (nucleotide exchange factor that controls RNA metabolism and transport, involves in yeast pheromone response pathway, and required for mRNA and ribosome nuclear export) and UTP14 (a component of the small subunit (SSU) processome that is required for the maturation of the pre-18S rRNA) genes. After 30 min from the osmotic stress, both genes showed time-courses of protein abundance that were quite different from those of mRNA. Further studies are also warranted to deepen understanding of such relationships between mRNA and protein abundance that were not focused in the present study.
Fig. 12.
Examples in which protein abundance could not be simply explained by the changes in mRNA. Upper: SRM1 gene; Lower: UTP14 gene.
We confirmed robustness of the analysis framework by applying it to another genome-wide time-series data in mammalian cells responding to stress of the endoplasmic reticulum (ER) and identifying the cytoskeleton-related gene cluster: keratin 18 (KRT18), keratin 17 (KRT17), and mitotic spindle positioning (MISP). Time courses of mRNA and protein abundance changes for these three genes (Fig. 10), indicated highly similar increase until 8 h after the stress in which the previous study [4] reported enrichment of mRNA expression changes of genes for apoptosis. It was reported that the keratin 8/18 intermediate filaments are required for the apoptosis-promoting function of eIF3k (the subunit k of eukaryotic initiation factor 3) [29]. In apoptotic cells, eIF3k colocalizes with keratin 8/18-containing inclusions and promotes the release of active caspase 3 from the insoluble compartment via a keratin 8/18-dependent manner. Studies in keratin 17-null mice uncovered several roles including resistance to TNFα-induced apoptosis [30]. MISP functions as a single actin-binding effector of cell morphology [31] and could be related to the morphological modifications of the apoptotic cell. The results suggest a concerted role of these genes in apoptosis after the ER stress in mammalian cells.
We inferred the extent of the time lag between mRNA and protein abundance changes, using an algorithm that was originally proposed to assess staggered relationships between the mRNA expression of pairs of genes in gene-regulation network analysis [10]. In this study, we used the algorithm for gene stratification by considering the extent of inferred time lags between mRNA and protein abundance changes. Namely, we applied the method to data of mRNA and protein abundance of the same gene rather than those of different genes [10].
We tested the statistical significance of the correlation between the time series of mRNA and protein abundance by using a permutation test, with Spearman's rank correlation as a test statistic, as was done in the previous study [7], in order to take natural correlations among repeated measures in a time series [14] into account. We assessed this permutation test by a simulation and confirmed that the type-I error rate was only slightly higher than 0.05. We conducted the permutation test for each stratum identified by gene clustering rather than for each gene, which increased the sample size and statistical power (note that any statistical test at the gene level was impossible because data were available only at the five time points).
Based on the extension, utilization, and improvement of the analysis methods, the present study provides a framework to study specific relationships between the mRNAs and their proteins in a cell, the details for which up to now have been controversial [1, 3]. Our framework provides a basis for identifying the kinds of genes or biological gene groups that have significant correlation between mRNA and protein abundance after accounting for potential time delays.
Declarations
Author contribution statement
Shimpei Morimoto, Koji Yahara: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank Dr. Nathalie Selevsek at ETH Zürich for providing us with the genome-wide time-series data. The computational calculations were performed at the Human Genome Center at the Institute of Medical Science (the University of Tokyo). We would like to thank Editage (www.editage.jp) for English language editing.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
The proteome (ProtSWATH_testResult_final.xlsx) and transcriptome (Gene2011_testResult_final.xlsx) data that were combined and analyzed in a previous study (detailed in Section 2.1 in Materials and methods).
The proteome and transcriptome data that were analyzed in a previous study (detailed in Section 2.6 in Materials and methods).
References
- 1.Jovanovic M., Rooney M.S., Mertins P., Przybylski D., Chevrier N., Satija R. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347(6226):1259038. doi: 10.1126/science.1259038. PubMed PMID: 25745177; PubMed Central PMCID: PMCPMC4506746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13(4):227–232. doi: 10.1038/nrg3185. PubMed PMID: 22411467; PubMed Central PMCID: PMCPMC3654667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165(3):535–550. doi: 10.1016/j.cell.2016.03.014. PubMed PMID: 27104977. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Z., Teo G., Krueger S., Rock T.M., Koh H.W., Choi H. Differential dynamics of the mammalian mRNA and protein expression response to misfolding stress. Mol. Syst. Biol. 2016;12(1):855. doi: 10.15252/msb.20156423. PubMed PMID: 26792871; PubMed Central PMCID: PMCPMC4731011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee M.V., Topper S.E., Hubler S.L., Hose J., Wenger C.D., Coon J.J. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 2011;7:514. doi: 10.1038/msb.2011.48. PubMed PMID: 21772262; PubMed Central PMCID: PMCPMC3159980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fournier M.L., Paulson A., Pavelka N., Mosley A.L., Gaudenz K., Bradford W.D. Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol. Cell. Proteom. 2010;9(2):271–284. doi: 10.1074/mcp.M900415-MCP200. PubMed PMID: 19955083; PubMed Central PMCID: PMCPMC2830839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H., Wang Q., Pape U.J., Shen B., Huang J., Wu B. Systematic investigation of global coordination among mRNA and protein in cellular society. BMC Genom. 2010;11:364. doi: 10.1186/1471-2164-11-364. PubMed PMID: 20529381; PubMed Central PMCID: PMCPMC2900266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selevsek N., Chang C.Y., Gillet L.C., Navarro P., Bernhardt O.M., Reiter L. Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry. Mol. Cell. Proteom. 2015;14(3):739–749. doi: 10.1074/mcp.M113.035550. PubMed PMID: 25561506; PubMed Central PMCID: PMCPMC4349991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teo G., Vogel C., Ghosh D., Kim S., Choi H. PECA: a novel statistical tool for deconvoluting time-dependent gene expression regulation. J. Proteome Res. 2014;13(1):29–37. doi: 10.1021/pr400855q. PubMed PMID: 24229407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian J., Dolled-Filhart M., Lin J., Yu H., Gerstein M. Beyond synexpression relationships: local clustering of time-shifted and inverted gene expression profiles identifies new, biologically relevant interactions. J. Mol. Biol. 2001;314(5):1053–1066. doi: 10.1006/jmbi.2000.5219. PubMed PMID: 11743722. [DOI] [PubMed] [Google Scholar]
- 11.Gillet L.C., Navarro P., Tate S., Rost H., Selevsek N., Reiter L. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012;11(6) doi: 10.1074/mcp.O111.016717. O111.016717. PubMed PMID: 22261725; PubMed Central PMCID: PMCPMC3433915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 13.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51(3):492–508. doi: 10.1080/10635150290069913. PubMed PMID: 12079646. [DOI] [PubMed] [Google Scholar]
- 14.Yule G.U. Why do we sometimes get nonsense-correlations between Time-Series?–a study in sampling and the nature of time-series. J. R. Stat. Soc. 1926;89(1):1–63. [Google Scholar]
- 15.Hanssum A., Zhong Z., Rousseau A., Krzyzosiak A., Sigurdardottir A., Bertolotti A. An inducible chaperone adapts proteasome assembly to stress. Mol. Cell. 2014;55(4):566–577. doi: 10.1016/j.molcel.2014.06.017. PubMed PMID: 25042801; PubMed Central PMCID: PMCPMC4148588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rousseau A., Bertolotti A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 2016;536(7615):184–189. doi: 10.1038/nature18943. PubMed PMID: 27462806; PubMed Central PMCID: PMCPMC4990136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt M., Finley D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta. 2014;1843(1):13–25. doi: 10.1016/j.bbamcr.2013.08.012. PubMed PMID: 23994620; PubMed Central PMCID: PMCPMC3858528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbert M., Rossi G., Brennwald P. The yeast par-1 homologs kin1 and kin2 show genetic and physical interactions with components of the exocytic machinery. Mol. Biol. Cell. 2005;16(2):532–549. doi: 10.1091/mbc.E04-07-0549. PubMed PMID: 15563607; PubMed Central PMCID: PMCPMC545889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anshu A., Mannan M.A., Chakraborty A., Chakrabarti S., Dey M. A novel role for protein kinase Kin2 in regulating HAC1 mRNA translocation, splicing, and translation. Mol. Cell Biol. 2015;35(1):199–210. doi: 10.1128/MCB.00981-14. PubMed PMID: 25348718; PubMed Central PMCID: PMCPMC4295377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cadou A., Couturier A., Le Goff C., Soto T., Miklos I., Sipiczki M. Kin1 is a plasma membrane-associated kinase that regulates the cell surface in fission yeast. Mol. Microbiol. 2010;77(5):1186–1202. doi: 10.1111/j.1365-2958.2010.07281.x. PubMed PMID: 20624220. [DOI] [PubMed] [Google Scholar]
- 21.Yuan S.M., Nie W.C., He F., Jia Z.W., Gao X.D. Kin2, the budding yeast ortholog of animal MARK/PAR-1 kinases, localizes to the sites of polarized growth and may regulate septin organization and the cell wall. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0153992. PubMed PMID: 27096577; PubMed Central PMCID: PMCPMC4838231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chantranupong L., Sabatini D.M. Cell biology: the TORC1 pathway to protein destruction. Nature. 2016;536(7615):155–156. doi: 10.1038/nature18919. PubMed PMID: 27462809. [DOI] [PubMed] [Google Scholar]
- 23.Cox J.S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87(3):391–404. doi: 10.1016/s0092-8674(00)81360-4. PubMed PMID: 8898193. [DOI] [PubMed] [Google Scholar]
- 24.Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1(9):803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. PubMed PMID: 9077435. [DOI] [PubMed] [Google Scholar]
- 25.Gething M.J., Sambrook J. Protein folding in the cell. Nature. 1992;355(6355):33–45. doi: 10.1038/355033a0. PubMed PMID: 1731198. [DOI] [PubMed] [Google Scholar]
- 26.Shamu C.E., Cox J.S., Walter P. The unfolded-protein-response pathway in yeast. Trends Cell Biol. 1994;4(2):56–60. doi: 10.1016/0962-8924(94)90011-6. PubMed PMID: 14731868. [DOI] [PubMed] [Google Scholar]
- 27.Chapman R.E., Walter P. Translational attenuation mediated by an mRNA intron. Curr. Biol. 1997;7(11):850–859. doi: 10.1016/s0960-9822(06)00373-3. PubMed PMID: 9382810. [DOI] [PubMed] [Google Scholar]
- 28.Aragon T., van Anken E., Pincus D., Serafimova I.M., Korennykh A.V., Rubio C.A. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457(7230):736–740. doi: 10.1038/nature07641. PubMed PMID: 19079237; PubMed Central PMCID: PMCPMC2768538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y.M., Chen Y.R., Lin J.R., Wang W.J., Inoko A., Inagaki M. eIF3k regulates apoptosis in epithelial cells by releasing caspase 3 from keratin-containing inclusions. J. Cell Sci. 2008;121(Pt 14):2382–2393. doi: 10.1242/jcs.021394. PubMed PMID: 18577580. [DOI] [PubMed] [Google Scholar]
- 30.Pan X., Kane L.A., Van Eyk J.E., Coulombe P.A. Type I keratin 17 protein is phosphorylated on serine 44 by p90 ribosomal protein S6 kinase 1 (RSK1) in a growth- and stress-dependent fashion. J. Biol. Chem. 2011;286(49):42403–42413. doi: 10.1074/jbc.M111.302042. PubMed PMID: 22006917; PubMed Central PMCID: PMCPMC3234953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumeta M., Gilmore J.L., Umeshima H., Ishikawa M., Kitajiri S., Horigome T. Caprice/MISP is a novel F-actin bundling protein critical for actin-based cytoskeletal reorganizations. Genes Cells. 2014;19(4):338–349. doi: 10.1111/gtc.12131. PubMed PMID: 24475924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The proteome (ProtSWATH_testResult_final.xlsx) and transcriptome (Gene2011_testResult_final.xlsx) data that were combined and analyzed in a previous study (detailed in Section 2.1 in Materials and methods).
The proteome and transcriptome data that were analyzed in a previous study (detailed in Section 2.6 in Materials and methods).