Abstract
Lipoprotein lipase (LPL) is widely linked to lipid and lipoprotein metabolism, but its effects on coronary artery disease (CAD) are not clearly elucidated. The aim of the present study was to clarify the association between LPL gene polymorphisms and CAD susceptibility. The pooled odds ratio (OR) and 95% confidence interval (CI) were calculated to estimate the strength of the relationship between LPL gene polymorphisms and CAD risk. Comprehensive electronic databases, including PubMed, EMBASE, Web of Science, and the Cochrane Library, were systematically searched. A total of 45 records containing 80 eligible studies were analyzed. The results indicated an increased risk between the LPL D9N polymorphism and susceptibility to CAD in the dominant genetic model (AA + GA vs. GG: OR = 1.46, 95% CI = 1.14–1.87), whereas the LPL HindIII polymorphism showed a protective effect against CAD under all tested models (GG + GT vs. TT: OR = 0.85, 95% CI = 0.75–0.97; GG vs. TT + TG: OR = 0.62, 95% CI = 0.47–0.83; G vs. T: OR = 0.81, 95% CI = 0.71–0.92). No significant association was identified for the LPL N291S and PvuII polymorphisms. Stratification analysis by ethnicity suggested a significant correlation between the LPL S447X polymorphism and CAD susceptibility in Caucasians under the dominant and allele genetic models. In summary, our meta-analysis indicated that the LPL D9N polymorphism was associated with an increased risk of CAD, whereas the S447X and HindIII polymorphisms showed protective effects. There was no association observed between the N291S and PvuII polymorphisms and CAD risk.
Keywords: coronary artery disease, gene polymorphisms, lipoprotein lipase, meta-analysis
Introduction
Coronary artery disease (CAD) is a complex multifactorial disease and a leading cause of morbidity and mortality worldwide [1]. Although genetic and environmental factors have been widely implicated in the mechanisms underlying the pathogenesis of CAD, these potential factors remain an area of active investigation [2]. Atherosclerosis is the underlying cause of CAD, which is primarily characterized by excessive lipid deposition in the endothelium of the vascular tree walls [3]. Individuals with aberrant lipid and lipoprotein metabolism, including elevated levels of triglyceride (TG), cholesterol (TC), and low-density lipoprotein cholesterol (LDL-C) and decreased levels of high-density lipoprotein cholesterol (HDL-C), are more inclined to the development of CAD [4]. Genome-wide association studies (GWAS) have also identified nearly 150 loci linked to plasma lipid traits, and some of these loci are associated with altered lipoprotein lipase (LPL) gene expression [5]. Furthermore, several studies have demonstrated a causal link between triglyceride-rich lipoproteins (TRLs) and CAD, with variants in several crucial genes involved in TRLs metabolism, including LPL and its regulators [2,6]. In the past decades, numerous studies have reported that the LPL gene variants directly affect abnormal lipid and lipoprotein metabolism and its influence on the risk of CAD [7–9]. However, the underlying mechanisms that mediate these effects remain poorly elucidated.
LPL is a glycoprotein containing 448 amino acids, which is synthesized and secreted by various tissues, such as adipose tissue, myocardium, and skeletal muscle [10]. As an important component in TRL metabolism, LPL binds to the capillary endothelium and primarily hydrolyzes TGs in circulating TRLs, chylomicrons (CM), and very-low-density lipoproteins (VLDL), providing fatty acids for the energy requirements of the heart and skeletal muscle and for storage [5,10].
The LPL gene maps to chromosome 8p22, and over 100 various mutations have been identified [11,12]. Several genetic variants in the LPL gene have been reported to be associated with CAD susceptibility [13–15]. However, the results were conflicted, and no general agreements existed between them. For example, the D9N (rs1801177, G to A mutations) and N291S (rs268, A to G mutations) polymorphisms, which both result in partial defects in LPL catalytic function, are reported to be associated with an increased risk of CAD [16–18]. Similarly, the HindIII (rs320, T to G mutations) and PvuII (rs285, C to T mutations) variant sites (located on introns 8 and 6 respectively), which are related to profound alterations in plasma lipids, also seemed to be associated with CAD [9,14]. However, other studies did not confirm these results [19–21]. Meanwhile, several gain-of-function LPL variants, such as the S447X (rs328) polymorphism, which lead to the transition of Serine (S) to a stop codon (X) at codon position 447, result in reduced TG levels and an overall favorable lipid profile [5]. In addition, certain studies demonstrated that carriers of the X447 allele are protected against CAD [22–24], while other studies drew the opposite conclusion [13,25].
To confirm the correlation existed between the LPL gene polymorphisms (HindIII, S447X, N291S, D9N, and PvuII) and CAD, we performed this meta-analysis by pooling all eligible studies to calculate the estimate of overall CAD risk.
Methods and materials
Literature search strategy
We performed the present study according to the MOOSE (Meta-analysis of Observational Studies in Epidemiology) guidelines for meta-analysis of observational studies (Supplementary Table S1) [26]. The literature search was performed by two authors (W.-Q.M. and Y.Z.). PubMed, Web of Science, EMBASE, and the Cochrane Library were systematically searched, and the time period for references searching was from the first available article to September 2017. The following search terms were applied: (“lipoprotein lipase” or “LPL” or “N291S” or “S477X” or “D9N” or “HindIII” or “PvuII”) and (“genetic polymorphisms” or “mutation” or “variant” or “polymorphism”) and (“coronary artery disease” or “coronary heart disease” or “atherosclerosis” or “acute coronary syndrome” or “angina” or “myocardial infarction”). Handsearching was also carried out to find potential relevant records.
Inclusion and exclusion criteria
The following criteria were applied for reference selection: (1) studies on the evaluation of the LPL gene polymorphisms (HindIII, S477X, D9N, N291S, and PvuII) and CAD susceptibility; (2) total CAD cases were documented by angiographic evidence of at least 50% stenosis of one major coronary vessel, myocardial infarction, angina, a history of prior angioplasty, or coronary artery bypass surgery; (3) the data in the reference were sufficient for the present estimation, such as the total number of cases and controls, distribution of genotypes or other relevant information; and (4) the language was limited to English. Studies were excluded if they met any of the following criteria: (1) non-English record; (2) abstracts, letters to the editor, reviews, case-only studies, meta-analysis, and animal studies; and (3) study with useless or insufficient data and multiple publications that reported the same or overlapping population information.
Data extraction
Data abstraction was independently performed by two investigators (W.-Q.M. and X.-Q.H.), and disagreements about study selection were discussed and resolved by a third investigator (N.-F.L.). The following information was extracted from each included article: author, publication date, ethnicity, total number of cases and controls, country, sources of controls, genotyping methods, genotype frequency in cases and controls, and Hardy–Weinberg equilibrium (HWE) in the controls.
Quality assessment
The Newcastle–Ottawa scale (NOS) was applied in the quality assessment [27]. The validated quality assessment instrument was composed of the following three parameters of quality: selection, comparability, and exposure assessment. NOS scores ranged from zero to nine. Studies with an NOS score of five or greater were considered moderate to high quality studies, whereas those with an NOS score of less than five were considered low quality.
Statistics analysis
Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were applied to estimate the strength of association between the LPL gene polymorphisms and CAD susceptibility. The dominant, recessive, and allele genetic models were applied to assess the correlation between the LPL HindIII, PvuII, and S477X gene polymorphisms and CAD risk. Only the dominant genetic model was applied for N291S and D9N, due to the low number of minor homozygotes. The Cochrane Q-test and index (I2) were calculated to evaluate the heterogeneity within studies. P-value < 0.1 in the Q-test or I2 > 50% indicated significant heterogeneity. According to the strength of heterogeneity among studies, the fixed- or random-effects model was applied to calculate the OR and the corresponding 95% CI. The Z-test was used to determine the significance of overall ORs. Subgroup analyses, which were based on ethnicity (Asians and Caucasians) and sample size (studies with more than 500 subjects were categorized as “large,” and studies with less 500 subjects were categorized as “small”), were applied to detect sources of heterogeneity. In addition, the influence of sample sizes on the overall risk estimation was assessed by a cumulative meta-analysis [28]. A sensitivity analysis was performed to assess the stability of the individual studies. Possible publication bias was assessed using funnel plots and the Egger linear regression test. All calculations were performed and graphs were made with Review Manager v5.2 (The Cochrane Collaboration, Oxford, U.K.) and Stata 12.0 (Stata Corporation, College Station, Texas, U.S.A.).
Results
Selection and characteristics of studies
A total of 958 articles were acquired after initial searching. Among them, 712 duplicate articles were excluded, and 160 articles were excluded for ineligibility after screening the titles and abstracts. In addition, 41 articles were excluded because of insufficient data, reviews, meta-analyses, or conference abstracts. Finally, 45 articles containing 80 eligible studies were included in this meta-analysis [7–9,13–25,29–57]. The flow chart of the retrieved and excluded studies with specifications of reasons is summarized in Figure 1.
Figure 1. Flow diagram of the study selection process.
The characteristics of the studies included in the meta-analysis are shown in Supplementary Table S2. Among 80 eligible studies, 18 studies, containing 5532 cases and 4813 controls, correlated the LPL HindIII polymorphism with susceptibility to CAD. Twenty-seven studies, involving 6959 cases and 9400 controls, focused on the relationship between the LPL S447X polymorphism and susceptibility to CAD. Eleven studies, including 9272 cases and 15,074 controls, focused on the relationship between the LPL N291S polymorphism and susceptibility to CAD. Eight studies, involving 2583 cases and 2525 controls, focused on the relationship between the LPL D9N polymorphism and susceptibility to CAD, and the remaining 16 studies, involving 7831 cases and 5966 controls, concerned the LPL PvuII polymorphism. The countries in which these studies occurred included the U.S.A., U.K., France, Brazil, China, Finland, and others. HWE had been applied for all polymorphisms in the controls. The quality of these enrolled studies was evaluated using the NOS quality scale (Supplementary Table S3).
Association between the LPL HindIII polymorphism and susceptibility to CAD
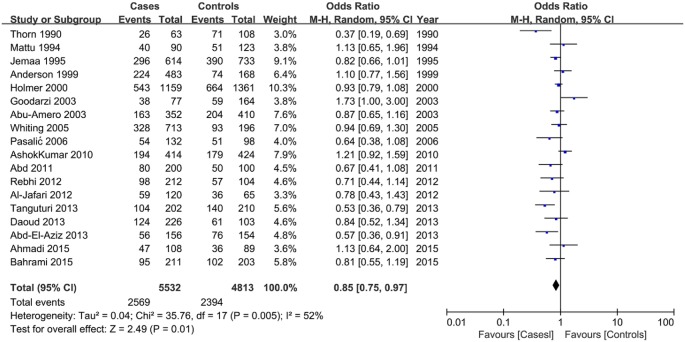
In all study subjects, the results indicated a reduced risk of CAD susceptibility associated with the LPL HindIII polymorphism in all tested genetic models (GG + GT vs. TT: OR = 0.85, 95% CI = 0.75–0.97; GG vs. TT + TG: OR = 0.62, 95% CI = 0.47–0.83; G vs. T: OR = 0.81, 95% CI = 0.71–0.92) with some evidence of interstudy heterogeneity (Table 1; Figure 2). Stratification analysis by ethnicity and sample size indicated a significant association between the HindIII polymorphism and CAD susceptibility in Caucasians and small sample size under all tested models (Table 1; Supplementary Figures S1 and S2).
Table 1. Summary of odds ratios (95% CI) in the analysis of the association between the LPL HindIII polymorphism and CAD susceptibility.
| Genetic model | Overall and subgroups | N | Test of association | Test of heterogeneity | |||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | PHeterogeneity | I2 (%) | |||
| GG + GT vs. TT | Overall | 18 | 0.85 | 0.75,0.97 | 0.010 | 0.005 | 52% |
| Asians | 7 | 0.86 | 0.70,1.07 | 0.190 | 0.050 | 52% | |
| Caucasians | 9 | 0.81 | 0.69,0.96 | 0.010 | 0.040 | 51% | |
| Large sample | 6 | 0.94 | 0.85,1.05 | 0.300 | 0.320 | 15% | |
| Small sample | 12 | 0.76 | 0.62,0.94 | 0.010 | 0.020 | 53% | |
| GG vs. TT + TG | Overall | 18 | 0.62 | 0.47,0.83 | 0.001 | 0.000 | 67% |
| Asians | 7 | 0.67 | 0.43,1.06 | 0.090 | 0.004 | 69% | |
| Caucasians | 9 | 0.58 | 0.38,0.88 | 0.010 | 0.000 | 71% | |
| Large sample | 6 | 0.82 | 0.60,1.12 | 0.220 | 0.030 | 60% | |
| Small sample | 12 | 0.50 | 0.34,0.75 | 0.000 | 0.006 | 58% | |
| G vs. T | Overall | 18 | 0.81 | 0.71,0.92 | 0.001 | 0.000 | 72% |
| Asians | 7 | 0.82 | 0.65,1.05 | 0.110 | 0.000 | 77% | |
| Caucasians | 9 | 0.78 | 0.66,0.92 | 0.003 | 0.001 | 69% | |
| Large sample | 6 | 0.94 | 0.85,1.04 | 0.200 | 0.180 | 35% | |
| Small sample | 12 | 0.73 | 0.59,0.89 | 0.002 | 0.000 | 70% | |
Abbreviations: CAD, coronary artery disease; CI, confidence interval; LPL, lipoprotein lipase; N, number of studies; OR, odds ratio; P-Value, P value for association; P Heterogeneity, P value for heterogeneity.
Figure 2. Forest plot of odds ratios for the association between the LPL HindIII polymorphism and CAD risk under dominant genetic model (GG + GT vs. TT).
Association between the LPL S477X polymorphism and susceptibility to CAD
No significant association was observed in any genetic model between the S477X polymorphism and CAD risk in the overall meta-analysis, and there was some evidence of interstudy heterogeneity (Table 2; Figure 3). The subgroup analysis stratified by ethnicity indicated that the S477X polymorphism was significantly associated with CAD risk for Caucasians, but not Asians, under the dominant and allele genetic models (GG + GC vs. CC: OR = 0.77, 95% CI = 0.64–0.93; GG vs. GC + CC: OR = 0.83, 95% CI = 0.72–0.94), with a reduction in interstudy heterogeneity (Table 2; Supplementary Figure S3). Stratification by sample size indicated that large sample size, but not small sample size, showed a reduced risk of CAD susceptibility associated with the S447X polymorphism under the recessive genetic model (GG vs. GC + CC: OR = 0.55, 95% CI = 0.35–0.86) (Table 2; Supplementary Figure S4).
Table 2. Summary of odds ratios (95% CI) in the analysis of the association between the LPL S447X polymorphism and CAD susceptibility.
| Genetic model | Overall and subgroups | N | Test of association | Test of heterogeneity | |||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | P Heterogeneity | I2 (%) | |||
| GG + GC vs. CC | Overall | 27 | 0.87 | 0.73,1.03 | 0.100 | 0.000 | 68% |
| Asians | 8 | 1.08 | 0.71,1.65 | 0.730 | 0.000 | 84% | |
| Caucasians | 16 | 0.77 | 0.64,0.93 | 0.008 | 0.007 | 52% | |
| Large sample | 9 | 0.87 | 0.75,1.00 | 0.050 | 0.080 | 44% | |
| Small sample | 18 | 0.87 | 0.62,1.22 | 0.430 | 0.000 | 74% | |
| GG vs. GC + CC | Overall | 19 | 1.00 | 0.60,1.68 | 1.000 | 0.040 | 40% |
| Asians | 6 | 0.90 | 0.30,2.69 | 0.850 | 0.002 | 74% | |
| Caucasians | 11 | 0.78 | 0.45,1.35 | 0.370 | 0.730 | 0% | |
| Large sample | 6 | 0.55 | 0.35,0.86 | 0.009 | 0.950 | 0% | |
| Small sample | 13 | 1.59 | 0.81,3.11 | 0.180 | 0.200 | 24% | |
| G vs. C | Overall | 21 | 0.94 | 0.77,1.15 | 0.540 | 0.000 | 76% |
| Asians | 8 | 1.11 | 0.72,1.72 | 0.630 | 0.000 | 88% | |
| Caucasians | 11 | 0.83 | 0.72,0.94 | 0.005 | 0.020 | 53% | |
| Large sample | 6 | 0.87 | 0.74,1.02 | 0.080 | 0.080 | 50% | |
| Small sample | 15 | 0.97 | 0.67,1.41 | 0.880 | 0.000 | 80% | |
Abbreviations: CAD, coronary artery disease; CI, confidence interval; LPL, lipoprotein lipase; N, number of studies; OR, odds ratio; P-Value, P value for association; P Heterogeneity, P value for heterogeneity.
Figure 3. Forest plot of odds ratios for the association between the LPL S447X polymorphism and CAD risk under the dominant genetic model (GG + GC vs. CC).
Association between the LPL N291S and D9N gene polymorphisms and susceptibility to CAD
Because of the low number of minor homozygotes, only the dominant genetic model was applied to the N291S and D9N polymorphisms. An increased risk of CAD susceptibility was associated with the D9N polymorphism under the dominant genetic model (AA + GA vs. GG: OR = 1.46, 95% CI = 1.14–1.87) with low interstudy heterogeneity (Table 3; Figure 4). No significant association was observed between the N291S polymorphism and CAD risk (Table 3; Figure 4). When we conducted subgroup analyses by ethnicity and sample size, the same significant association was observed in large sample size of the D9N polymorphism (Table 3; Supplementary Figure S5). However, no significant association was observed between CAD risk and the N291S polymorphism in the subgroup analysis (Table 3; Supplementary Figure S6).
Table 3. Summary of odds ratios (95% CI) in the analysis of the association between the LPL N291S and D9N polymorphisms and CAD susceptibility.
| Genetic model | Overall and subgroups | N | Test of association | Test of heterogeneity | |||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | P Heterogeneity | I2 (%) | |||
| N291S | |||||||
| GG + GA vs. AA | Overall | 11 | 1.11 | 0.97,1.28 | 0.130 | 0.420 | 3% |
| Asians | 1 | 1.54 | 0.89,2.68 | 0.120 | N/A | N/A | |
| Caucasians | 8 | 1.10 | 0.95,1.27 | 0.210 | 0.300 | 16% | |
| Large sample | 7 | 1.13 | 0.98,1.30 | 0.100 | 0.320 | 15% | |
| Small sample | 4 | 0.96 | 0.57,1.60 | 0.860 | 0.430 | 0% | |
| D9N | |||||||
| AA + GA vs. GG | Overall | 8 | 1.46 | 1.14,1.87 | 0.002 | 0.360 | 9% |
| Asians | 1 | 0.41 | 0.05,3.73 | 0.430 | N/A | N/A | |
| Caucasians | 4 | 1.47 | 1.00,2.14 | 0.050 | 0.200 | 36% | |
| Large sample | 4 | 1.49 | 1.03,2.15 | 0.040 | 0.180 | 38% | |
| Small sample | 4 | 0.94 | 0.42,2.10 | 0.890 | 0.670 | 0% | |
Abbreviations: CAD, coronary artery disease; CI, confidence interval; LPL, lipoprotein lipase; N, number of studies; N/A, not applicable; OR, odds ratio; P-Value, P value for association; P Heterogeneity, P value for heterogeneity.
Figure 4. Forest plot of odds ratios for the association of polymorphisms in LPL N291S and D9N and susceptibility to CAD.
(A) The N291S polymorphism under the dominant genetic model (GG + GA vs. AA). (B) The D9N polymorphism under the dominant genetic model (AA+GA vs. GG).
Association between the LPL PvuII polymorphism and susceptibility to CAD
No significant associations were observed between the LPL PvuII polymorphism and CAD susceptibility in any genetic model (Table 4; Figure 5). This was also the case in the subgroup analysis (Table 4; Supplementary Figures S7 and S8).
Table 4. Summary of odds ratios (95% CI) in the analysis of the association between the LPL PvuII polymorphism and CAD susceptibility.
| Genetic model | Overall and subgroups | N | Test of association | Test of heterogeneity | |||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | PHeterogeneity | I2 (%) | |||
| TT + CT vs. CC | Overall | 16 | 1.00 | 0.92,1.08 | 0.920 | 0.450 | 0% |
| Asians | 4 | 1.09 | 0.90,1.33 | 0.360 | 0.660 | 0% | |
| Caucasians | 11 | 0.99 | 0.91,1.08 | 0.810 | 0.480 | 0% | |
| Large sample | 4 | 1.03 | 0.87,1.23 | 0.700 | 0.080 | 55% | |
| Small sample | 12 | 0.92 | 0.78,1.09 | 0.320 | 0.790 | 0% | |
| TT vs. CC + CT | Overall | 16 | 0.90 | 0.78,1.04 | 0.150 | 0.100 | 32% |
| Asians | 4 | 0.95 | 0.74,1.22 | 0.670 | 0.670 | 0% | |
| Caucasians | 11 | 0.85 | 0.69,1.05 | 0.120 | 0.030 | 51% | |
| Large sample | 4 | 0.98 | 0.82,1.16 | 0.780 | 0.140 | 45% | |
| Small sample | 12 | 0.82 | 0.68,1.00 | 0.040 | 0.290 | 16% | |
| T vs. C | Overall | 16 | 0.99 | 0.94,1.04 | 0.670 | 0.200 | 22% |
| Asians | 4 | 1.03 | 0.90,1.17 | 0.680 | 0.490 | 0% | |
| Caucasians | 11 | 0.94 | 0.84,1.04 | 0.210 | 0.110 | 37% | |
| Large sample | 4 | 1.01 | 0.89,1.14 | 0.900 | 0.040 | 65% | |
| Small sample | 12 | 0.90 | 0.81,1.01 | 0.060 | 0.780 | 0% | |
Abbreviations: CAD, coronary artery disease; CI, confidence interval; LPL, lipoprotein lipase; N, number of studies; OR, odds ratio; P-Value, P value for association; P Heterogeneity, P value for heterogeneity.
Figure 5. Forest plot of odds ratios for the association between the LPL PvuII polymorphism and CAD risk under the dominant model (TT + CT vs. CC).
Cumulative analysis
For the LPL HindIII and D9N polymorphisms, the cumulative meta-analysis showed that as publication year increased, the CI became increasingly narrower, and statistical significance was more common. The association between the LPL S447X polymorphism and CAD risk appeared to fluctuate with the number of studies accumulated. For the LPL N291S and PvuII polymorphisms, no significant association was observed with the number of studies accumulated (Figure 6).
Figure 6. Forest plots of the cumulative odds ratio for the association between the LPL gene polymorphisms and CAD risk under the dominant genetic model.
(A) HindIII polymorphism; (B) S447X polymorphism; (C) N291S polymorphism; (D) D9N polymorphism; (E) PvuII polymorphism.
Heterogeneity and sensitivity analysis
The heterogeneity within each study in each comparison is shown in Tables 1–4. The influence of each study on the overall meta-analysis was evaluated by deleting one study at a time. The results indicated that no individual study influenced the pooled OR significantly (Supplementary Figure S9).
Publication bias
The Funnel plot and Egger’s regression test were applied to assess the publication bias of the included studies. The results indicated that the distribution of the included studies on the funnel plot appeared roughly symmetrical (Figure 7). The results of Egger’s regression test are also presented under the dominant models (HindIII: t = −1.23, P=0.237; S477X: t = −3.12, P=0.005; N291S: t = −0.96, P=0.363; D9N: t = −1.62, P=0.157; PvuII: t = −1.05, P=0.311) (Supplementary Figure S10).
Figure 7. Funnel plot of publication bias for the association between LPL gene polymorphisms and susceptibility to CAD under dominant models.
(A) Hind polymorphism; (B) S447X polymorphism; (C) N291S polymorphism; (D) D9N polymorphism; (E) PvuII polymorphism.
Discussion
Genetic variations in the LPL gene could influence lipid transport and metabolism and could consequently modulate an individual’s susceptibility to atherosclerosis. However, it is difficult to draw a definite conclusion for whether LPL is a proatherosclerotic or antiatherosclerotic factor, since the effects of LPL partly depend on its locations and activity [58]. The enzyme, when expressed in adipose tissue, heart, and skeletal muscle, has been regarded as an antiatherosclerotic factor by reducing atherogenic lipoproteins or increasing HDL, whereas the effect of LPL on the biology of arterial wall seems to be atherogenic by accelerating lipid accumulation [58,59]. Several lines of evidence also suggest that LPL activity is higher in atherosclerotic arteries compared with normal arteries [10,60]. Increased plasma LPL activity could alter lipid traits, such as decreasing TG and increasing HDL levels, generating a profile associated with protection against atherosclerosis, while the down-regulation of LPL gene expression has been shown to play an opposite role [61,62].
Although numerous studies have investigated the correlation between LPL and CAD risk in the past several decades, no definite conclusions have been reached regarding gene polymorphisms. This meta-analysis has combined and reanalyzed individual participant data from 80 eligible studies of the effect of LPL gene polymorphisms on CAD incidence. In our study, all of the results revealed that three LPL gene variants (Hind III, S447X, and D9N) were associated with CAD susceptibility. When Asians or Caucasians were analyzed independently, the heterogeneity of the population tended to be weaker, and the subgroup analysis indicated that S447X polymorphism decreased CAD risk in Caucasians. On the other hand, the stratified analysis by ethnicity for the S447X polymorphism was successfully applied to relieve the heterogeneity bias in the polymorphism analysis within Caucasians, suggesting that ethnicity may potentially be the source of the heterogeneity. In addition, it is worth noting that the P value of the Egger’s regression test for the S447X polymorphism was less than 0.05, which indicated that publication bias likely existed; however, the funnel plot appeared roughly symmetrical, and the sensitivity analysis indicated the stability of the results. Consequently, future studies are warranted to validate our conclusion.
Although several relevant meta-analyses have been published, our study had certain specific advantages [63–65]. Compared with other studies, we incorporated more eligible articles, conducted quality assessment, and performed a comprehensive analysis, whereas previous studies primarily focused on the plasma levels of lipids and lipoproteins, or they only analyzed a single gene variant in the meta-analysis. Furthermore, in the present study, a cumulative meta-analysis was performed to assess the pattern of the evidence accumulated over time.
Several limitations in our study should also be addressed. First, some genetic models displayed high heterogeneity, although subgroup analysis was performed to detect the sources of this heterogeneity. Second, the ethnic distribution of included studies was primarily Asians and Caucasians. Racial bias may exist, and the conclusions may not be applicable to other races. Third, we searched and collected articles in English from four comprehensive electronic databases, including PubMed, Web of Science, EMBASE, and Cochrane database. Several publications related to this topic written in other languages might have been ignored. Thus, publication bias likely existed. However, the articles included in these four databases are more authoritative and more convenient for readers compared with the original literature.
In summary, this updated meta-analysis suggested that the LPL D9N polymorphism was associated with the increased risk of CAD, whereas the LPL HindIII and S447X polymorphisms showed protective effects against CAD. No associations were observed between the LPL N291S and PvuII polymorphisms and susceptibility to CAD.
Supporting information
Supplementary Table 1. MOOSE checklist for meta-analysis of observational studies.
Supplementary Table 2. Characteristics of the individual studies included in the meta-analysis.
Supplementary Table 3. Methodological quality of the selected studies according to the Newcastle-Ottawa Scale.
supplementary Figure 1.
Stratified analysis based on ethnicity for the association between the LPL HindIII polymorphism and CAD risk using dominant genetic model (GG+GT vs. TT).
supplementary Figure 2.
Stratified analysis based on sample size for the association between the LPL HindIII polymorphism and CAD risk using dominant genetic model (GG+GT vs. TT).
supplementary Figure 3.
Stratified analysis based on ethnicity for the association between the LPL S447X polymorphism and CAD risk using dominant genetic model (GG+GC vs. CC).
supplementary Figure 4.
Stratified analysis based on sample size for the association between the LPL S447X polymorphism and CAD risk using dominant genetic model (GG+GC vs. CC).
supplementary Figure 5.
Stratified analysis based on sample size for the association between the LPL D9N polymorphism and CAD risk using dominant genetic model (AA+GA vs. GG).
supplementary Figure 6.
Stratified analysis based on sample size for the association between the LPL N291S polymorphism and CAD risk using dominant genetic model (GG+GA vs. AA).
supplementary Figure 7.
Stratified analysis based on ethnicity for the association between the LPL PvuII polymorphism and CAD risk using dominant genetic model (TT+CT vs. CC).
supplementary Figure 8.
Stratified analysis based on sample size for the association between the LPL PvuII polymorphism and CAD risk using dominant genetic model (TT+CT vs. CC).
supplementary Figure 9.
Egger’s regression test of publication bias for the association between the LPL gene polymorphisms and susceptibility to CAD. (a). HindIII polymorphism; (b). S447X polymorphism; (c). N291S polymorphism; (d). D9N polymorphism; (e). PvuII polymorphism.
supplementary Figure 10.
Sensitivity analysis on the correlation between the LPL gene polymorphisms and susceptibility to CAD. (a). sensitivity analysis for HindIII and CAD risk; (b). sensitivity analysis for S447X and CAD risk; (c). sensitivity analysis for N291S and CAD risk; (d). sensitivity analysis for D9N and CAD risk; (e). sensitivity analysis for PvuII and CAD risk;
Abbreviations
- CAD
coronary artery disease
- CM
chylomicrons
- CI
confidence interval
- GWAS
genome-wide association studies
- HDL-C
high-density lipoprotein cholesterol
- HWE
Hardy–Weinberg equilibrium
- LDL-C
low-density lipoprotein cholesterol
- LPL
lipoprotein lipase
- NOS
Newcastle–Ottawa scale
- OR
odds ratio
- TC
cholesterol
- TG
triglyceride
- TRL
triglyceride-rich lipoprotein
- VLDL
very-low-density lipoprotein
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by National Nature Science Foundation of China (No. 81770451).
Author Contribution
W.-Q.M. and N.-F.L. conceived and designed the study. X.-Q.H., Y.W., and N.-F.L. performed in data collection and management. Y.Z. performed in data analysis. W.-Q.M. and N.-F.L. wrote the paper. All the authors reviewed the manuscript.
References
- 1.Mozaffarian D., Benjamin E.J., Go A.S., Arnett D.K., Blaha M.J., Cushman M. et al. (2016) Heart disease and stroke statistics-2016 update: a report from the American heart association. Circulation 133, e38–e360 10.1161/CIR.0000000000000350 [DOI] [PubMed] [Google Scholar]
- 2.Lanktree M.B. and Hegele R.A. (2009) Gene-gene and gene-environment interactions: new insights into the prevention, detection and management of coronary artery disease. Genome Med. 1, 28 10.1186/gm28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakashima Y., Fujii H., Sumiyoshi S., Wight T.N. and Sueishi K. (2007) Early human atherosclerosis: accumulation of lipid and proteoglycans in intimal thickenings followed by macrophage infiltration. Arterioscler. Thromb. Vasc. Biol. 27, 1159–1165 10.1161/ATVBAHA.106.134080 [DOI] [PubMed] [Google Scholar]
- 4.Patsch J.R., Miesenbock G., Hopferwieser T., Muhlberger V., Knapp E., Dunn J.K. et al. (1992) Relation of triglyceride metabolism and coronary artery disease. Studies in the postprandial state. Arterioscler. Thromb. 12, 1336–1345 10.1161/01.ATV.12.11.1336 [DOI] [PubMed] [Google Scholar]
- 5.Bauer R.C., Khetarpal S.A., Hand N.J. and Rader D.J. (2016) Therapeutic targets of triglyceride metabolism as informed by human genetics. Trends Mol. Med. 22, 328–340 10.1016/j.molmed.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 6.Toth P.P. (2016) Triglyceride-rich lipoproteins as a causal factor for cardiovascular disease. Vasc. Health Risk Manag. 12, 171–183 10.2147/VHRM.S104369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorn J., Chamberlain J.C., Alcolado J.C., Oka K., Chan L., Stocks J. et al. (1990) Lipoprotein and hepatic lipase gene variants in coronary atherosclerosis. Atherosclerosis 85, 55–60 10.1016/0021-9150(90)90182-I [DOI] [PubMed] [Google Scholar]
- 8.Peacock R.E., Hamstenb A. and Nilsson-Ehle P. Humphries S.E. (1992) Associations between lipoprotein lipase gene polymorphisms and plasma correlations of lipids, lipoproteins and lipase activities in young myocardial infarction survivors and age-matched healthy individuals from Sweden. Atherosclerosis 97, 171–185 10.1016/0021-9150(92)90130-9 [DOI] [PubMed] [Google Scholar]
- 9.Wang X.L., McCredie R.M. and Wilcken D.E. (1996) Common DNA polymorphisms at the lipoprotein lipase gene: Association with severity of coronary artery disease and diabetes. Circulation 93, 1339–1345 10.1161/01.CIR.93.7.1339 [DOI] [PubMed] [Google Scholar]
- 10.Goldberg I.J. (1996) Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J. Lipid Res. 37, 693–707 [PubMed] [Google Scholar]
- 11.Sparkes R.S., Zollman S., Klisak I., Kirchgessner T.G., Komaromy M.C., Mohandas T. et al. (1987) Human genes involved in lipolysis of plasma lipoproteins: mapping of loci for lipoprotein lipase to 8p22 and hepatic lipase to 15q21. Genomics 1, 138–144 10.1016/0888-7543(87)90005-X [DOI] [PubMed] [Google Scholar]
- 12.Murthy V.J.P. and Gagne C. (1996) Molecular pathobiology of the human lipoprotein lipase gene. Pharmacol. Ther. 70, 101–135 10.1016/0163-7258(96)00005-8 [DOI] [PubMed] [Google Scholar]
- 13.Ahmadi Z., Senemar S., Toosi S. and Radmanesh S. (2015) The association of lipoprotein lipase genes, HindIII and S447X polymorphisms with coronary artery disease in Shiraz city. J. Cardiovasc. Thorac. Res. 7, 63–67 10.15171/jcvtr.2015.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rebhi L., Kchok K., Omezzine A., Kacem S., Rejeb J., Ben H.I. et al. (2012) Six lipoprotein lipase gene polymorphisms, lipid profile and coronary stenosis in a Tunisian population. Mol. Biol. Rep. 39, 9893–9901 10.1007/s11033-012-1856-9 [DOI] [PubMed] [Google Scholar]
- 15.Al-Jafari A.A., Daoud M.S., Mobeirek A.F. and Anazi M.S.A. (2012) DNA polymorphisms of the lipoprotein lipase gene and their association with coronary artery disease in the Saudi population. Int. J. Mol. Sci. 13, 7559–7574 10.3390/ijms13067559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izar M.C., Helfenstein T., Ihara S.S., Relvas W.G., Santos A.O., Fischer S.C. et al. (2009) Association of lipoprotein lipase D9N polymorphism with myocardial infarction in type 2 diabetes: the genetics, outcomes, and lipids in type 2 diabetes (GOLD) study. Atherosclerosis 204, 165–170 10.1016/j.atherosclerosis.2008.08.006 [DOI] [PubMed] [Google Scholar]
- 17.van Bockxmeer F.M., Liu Q., Mamotte C. and Taylor R. (2001) Lipoprotein lipase D9N, N291S and S447X polymorphisms: their influence on premature coronary heart disease and plasma lipids. Atherosclerosis 157, 123–129 10.1016/S0021-9150(00)00717-6 [DOI] [PubMed] [Google Scholar]
- 18.Abdel Hamid M.M., Ahmed S., Salah A., Tyrab E.M., Yahia L.M., Elbashir E.A. et al. (2015) Association of lipoprotein lipase gene with coronary heart disease in Sudanese population. J. Epidemiol. Glob. Health 5, 405–407 10.1016/j.jegh.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahrami M., Barati H., Jahani M.M., Fatemi A., Sharifi Z., Eydi A. et al. (2015) Lipoprotein lipase gene variants: Association with acute myocardial infarction and lipid profiles. Egypt. J. Med. Hum. Genet. 16, 327–332 10.1016/j.ejmhg.2015.04.001 [DOI] [Google Scholar]
- 20.Georgiev A., Panov S. and Sadikario S. (2008) Association of PVUII polymorphism in the lipoprotein lipase gene with the coronary artery disease in Macedonian population. Prilozi 29, 213–225 [PubMed] [Google Scholar]
- 21.Duman B.S., Turkoglu C., Akpınar B., Guden M., Vertii A., Dak E. et al. (2004) Lipoprotein lipase gene polymorphism and lipid profile in coronary artery disease. Arch. Pathol. Lab. Med. 128, 869–874 [DOI] [PubMed] [Google Scholar]
- 22.Agirbasli M., Eren F., Sumerkan M.C. and Agirbasli D. (2011) The S447X variant of lipoprotein lipase gene is inversely associated with severity of coronary artery disease. Heart Vessels 26, 457–463 10.1007/s00380-010-0077-1 [DOI] [PubMed] [Google Scholar]
- 23.Aydogan H.Y., Isbİr S., Kurnaz O., Gormus U. and Isbİr T. (2009) Associations of lipoprotein lipase S447X and apolipoprotein E genotypes with low-density lipoprotein subfractions in Turkish patients with coronary artery disease. In Vivo 23, 155–161 [PubMed] [Google Scholar]
- 24.Gagné S.E., Larson M.G., Pimstone S.N., Schaefer E.J., Kastelein J.J., Wilson P.W. et al. (1999) A common truncation variant of lipoprotein lipase (Ser447X) confers protection against coronary heart disease: the Framingham Offspring Study. Clin. Genet. 55, 450–454 10.1034/j.1399-0004.1999.550609.x [DOI] [PubMed] [Google Scholar]
- 25.Almeida K.A., Strunz C.M., Maranhão R.C. and Mansur A.P. (2007) The S447X polymorphism of lipoprotein lipase: effect on the incidence of premature coronary disease and on plasma lipids. Arq. Bras. Cardiol. 88, 297–303 [DOI] [PubMed] [Google Scholar]
- 26.Stroup D.F., Berlin J.A., Morton S.C. and Olkin I. (2000) Meta-analysis of observational studies in epidemiology. J. Am. Med. Assoc. 283, 2008–2012 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 27.Stang A. (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 28.Rotondi M.A. and Bull S.B. (2012) Cumulative meta-analysis for genetic association: when is a new study worthwhile? Hum. Hered. 74, 61–70 10.1159/000345604 [DOI] [PubMed] [Google Scholar]
- 29.Mattu R.K., Needham E.W., Morgan R., Rees A., Hackshaw A.K., Stocks J. et al. (1994) DNA variants at the LPL gene locus associate with angiographically defined severity of atherosclerosis and serum lipoprotein levels in a Welsh population. Arterioscler. Thromb. 14, 1090–1097 10.1161/01.ATV.14.7.1090 [DOI] [PubMed] [Google Scholar]
- 30.Jemaa R., Fumeron F., Pokier O., Kerf L., Evans A., Ameiler D. et al. (1995) Lipoprotein lipase gene polymorphisms: associations with myocardial infarction and lipoprotein levels, the ECTIM study. J. Lipid Res. 36, 2141–2146 [PubMed] [Google Scholar]
- 31.Zhang Q., Cavanna J., Winkelman B.R., Shine B., Gross W., Marz W. et al. (1995) Common genetic variants of lipoprotein lipase that relate to lipid transport in patients with premature coronary artery disease. Clin. Genet. 48, 293–298 10.1111/j.1399-0004.1995.tb04112.x [DOI] [PubMed] [Google Scholar]
- 32.Wittrup H.H., Tybjaerg-Hansen A., Abildgaard S., Steffensen R., Schnohr P. and Nordestgaard B.G. (1997) A common substitution (Asn291Ser) in lipoprotein lipase is associated with increased risk of ischemic heart disease. J. Clin. Invest. 99, 1606–1613 10.1172/JCI119323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stepanov V.A., Puzyrev V.P., Karpov R.S. and Kutmin A.I. (1998) Genetic markers in coronary artery disease in a Russian population. Hum. Biol. 7, 47–57 [PubMed] [Google Scholar]
- 34.Anderson J.L., King G.J., Bair T.L., Elmer S.P., Muhlestein J.B., Habashi J. et al. (1999) Association of lipoprotein lipase gene polymorphisms with coronary artery disease. J. Am. Coll. Cardiol. 33, 1013–1020 10.1016/S0735-1097(98)00677-9 [DOI] [PubMed] [Google Scholar]
- 35.Arca M., Campagna F., Montali A., Barillà F., Mangieri E., Tanzilli G. et al. (2000) The common mutations in the lipoprotein lipase gene in Italy: effects on plasma lipids and angiographically assessed coronary atherosclerosis. Clin. Genet. 58, 369–374 10.1034/j.1399-0004.2000.580507.x [DOI] [PubMed] [Google Scholar]
- 36.Holmer S.R., Hengstenberg C., Mayer B., Döring A., Löwel H., Engel S. et al. (2000) Lipoprotein lipase gene polymorphism, cholesterol subfractions and myocardial infarction in large samples of the general population. Cardiovasc. Res. 47, 806–812 10.1016/S0008-6363(00)00131-0 [DOI] [PubMed] [Google Scholar]
- 37.Moennig G., Wiebusch H., Enbergs A., Dorszewski A., Kerber S., Schulte H. et al. (2000) Detection of missense mutations in the genes for lipoprotein lipase and hepatic triglyceride lipase in patients with dyslipidemia undergoing coronary angiography. Atherosclerosis 149, 395–401 10.1016/S0021-9150(99)00330-5 [DOI] [PubMed] [Google Scholar]
- 38.Myllykangas L., Polvikoski T., Sulkava R., Notkola I.L., Rastas S., Verkkoniemi A. et al. (2001) Association of lipoprotein lipase Ser447Ter polymorphism with brain infarction: a population-based neuropathological study. Ann. Med. 33, 486–492 10.3109/07853890109002098 [DOI] [PubMed] [Google Scholar]
- 39.Sawano M., Watanabe Y., Ohmura H., Shimada K., Daida H., Mokuno H. et al. (2001) Potentially protective effects of the Ser447-Ter mutation of the lipoprotein lipase gene against the development of coronary artery disease in Japanese subjects via a beneficial lipid profile. Jpn. Circ. J. 65, 310–314 10.1253/jcj.65.310 [DOI] [PubMed] [Google Scholar]
- 40.Abu-Amero K.K., Wyngaard C.A., Al-Boudari O.M., Kambouris M. and Dzimiri N. (2003) Lack of association of lipoprotein lipase gene polymorphismswith coronary artery disease in the Saudi Arab population. Arch. Pathol. Lab. Med. 127, 597–600 [DOI] [PubMed] [Google Scholar]
- 41.Goodarzi M.O., Guo X., Taylor K.D., Quinones M.J., Samayoa C., Yang H. et al. (2003) Determination and use of haplotypes: ethnic comparison and association of the lipoprotein lipase gene and coronary artery disease in Mexican-Americans. Genet. Med. 5, 322–327 10.1097/01.GIM.0000076971.55421.AD [DOI] [PubMed] [Google Scholar]
- 42.Isbir T., Yilmaz H., Agachan B. and Karaali Z.E. (2003) Cholesterol ester transfer protein, apolipoprotein E and lipoprotein lipase genotypes in patients with coronary artery disease in the Turkish population. Clin. Genet. 64, 228–234 10.1034/j.1399-0004.2003.00137.x [DOI] [PubMed] [Google Scholar]
- 43.Keavney B., Palmer A., Parish S., Clark S., Youngman L., Danesh J. et al. (2004) Lipid-related genes and myocardial infarction in 4685 cases and 3460 controls: discrepancies between genotype, blood lipid concentrations, and coronary disease risk. Int. J. Epidemiol. 33, 1002–1013 10.1093/ije/dyh275 [DOI] [PubMed] [Google Scholar]
- 44.Tobin M.D., Braund P.S., Burton P.R., Thompson J.R., Steeds R., Channer K. et al. (2004) Genotypes and haplotypes predisposing to myocardial infarction: a multilocus case-control study. Eur. Heart J. 25, 459–467 10.1016/j.ehj.2003.11.014 [DOI] [PubMed] [Google Scholar]
- 45.Whiting B.M., Anderson J.L., Muhlestein J.B., Horne B.D., Bair T.L., Pearson R.R. et al. (2005) Candidate gene susceptibility variants predict intermediate end points but not angiographic coronary artery disease. Am. Heart J. 150, 243–250 10.1016/j.ahj.2004.08.034 [DOI] [PubMed] [Google Scholar]
- 46.Baum L., Ng H.K., Wong K.S., Tomlinson B., Rainer T.H., Chen X. et al. (2006) Associations of apolipoprotein E exon 4 and lipoprotein lipase S447X polymorphisms with acute ischemic stroke and myocardial infarction. Clin. Chem. Lab. Med. 44, 274–281 10.1515/CCLM.2006.047 [DOI] [PubMed] [Google Scholar]
- 47.Pasalić D., Ferencak G., Grsković B., Sesto M. and Stavljenić-Rukavina A. (2006) Association of two genetic variations of lipoprotein lipase, S447X and Hind III, with coronary artery disease and hypertriglyceridemia. Coll. Antropol. 30, 549–554 [PubMed] [Google Scholar]
- 48.Yamada Y., Matsuo H., Segawa T., Watanabe S., Kato K., Hibino T. et al. (2006) Assessment of genetic risk for myocardial infarction. Thromb. Haemost. 96, 220–227 [PubMed] [Google Scholar]
- 49.Ak K., Isbir S., Tekeli A., Ergen A., Atalan N., Dogan S. et al. (2007) Presence of lipoprotein lipase S447X stop codon affects the magnitude of interleukin 8 release after cardiac surgery with cardiopulmonary bypass. J. Thorac. Cardiovasc. Surg. 134, 477–483 10.1016/j.jtcvs.2007.03.017 [DOI] [PubMed] [Google Scholar]
- 50.AshokKumar M., Veera Subhashini N.G., Kanthimathi S., SaiBabu R., Ramesh A., Cherian KM. et al. (2010) Associations for lipoprotein lipase and peroxisome proliferator-activated receptor-gamma gene and coronary artery disease in an Indian population. Arch. Med. Res. 41, 19–25 10.1016/j.arcmed.2010.01.005 [DOI] [PubMed] [Google Scholar]
- 51.Bhanushali A.A. and Das B.R. (2010) Genetic variants at the APOE, lipoprotein lipase (LPL), cholesteryl ester transfer protein (CETP), and endothelial nitric oxide (eNOS) genes and coronary artery disease (CAD): CETP Taq1 B2B2 associates with lower risk of CAD in Asian Indians. J. Community Genet. 1, 55–62 10.1007/s12687-010-0005-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abd El-Aziz T.A., Mohamed R.H. and Hashem R.M. (2011) Association of lipoprotein lipase and apolipoprotein C-III genes polymorphism with acute myocardial infarction in diabetic patients. Mol. Cell. Biochem. 354, 141–150 10.1007/s11010-011-0813-6 [DOI] [PubMed] [Google Scholar]
- 53.Tripathi R., Agarwal S. and Ramesh V. (2011) Lipoprotein lipase (A1127G) gene polymorphism: a case–control association study. Biochem. Genet. 49, 587–591 10.1007/s10528-011-9433-9 [DOI] [PubMed] [Google Scholar]
- 54.Abd-El-Aziz T.A., Mohamed R.H. and El-Shal A.S. (2013) Synergistic effect between lipoprotein lipase and apolipoprotein C3 genes in determining the severity of coronary artery disease. J. Cardiovasc. Transl. Res. 6, 430–435 10.1007/s12265-013-9446-3 [DOI] [PubMed] [Google Scholar]
- 55.Daoud M.S., Ataya F.S., Fouad D., Alhazzani A., Shehata A.I., Al-Jafari A.A. et al. (2013) Associations of three lipoprotein lipase gene polymorphisms, lipid profiles and coronary artery disease. Biomed. Rep. 1, 573–582 10.3892/br.2013.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanguturi P.R., Pullareddy B., Rama Krishna B.S. and Murthy D.K. (2013) Lipoprotein lipase gene HindIII polymorphism and risk of myocardial infarction in South Indian population. Indian Heart J. 65, 653–657 10.1016/j.ihj.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferencak G., Pasalić D., Grsković B., Cheng S., Fijal B., Sesto M. et al. (2003) Lipoprotein lipase gene polymorphisms in croatian patients with coronary artery disease. Clin. Chem. Lab. Med. 41, 541–546 10.1515/CCLM.2003.082 [DOI] [PubMed] [Google Scholar]
- 58.Li Y., He P.P., Zhang D.W., Zheng X.L., Cayabyab F.S., Yin W.D. et al. (2014) Lipoprotein lipase: from gene to atherosclerosis. Atherosclerosis 237, 597–608 10.1016/j.atherosclerosis.2014.10.016 [DOI] [PubMed] [Google Scholar]
- 59.Clee S.M., Bissada N., Miao F., Miao L., Marais A.D., Henderson H.E. et al. (2000) Hayden plasma and vessel wall lipoprotein lipase have different roles in atherosclerosis. J. Lipid Res. 41, 521–531 [PubMed] [Google Scholar]
- 60.Preiss-Landl K., Zimmermann R., Hämmerle G. and Zechner R. (2002) Lipoprotein lipase: the regulation of tissue specific expression and its role in lipid and energy metabolism. Curr. Opin. Lipidol. 13, 471–481 10.1097/00041433-200210000-00002 [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y., Xie W., Li L., Zhang M., Cheng H.P., Gong D. et al. (2016) MicroRNA-27 prevents atherosclerosis by suppressing lipoprotein lipase-induced lipid accumulation and inflammatory response in apolipoprotein E knockout mice. PLoS One 11, e0157085 10.1371/journal.pone.0157085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ani A., Ani M., Moshtaghie A.A. and Ahmadvand H. (2010) Effect of titanium on lipoprotein lipase activity in vivo and in vitro. J. Trace Elem. Med. Biol. 24, 95–98 10.1016/j.jtemb.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 63.Sagoo G.S., Tatt I., Salanti G., Butterworth A.S., Sarwar N., van Maarle M. et al. (2008) Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: a HuGE association review and meta-analysis. Am. J. Epidemiol. 168, 1233–1246 10.1093/aje/kwn235 [DOI] [PubMed] [Google Scholar]
- 64.Wittrup H., Tybjaerg-Hansen A. and Nordestgaard B. (1999) Lipoprotein lipase mutations, plasma lipids and lipoproteins, and risk of ischemic heart disease. Circulation 99, 2901–2907 10.1161/01.CIR.99.22.2901 [DOI] [PubMed] [Google Scholar]
- 65.Cagatay P., Susleyici-Duman B. and Ciftcic C. (2007) Lipoprotein lipase gene PvuII polymorphism serum lipids and risk for coronary artery disease: meta-analysis. Dis. Markers 23, 161–166 10.1155/2007/863712 [DOI] [PMC free article] [PubMed] [Google Scholar]