Summary
Perturbations in stress granule (SG) dynamics may be at the core of amyotrophic lateral sclerosis (ALS). Since SGs are membraneless compartments, modeling their dynamics in human motor neurons has been challenging, thus hindering the identification of effective therapeutics. Here, we report the generation of isogenic induced pluripotent stem cells carrying wild-type and P525L FUS-eGFP. We demonstrate that FUS-eGFP is recruited into SGs and that P525L profoundly alters their dynamics. With a screening campaign, we demonstrate that PI3K/AKT/mTOR pathway inhibition increases autophagy and ameliorates SG phenotypes linked to P525L FUS by reducing FUS-eGFP recruitment into SGs. Using a Drosophila model of FUS-ALS, we corroborate that induction of autophagy significantly increases survival. Finally, by screening clinically approved drugs for their ability to ameliorate FUS SG phenotypes, we identify a number of brain-penetrant anti-depressants and anti-psychotics that also induce autophagy. These drugs could be repurposed as potential ALS treatments.
Keywords: amyotrophic lateral sclerosis, induced pluripotent stem cells, FUS, stress granules, autophagy, gene editing, CRISPR/Cas9n
Highlights
-
•
Generation of isogenic WT and P525L FUS-eGFP reporter iPSCs
-
•
P525L FUS-eGFP SGs are more numerous, more intense, and larger than WT
-
•
Increasing PI3K/AKT/mTOR-regulated autophagy reduces FUS-eGFP recruitment to SGs
-
•
Brain-penetrant drugs that induce autophagy ameliorate the FUS SG phenotype
Sterneckert and colleagues generate isogenic FUS-eGFP reporter iPSCs that enable the identification of stress granule (SG) phenotypes specifically induced by the ALS mutation FUS P525L. Compound screening shows that modulation of the PI3K/AKT/mTOR pathway regulating autophagy ameliorates SG phenotypes. A second screen identifies similarly acting brain-penetrant US FDA-approved drugs that could be repurposed to treat ALS.
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are neurodegenerative diseases characterized by progressive neurodegeneration and protein aggregation. In ALS, upper and lower motor neurons (MNs) degenerate, causing paralysis and death. In FTD, cerebral neurons degenerate to cause early-onset dementia. Although riluzole and edaravone are used for treating ALS, they only partially slow disease progression (Sawada, 2017, Writing Group and Edaravone (MCI-186) ALS 19 Study Group, 2017). Similarly, no medications specifically prevent or effectively slow FTD, thus effective therapies are urgently needed.
Genetics and neuropathology link ALS and FTD. About 10% of ALS cases are familial (fALS) and mostly caused by autosomal dominant genetic mutations. As the third most common monogenetic form of ALS is associated with FUS mutations, FUS has aroused considerable interest among researchers. In its wild-type (WT) form, the FUS protein interacts with components of the splicing machinery, binds and processes RNA, and shuttles between nucleus and cytoplasm for mRNA export and stress granule (SG) formation (Dormann et al., 2010). Mutations causing FUS-ALS primarily occur in two domains: the prion-like domain and the nuclear localization signal (NLS). NLS mutations, including P525L, which results in a severe early-onset form of ALS (Conte et al., 2012), cause aberrant localization of FUS to the cytoplasm (Dormann et al., 2010). Aggregates of FUS are a hallmark of FUS-ALS, but the dynamics of their formation are poorly understood. Significantly, cytoplasmic FUS aggregates are also found in FTD patients (Neumann et al., 2009), most of whom lack FUS mutations, demonstrating that disruption of FUS nuclear transport can occur even in the absence of mutations, perhaps by FUS protein misfolding.
Recent evidence suggests that SGs (non-membrane-bound compartments of regulated protein aggregation upon stress) may act as nucleation centers for the formation of insoluble aggregates in pathological conditions. FUS is recruited to SGs by its prion-like domain, and mutations in this domain promote FUS aggregation, resulting in liquid-to-solid phase transition. Other ALS-associated proteins behave similarly to FUS, including TAR DNA binding protein (TARDBP/TDP43), Ataxin2 (ATX2), TATA-box binding protein-associated factor 15 (TAF15), Ewing sarcoma RNA binding protein 1 (EWSR1), Heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1), and Heterogeneous nuclear ribonucleoprotein A2B1 (HNRNPA2B1). These contain prion domains, can be recruited to SGs, and are prone to protein aggregation when mutated. Similar to FUS, genetics and neuropathology studies show that TDP43, EWSR1, and TAF15 are linked to FTD (Mackenzie et al., 2010, Neumann et al., 2011), reinforcing the observation that ALS and FTD are connected.
Misregulation of autophagy also plays a critical role in ALS/FTD pathogenesis since disease-causing mutations were identified in many proteins that regulate this system, including Sequestosome 1 (SQSTM1/P62), valosin-containing protein (VCP), Optineurin (OPTN), TANK binding kinase 1 (TBK1), and chromosome 9 open reading frame 72 (C9ORF72). In addition, proteins such as heat-shock protein B8 (HSPB8) play a dual role in SG disassembly and promotion of autophagic removal of misfolded proteins (Ganassi et al., 2016), suggesting a link between autophagy and SG dynamics. Thus, compounds inducing autophagy might protect neurons from FUS pathology. However, the mechanism linking autophagy, SGs, and ALS pathogenesis is unclear, significantly impeding the identification of effective drugs for ALS/FTD.
The inability to directly probe patient MNs has hampered ALS research, and studying SGs in human neurons has been challenging due to their lack of membrane. Induced pluripotent stem cells (iPSCs), capable of immortal self-renewal and differentiation into specialized cell types, represent an exciting tool to recapitulate the disease and study the intricate biology of neurons. Multiple groups reported the generation of iPSCs from ALS-FUS patients (Japtok et al., 2015, Lenzi et al., 2015) and showed FUS to be recruited to SGs in mutant iPSC-derived MNs, with cytoplasmic aggregates manifesting upon increased neuronal aging (Japtok et al., 2015). However, the ability to monitor FUS and SG dynamics in human neurons in real time was still lacking.
Here, we report the generation of WT and P525L FUS-eGFP reporter iPSC lines using CRISPR/Cas9n-mediated gene editing. We confirm that the P525L mutation in the NLS domain causes mislocalization of FUS-eGFP protein to the cytoplasm and that FUS-eGFP is incorporated into SGs upon oxidative stress induction via arsenite treatment (Japtok et al., 2015). Even without stress, P525L FUS-GFP spontaneously formed puncta, particularly in cells expressing a variant of FUS-eGFP leading to exacerbated cytoplasmic mislocalization, and which we demonstrate were subcellular structures distinct from SGs. Although WT and mutant FUS-eGFP proteins are both recruited to SGs in our system, we show that SGs in iPSCs and iPSC-derived neuronal cultures with P525L FUS-eGFP contained more FUS-eGFP. With a phenotypic screening campaign, we identified inhibitors of the PI3K/AKT/mTOR pathway that reduced FUS-eGFP recruitment to iPSC SGs. Rapamycin, a well-characterized mTOR inhibitor and autophagy inducer, rescued the P525L FUS-linked SG phenotype in iPSCs and iPSC-derived neurons, although the response in neuronal cultures was delayed and weaker compared with iPSCs. The potent and direct ATP competitor torkinib rapidly ameliorated FUS SG phenotypes in neurons more efficiently. Using a Drosophila model of FUS-ALS, we further validated that autophagy induction significantly increases survival. As proof of principle, we further screened a library of clinically approved drugs and found that anti-depressants and anti-psychotics known to also induce autophagy ameliorated the FUS SG phenotype, suggesting they could be repurposed as ALS treatments. We conclude that the combination of isogenic reporter iPSCs with compound screening is a powerful tool to identify novel ALS drugs.
Results
Generation of WT and P525L FUS-eGFP Reporter iPSC Lines
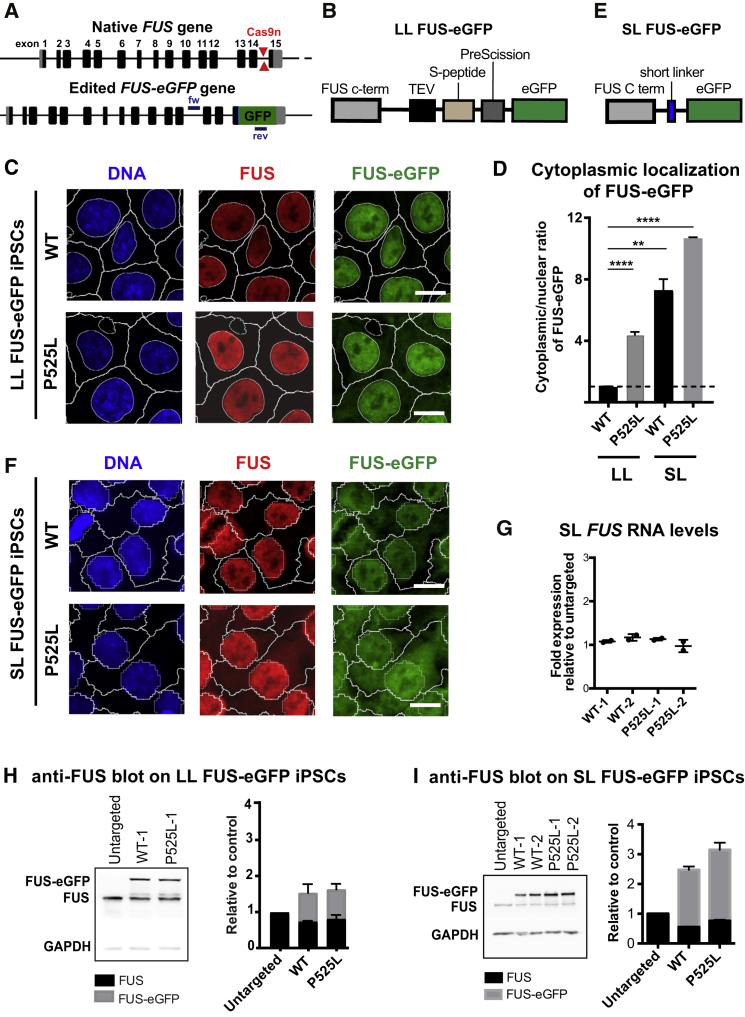
To study the molecular mechanisms of FUS-ALS, we generated isogenic FUS-eGFP reporter lines (Figure 1A). For this, we used WT iPSC lines that had been previously derived and characterized from healthy donors (details in Supplemental Information). Cells were transiently transfected for CRISPR-mediated gene editing with a plasmid containing Cas9n and two single guide RNAs to introduce two opposing nicks within the last intron of the FUS gene (Figures 1A and S1A). Simultaneously, we provided a donor construct (Figure S1A) containing a long and well-characterized linker, which would prevent the eGFP tag from interfering with FUS protein folding (Figure 1B). One of the donor constructs included C1679T, which encodes the P525L mutation known to cause a severe form of FUS-fALS. Genotyping showed targeted integration of the eGFP reporter sequence (Figure S1B). Sequencing of the edited region confirmed the presence of the P525L mutation in mutant lines and excluded incorporation of unwanted mutations at the FUS-eGFP junction site in both WT and mutant lines (Figure S1C). Immunostaining proved that gene-edited iPSCs maintained expression of the pluripotency markers OCT4, SOX2, and SSEA4 (Figure S1F). Finally, metaphase spread analysis showed euploidy (Figure S2).
Figure 1.
Generation of FUS-eGFP Reporter iPSCs
(A–F) Examples of FUS gene before and after gene editing to insert eGFP (A). Cas9n cut sites are shown with locations of primers used for genotyping. Schematic representation of the linkers used to knock in eGFP in LL (B) and SL (E) iPSC lines. Confocal micrographs showing FUS localization in WT and P525L FUS-eGFP cells for LL (C) and SL (F). Nuclear and cytoplasmic segmentation is displayed. Scale bar, 10 μm. (D) Quantification of the FUS-eGFP cytoplasmic-nuclear ratio in WT and P525L FUS-eGFP LL and SL iPSCs. All values are expressed as fold relative to LL WT FUS-eGFP iPSCs. Data represent results from three independent measurements. Error bars indicate SD. ∗∗p < 0.01 and ∗∗∗∗p < 0.0001.
(G–I) qRT-PCR showing total FUS RNA levels (G). WB and relative quantification using an anti-FUS antibody showing expression of FUS and FUS-eGFP in LL (H) and SL (I) iPSCs. Results are shown for two different WT and P525L lines as biological replicates and were reproduced in three independent experiments. Error bars indicate SD.
P525L lies in the NLS of FUS protein, and ALS mutations in the NLS domain were reported to cause FUS protein mislocalization to the cytoplasm (Dormann et al., 2010). Thus, we used fluorescence microscopy to examine the localization of WT and P525L FUS-eGFP. WT FUS-eGFP primarily localized to the nucleus (Figure 1C). Consistent with previous reports, P525L FUS-eGFP had increased cytoplasmic localization, with substantial FUS-eGFP still detected in the nucleus (Figure 1C). Quantification of the cytoplasmic-nuclear ratio revealed a 4-fold increase of cytoplasmic FUS-eGFP in P525L iPSC lines compared with WT (Figure 1D).
Because FTD is associated with cytoplasmic aggregation of WT FUS, we speculated that further impairing the nuclear localization of FUS may exacerbate disease pathology, which could be a considerable advantage for disease modeling considering the typically late onset of ALS and FTD. For this reason, we generated a second set of iPSC reporter lines carrying a shorter linker stretch (Figure 1E), which we speculated would increase aberrant cytoplasmic localization by disrupting the interaction with nuclear transport receptors such as Transportin and/or interfering with FUS folding, particularly considering proximity to the adjacent NLS. Genotyping and sequencing demonstrated successful editing. Immunostaining showed maintenance of pluripotent markers, and metaphase spread analysis confirmed euploidy (Figures S1D–S1F and S2). Confocal fluorescence microscopy followed by quantification indicated that short linker (SL) cells, unlike long linker (LL), had cytoplasmic WT FUS-eGFP (Figure 1F). Since FUS was WT, this was likely due to disrupted function of the FUS NLS. Consistent with the results from LL cells, P525L-eGFP in SL cells showed higher levels of cytoplasmic mislocalization (Figures 1D and 1F).
We further characterized our FUS-eGFP reporters to understand the mechanism of increased cytoplasmic FUS-eGFP localization. qRT-PCR demonstrated that knocking in the eGFP sequence in SL cells did not alter FUS expression levels (Figure 1G). Western blot (WB) analysis confirmed production of the FUS-eGFP fusion protein in LL and SL reporter lines. Blots probed with anti-FUS antibody revealed two bands of 75 and 100 kDa, corresponding to the native and the fused protein, respectively (Figures 1H and 1I), which demonstrated heterozygosity for the reporter sequence. Blots probed with anti-eGFP antibody showed one band at about 100 kDa, consistent with the FUS-eGFP fusion protein (Figures S3A and S3B). Quantification revealed reduced turnover of the FUS-eGFP protein in SL reporter cells, especially in P525L, where FUS-eGFP increased by about 1.2-fold compared with the WT counterpart (Figure 1I). Because FUS RNA levels were unchanged, this suggested that the SL eGFP tag as well as the P525L mutation affects protein turnover by reducing its clearance. Flow cytometry demonstrated that eGFP was uniformly expressed by WT and P525L SL FUS-eGFP cells in contrast to untargeted parental iPSCs, and median fluorescence intensity confirmed increased signal in mutant SL cells (Figure S3C). These trends were not evident in LL FUS-eGFP lines (Figures 1H and S3A).
P525L Alters FUS-eGFP SG Dynamics
Since FUS was reported to be recruited into SGs, we tested this in our iPSC models. iPSCs were treated with 0.5 mM sodium arsenite for 1 hr and immunostained for eIF3 and TIAR as eukaryotic SG markers (Figures 2A, S3D, and S3E). In SL lines, >95% of WT and P525L FUS-eGFP subcellular structures co-localized with eIF3 after arsenite treatment, confirming FUS-eGFP recruitment (Figures 2A and 2B). LL reporter lines behaved similarly, but FUS-eGFP was recruited less abundantly than in SL iPSCs, particularly in WT cells, where FUS-eGFP-positive SGs were almost imperceptible (Figure S3D). These data suggest that SGs preferentially recruit FUS-eGFP mislocalized to the cytoplasm, for example, by the ALS mutation P525L or by disrupted interaction with nuclear transport proteins due to impaired NLS function.
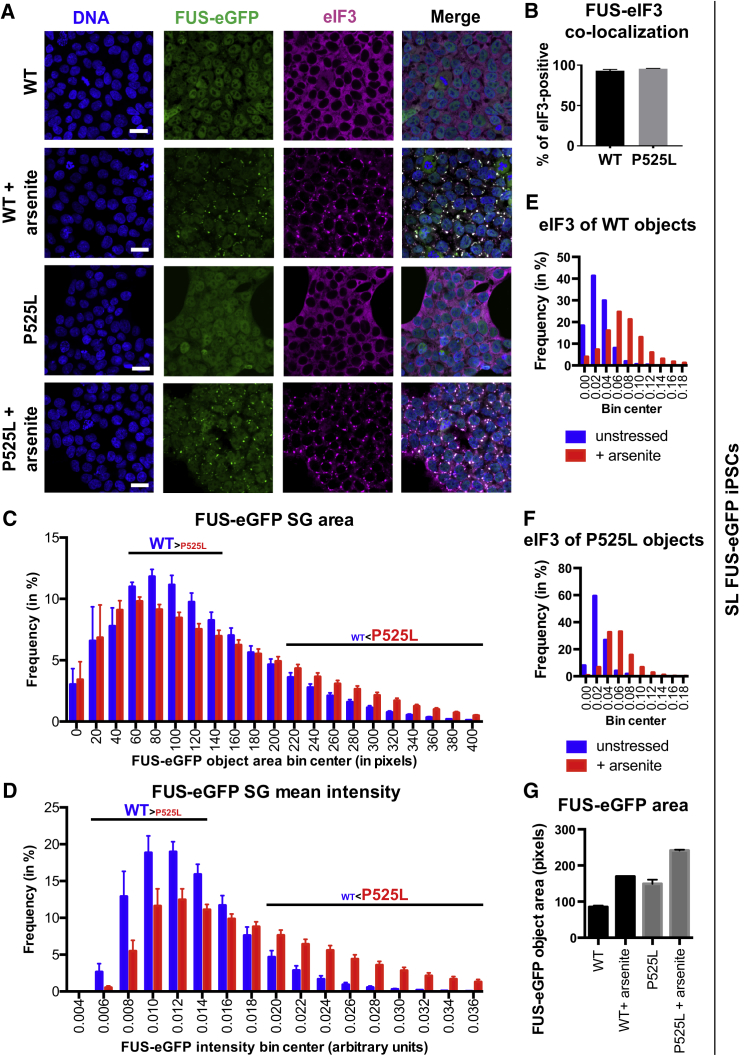
Figure 2.
Characterization of Stress Granules in WT and P525L FUS-eGFP SL iPSCs
(A) Confocal micrographs demonstrating that iPSCs form FUS-eGFP SGs that co-localize with eIF3. Scale bar, 20 μm.
(B–G) Quantification of SGs positive for FUS-eGFP in WT and P525L iPSCs (B). Error bars indicate SD. Histograms are shown for the frequency of WT and P525L FUS-eGFP SG area (C) and mean intensity (D). In contrast to SGs, FUS-eGFP puncta do not co-localize with eIF3-positive structures (E and F) and display smaller size (G). Data represent results from three independent experiments.
Our FUS-eGFP reporter lines are particularly powerful because they enable monitoring endogenous FUS protein dynamics in real time. Yet comparing the effects of WT and mutant FUS on SG dynamics in LL reporter cells is extremely challenging due to reduced FUS-eGFP recruitment into SGs. In contrast, since SL mutant lines recapitulate a more exacerbated pathology, they may facilitate the detection of phenotypes that would be too subtle in LL lines. We also hypothesized that a treatment ameliorating a disease phenotype in SL cells would also reduce disease-associated phenotypes in the context of a less severe pathology, such as that of LL cells. Therefore, to better understand the effects of P525L FUS on SG dynamics compared with WT FUS, we primarily focused on SL cells and used LL lines for validation. Live-cell imaging confirmed P525L FUS-eGFP mislocalization and recruitment to SGs upon arsenite stress. When cells were heat shocked, P525L FUS-eGFP iPSCs formed SGs at a lower temperature (38.5°C) compared with WT iPSCs (40°C; Figure S4A), demonstrating that the P525L mutation alters SG dynamics.
To quantify SG dynamics, we developed a phenotypic imaging assay that segmented each FUS-eGFP object and assessed the following parameters: number of FUS-eGFP objects per cell, area, and mean FUS-eGFP intensity. We applied this assay to unstressed SL iPSCs with WT and P525L FUS-eGFP (Figure S4B). Without added stress, P525L FUS-eGFP iPSCs manifested significantly increased numbers of FUS-eGFP objects per cell compared with WT (Figure S4C; 1.83 versus 0.27 objects per cell; p = 6.1 × 10−5). P525L FUS-eGFP objects were significantly larger (Figure S4D; 27.26 versus 18.12 pixels; p = 0.000143) and had significantly increased average fluorescence intensity (Figure S4E; 0.0111versus 0.0087 a.u.; p = 1.82 × 10−5) compared with WT.
Subsequently, we quantified SGs in WT and P525L SL FUS-eGFP iPSCs treated with 0.5 mM sodium arsenite (Figure S4F). Since arsenite induced a diverse population of SGs, individual FUS-eGFP SGs were divided into bins according to area and intensity, and the relative frequency of each bin was compared between WT and P525L. The population of FUS-eGFP SGs with a relatively small area (bin centers 60 through 140 pixels) was significantly less frequent in P525L compared with WT (Figure 2C). In contrast, there was a significant increase in the frequency of FUS-eGFP SGs with a larger area (220–400 pixels). Both trends became more striking when comparing populations of FUS-eGFP SGs binned for mean intensity (Figure 2D). Together, these data demonstrate that P525L leads to aberrant recruitment of FUS protein to SGs, which could be seeding aggregate formation and cause MN degeneration in FUS-ALS.
We investigated whether FUS-eGFP objects that formed spontaneously without arsenite in SL iPSCs were SGs or, if not, whether they had different characteristics from SGs. Thus, we performed immunostaining for the SG marker eIF3 and quantified its intensity within FUS-eGFP objects with or without arsenite. Histogram analysis showed that WT (Figure 2E) and P525L (Figure 2F) FUS-eGFP objects had a significantly lower eIF3 intensity compared with FUS-eGFP-positive SGs induced by arsenite. In addition, FUS-eGFP objects that formed spontaneously in the absence of arsenite (which we term “puncta” to distinguish them from arsenite-induced SGs) were much smaller compared with SGs. FUS-eGFP puncta were also detected in LL P525L FUS-eGFP iPSCs, although less frequently (Figure S4B).
Inhibition of PI3K/AKT/mTOR Pathway Ameliorates Aberrant SG Dynamics
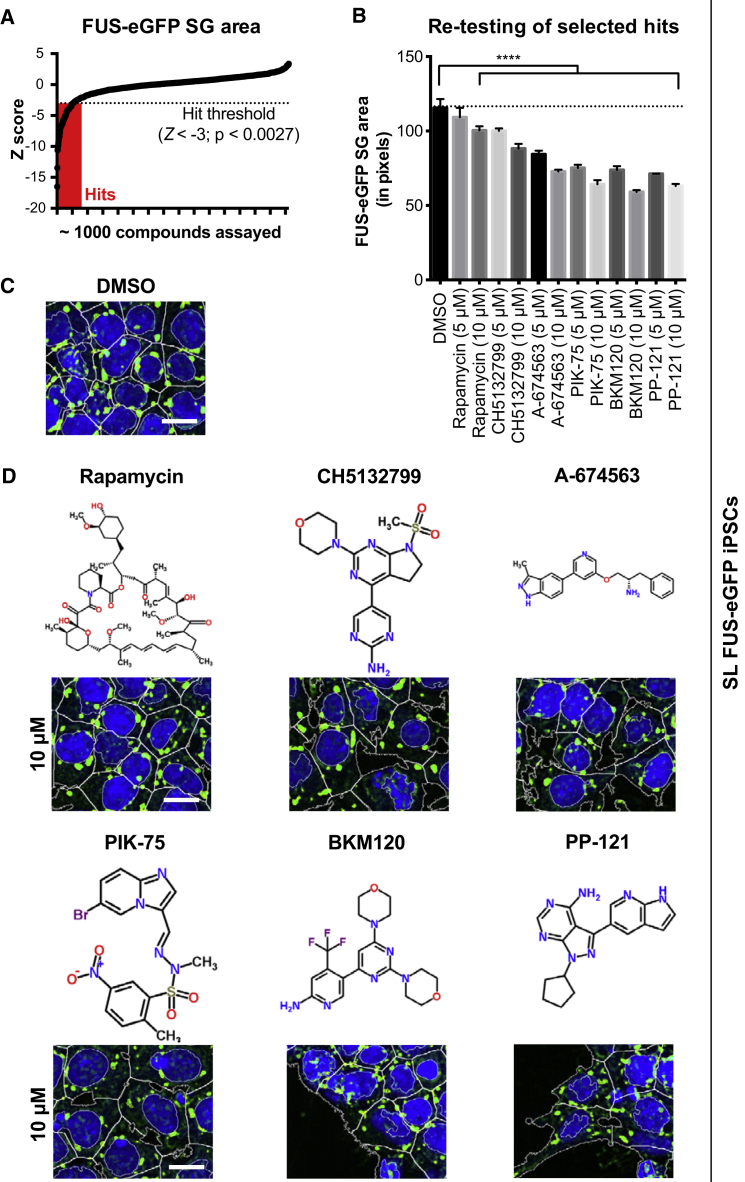
Compounds that modulate FUS SGs could represent novel therapeutics for ALS. Exploiting the scalability of our FUS-eGFP reporter lines, we performed a screening campaign of ∼1,000 small molecules against known targets to investigate the molecular mechanisms regulating FUS SG dynamics in SL P525L FUS-eGFP iPSCs. Cells were treated with each compound for 1 hr followed by addition of arsenite in the presence of the compound for an additional hour. Sixty-nine compounds showed significant reduction of the average FUS-eGFP area with a Z score < −3, giving a hit rate of about 6.1% (Figure 3A). Among these were 13 compounds that inhibit PI3K, AKT, and mTOR, a signaling pathway known to regulate autophagy, suggesting that inhibition of this pathway decreases FUS SG size in P525L iPSCs, making them more similar to their WT counterparts. To confirm these results, we validated five of these hits at two different concentrations (5 and 10 μM) and included rapamycin, a well-characterized mTOR inhibitor. Consistent with our primary screening results, each compound at 5 μM showed a significant reduction in FUS-eGFP SG area, and this reduction increased at 10 μM (Figures 3B–3D). We also performed a complete dose-response curve for rapamycin, which showed the strongest effects at 15 μM (data not shown).
Figure 3.
Identification of Compounds Reducing P525L FUS-eGFP SG Area in SL iPSCs
(A) Results for ∼1,000 compounds on P525L FUS-eGFP SG area arranged in ascending order. Compounds were considered as hits when Z < −3.
(B–D) Validation of selected compounds at 5 and 10 μM for their effects on P525L FUS-eGFP SG area (B). Each time, images were acquired from at least six positions/well from three different wells per line. Error bars indicate SEM. ∗∗∗∗p < 0.0001. Example micrographs are shown for DMSO control (C) and selected hit compounds (D). Chemical structures for the selected compounds were downloaded from ChemSpider. Scale bar, 10 μm.
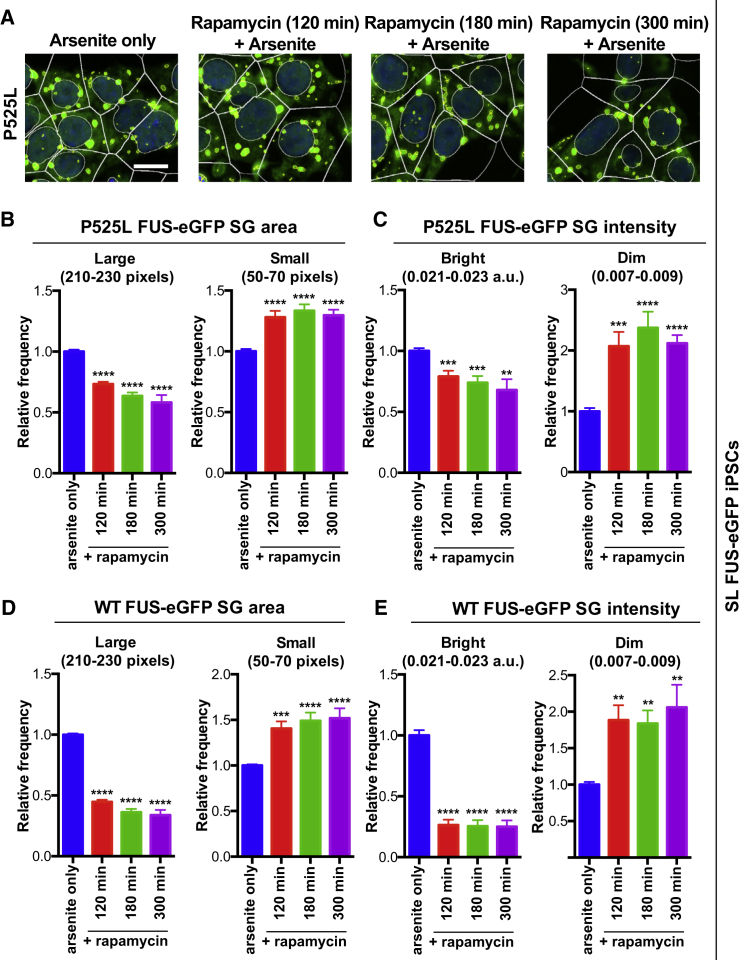
We then treated SL P525L FUS-eGFP iPSCs with 15 μM rapamycin for 120, 180, and 300 min with 0.5 mM sodium arsenite being included in the last 1 hr. FUS SGs were analyzed as populations; individual SGs were segmented (Figure 4A) and binned according to FUS-eGFP SG area or mean intensity. The frequency of large FUS-eGFP-containing SGs was significantly decreased by rapamycin, while the population of small FUS-eGFP SGs was significantly increased over time (Figure 4B). FUS-eGFP SG intensity behaved similarly (Figure 4C). These data demonstrate that mTOR inhibition reduces size and fluorescence intensity of the population of P525L FUS-eGFP SGs.
Figure 4.
Rapamycin Treatment Ameliorates P525L FUS-eGFP SG Pathology in SL iPSCs
(A) Confocal micrographs of P525L FUS-eGFP iPSCs treated with 15 μM rapamycin for time indicated. Arsenite was added for the final 1 hr. Scale bar, 10 μm. Individual FUS-eGFP drops were segmented and quantified by binning. The relative frequency of each bin was compared with 1 hr of arsenite only.
(B) Rapamycin treatment reduces P525L FUS-eGFP droplet area.
(C) Rapamycin treatment reduces P525L FUS-eGFP SG intensity.
(D and E) WT iPSCs respond more promptly to rapamycin than P525L. (D) Droplet area. (E) Intensity. n = 3 independent experiments, and in each experiment images were acquired from at least six positions/well from four different wells per line.
Error bars indicate SEM. ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Next, we quantified the effects of rapamycin on WT SL FUS-eGFP SGs and compared them with P525L. As with P525L, the frequency of large and bright WT FUS-eGFP SGs significantly decreased upon rapamycin treatment, while the frequency of small and dim SGs significantly increased (Figures 4D and 4E). Although rapamycin induced a comparable improvement, the dynamics of this transition were clearly altered, as WT FUS-eGFP iPSCs had a more rapid decrease in size and intensity. We speculate that mTOR inhibition may require more time to have an effect in P525L cells because, in addition to forming larger and more intense FUS-eGFP-containing objects compared with WT upon arsenite treatment (Figures 2C and 2D), P525L FUS-eGFP accumulates in puncta even when unstressed (Figures S4B–S4E).
We further validated the effects of mTOR inhibition using LL P525L FUS-eGFP reporter iPSCs. Cells were treated with 15 μM rapamycin for 120 min with 0.5 mM sodium arsenite being added for the last 1 hr (Figure S5A). Compared with arsenite only, P525L FUS-eGFP SGs were significantly smaller in the presence of rapamycin (Figure S5B) also in LL reporter lines, demonstrating that mTOR inhibition ameliorates the FUS SG phenotype independent of the linker.
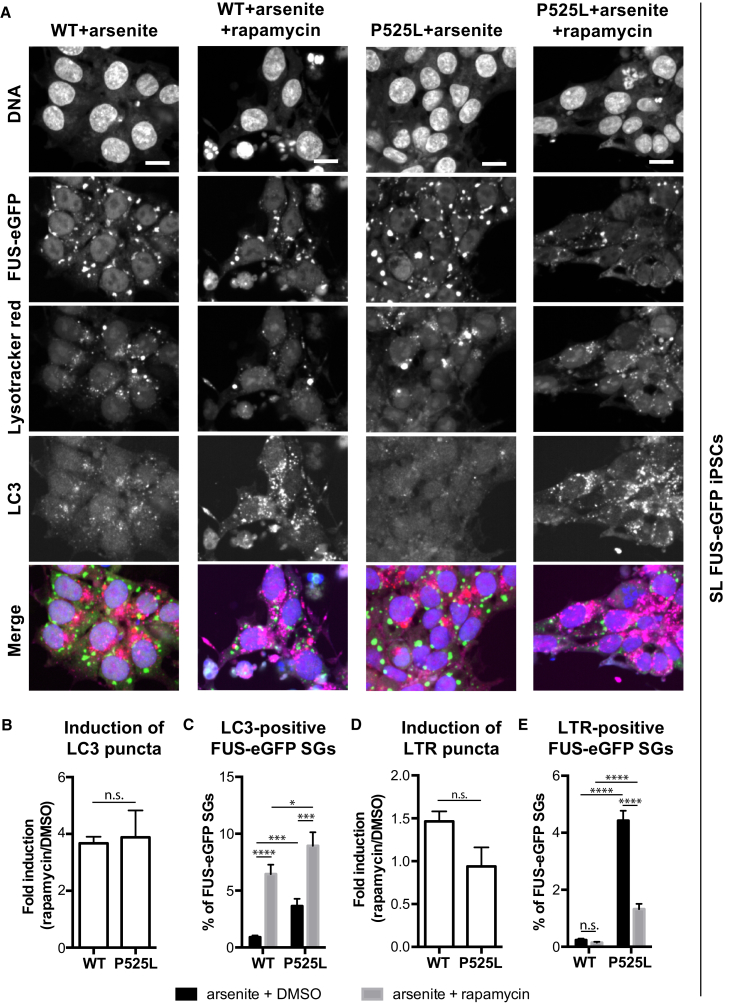
We then assessed autophagy. WB analysis of iPSC lysates showed strong LC3-II induction in the presence of 15 μM rapamycin within a short incubation period (Figure S5C). We analyzed autophagosome and lysosome distribution using LC3 immunostaining in combination with LysoTracker red (LTR; Figure 5A). We observed LC3 puncta in WT and P525L SL iPSCs in the presence of arsenite treatment, with greater numbers in WT cells (Figure 5B). Consistent with one report (Ganassi et al., 2016), we found less than 1% of FUS-eGFP SGs immunostained positive for LC3 in the presence of arsenite (Figure 5C). As expected, rapamycin induced a significant increase in the number of LC3 puncta per cell as well as a slight increase in LC3-positive FUS-eGFP SGs. Yet, the overall percentage of LC3-positive FUS-eGFP-containing SGs was less than 10%, which is too low to explain the effects of rapamycin on the SG dynamics described above.
Figure 5.
Induction of Autophagy Rescues P525L FUS-eGFP SG Pathology in iPSCs
(A) Confocal micrographs of WT and P525L FUS-eGFP SL iPSCs treated with 15 μM rapamycin for 5 hr. Arsenite was added for the final 1 hr. LysoTracker red labels lysosomes, LC3 immunostaining highlights autophagosomes. Scale bar, 10 μm.
(B) Two hours of rapamycin increases LC3 puncta per cell relative to DMSO.
(C) Analysis of the frequency of LC3-positive FUS-eGFP SGs after 5 hr of rapamycin.
(D) Increased frequency of LTR-positive puncta per cell upon rapamycin treatment for 2 hr relative to DMSO.
(E) Analysis of the frequency of LTR-positive FUS-eGFP SGs after rapamycin treatment for 5 hr. All experiments used arsenite for 1 hr. n = 3 independent experiments, and images were acquired from at least six positions/well from four different wells per line.
Error bars indicate SEM. ∗p < 0.05, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Rapamycin treatment also significantly increased the number of lysosomes, measured as LTR-positive organelles. However, no significant difference in the number of lysosomes was observed between WT and P525L with or without arsenite (Figure 5D). Because eGFP is sensitive to acidic conditions (Kneen et al., 1998), it should not co-localize with LTR when properly incorporated in acidic organelles for degradation. Accordingly, with arsenite only, almost no co-localization was observed between WT SL FUS-eGFP and LTR, which was even further reduced by rapamycin treatment (Figure 5E). In contrast, we observed a significant increase in the percentage of P525L FUS-eGFP SGs positive for LTR compared with WT. Again, the overall percentage of P525L FUS-eGFP was less than 5% and too low to fully explain the observed effects of rapamycin on SG dynamics as dependent on FUS SG degradation by autophagy. Nevertheless, this co-localization suggested impaired lysosomal degradation of P525L FUS-eGFP protein. Similarly, rapamycin significantly reduced the percentage of P525L FUS-eGFP SGs positive for LTR, but not to the same level as WT. Together, these data suggest that rapamycin may ameliorate FUS SG dynamics by degrading FUS proteins that become aberrantly incorporated into SGs, such as mislocalized cytoplasmic P525L FUS-eGFP, rather than inducing the degradation of aberrant FUS SGs.
Rapamycin and Torkinib Ameliorate FUS SG Phenotypes in iPSC-Derived Neurons
ALS is an MN disease so we extended our analysis from iPSCs to iPSC-derived neurons. WT and P525L FUS-eGFP iPSCs were differentiated into a mixed population of neuronal cells containing MNs using a two-step protocol (Figure S6A); iPSCs were differentiated into an intermediate population of expandable neural progenitor cells (smNPCs) followed by MN specification. Immunostaining demonstrated that WT and P525L smNPCs expressed the neural progenitor markers NESTIN and SOX1 (Figure S6B), while smNPC-derived neurons expressed MAP2, SMI32, ISLET1, and HB9 (Figure S6C). Quantification of ISLET1 and HB9 positivity, which mark human MNs (Stifani, 2014), revealed that approximately 10%–15% of cultured cells were MNs (Figures S6D and S6E); no difference was observed in differentiation efficiency between WT and P525L.
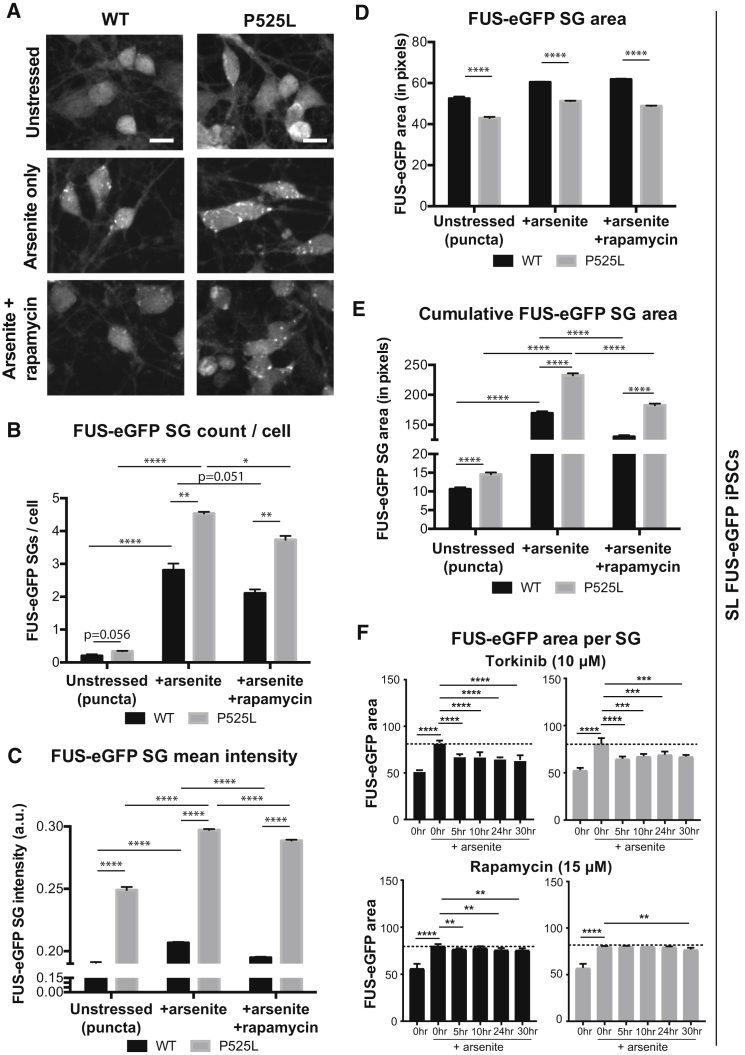
We applied our quantitative imaging assay to assess FUS-eGFP in SL iPSC-derived neuronal cultures containing MNs (Figure 6A). Without any stress, we observed more FUS-eGFP puncta per cell in P525L relative to WT (Figure 6B). Although statistical significance was not reached (p = 0.056), this was likely due to the technical challenge in quantifying objects in neuronal cell bodies, which are much smaller and more compact compared with iPSCs. When cultures of iPSC-derived neurons were treated with 0.5 mM arsenite for 1 hr, the number of FUS-eGFP objects per cell significantly increased. We detected significantly more FUS-eGFP SGs per cell and increased mean intensity in P525L neurons compared with WT (Figures 6B and 6C). In contrast to iPSCs, the area of FUS-eGFP SGs in neuronal cultures was significantly decreased in P525L compared with WT (Figure 6D). We speculated that the increased number of SGs per cell in P525L neurons may indicate that SGs are more fragmented, which predicted that the cumulative FUS-eGFP SG area within each cell would increase in P525L neurons compared with WT. To test this, we quantified the total area of FUS-eGFP SGs per cell (Figure 6E) and confirmed our hypothesis (Figure 6E). Rapamycin treatment ameliorated the phenotype for all tested parameters, although P525L cells were never fully rescued to WT levels. Because rapamycin induced a significant decrease in FUS-eGFP SG count per cell, mean intensity, and cumulative area in neurons, this suggests an important role for autophagy (Figures 6B–6E).
Figure 6.
Rapamycin Rescues P525L FUS-eGFP SG Pathology in SL iPSC-Derived Neurons
(A) Fluorescence confocal micrographs of P525L FUS-eGFP iPSC-derived neurons treated with 15 μM rapamycin in the presence of arsenite stress. Scale bar, 10 μm. Individual FUS-eGFP objects were identified and quantified. n = 3.
(B) Neurons with P525L FUS-eGFP have significantly more SGs per cell compared with WT and rapamycin reduces overall numbers.
(C) Neurons with P525L FUS-eGFP have significantly brighter SGs compared with WT. Rapamycin significantly decreases the brightness of WT and P525L FUS-eGFP SGs.
(D and E) P525L FUS-eGFP SGs in iPSC-derived neurons are significantly smaller compared with WT (D). However, total area of FUS-eGFP SGs per cell is significantly increased in neurons with P525L (E).
(F) Comparison of torkinib and rapamycin effect on neuronal SG area. Torkinib (top) significantly reduces SG area in both WT (left) and P525L (right) FUS-eGFP neurons after 5 hr of treatment. Rapamycin (bottom) is effective at the tested concentration only upon 24 hr treatment in P525L neurons (right). Effects on SG area appear earlier in WT cells (left), but are not as strong as under torkinib treatment.
Error bars indicate SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Autophagy was assessed in differentiated neurons using WB analysis for the conversion of soluble LC3-I to lipid-bound LC3-II. Here, we observed a clear increase in LC3-II in the presence of 15 μM rapamycin after 24 hr of treatment (Figure S5D). However, we found that LC3-II increased in neurons much more slowly than in iPSCs. Since rapamycin inhibits mTOR indirectly, differences in the response may exist between cells. For this reason, we also tested torkinib, which directly and selectively inhibits mTOR by competing with ATP. When neurons were treated with 10 μM torkinib for a time frame of 5–30 hr, SL FUS-eGFP SG area (Figure 6F) and intensity (data not shown) of both WT and P525L FUS-eGFP neurons were significantly reduced even upon the shortest treatment tested, i.e., 5 hr. Therefore, compared with rapamycin treatment performed under the same conditions (Figure 6F), torkinib proved more efficacious, which was also confirmed in LL FUS-eGFP neuronal cultures (Figure S7E). These results corroborated the link between autophagy and SG dynamics and highlighted the importance of studying ALS pathology in disease-relevant models, as the same compound may produce different effects in different cell types.
Stimulation of Autophagy Improves Neuronal Survival
We assessed if induction of autophagy, which reduced FUS-eGFP recruitment into SGs, would decrease neurodegeneration. We first immunostained neuronal cultures for cleaved caspase-3 (CC3), which labels apoptotic cells, in the presence or absence of rapamycin. For quantification, we used an unbiased approach based on setting different thresholds of CC3 intensity to define cells as “positive” for the apoptotic marker. We found that rapamycin significantly reduced the percentage of CC3-positive P525L SL FUS-eGFP neurons regardless of the selected threshold (Figures S7A and S7B). Next, we investigated changes in neuronal morphology by immunostaining neurons for TUBB3 followed by quantification of neurite branch length. Branch length is calculated as the connection between two crossing points. Thus, short branch lengths identify denser networks, which are formed by healthy neurons. Notably, rapamycin increased the number of short branch segments (Figure S7C). Together, these data show that autophagy stimulation reduces apoptosis in cultures of differentiated neurons and promotes healthy neuronal branching, consistent with improved survival.
Activation of Autophagy Rescues a Drosophila Model of FUS-ALS
We assessed the efficacy of inducing autophagy in vivo using a transgenic animal model of FUS-ALS. We employed a Drosophila model of ALS in which the neuronal ELAV-GS (gene switch) drove conditional expression of WT and P525L FUS throughout the CNS in adult animals. Consistent with previous reports (Daigle et al., 2013, Lanson et al., 2011), overexpression of WT and P525L FUS significantly truncated the animals' lifespan compared with non-transgenic controls (Figure S7D). To induce autophagy, UAS-Atg1 was expressed using the ELAV-GS driver. Atg1 is a direct target of mTOR and positively regulates the initiation of autophagy (Nakatogawa et al., 2009). As per our small-molecule results, Atg1 overexpression significantly extended the lifespan of transgenic flies expressing WT and P525L FUS (p < 0.0001; Figure S7D).
To further test the effects of autophagy stimulation on FUS-ALS pathology, we looked at retinal degeneration in our Drosophila model. We expressed WT or a disease-causing mutation in FUS in the fly eyes using the GMR-gal4 driver and found that overexpression of FUS led to external degeneration as evident from necrotic patches, loss of ommatidial organization, and depigmentation (Figure S7E). When atg1 was ectopically expressed, the eye degeneration phenotype was suppressed (Figures S7E and S7F). These results came from Drosophila models of FUS-ALS carrying other mutations than P525L, indicating that inducing autophagy may be an effective strategy for multiple ALS subtypes.
Clinically Approved Anti-depressants and Anti-psychotics Ameliorate SG Phenotypes
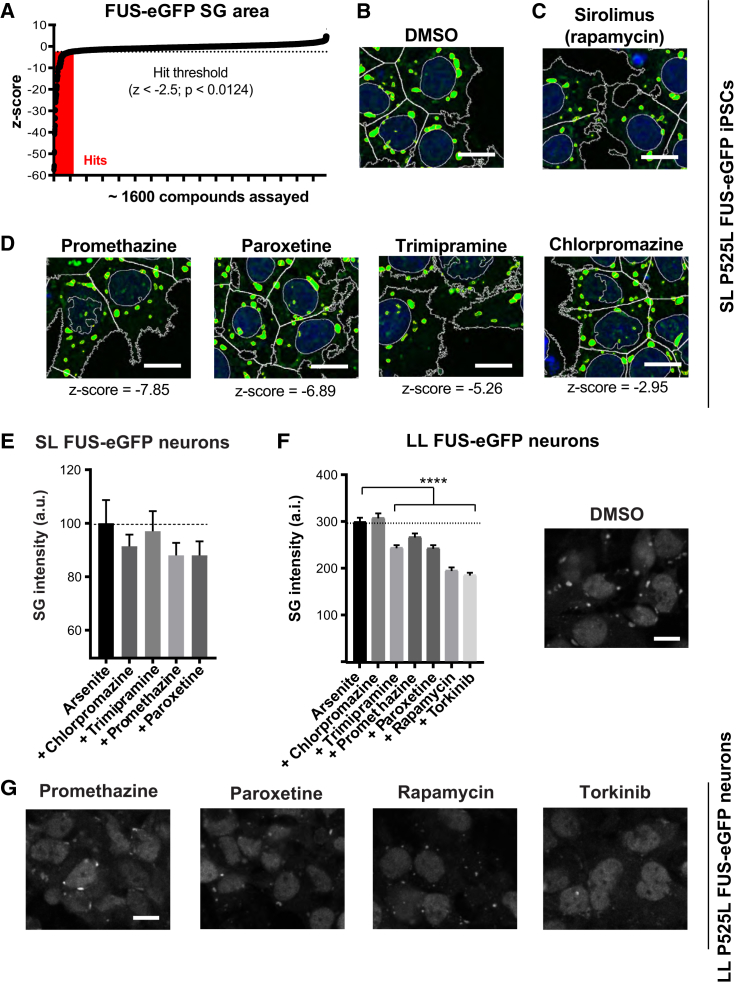
We aimed to identify clinically approved drugs that could reduce FUS-ALS SG phenotypes. SL P525L FUS-eGFP iPSCs were treated for 1 hr with ∼1,600 US Drug Collection MicroSource Discovery compounds at 10 μM each, followed by addition of 0.5 mM arsenite for 1 hr (Figure 7A). FUS-eGFP SG area was quantified and compared with controls. With −2.5 as a Z score threshold (p < 0.0124), 70 hit compounds were identified, including sirolimus (rapamycin), which corroborated our earlier results (Figures 7B and 7C).
Figure 7.
Identification of Drugs for Repurposing
(A–D) Chart of Z scores from drug screen identifying US FDA-approved drugs able to reduce SG area (A). Representative images are shown for DMSO control in (B), sirolimus in (C), and selected anti-depressants and anti-psychotics in (D). Scale bars, 10 μm.
(E) SL P525L FUS-eGFP neurons treated with chlorpromazine, paroxetine, promethazine, and trimipramine show a slight reduction in FUS SG intensity. Error bars represent SEM.
(F and G) LL P525L FUS-eGFP neurons treated with the indicated compounds show a reduction in FUS SG intensity (F), which is significant for paroxetine, promethazine, rapamycin, and torkinib as displayed in the respective confocal micrographs (G). Scale bar, 10 μm. n = 3 independent experiments and in each experiment, images were acquired from three positions/well from three different wells per line. Error bars represent SEM. ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Since MN cell bodies and dendrites are protected by the blood-brain barrier, brain-penetrant drugs would be ideal for treating ALS. Among the hits, we found a number of brain-penetrant anti-psychotics/depressants that reduced the area of P525L FUS-eGFP SGs induced by arsenite (Figure 7D). These drug hits, including paroxetine, promethazine, trimipramine, penfluridol, imipramine, and chlorpromazine, were reported to induce autophagy (Tsvetkov et al., 2010). Paroxetine and promethazine showed a trend toward ameliorating arsenite-induced FUS-eGFP SG phenotypes in SL P525L neurons (Figure 7E), although the effect did not reach statistical significance. When paroxetine, promethazine, and trimipramine were tested in the context of a milder and more physiological phenotype, such as that of P525L LL neurons, they produced a highly significant reduction of FUS-eGFP SG intensity (Figures 7F and 7G) and size (data not shown).
Discussion
Multiple proteins that are recruited to SGs, including FUS, were shown to cause ALS when mutated. Thus, SGs are hypothesized to be crucibles of FUS-ALS pathology (Li et al., 2013). Because many of these same proteins implicated in ALS also form aggregates in FTD, ALS and FTD are considered related. FUS is particularly interesting because, although aggregates are found in a subset of FTD patients, these patients almost never harbor FUS mutations, which, instead, are specifically associated with ALS. Understanding how improperly localized WT and/or mutated FUS protein affects SG dynamics is critical for developing therapeutics.
As SGs are membraneless compartments, investigating their behavior poses challenges. Here, we reported the first real-time analysis of endogenous SG dynamics in iPSCs and differentiated cultures of neurons including MNs. We generated isogenic FUS-eGFP iPSC reporter lines using CRISPR/Cas9n gene editing to knock in the ALS-causing mutation P525L as well as a GFP tag at the C terminus of the endogenous FUS protein. This approach is powerful to investigate the effects of specific genetic traits, since isogenic lines avoid variance caused by polymorphisms in the genetic background as well as overexpression artifacts.
Since cytoplasmic mislocalization of WT and mutant FUS protein are both linked to neurodegeneration, we generated two different types of reporters. An LL FUS-eGFP enabled monitoring native FUS protein, entirely nuclear in WT reporter cells. When these were treated with arsenite, SGs contained very weak levels of FUS-eGFP. Augmenting the levels of cytoplasmic FUS-eGFP by knocking in either the P525L mutation or an SL increased recruitment of FUS-eGFP to SGs in the presence of arsenite. Together, this evidence suggests that SGs preferentially recruit cytoplasmic FUS, which can be increased through diverse mechanisms, consistent with differences in the genetics of FUS-FTD and FUS-ALS. SL P525L FUS-eGFP cells showed the most severe phenotype and spontaneously formed subcellular structures under unstressed conditions. In contrast to arsenite-induced SGs, these objects were eIF3 negative and relatively small. To distinguish these objects from SGs, we termed them puncta, and we speculate that FUS aggregates may also arise from puncta independent of canonical SG formation.
It has been demonstrated that misfolded proteins as well as mutant FUS protein accumulate in SGs, which may be linked to aggregation (Ganassi et al., 2016, Patel et al., 2015). Consistent with this, we found that SGs showed greater recruitment of P525L FUS-eGFP compared with WT, leading to aberrant FUS SG dynamics. The life cycle of SGs can be broken down into multiple steps (Protter and Parker, 2016). First, nucleation occurs in the presence of oligomeric assemblies of messenger ribonucleoproteins (mRNPs). Next, the nucleated states increase in size by the addition of further mRNPs. In mammals, a third step is the merging of smaller SGs to form higher-order assemblies via mechanisms relying on microtubule-dependent transport. In iPSCs with mutant FUS-eGFP, we observed formation of SGs more rapidly compared with WT, as well as larger SGs. These observations suggest that in iPSCs, the sequence of steps is followed, but that mutant cells form large-sized, later-stage SGs faster than WT due to their intrinsic inclination to coalesce in aggregates caused by P525L-induced FUS mislocalization. Interestingly, while total FUS SG area was increased in neurons with mutant FUS, neuronal SGs were more fragmented compared with WT, suggesting a significantly decreased efficiency in coalescence, the last step of SG formation dependent on microtubule transport. Consistent with this, RNPs containing FUS are transported in MN axons (Schoen et al., 2015), and disruptions in microtubule transport are linked to ALS pathogenesis (Munch et al., 2004).
iPSCs' ability to limitlessly self-renew makes them ideal for drug screening. Thus, we used our FUS-eGFP reporter lines to identify small molecules with known targets, as well as approved drugs, that ameliorated SG phenotypes induced by P525L. Inhibition of the PI3K/AKT/mTOR pathway at different levels decreased FUS-eGFP SG numbers, area, and intensity, resembling WT conditions. The mTOR pathway is notoriously associated with a plethora of biological functions, including cell growth, proliferation, and survival. However, because mTOR kinase is a master regulator of autophagy induction, and defects in autophagy are prominent in diverse disease states such as neurodegenerative diseases, we explored the possibility of a mechanistic link between autophagy and SG dynamics. In contrast to one report (Ryu et al., 2014), we observed that only a small fraction of WT or P525L FUS-eGFP SGs were positive for LC3, indicating that very few SGs are degraded by autophagy. Similarly, Buchan et al. reported that disruption of autophagy increased SG formation, but this was linked to decreased SG clearance, resulting in their accumulation (Buchan et al., 2013). In contrast, our findings extend the results of Ganassi et al. (2016) by showing that triggering autophagy prior to SG induction ameliorates phenotypes in a dose- and time-dependent manner, without a significant fraction of SGs undergoing autophagy. We speculate this may be due to increased clearance of soluble mislocalized FUS proteins that would otherwise be incorporated into SGs. Further experiments are necessary to better characterize the interplay between autophagy and SG dynamics. Our observation that mTOR inhibition decreased the levels of CC3 and increased neuronal network arborization enables these mechanistic studies to be linked to human neurodegeneration.
Direct inhibition of mTOR proved beneficial in our cell-based experimental setup. However, rapamycin (or similarly, torkinib) administration is unlikely to be a viable clinical strategy for ALS because it is a potent immunosuppressant. Previously, rapamycin was shown to exacerbate ALS pathology in a mouse model of ALS, and it was beneficial only when lymphocytes were depleted (Staats et al., 2013). Thus, we also screened a library of clinically approved drugs to identify other potentially interesting compounds. We found multiple anti-depressants and anti-psychotics, whose mechanism of action was suggested to be independent of mTOR (Park et al., 2014, Tsvetkov et al., 2010). At least one of these drugs, paroxetine, was shown to ameliorate Huntington's and Alzheimer's pathology in mice, corroborating the idea that modulation of autophagy is beneficial in the context of neurodegenerative diseases. The identified drugs are brain penetrant and their effects well characterized. Importantly, promethazine, and chlorpromazine share the same chemical scaffold, which is directly related to the tricyclic structure of trimipramine and imipramine. This could be an ideal starting point for medicinal chemistry to optimize a novel compound that specifically ameliorates FUS pathology in ALS patients. It is important to note that increasing autophagy protected neurons against mutant TDP-43, suggesting that these compounds might be beneficial for multiple forms of ALS (Barmada et al., 2014). However, because our data link induction of autophagy to neuroprotection, it will may be necessary to administer these treatments when MNs are still functionally attached to skeletal muscle, i.e., upon diagnosis, and earlier diagnosis may be required, for instance, using gene sequencing, before symptom onset.
Experimental Procedures
Gene Editing
Dr. Boris Greber donated the guide RNA expression vector. Donor vectors were synthesized (Integrated DNA Technologies). iPSCs used in this project were previously generated (see Supplemental Information). A 1:1 mix of donor construct and guide RNA was used to transfect 175,000 iPSCs with FuGENE HD Transfection Reagent (Promega). Neural progenitor cells and neurons were derived as previously described (Reinhardt et al., 2013).
Immunocytochemistry Staining
Cells were fixed for 20 min with 4% paraformaldehyde (EM Sciences). Permeabilization and blocking were performed using 0.1% Triton X-100 (Sigma), 10% fetal calf serum (GE Healthcare), and 1% BSA in PBS (Lonza). Primary antibodies were applied overnight at 4°C. Secondary antibodies were applied for 1 hr at room temperature.
Compound Screening Protocol
iPSCs were seeded onto Greiner μClear 384-well plates. Compounds were added the next day at a final concentration of 10 μM. After 1 hr, cells were stressed with 500 μM sodium arsenite. Subsequently, cells were fixed with 3.7% formaldehyde and stained with Hoechst and CellMask blue. Plates were imaged with Yokogawa Cell Voyager CV7000 at 40× magnification.
Ethical Approval
All procedures involving human participants were performed in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Author Contributions
Conceptualization, S.A., M.B., A.H., A.A.H., and J.L.S.; Investigation, L.M., I.P., J.J., P.R., A.J., C.A., H.O.L., C.M., E.K., L.R., M.E.C., K.H., B.K., and A.P.; Writing, L.M. and J.L.S.; Project Administration, S.A., M.B., A.H., U.P., A.A.H., and J.L.S.
Acknowledgments
We gratefully acknowledge financial support from the Deutsche Forschungsgemeinschaft (DFG) and the CRTD/TUD. This work was financed by DFG Research Center (DFG FZ 111) and Cluster of Excellence (DFG EXC 168), including a seed grant. L.M. was sponsored by the CRTD and the Hans and Ilse Breuer Stiftung. This work was additionally supported by the CRTD Light Microscopy and FACS facilities. The Sterneckert lab is supported by the European Union's Horizon 2020 research and innovation program (643417) and the Bundesministerium für Bildung und Forschung (01ED1601B). This is an EU Joint Programme – Neurodegenerative Disease Research (JPND) project. The project is supported by the following funding organizations under the aegis of JPND—www.jpnd.eu: Germany, Bundesministerium für Bildung und Forschung; Israel, Ministry of Health; Italy, Ministero dell'Istruzione dell'Università e della Ricerca; Sweden, Swedish Research Council; and Switzerland, Swiss National Science Foundation. In addition, this work was supported by the Robert Packard Center for ALS at Johns Hopkins (to J.L.S. and U.P.). U.P. was supported by the USA National Institutes of Health (R21NS094921, R01NS081303, R21NS100055, R21NS098379, R21NS094921) and the Muscular Dystrophy Association. We thank Bill Skarnes for iPSC lines. This work was supported, in part, by the Deutsche Gesellschaft für Muskelerkrankungen (He2/2), the NOMIS Foundation, and the Helmholtz Virtual Institute RNA dysmetabolism in ALS and FTD (VH-VI-510) to A.H.
Published: January 18, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and seven figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.12.018.
Supplemental Information
References
- Barmada S.J., Serio A., Arjun A., Bilican B., Daub A., Ando D.M., Tsvetkov A., Pleiss M., Li X., Peisach D. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat. Chem. Biol. 2014;10:677–685. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., Kolaitis R.M., Taylor J.P., Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte A., Lattante S., Zollino M., Marangi G., Luigetti M., Del Grande A., Servidei S., Trombetta F., Sabatelli M. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul. Disord. 2012;22:73–75. doi: 10.1016/j.nmd.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Daigle J.G., Lanson N.A., Jr., Smith R.B., Casci I., Maltare A., Monaghan J., Nichols C.D., Kryndushkin D., Shewmaker F., Pandey U.B. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum. Mol. Genet. 2013;22:1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M.E., Mackenzie I.R., Capell A., Schmid B. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M., Mateju D., Bigi I., Mediani L., Poser I., Lee H.O., Seguin S.J., Morelli F.F., Vinet J., Leo G. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol. Cell. 2016;63:796–810. doi: 10.1016/j.molcel.2016.07.021. [DOI] [PubMed] [Google Scholar]
- Japtok J., Lojewski X., Naumann M., Klingenstein M., Reinhardt P., Sterneckert J., Putz S., Demestre M., Boeckers T.M., Ludolph A.C. Stepwise acquirement of hallmark neuropathology in FUS-ALS iPSC models depends on mutation type and neuronal aging. Neurobiol. Dis. 2015;82:420–429. doi: 10.1016/j.nbd.2015.07.017. [DOI] [PubMed] [Google Scholar]
- Kneen M., Farinas J., Li Y.X., Verkman A.S. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanson N.A., Jr., Maltare A., King H., Smith R., Kim J.H., Taylor J.P., Lloyd T.E., Pandey U.B. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum. Mol. Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi J., De Santis R., de Turris V., Morlando M., Laneve P., Calvo A., Caliendo V., Chio A., Rosa A., Bozzoni I. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Dis. Model. Mech. 2015;8:755–766. doi: 10.1242/dmm.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.R., King O.D., Shorter J., Gitler A.D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie I.R., Rademakers R., Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- Munch C., Sedlmeier R., Meyer T., Homberg V., Sperfeld A.D., Kurt A., Prudlo J., Peraus G., Hanemann C.O., Stumm G. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 2004;63:724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Neumann M., Bentmann E., Dormann D., Jawaid A., DeJesus-Hernandez M., Ansorge O., Roeber S., Kretzschmar H.A., Munoz D.G., Kusaka H. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Rademakers R., Roeber S., Baker M., Kretzschmar H.A., Mackenzie I.R. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.W., Lee J.G., Seo M.K., Lee C.H., Cho H.Y., Lee B.J., Seol W., Kim Y.H. Differential effects of antidepressant drugs on mTOR signalling in rat hippocampal neurons. Int. J. Neuropsychopharmacol. 2014;17:1831–1846. doi: 10.1017/S1461145714000534. [DOI] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Protter D.S., Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P., Glatza M., Hemmer K., Tsytsyura Y., Thiel C.S., Hoing S., Moritz S., Parga J.A., Wagner L., Bruder J.M. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One. 2013;8:e59252. doi: 10.1371/journal.pone.0059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H.H., Jun M.H., Min K.J., Jang D.J., Lee Y.S., Kim H.K., Lee J.A. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol. Aging. 2014;35:2822–2831. doi: 10.1016/j.neurobiolaging.2014.07.026. [DOI] [PubMed] [Google Scholar]
- Sawada H. Clinical efficacy of edaravone for the treatment of amyotrophic lateral sclerosis. Expert Opin. Pharmacother. 2017;18:735–738. doi: 10.1080/14656566.2017.1319937. [DOI] [PubMed] [Google Scholar]
- Schoen M., Reichel J.M., Demestre M., Putz S., Deshpande D., Proepper C., Liebau S., Schmeisser M.J., Ludolph A.C., Michaelis J. Super-resolution microscopy reveals presynaptic localization of the ALS/FTD related protein FUS in hippocampal neurons. Front. Cell. Neurosci. 2015;9:496. doi: 10.3389/fncel.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats K.A., Hernandez S., Schonefeldt S., Bento-Abreu A., Dooley J., Van Damme P., Liston A., Robberecht W., Van Den Bosch L. Rapamycin increases survival in ALS mice lacking mature lymphocytes. Mol. Neurodegener. 2013;8:31. doi: 10.1186/1750-1326-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifani N. Motor neurons and the generation of spinal motor neuron diversity. Front. Cell. Neurosci. 2014;8:293. doi: 10.3389/fncel.2014.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov A.S., Miller J., Arrasate M., Wong J.S., Pleiss M.A., Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc. Natl. Acad. Sci. USA. 2010;107:16982–16987. doi: 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Writing Group. Edaravone (MCI-186) ALS 19 Study Group Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017;16:505–512. doi: 10.1016/S1474-4422(17)30115-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.