Abstract
Imaging flow cytometry has been applied to address questions in infection biology, in particular, infections induced by intracellular pathogens. This methodology, which utilizes specialized analytic software makes it possible to analyze hundreds of quantified features for hundreds of thousands of individual cellular or subcellular events in a single experiment. Imaging flow cytometry analysis of host cell-pathogen interaction can thus quantitatively addresses a variety of biological questions related to intracellular infection, including cell counting, internalization score, and subcellular patterns of co-localization. Here, we provide an overview of recent achievements in the use of fluorescently labeled prokaryotic or eukaryotic pathogens in human cellular infections in analysis of host-pathogen interactions. Specifically, we give examples of Imagestream-based analysis of cell lines infected with Toxoplasma gondii or Mycobacterium tuberculosis. Furthermore, we illustrate the capabilities of imaging flow cytometry using a combination of standard IDEAS™ software and the more recently developed Feature Finder algorithm, which is capable of identifying statistically significant differences between researcher-defined image galleries. We argue that the combination of imaging flow cytometry with these software platforms provides a powerful new approach to understanding host control of intracellular pathogens.
Keywords: imaging flow cytometry, fluorescent protein, intracellular pathogen, Mycobacteria tuberculosis, Toxoplasma gondii, Feature Finder, cellular heterogeneity, colocalization, phagosome maturation, Rab5, Rab7
Graphical abstract
1. Introduction
Intracellular bacteria and protozoans include symbionts and pathogens that live in the cytoplasm, cytoplasmic vacuoles or nuclei of host eukaryotic cells, which are integral to the biology, pathology and evolution of infection in their hosts [1]. Among them are major infectious agents of humans such as Mycobacterium tuberculosis (MTb), and other intracellular pathogens, where humans serve as the intermediate host, such as Toxoplasma gondii (T.gondii). The molecular mechanisms that allow these pathogens to invade intracellular spaces invasion and manipulate host cell machinery are critical to understand host control of infection and the unique niches that support for developmental cycles of invading parasites.
Parameters that describe the interaction of intracellular pathogens with host cells are traditionally quantitated on a cellular population level. However, intracellular replication is heterogeneous, and infection of host cells with a clonal population of intracellular pathogen frequently results in variable numbers of bacterial or protozoan pathogens in each host cell [2–5]. Microscopic analysis reveals heterogeneity during intracellular pathogen internalization and intracellular replication by interaction of a variety of host- and pathogen-depending factors. To study host-pathogen interaction heterogeneity on the single cell level requires techniques capable of analyzing thousands of cellular images in a short period of time. Imaging flow cytometry (IFC) provides a very powerful tool for researching intracellular pathogens. First, IFC allows quantification of morphological cellular features as well as spatial distribution of numerous fluorescent markers in single cells and in heterogeneous cellular populations [6–7]. Second, IFC allows direct visualization of cells and fluorescently labeled pathogens and makes it possible to correlate acquired cellular images with events on bivariate dotplots (one dot for one cellular event). Third, IFC instruments such as the Imagestream platform (Amnis-Merck, USA) provide the opportunity of acquisition and quantitative analysis of tens and hundreds of thousands of cells in a short period of time (Inspire™ acquisition software and IDEAS™ analysis software (Amnis-Merck, USA) for Imagestream platform). Finally, in complex multiple-labeling imaging experiments, IFC allows quantitation and compensation for spectral crosstalk on a pixel-to-pixel basis using personal computer and specialized software. The use of fluorescent markers successfully incorporated by intracellular pathogens has proven to be an especially powerful tool in combination with IFC technology.
Notably, detection and quantification of fluorescent signals using IFC can be performed using live cells without fixation (with appropriate biosafety guidelines). In the emerging era of systemic and synthetic biology, IFC coupled with genetic-tagging technologies is becoming the powerful instrument to visualize dynamic components of the cellular system. We discuss recent advances in the use of fluorescent proteins for analysis of host-pathogen interaction markers and show examples of sophisticated IFC analysis of T.gondii and MTb internalization.
2. Use of external and internal fluorescent reporters for intracellular pathogens
Use of image-based platforms such as fluorescent (confocal) microscopy, intravital multiphoton microscopy and IFC (see Table 1 for comparison of IFC and other techniques for cell analysis) significantly expanded the understanding of complex diseases induced by intracellular pathogens For example:1) internalization and phagocytosis of intracellular pathogens; 2) differentiation intracellular forms of pathogens and survival inside phagocytes; 3) replication inside phagocytes and multiplication of pathogens(see ref. [8–9] and [6] for review).
Table 1.
Comparison of IFC and Other Common Techniques for Cell Analysis (directly modified from [6]).
| Feature | IFC | FC | Spectral FC* |
LSC** | High- Thoughput Fluorescent Microscopy |
Western Blotting |
|---|---|---|---|---|---|---|
| Light Scatter | Yes (side scatter) | Yes | Yes | Yes | No | No |
| Brightfield | Yes | No | No | Yes | Yes | No |
| Spectral Compensation required | Yes | Yes | No | No | No | No |
| Time-lapse (single-cell) | No | No | No | Yes | Yes | No |
| Time parameter (population analysis) | Yes | Yes | Yes | Yes | Yes | No |
| Morphological analysis | Yes | No | No | Yes | Yes | No |
| Speed of acquisition | Up to 5×103 events/sec | Up to 25×103 events/sec | Up to 5×104 events/sec | Up to 100 cells/sec | Rate depends on exposure time; parallel acquisition | Averaged data from 5×105–106 per slot |
| Size range | Up to 100 µ | Instrument- and nozzle-dependent (40 µ cell size limit-for 130 µ nozzle) | Up to 40 µ | Not limited | Not limited | Not limited |
| Signal spread error | Yes | Yes | No | Yes | No | N/A |
| Rare cell analysis (<0.01%) | Yes | Yes | No available publications | Capable of analyzing 1:104 cells | No | No |
| Capable to analyze cell heterogeneity | Yes | Yes | Yes | Yes | Yes | No |
Spectral FC- Sony Biotechnology Inc. instruments (SP6800 spectral analyser).
LSC – Compucyte Inc (now Thorlabs, Inc., USA) instruments.
2.1 Fluorescent protein palette for fluorescence-based techniques and IFC
Recent advances in the development of fluorescent sensors has increased and diversified the utility of fluorescent protein (FP) for analysis of intracellular parasites and bacteria in vitro and in vivo. In the case of parasites, green fluorescent protein (GFP) originally purified from the jellyfish (Aequoria victoria) has been successfully expressed in several protozoan intracellular parasites such as Leishmania species [10–12], Trypanosoma [13–14], Enatomoeba [15], Plasmodium [16] and Toxoplasma [17], and more recently as a dual combination with other fluorescent reporters (for example with mCherry) to follow the developmental stages of T. gondii [18]. FPs have been also successfully used for deciphering pathogenesis of bacterial intracellular infection (some examples are given in [19–24]). In these cases, the FP-tag sequence is fused to the DNA sequence of a known protein-of-interest in an expression vector that is translated as a fusion protein in the bacteria. The discovery of GFP homologues that emit not only in green light spectrum, but in all parts of the light spectrum, provided a significant boost in imaging host-pathogen research [25–26]. A particular interest is geared toward the novel group of red fluorescent proteins (RFPs). These RFPs-based sensors provide an advantage of reduced autofluorescence compared to GFP, and relatively low light scattering and minimal absorbance at the longer wavelengths [27–29] (summarized in Table 2). Also, near-infrared FPs are now emerging group due to high transparency of mammalian tissues and whole animals to light in 650 nm to 900 nm “optical window”and to low autofluorescence/light scattering advantage of this type of FP for in vivo molecular imaging [30].
Table 2.
Properties of FP proteins used in IFC research of intracellular pathogens
| Protein | Class | Ex.(n m) |
Em.(nm) | Brightness* | Oligomerization | IFC | Source | Refs |
|---|---|---|---|---|---|---|---|---|
| EGFP | Green | 488 | 507 | 1 | Weak dimer | Yes | Clontech | [3; 34; 36–41] |
| ZsGreenl | Green | 493 | 505 | 2.5 | Tetramer | Yes | [42] Clontech | [43] |
| EYFP | Yellow-Green | 514 | 527 | 1.5 | Weak dimer | Yes | Invitro-gen | [39] |
| tdTomato | Red | 554 | 581 | 2.8 | Tandem dimer | Yes | [31] | [43–45] |
| mCherry | Red | 587 | 610 | 0.47 | Monomer | Yes | [31] | [43–44;46] |
| DsRed-monomer | Red | 556 | 586 | 0.1 | Monomer | Yes | Clontech | [47] |
| RFP (tag) | Red | 555 | 584 | 1.41 | Monomer | Yes | Eurogen | [37] |
EGFP brightness equals 1.
Dimeric (d) and monomeric (m) RFP proteins such as dTomato, mOrange and mCherry have also been employed for labeling intracellular parasites. These proteins were chosen based on relatively high brightness and low maturation time (time from FP expression to the point where FP becomes functional) [31]. The tandem version of dimeric Tomato (tdTomato) is the brightest FP, but its usage is limited by a relatively large molecular size of tdTomato, which may interfere with fusion-protein packaging [32]. In general, under optimal cellular growth conditions the expression of monomeric FPs by bacterial or protozoan cells does not have a detectable effect on growth of transfected cells. However, adding a fluorescent tag may lead to toxicity in the case of high-copy-number FP expression plasmids for bacterial cells, when fusion proteins are expressed at a concentration that significantly exceeds expression of endogenous protein [33]. Another concern is that the introduction of a plasmid expressing FP to a bacterial cell may lead to a reduced intracellular infection [34].
Recently, a wide spectrum of FP-broad-range-expressing plasmids was developed for bacterial research [35]. Authors reported a set of 12 different FPs covering a wide range of excitation (ex.) and emission (em.) wavelengths (ex. 399–610 nm; em. 476–649 nm), and emphasized the importance of selecting appropriate FPs for specific fluorescence-based application such as confocal microscopy or molecular imaging. Thus, the authors recommended using E2-Crimson FP (ex./em. 611/646 nm), due to its low photobleaching, fast maturation and relatively high brightness for instruments with green-yellow (561 nm) and red laser (633–640 nm) excitation wavelengths. Furthermore, the authors also analyzed the usefulness of their plasmids for conventional flow cytometry and microscopy, as well as for in vivo imaging, but not for IFC.
In IFC technology relative brightness is one of the most important characteristics, and tdTomato excited at 561 nm and determined with 610/20 em. filter will be the primary candidate for intracellular pathogen research. Alternatively, mCherry monomeric protein with less brightness but suitable for 561 nm ex. is a popular choice at IFC applications. In the Table 2 we summarized existing publications on IFC exploiting different FP fusions, and the majority of publications reported using mCherry and tdTomato as well as the classic EGFP.
2.2 IFC in the studies of bacterial infection of host cells
Conventional flow cytometry analyses allowed facile visualization of bacterial association with target host cells. Cell-pathogen associations have been evaluated by i) labeling bacteria externally (with antibodies against bacterial cell membrane components conjugated with fluorochromes or fluorescent dye) or by ii) expressing FPs in bacterial cells and analyzing host cells infected with fluorescently labeled pathogen, via conventional flow cytometer. This latter method has been used by a number of researchers and correlated with standard colony forming units (CFU) assays to study host cell association with for example, the pathogens Chlamydia psittaci [48], Mycobacterium bovis [49] etc. A functional FP such as EGFP or mCherry can be expressed in diverse bacterial systems and have minimal effect on the interactions of pathogens with their respective host cells [20]. However, the quantification of intracellular particles and/or bacteria using traditional flow cytometry is complicated by extracellular bound bacteria that make it technically challenging to discriminate between intracellular and extracellular bacteria. See Table 3 for a summary of applications of IFC for intracellular pathogens research.
Table 3.
IFC analysis of intracellular bacterial and protozoan pathogens
| Intracellular parasite |
Fluorescent reporter |
IFC- platform |
Assays | IFC analysis (parameters) |
References |
|---|---|---|---|---|---|
| Burkholderia thailandensis | EGFP Baclighttm Green | IS-X Mark II | Growth curves; internalization; effect of inhibitor (wortmannin) | Max pixel fluorescence; Eroded intracellular and spot count masks | [34] |
| Cryptococcus neoformans | Alexa 488; CMDFA | IS-X | Phagocytosis; stress response; C.neoformans multiplication | [50–51] | |
| Brucella melitensis | mCherry | IS100 | Internalization | Eroded mask for erythrocytes (BF) from infected mice and 50% intensity mask for mCherry | [46] |
| Leishmania amazonensis | CFSE | IS-X | % infected cells | % positive cells; mean number of spots per infected activated cell; fluorescent intensity; max pixel of fluorescence | [52] |
| Leishmania donovani | DsRed2 | FlowSight | Quantitation of infected cells/L.donovani load; NFκB-p65 translocation in infected macrophages | MaxPixel vs. intensity, spot counting; similarity score | [47] |
| T.gondii | EGFP | IS100 | Quantitation and detection of T.gondii by size and fluorescence in cell line | Spot counting | [3] |
| T.gondii | ZsGreenl; mCherry | IS-X | Quantitation and detection of T.gondii by size and fluorescence in peritoneal excudate cellular subset | Multi-color staining and detection T.gondii by size and fluorescence; colocalization; | [53] |
| T.gondii | mCherry; tdTomato ZsGreenl | IS-X | Quantitation and detection of T.gondii by size and fluorescence in peritoneal excudate cellular subset;STAT6 nucleo-cytoplasmic translocation in infected cells | Multi-color staining and detection T.gondii by size and fluorescence; colocalization; similarity (DAPI/pSTAT6) | [43] |
| T.gondii | mCherry; tdTomato; CellTrace Violet | IS-X | Quantitation and detection of T.gondii in peritoneal excudate cellular subset | Multi-color staining and detection T.gondii by size and fluorescence; colocalization | [44] |
| T.gondii | tdTomato | IS-X | Detection of T.gondii in blood cells | 4-color staining of blood cells and detection of T.gondii by size and fluorescence | [45] |
| T.gondii | EGFP, EYFP | IS** | Quantitation and detection of T.gondii in peritoneal excudate subset | Multi-color staining and detection T.gondii by size and fluorescence | [39] |
| Salmonella enterica, serovar Thyphimurium | EGFP | IS-X | Internalization; counting bacterial load inside cell | Mean number of spots per infected cell; internalization score | [39] |
| Salmonella enterica, serovar Thyphimurium | EGFP, AB against O-antigen | IS** | Bacterial redox “stress” ; intracellular/extracellular bacteria | Fluorescent intensities; ratio parameters | [54] |
| Salmonella enterica | EGFP | IS-X | Quantification of Salmonella in infected B-cells | Fluorescent intensities | [54] |
| Plasmodium berghei | EGFP | IS-X | Size of merozoits | Size | [40] |
| Plasmodium | EGFP; RFP | IS-X | Interaction with erythrocytes | Colocalization of EGFP and PKH26 (similarity) | [37] |
| Plasmodium falciparum | Fluo-4AM, Hoechst | IS100 | Phenotypic screen for compounds destroying P.falciparum digestive vacuole | Colocalization Fluo-4-AM and DNA dye-stained P. falciparum | [56] |
| Plasmodium falciparum | EGFP | IS100 | Analysis of sorted EGFP-expressing P.falciparum | Fluorescent intensity; morphological features | [36] |
| Plasmodium falciparum | Hoechst | IS** | Surface area, sphericity | Diameter; perimeter; circularity; projected surface area loss | [57] |
| Plasmodium falciparum | Calcein-AM; AnV | IS100 | Microvesicles (MVs) quantitation by size | SSC, bright field and two fluorescent channels to quantitate MVs | [58] |
| Blastocystis spp. | CFSE, Hoechst | IS-X Mark II | Shape, size, granularity, nuclear arrangement | Software shape wizard; aspect ratio for CFSE and BF; circularity, granularity; DNA content | [59] |
| Aeromonas veronii* | Syto9 (bacteria); AnV; DRAQ5; PI; DHR; | IS-X Mark II | Phagocytosis; proliferation; apoptosis; NFκB translocation in infected host cells | Morphology; similarity score; fluorescent intensity | [60] |
| M.tuberculosis | EGFP; Liso Tracker Deep Red | IS-X Mark II | Phagocytosis;-phagosome maturation; intracellular replication | Spot counting; phagosome maturation score | [41] |
| M.tuberculosis | AB to CD4, IFNgamma, IL17 | IS** | Immunophenotyping of MTb-activated dendritic cells | Fluorescent Intensities | [61] |
| M.abscessus | mCherry; FLICA H2DCFDA | IS-X Mark II | Apoptotic/necrotic; FLICA/caspase 3/7 and H2DCFDA activated cells | Fluorescent Intensities | [62] |
| Entamoeba histolytica | PE-anti-TMK antibody | IS** | Co-localization; phagocytosis | Bright detail similarity score (co-localization between fluorescence in different channels) | [63] |
| Entamoeba histolytica | No fluorescent reporter | IS** | Shape | Bright field area and circularity score | [64] |
| Entamoeba histolytica | CMFDA; DID; live/dead violet dye | IS-X Mark II | Viability; internalization; amoebic trogocytosis | Fluorescent intensities; spot count; Max pixel; internalization score | [65] |
| Pneumocystis carinii | PE-CD11c | IS-100 | Immunophenotyping | Fluorescent intensities; Max Pixel values (discriminates punctate from background) | [66] |
| Legionella pneumophila | Polyclonal AB to L.p. | IS** | Detection of L.pneumophila in macrophages | Fluorescent intensities; (Legionella-positive punctate) | [67] |
| Francisella tularensis | Phalloidin, Fura Red | IS** | Ca++ production; actin polymerization | Fluorescent intensities | [68] |
| Brucella melitensis | mCherry | IS-100 | Detection of mCherry-tagged B.melitensis in red blood cells (free, adherent, overlapping) | Fluorescent intensities | [69] |
| Chlamydia pneumoniae | Cholera toxin-Al488 | IS** | Membrane fluidity and uniformity of lipid rafts in C.pneumoniae –infected cells | Lipid raft spot counting; homogeneity | [70] |
Aeromonas veronii is an intracellular pathogen of different eukaryotic hosts [71];
IS** model of Imagestream instrument is not specified.
The introduction of high-throughput fluorescence microscopy made tremendous progress in quantitation of image features using proprietary as well as open image analysis software such as ImageJ program (originally NIH Image)[72], and more recently CellProfiler platform [73]. IFC differs from the fluorescent microscopy since the first method utilizes images of individual cells or small groups of cells in the stream, while the microscopy deals with cells attached to the coverslip. In the IFC it is fairly easy to separate images of single cells or cell groups, while in microscopy such separation (based on image segmentation) is less feasible and is typically performed by investigator. However, the difficulties of proper segmentation of the images and the absence of an algorithm for the identification of statistically significant differences between series of images manually picked by the investigator continue to remain key challenges in the development of semi-automatic algorithms of image analysis in high-throughput microscopy.
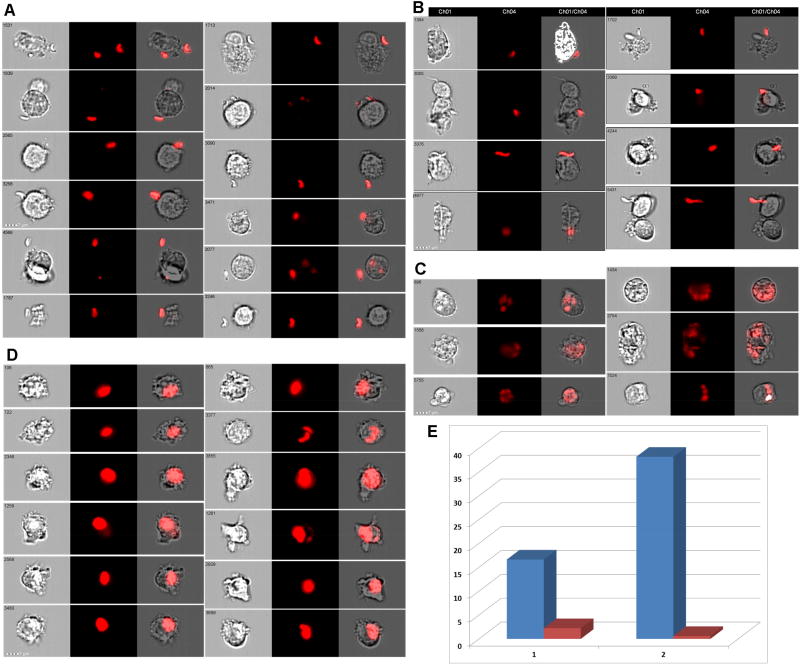
IFC analysis by Imagestream (Amnis-Merck, USA) employs powerful image analysis software (IDEAS™) with more than hundred fluorescence and morphology-based features. Recently, commercially available IDEAS™ software was upgraded with a Feature Finder algorithm (Fig.1) which utilizes the IFC advantage of single cell analysis. Furthermore, IDEAS™ Feature Finder algorithm provides a researcher with a scored table of statistically significant differences of multiple parameters derived from comparing two panels of manually picked image galleries. For example, Fig.1 illustrates, how to identify critical parameters differentiating T.gondii-infected cells inside large cellular terminal apoptotic “bubbles” from infected, but appearing morphologically normal cells, by comparing two sets of cellular images with and without large cellular “bubbles”.
Figure 1. Feature Finder algorithm implementation for T.gondii-infected THP-1 cells (defining of statistically significant feature for identification of cellular “bubbles”, containing mCherry-labeled Toxoplasma).
1A, B: Two image panels from experiment with T.gondii-infected THP-1 cells (hand-picked by investigator). Ch1 – bright field; Ch4 – mCherry fluorescence (mCherry-labeled T.gondii); Ch1/Ch4 –merged cellular and T.gondii images. Right – images show the intracellular T.gondii clustered in vacuoles inside the normal size cells. Left – images show the intracellular T.gondii in vacuoles inside swelling cells (cellular “bubbles”).
1C: Major step in Feature Finder algorithm – to define Ch1 (brightfield) and Ch4 (mCherry fluorescence) as channels of interest, and ALL software features as categories of interest. Alternatively, categories of interest can be narrowed to “Fluorescent Intensity”category etc. 1D: Feature Finder wizard results – “Delta Centroid XY” feature provides the best statistical significance for two analyzed galleries of images chosen to represent T.gondii inside “bubble-cells” and in appearing morphologically “normal” cells.
1E, G: “Delta Centroid XY” feature applied to two initially hand-picked galleries of images. Dotblot axises: height of image vs Delta Centroid XY.
1F, H: “Delta Centroid XY” feature applied to single, focused cellular population of acquired file. The populations corresponding to initial hand-picked galleries of images are automatically gated on dotplot.
2.3 IFC in research of protozoans expressing fluorescent proteins
IFC techniques with FPs have been used for different applications such as detection and quantitation of intracellular pathogens, phagocytosis, colocalisation with membrane dyes and subcellular organelle markers (summarized in Table 3). Specifically, FPs have been successfully expressed in several Toxoplasma, Plasmodium, Trypanosoma and Leishmania species [12]. Some of them produce FP only in replicative stages of the parasite living cycle; for example, epimastigotes and amastigotes of Trypanosoma cruzi [13], making FPs useful for the detection of proliferating parasites. Recently, functional and genetically modified parasite strains with dual expression of fluorescent tags under control of stage-specific promoters (BAG1-GFP; SAG1-mCherry) were developed for T. gondii [18]. However, in parasitic protozoans FP-based tagging is mostly restricted to aerobic protozoans or their oxygenated stages of the life cycle because of the oxygen requirement for the maturation of FPs. Recent alternative to GFP-protein fusion is tagging of parasites with biarsenical-tetracysteine FPs (either FISsH-EDT(2) or ReAsH-EDT(2)), which has been successfully used in a variety of cell models including Dictyostelium [74]. IFC techniques are using FPs for different applications such as detection and quantitation of intracellular pathogens, phagocytosis, colocalisation with membrane dyes and subcellular organelle markers (summarized in Table 3).
2.4 Fluorescent staining of intracellular pathogens with external reporters
Similar to fluorescent microscopy, IFC employs staining with specific antibodies to simultaneously immunophenotype host cells, identify intracellular pathogen and/or organelles, transcription factors and other subcellular components [54; 63; 66]. Though fluorescent reporters are commonly used to detect intracellular parasites, in the absence of the specific fluorescent tag, it is possible to follow parasites using membrane cell tracking dyes [52; 59]. A fluorescent membrane cell-tracking dye such as carboxyfluorescein diacetate succinimidyl ester (CFSE) has been extensively used in the last few years to quantitate mammalian cell divisions [75]. The same correlative approach of membrane dye (PKH26) dilution with cell division can be applied to measure the rate of bacterial replication in vitro through conventional flow cytometry [76]. For example, Gibson-Corley et al. [52] labeled Leishmania amazonensis promastigotes with CFSE dye and quantitated the number of parasite-positive spots in infected cells using an Imagestream instrument (Amnis-Merck, USA). Similarly, Yason and Tan [59] labeled protistan parasite Blastocystis with CFSE dye and described the shape, size, granularity and nuclear arrangement in a variety of morphological forms of Blastocystis by IFC technique. Furthermore, Dupont et al [44] used a combination of an external staining of T. gondii parasites with CellTrace Violet (pH sensitive dye) and internal fluorescent tag mCherry (pH independent) to follow the fate of T.gondii in vivo via conventional flow cytometry and high throughput IFC. This report emphasized the importance of dual fluorescent reporters to track the parasite and distinguish different ways for parasite to infect cells.
3. IFC applications in intracellular pathogen research
3.1 Quantification and internalization of intracellular pathogens
The standard approach to quantify intracellular pathogen burden is mainly cell culture-based, time-consuming and expensive (by for example colony forming unit (CFU) determination). Moreover, analysis is usually hampered by low infection rates (less than 2% in some models) [77] and may require complicated methods to monitor intracellular development such as quantitative RT-PCR (qRT-PCR) techniques [78–79]. Alternatively, counting of intracellular pathogens involves direct quantification of pathogen viability and development of microscopy applications. Light microscopic analysis of pathogen or pathogen-induced cellular inclusions can be carried out with a variety of dyes such as a Giemsa stain [80–82], or staining with acridine orange, RNA and DNA probes, or specific immunofluorescent staining. Light microscopy yields very good qualitative [82] and quantitative [89] results, however, it is limited by sampling of one-several hundreds of events and might be prone to subjective variations between observers.
Conventional flow cytometry offers a statistically robust alternative for the quantitation of thousands of fluorescently labelled parasites and bacteria [83], and has been intensively used during the last twenty years. It may employ the staining of intracellular pathogens with nonspecific nuclear dyes such as propidium iodide (PI) or acridine orange [84], immunofluorescent staining with specific antibodies to intracellular pathogens [85–86], and counts of fluorescently tagged bacteria and protozoans [87]. However, there are some constraints that limit the use of flow cytometry in the quantitation of intracellular pathogens. Specifically, it is possible, yet technically challenging to differentiate between attached and internalized parasites using conventional flow cytometry [87]. Another obstacle is cellular and parasite aggregation which could have a significant effect on counting results. Besides, flow cytometry cannot address the question on intracellular localization of pathogens that is important for studies on pathogens development.
The use of fluorescence-based methods such as fluorescent microscopy and/or conventional flow cytometry to study intracellular pathogens became a common approach with the introduction of intracellular pathogens expressing FPs [3; 74; 88]. The IFC offers more information than just counting viable intracellular parasites. It allows researcher to combine intracellular parasite counting with the quantitation of a multitude of subcellular morphological and fluorescence features (Table 3), as well as robust statistics. The good example provides “Similarity Score” - a log-transformed Pearson's correlation coefficient between the pixel values of two image pairs, used in IDEAS™ software to evaluate a degree of colocalisation [89]. This feature as well as IFC use of Rd value – shift in the statistical distribution between two populations (eg. “treated” vs. “untreated”) described in details by Maguire et al. [90]. This multi-parametric multi-color approach is actively pursued in parallel with IFC by high-content microscopy, generally using 10× and 20× magnification [91–92]. For example, Maudet et al [92] employed a fluorescence-based screen to study a library of miRNA mimics in the molecular interplay between intercellular Salmonella enterica GFP-expressing strain and host cells. The authors successfully used a high-content screening fluorescent microscope (Molecular Devices, USA) at ×20 magnification and classified cells positive and negative for Salmonella, depending on the total area of GFP-fluorescence in the cell. Furthermore, GFP fluorescent intensities in the cell were used as a surrogate marker for the amount of intracellular bacteria [92]. However, high-throughput microscopy approach is limited by analysis of adhesive cells growing as monolayers, when segmentation algorithm to define single cells prior to analysis is mandatory. Imagestream-based IFC analysis, on the other hand, can be applied to hundred thousands of single events without segmentation: either cells (growing in suspension or detached), cell clusters, or cell fragments (one dot on dotplot corresponds to single event and vice versa) at ×60 magnification.
3.1.1 Quantification and internalization of Toxoplasma gondii
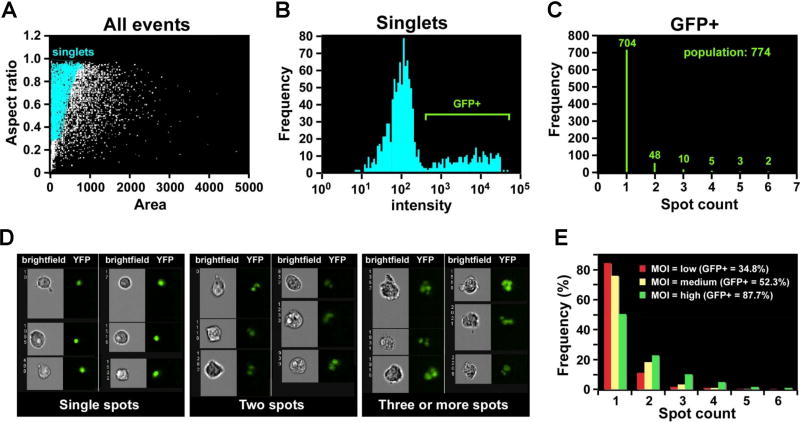
For the determination of intracellular pathogens ingested in each cell, IFC offers spot count (Fig.2) and internalization features. The spot count approach to quantitate intracellular pathogens by IFC can be illustrated by this figure adopted from Muskavitch et al. [2] (Fig. 2). We used the IDEAS™ software Spot count feature to calculate the number of independent green vesicles (corresponding to GFP+ Toxoplasma inclusions) determined in each single cell [2]. More sophisticated IFC analysis based on an internalization score or spot count feature includes the development of a mask based on a BrightField cell surface. The mask is eroded by 3–5 pixels to exclude the cell membrane and signals associated with the cell surface. The resulting Eroded mask is applied to a fluorescence channel (FP channel: for example, GFP or mCherry) and the internalization score is then calculated using the Internalization feature provided by IDEAS™ software (Amnis-Merck, USA). Internalization is represented as a log-scaled ratio between fluorescent intensity in the intracellular space versus fluorescent intensity of the entire cell. Cells that internalized FP-labeled intracellular pathogen typically have positive scores, whereas cells with parasites only attached to the plasma membrane have negative scores. Furthermore, co-localization of internalized intracellular parasites with intracellular compartments (for example, lysosomes or endosomes) can be calculated using the Bright Detail Similarity (BDS) feature. This feature quantifies the correlation between the bright details of the GFP+ Toxoplasma and fluorescent organelle markers. High values of similarity scores indicate that the fluorescence of intracellular pathogen and organelle marker colocalises in the same place.
Figure 2. Quantification of internalized GFP-labeled T.gondii using Spot feature of Imagestream (reprinted from [2] with permission from Bentham Science Publishers).
2A: Single-cell population defined by Area/Aspect ratio dotplot;
2B: Histogram of fluorescent intensity (GFP), gated on GFP+ cells;
2C: Histogram of Spot feature (IDEAS™, Amnis-Merck, USA) applied to GFP+, single, focused cells. Represents a quantitative distribution of cells with T.gondii-inclusions (distribution of GFP+-spots inside T.gondii-infected cells).
2D: Representative image galleries of cells, infected with T.gondii (cells with only one, two and three or more inclusions (spots)).
2E: Histogram of Spot Feature distribution (IDEAS™, Amnis-Merck, USA) representing GFP+ T.gondii inclusions in the infected cells after different multiplicity of infection (MOI).
The application of Eroded mask to T. gondii-infected cells (THP-1 cell line) allows to exclude from quantitation attached but non-internalized parasites, which otherwise will compromise counting (Fig.3 A, B). In our IFC experiments invasion of T.gondii parasites led to single and/or multiple inclusions in the infected cells (Fig. C, D), and had retained sensitivity to the actin polymerization inhibitor cytochalasin B (CytB) (Fig. 3E). In the presence of CytB (inhibits formation of actin filaments network), the percentage of invaded parasites was reduced to less than 5%. These results are consistent with previous publication where penetration of T. gondii was shown to be actin-dependent [93–95].
Figure 3. Quantification of internalized T.gondii.
3A, B: Representative image galleries of cells, which did not internalized T.gondii (mCherry-labeled T.gondii attached or not attached, but located close to cells).
3C, D: Representative image galleries of cells with multiple (C) or single (D) T.gondii inclusions.
3E: Bar graph showing % of THP-1 cells infected with mCherry+ T.gondii cells (defined with a help of Eroded (3 pixel) mask). Blue: % of T.gondii-positive cells without treatment with inhibitor; Red: % of T.gondii-positive cells 24 hours after treatment with CytB (10 µM).
3.1.2 Internalization and quantification of MTb
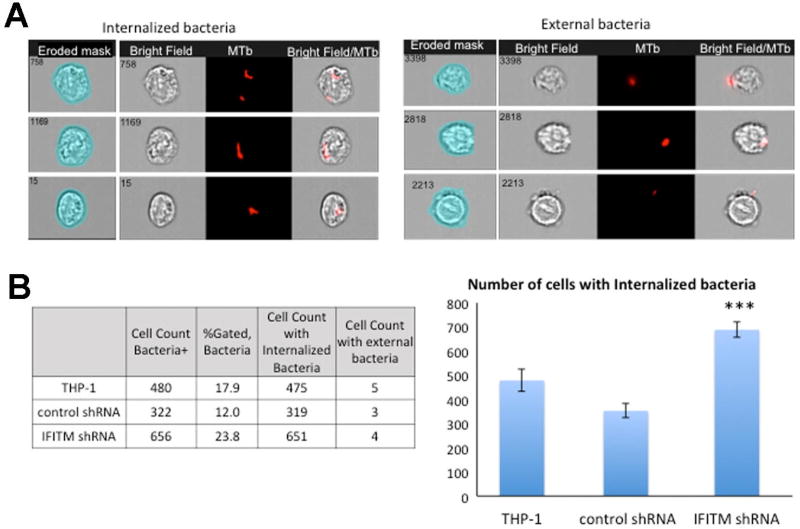
To understand how intracellular pathogen such as MTb survives inside potentially hostile cells, such as macrophages, it is especially important to understand the processes allowing pathogen internalization, interaction with cellular host factors and, in case of MTb, phagosome maturation. Recently, Ranjbar et al. demonstrated that the interferon (IFN)-induced transmembrane (IFITM) proteins, particularly IFITM3 is a host restriction factor for MTb [90]. As part of this study, the group used IS-X Mark II (Amnis-Merck, USA) and performed a quantitative analysis investigating the impact of knockdown of IFITM1–3, or the impact of overexpression of IFITM3 on MTb growth. Using the IDEAS™-guided algorithm, the study distinguished between internalized and non-internalized MTb bacteria (IDEAS™, Amnis-Merck, USA). An example of this approach is presented in Fig. 4A, which shows quantification of internalized TB bacteria and external bacteria binding to the cell membrane (Fig. 4A; the 4-pixel eroded mask is shown on the left for both panels).
Figure 4. Quantification of internalized mCherry labeled MTb bacteria.
4A: Left panel - Representative images of internalized MTb-mCherry acquired on IS-X Mark II.
4A: Right panel - external bacteria. Images shown from left to right : BF with erode mask (4 pixels), BF indicating cellular outline, H37Rv-mCherry+-MTb bacilli (Red), merged image of MTb-mCherry fluorescence and BF showing intracellular or external localisation of MTb-mCherry.
4B: Left - Chart showing wild type THP-1 cells, control shRNA cells, and IFITM shRNA cells lines number of cells being infected with mCherry-MTb, cells, internalized and external bacteria.
4B: Right -Bar graphs show the number of infected cells in all three cell lines, IFITM shRNA cells had significantly higher number of infected cells compared to control shRNA (p=0.0001) and wild type THP-1 (p=0.002). Histograms show the results of three separate experiments.
About 1% of cells in which bacteria were binding to the cell membrane were excluded from our analysis based on internalization mask (Fig. 4B, Table). The percentage of infected cells was comparable to results of flow cytometry analysis performed previously [90]. IFITM shRNA cell cultures presented significantly higher infected cells with H37Rv-mCherry compared to the control (see Figure 4A based on data presented in Ranjbar et al. [90]). These results were consistent with conventional CFU assay, which showed a significant increase (p<0.01) in MTb infection at days 2 and 3 post-infection in IFITM shRNA cells compared to control shRNA cells [90]. Notably, the IFC assay also allows capturing numerous images from infected cells that can be used for further analysis.
3.2 Subcellular characterization of intracellular pathogens by IFC
MTb has developed elegant strategies to avoid both the innate and adaptive immune responses [96–97]. One of the mechanisms used by MTb to survive in macrophages is manipulation of host phagosome/endosome pathways and inhibition of phagosome-lysosome fusion [98]. One cause of the phagosomal arrest is attributed to altered Rab7 function [99–102] coupled with retention of Rab5 (an early endosome marker) [99].
3.2.1 MTb localization in early endosome or late endosome compartments
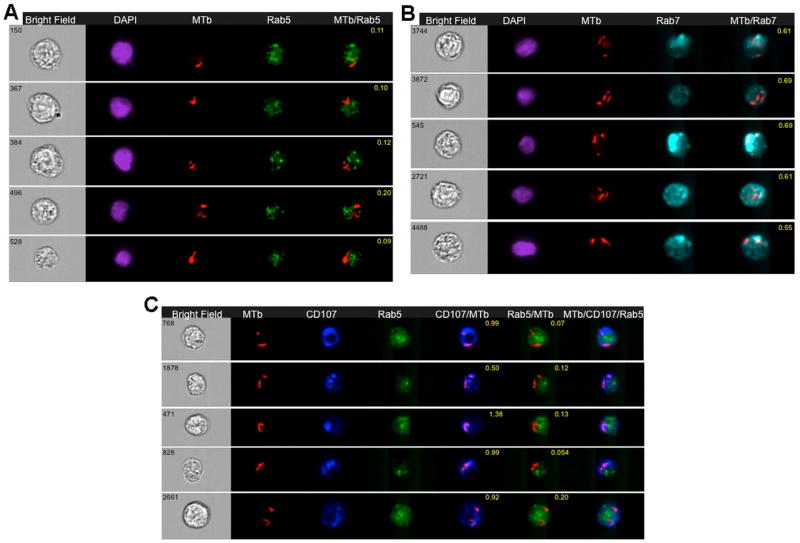
Since IFC is capable of quantitating co-localization of intracellular parasites with different subcellular compartments [63], this technology was used to show that internalized bacilli were restricted to early endosome or late endosome compartments co-localized with IFITM3 [90]. In the study by Ranjbar et al., IFC showed that that internalized bacilli were restricted to early endosome or late endosome compartments co-localized with IFITM3 [90].
Here, we performed the same experiment in THP-1 cells that did not contain overexpressed tagged IFITM3, and MTb-mCherry-infected cells were acquired by IS-X Mark II (Amnis-Merck, USA). Results were then analyzed with IDEAS™ software (Amnis-Merck, USA) as described above in section 3.1.2. Internalized bacteria were selected and followed by the co-localization wizard to attribute bacteria to early or late endosomal compartments. The IDEAS™ feature Bright Detail Similarity (BDS) was used to measure co-localization between two signals. A BDS score of 2 or greater represents a high degree of overlap. Representative images of co-localization of MTb-mCherry bacilli within the early (Rab5 positive) endosomal compartment and in the late (Rab7 positive) endosomal compartment are shown respectively in Fig. 5A and Fig. 5B. BDS coefficients are indicated in the merged images.
Figure 5. Detection of H37Rv-mCherry MTb in early and/or late phagosomal compartments of infected human THP-1 cells.
Wild type THP-1 cell were infected with H37Rv-mCherry MTb for 24 hours, fixed in 4% PFA, and stained with AB to Rab5 or Rab7 or Rab5 and CD107a and acquired on IS-X Mark II, as described in Ranjbar et al [90].
5A: Representative images of internalized MTb-mCherry in wild type THP-1 cells 24 hr after infection. Cells were labeled with Rab5 antibody and stained with DAPI as above. Images shown from left to right: BF image (Grey), DAPI (Purple), MTb-mCherry fluorescence (Red), Rab5 (Green), and a merged image showing colocalisation of MTb-mCherry fluorescence and Rab5. BF similarity coefficient values are shown in yellow font.
5B: Representative images of internalized MTb-mChery in wild type THP-1 cells that were labeled with Rab7 and stained with DAPI as above. From left to right: BF image (Grey), DAPI (Purple), MTb-mCherry fluorescence (Red), Rab 7 (Blue), and a merged image of MTb-mCherry fluorescence and Rab7 with BF similarity coefficient shown in yellow font.
5C: Representative images of internalized MTb-mCherry in wild type THP-1 cells infected with MTb-mCherry and labeled with Rab5 and CD107a (LAMP1) as above. From left to right: BF image (Grey), MTb-mCherry fluorescence (Red), CD107a (Dark blue), Rab5 (Green), and a merged image of MTb-mCherry fluorescence and CD107a with BF similarity coefficient in yellow font, a merged image of MTb-mCherry fluorescence and Rab5, and a merged image of MTb-mCherry fluorescence, CD107a and Rab5.
To further characterize the MTb localization in infected cells that were labeled with Rab5, the cells were also labeled with LAMP1 (CD107 conjugated antibody, lysosome marker). As shown in a new analysis presented in Fig. 5, which recapitulates work in Ranjbar et al [90], H37Rv-mCherry bacilli were co-localized to a greater degree at LAMP1/CD107 late endosome/lysosome and Rab7 associated late phagosome compartment as compared to the Rab5 associated early phagosome (Fig. 5C). Thus the study by Ranjbar et al., demonstrated the ability of IFC to produce a statistically robust evaluation of colocalization of MTb with various cellular compartments and to compare the trafficking of MTb in cells where IFITM1–3 had been knocked down [90].
The strategies to analyze phagosome maturation in infected by MTb cells were further expanded by Johansson et al [41]. In order to develop small-scale screening strategy for chemical compounds that enhance phagosome maturation in MTb-infected cells, the group used fixable (CD63) and non-fixable (LysoTracker Deep Red) lysosome markers, and introduced custom-designed “phagosome-maturation score” [41]. Essentially, the analysis strategy was to quantify an extent to which the lysosomal marker is enriched in phagosome comparing with the whole body of the cell. The use of “phagosome-maturation score” (ratio of mean pixels values) is based on the mean rather than the total intensity of lysosomal marker and therefore is less sensitive to the size of phagosome and cell comparing with standard approach based on the total intensity of lysosomal marker in the cell [41]. To validate the new approach authors imported images obtained using IS-X and quantitated phagosome maturation by techniques well-established in microscopy analysis [104–105].
Despite several decades of intensified research, the mechanisms involved in the protective immune response against MTb are not well understood. Use of multi-parametric and statistically robust IFC, and providing morphometric and fluorescence-based assessment of colocalization of MTb with different subcellular compartments will be beneficial in studying host-pathogen interactions in MTb infection and disease progression.
3.3 IFC and host-pathogen communication studies
IFC has also been used to study the dynamic of interaction between intracellular parasites and host cells. Bargieri et al. [37] quantified the invasion of PKH26-stained erythrocytes with GFP+ merozoites of P.berghei using co-localization and similarity scores to evaluate the efficiency of internalization of wild type and AMA1-deficient parasites. The decrease in invasion efficiency of AMA1-deficient merozoites was associated with altered adhesion to host cells [37]. The high sensitivity of Imagestream instruments allowed efficient detection of extracellular vesicles (EVs) in the wide size range [106–107]. Furthermore, we performed quantitative measurement of release of EVs of 100–400 nm size from P. falciparum schizont-stage parasites before egress [58], and it was possible to demonstrate that EVs originating from P. falciparum-infected red blood cells act as intercellular communicators and induce commitment of malaria parasites toward sexual differentiation. EVs released by infected cells could have multiple roles during infection [108–111]. Intracellular pathogen antigens may be transported by EVs or internalized by dendritic cells (DC), and this might amplify the cellular immune response [108]. EVs production is an universal cellular feature [112], and considered to be the major way to communicate and synchronize development and fate of intracellular pathogens. The application of IFC technology for research of EVs production by infected cells will further expand our understanding of intracellular pathogens.
3.4 IFC as a method to study heterogeneity and fate of intracellular pathogens
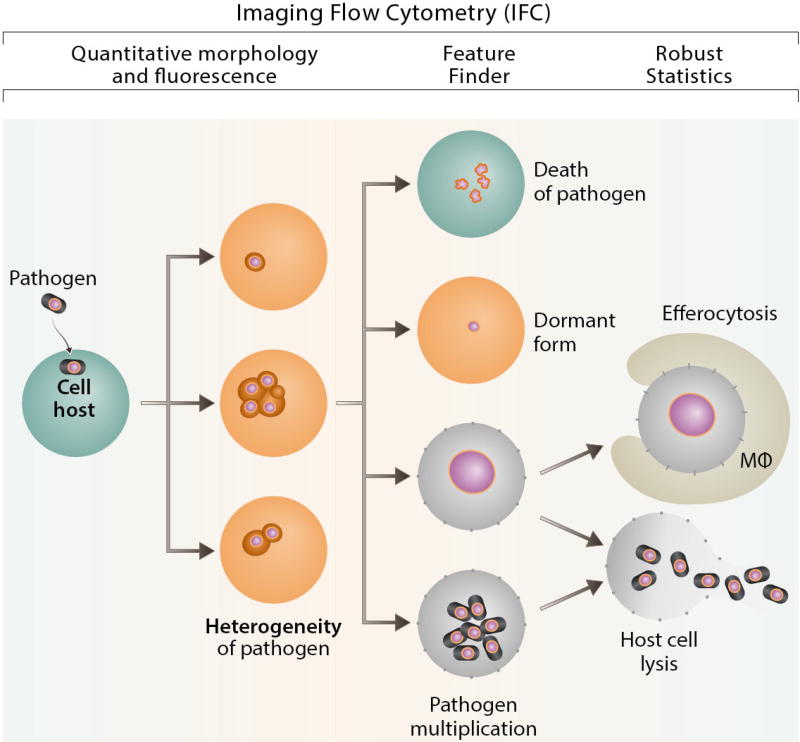
The fate of an intracellular pathogen after cell invasion depends on a variety of factors including type of the cell and/or pathogen, metabolic state and cell cycle stage of host cell (eg. professional phagocyte; endothelial cell, etc). Epigenetic factors can generate differentiation within genetically identical single cells of a clonal population [113] leading to the different fate of invading pathogen. Until recently, the technique of choice to study heterogeneity of intracellular pathogens and their fate in a clonal cellular population was flow cytometry supplemented by different variants of microscopy [114]. However, the statistically robust IFC technology platform allows quantification of a multitude of morphological pathogen forms present at the same time in the different subcellular compartments in a clonal cell line population leading to: 1) death of the pathogen; 2) long-term survival as a dormant form; 3) programmed cell death of the cellular host and efferocytosis; 4) proliferation inside phagocytes and multiplication of pathogen; or 5) death of pathogen (Fig. 6).
Figure 6. IFC in intracellular pathogen studies.
IFC allows to quantitate morphology and fluorescence, provides robust statistics, colocalization quantitation and Feature Finder capabilities.
3.4 IFC in drug discovery
In view of the recent spreading of intracellular pathogens showing resistance to the existing drugs [115–117], there is an urgent need to develop new, safe, and affordable drugs [118–119]. While several new chemicals were identified recently, they were primarily developed towards extracellular form of pathogens [120–121]. The biology of different parasite stages is important for research of intracellular pathogen response to drugs [122–124]. When comparing drug activity against Leishmania in vivo with drug activity against cultured parasites and intracellular forms in vitro, a correlation is better with the in vitro assays performed with drug screens using intracellular forms (amastigotes) vs. extracellular forms (promastigotes) [125]. However, the use of macrophage cell lines as host for intracellular parasites is problematic, since chemicals used to induce differentiation of macrophages in vitro modulate macrophage drug sensitivity [126–127].
Recent studies have suggested that IFC will be extremely helpful in screening for new drugs against intracellular pathogens. Recently, Lee et al. [56; 128] provided a detailed description of phenotype-based IFC assays to analyze a library of chemicals disrupting the integrity of the digestive vacuole of Plasmodium falciparum; these positive results of drug screening were further validated by confocal microscopy. It should be noted that these studies used colocalization of Fluo-4-AM and DNA dye-stained P. falciparum by IFC and demonstrated the capabilities of the method.
4. Perspectives and Challenges
We have described here data showing that IFC technology is a dependable and statistically robust platform for intracellular pathogen studies. Use of IFC and other image-based technologies in studying of host-pathogen interactions, has expanded our understanding of bacterial and parasite disease pathogenesis. Recent advances in red-shifted fluorescent protein development, advantageous due their low autofluorescence and light scattering, formed a basis for multiparameter and multi-color IFC. With the introduction of the Feature Finder algorithm, the IFC will further advance the field of imaging analysis. The availability and high sensitivity of Imagestream instruments also provides powerful tool for the analysis of intracellular pathogen heterogeneity.
By allowing an examination of pathogen morphology and subcellular localization and co-localization of pathogens with cytoplasmic organelles, along with statistical robustness of image analysis based on multi-parametric software (more than hundred morphological and fluorescent parameters can be combined with sequentially built customized masks), IFC gives novel opportunities in the analysis of the life cycle of pathogens and details of host-pathogen interactions.
The obvious limitation of IFC is the absence of a cell sorting option on the current instrumentation, which we expect will be overcome in the next years. The second limitation is its spatial resolution that is several fold less as compared to the fluorescent microscopy. However, introduction of the MultiMag option with 60× objective for Imagestream instrument results in a 0.3 µm pixel size sensitivity and along with the extended depth of focus option, makes this difference less significant.
Another challenge in the use of IFC is related to the increasing size of compounds libraries used in drug discovery for intracellular pathogens which have rigorous demands with respect to assay time and robustness. Future perspectives on drug screening may require compatibility of the IFC instrument with multi-well plate format (96–384 wells).
Finally, the development of IFC techniques and their recent applications to the study of intracellular pathogens at the single cell and population levels greatly enhances our understanding of the physiology of intracellular pathogens, and of the host-pathogen interface.
Highlights.
-
-
Imaging flow cytometry (IFC) characterize hundreds thousands of cellular images using hundreds of measurements of morphological and fluorescence cellular features.
-
-
Complex morphology of host-intracellular parasite interaction as well as standard cellular features (counts, size, shape) can be quantified in statistically robust manner using IFC.
-
-
Feature Finder algorithm is useful in defining statistically significant differences in morphological and fluorescent features of cells infected with T. gondii cells.
-
-
Imaging flow cytometry technique revealed that Mycobacteria tuberculosis internalized bacilli were co-localized to a greater degree at late endosome/lysosome CD107 and late endosome Rab7 compartment than Rab5 early endosome compartment.
Acknowledgments
This work was supported by grants from MES of Republic of Kazakhstan PI NURIS 055 project #100/14 to N.S.B., from the NIH (1R01AI117929-01A1) and the Annenberg Foundation to A.E.G., and by RFBR # 13-04-40189-H to I.A.V. We thank Sabrina Hawthorn and Rich De Marco of Amnis-Merck and Kenneth Ketman from PCMM for their generous help and advice with Imagestream instrumentation.
Abbreviations
- An5
annexin V
- APC
antigen-presenting cells
- BDS
Bright detail similarity
- BF
bright field
- CFSE
carboxyfluoresceinsuccinimidyl ester
- CFU
colony-forming unit
- CMDFA
5-chloromethylfluoresceinDiacetate
- DAPI
(4',6-diamidino-2-phenylindole)
- DHR
dihydrorhodamine
- DMEM
Dulbecco’s modified Eagle medium
- FP
fluorescent protein
- EGFP
enhanced green fluorescent protein
- em.
emission
- EVs
extracellular vesicles
- EYFP
enhanced yellow fluorescent protein
- ex.
excitation
- H2DCFDA
2',7'-dichlorodihydrofluorescein diacetate
- IFC
imaging flow cytometry
- IFITM
interferon-induced transmembrane proteins
- IS100
Imagestream 100
- IS-X
ImagestreamX
- IS-X MK
ImagestreamX MarkII
- LAMP1
lysosomal-associated membrane protein-1
- MTb
Mycobacterium tuberculosis
- MVs
microvesicles
- OD
optical density
- PBS
phosphate-buffered saline
- PCD
programmed cell death
- PE
phycoerythrin
- PI
propidium iodide
- qRT-PCR
quantitative RT-PCR
- RFP
red fluorescent protein
- shRNA
short hairpin RNA
- SI
staining index
- SSC
side scatter
- TNK
transmembrane kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fuchs TM, Eisenreich W, Heesemann J, Goebel W. Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra- and intracellular habitats. FEMS Microbiol. Rev. 2012;36:435–462. doi: 10.1111/j.1574-6976.2011.00301.x. [DOI] [PubMed] [Google Scholar]
- 2.Muskavitch MA, Barteneva N, Gubbels MJ. Chemogenomics and parasitology: small molecules and cell-based assays to study infectious processes. Comb. Chem. High Throughput Screen. 2008;11:624–646. doi: 10.2174/138620708785739989. [DOI] [PubMed] [Google Scholar]
- 3.Helaine S, Thompson JA, Watson KG, Liu M, Boyle C, Holden DW. Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. USA. 2010;107:3746–3751. doi: 10.1073/pnas.1000041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toma C, Okura N, Takayama C, Suzuki T. Characteristic features of intracellular pathogenic Leptospira in infected murine macrophages. Cell. Microbiol. 2011;13:1783–1792. doi: 10.1111/j.1462-5822.2011.01660.x. [DOI] [PubMed] [Google Scholar]
- 5.Bailo N, Cosson P, Charette SJ, Paquet VE, Doublet P, Letourneur F. Defective lysosome maturation and Legionella pneumophila replication in Dictostelium cells mutant for the Arf GAP ACAP-A. J. Cell Sci. 2014;127:4702–4713. doi: 10.1242/jcs.154559. [DOI] [PubMed] [Google Scholar]
- 6.Barteneva NS, Fasler-Kan E, Vorobjev IA. Imaging flow cytometry: coping with heterogeneity in biological systems. J. Histochem. Cytochem. 2012;60:723–733. doi: 10.1369/0022155412453052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basiji DA. Principles of Amnis imaging flow cytometry. In: Barteneva Natasha S, Vorobjev Ivan A., editors. Imaging Flow Cytometry: methods and protocols. Humana Press; New York: 2016. pp. 13–21. [DOI] [PubMed] [Google Scholar]
- 8.Ha DS, Schwarz JK, Turco SJ, Beverley SM. Use of the green fluorescent protein as a marker in transfected Leishmania. Mol. Biochem. Parasitol. 1996;77:57–64. doi: 10.1016/0166-6851(96)02580-7. [DOI] [PubMed] [Google Scholar]
- 9.Coombes JL, Robey EA. Dynamic imaging of host-pathogen interactions in vivo. Nat. Rev. Immunol. 2010;10:353–364. doi: 10.1038/nri2746. [DOI] [PubMed] [Google Scholar]
- 10.Florentino PT, Real F, Bonfim-Melo A, Orikaza CM, Fereira ER, Pessoa CC, Lima BR, Sasso GR, Mortara RA. An historical perspective on how advances in microscopic imaging contributed to understanding the Leishmania spp. and Trypanosoma cruzi host-parasite relationship. Biomed. Res. Int. 2014:565291. doi: 10.1155/2014/565291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fumarola L, Spinelli R, Brandonisio O. In vitro assays for evaluation of drug activity against Leishmania spp. Res. Microbiol. 2004;155:224–230. doi: 10.1016/j.resmic.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Bolhassani A, Taheri T, Taslimi Y, Zamanilui S, Zahedifard F, Seyed N, Torkashvand F, Vaziri B, Rafati S. Fluorescent Leishmania species: development of stable GFP expression and its application for in vitro and in vivo studies. Exp. Parasitol. 2011;127:637–645. doi: 10.1016/j.exppara.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Kessler RL, Gradia DF, Rampazzo RCP, Lourenco EE, Fidencio NJ, Manhaes L, Probst CM, Avila AR, Fragoso SP. Stage-regulated GFP expression in Trypanosoma cruzi: applications from host-parasite interactions to drug screening. PLoS One. 2013;8:e67441. doi: 10.1371/journal.pone.0067441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padmanabhan PK, Polidoro RB, Barteneva NS, Gazzinelli RT, Burleigh BA. Transient transfection and expression of foreign and endogenous genes in the intracellular stages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 2014;198:100–103. doi: 10.1016/j.molbiopara.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Ramakrishnan G, Rogers J, Mann BJ, Petri WA. New tools for genetic analysis of Entamoeba histolytica: blasticidin S deaminase and green fluorescence protein. Parasitol. Int. 2001;50:47–50. doi: 10.1016/s1383-5769(00)00067-2. [DOI] [PubMed] [Google Scholar]
- 16.Sultan AA, Thathy V, Nussenzweig V, Menard R. Green fluorescent protein as a marker in Plasmodium berghei transformation. Infect. Immun. 1999;67:2602–2606. doi: 10.1128/iai.67.5.2602-2606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Striepen B, He CY, Matrajt M, Soldati D, Roos DS. Expression, selection, and organellar targeting of the green fluorescent protein in Toxoplasma gondii. Mol. Biochem. Parasitol. 1998;92:325–338. doi: 10.1016/s0166-6851(98)00011-5. [DOI] [PubMed] [Google Scholar]
- 18.Paredes-Santos TC, Tomita T, Yan Fen M, de Souza W, Vommaro RC, Weiss LM. Development of dual fluorescent stage specific reporter strain of Toxoplasma gondii to follow tachyzoite and bradyzoite development in vitro and in vivo. Microb. Inf. 2016;18:39–47. doi: 10.1016/j.micinf.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker AE, Bermudez LE. Expression of the green fluorescent protein (GFP) in Mycobacterium avium as a tool to study the interaction between Mycobacteria and host cells. Microb. Pathog. 1997;22:193–198. doi: 10.1006/mpat.1996.0106. [DOI] [PubMed] [Google Scholar]
- 20.Valdivia RH, Hromockyj AE, Monack D, Ramakrishnan L, Falkow S. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]
- 21.Valdivia RH, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 22.Bumann D. In vivo visualization of bacterial colonization, antigen expression, and specific T-cell induction following oral administration of live recombinant Salmonella enterica serovar Typhimurium. Infect. Immun. 2001;69:4618–4626. doi: 10.1128/IAI.69.7.4618-4626.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norris MH, Kang Y, Wilcox B, Hoang TT. Stable, site-specific fluorescent tagging constructs optimized for Burkholderia species. Appl. Environ. Microb. 2010;76:7635–7640. doi: 10.1128/AEM.01188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lathrop SK, Binder KA, Starr T, Cooper KG, Chong A, Carmody AB, Steele-Mortimer O. Replication of Salmonella enterica serovar Typhimurium in human monocyte-derived macrophages. Infect. Immun. 2015;83:2661–2671. doi: 10.1128/IAI.00033-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvo-Alvarez E, Stamatakis K, Punzon C, Alvarez-Velilla R, Teieria A, Escudero-Martinez JM, Perez-Perteio Y, Fresno M, Balana-Fouce R, Reguera RM. Infrared fluorescent imaging as a potential tool for in vitro, ex vivo and in vivo models of viscereal leishmaniasis. PLoS Negl. Trop. Dis. 2015;9:e0003666. doi: 10.1371/journal.pntd.0003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santi-Rocca J, Chenouard N, Fort C, Lagache T, Olivo-Marin J-C, Bastin P. Imaging intraflagellar transport in trypanosomes. Methods Cell Biol. 2015;127:487–508. doi: 10.1016/bs.mcb.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Wu B, Piatkevich KD, Lionnet T, Singer RH, Verkhusha VV. Modern fluorescent proteins and imaging technologies to study gene expression, nuclear localization, and dynamics. Curr. Opin. Cell Biol. 2011;23:310–317. doi: 10.1016/j.ceb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singer JT, Phennicie RT, Sullivan MJ, Porter LA, Shaffer VJ, Kim CH. Broad-host-range plasmids for red fluorescent protein labeling of gram-negative bacteria for use in the zebrafish model system. Appl. Environ. Microbiol. 2010;76:3467–3464. doi: 10.1128/AEM.01679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vacchina P, Morales MA. In vitro screening test using Leishmania promastigotes stably expressing mCherry protein. Antimicrob. Agents Chemother. 2014;58:1825–1828. doi: 10.1128/AAC.02224-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvo-Alvarez E, Stamatakis K, Punzon C, Alvarez-Vellila R, Tejeria A, Escudero-Martinez JM, Perez-Pertejo Y, Fresno M, Balana-Fouce R, Reguera RM. Infrared fluorescent imaging as a potent tool for in vitro, ex vivo and in vivo models of visceral leishmaniasis. PLoS Negl. Trop. Dis. 2015;9:e0003666. doi: 10.1371/journal.pntd.0003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 32.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shemiakina II, Ermakova GV, Cranfill PJ, Baird MA, Evans RA, Souslova EA, et al. A monomeric red fluorescent protein with low cytotoxicity. Nat. Commun. 2012;3:1204. doi: 10.1038/ncomms2208. [DOI] [PubMed] [Google Scholar]
- 34.Jenner D, Ducker C, Clark G, Prior J, Rowland CA. Using multispectral imaging flow cytometry to assess in vitro intracellular Burkholderia thailandensis infection model. Cytometry A. 2016;89:328–337. doi: 10.1002/cyto.a.22809. [DOI] [PubMed] [Google Scholar]
- 35.Barbier M, Damron FH. Rainbow vectors for broad-range bacterial fluorescence labeling. PLoS ONE. 2016;11:e0146827. doi: 10.1371/journal.pone.0146827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vorobjev IA, Buchholz K, Prabhat P, Ketman K, Egan ES, Marti M, Duraisingh MT, Barteneva NS. Optimization of flow cytometric detection and cell sorting of transgenic Plasmodium parasites using interchangeable optical filters. Malar. J. 2012;11:312. doi: 10.1186/1475-2875-11-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bargieri DY, Andenmatten N, Lagal V, Thiberge S, Whitelaw JA, Tardieux I, Meissner M, Menard R. Apical membrane antigen 1 mediates apicomplexan parasite attachment but is dispensable for host cell invasion. Nat. Comm. 2013;4:2552. doi: 10.1038/ncomms3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phanse Y, Ramer-Tait AE, Friend SL, Carillo-Conde B, Lueth P, Oster CJ, Phillips GJ, Narsimhan B, Wannemuehier MJ, Bellaire BH. Analyzing cellular internalization of nanoparticles and bacteria by multi-spectral imaging flow cytometry. J. Vis. Exp. 2012;64:e3884. doi: 10.3791/3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagal V, Dinis M, Canella D, Bargleri D, Gonzalez V, Andenmatten N, Meissner M, Tardleux I. AMA1-deficient Toxoplasma gondii parasites transiently colonize mice and trigger an innate immune response that leads to long-lasting protective immunity. Inf. Immun. 2015;83:2475–2486. doi: 10.1128/IAI.02606-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin J-W, Spaccapelo R, Schwarzer E, Sajid M, Annoura T, Deroost K, et al. Replication of Plasmodium in reticulocytes can occur without hemozoin formation, resulting in chloroquine resistance. J. Exp. Med. 2015;212:893–903. doi: 10.1084/jem.20141731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansson J, Karlsson A, Bylund J, Welin A. Phagocyte interactions with Mycobacterium tuberculosis – simultaneous analysis of phagocytosis, phagosome maturation and intracellular replication by imaging flow cytometry. J. Immunol. Methods. 2015;427:73–84. doi: 10.1016/j.jim.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotech. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 43.Christian DA, Koshy AA, Reuter MA, Betts MR, Boothroyd JC, Hunter CA. Use of transgenic parasites and host reporters to dissect events that promote interleukin-12 production during toxoplasmosis. Inf. Immun. 2014;82:4056–4067. doi: 10.1128/IAI.01643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupont CD, Christian DA, Selleck EM, Pepper M, Leney-Greene M, Pritchard GH, et al. Parasite fate and involvement of infected cells in the induction of CD4+ and CD8+ T cell responses to Toxoplasma gondii. PLoS Pathogens. 2014;10:e1004047. doi: 10.1371/journal.ppat.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konradt C, Ueno N, Christian DA, Delong JH, Pritchard GH, Herz J, et al. Endothelial cells are a replicative niche for entry of Toxoplasma gondii to the central nervous system. Nature Microb. 2016;1:16001. doi: 10.1038/nmicrobiol.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vltry M-A, Mambres DH, Deghelt M, Hack K, Machelart A, et al. Brucella melitensis invades murine erythrocytes during infection. Inf. Immun. 2014;82:3927–3938. doi: 10.1128/IAI.01779-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terrazas C, Oghumu S, Varikuti S, Martinez-Saucedo D, Beverley SM, Satoskar AR. Uncovering Leishmania-macrophage interplay using imaging flow cytometry. J. Immunol. Methods. 2015;423:93–98. doi: 10.1016/j.jim.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutierrez-Martin CB, Ojcius DM, Hsia R, Hellio R, Bavoil PM, Dautry-Varsat A. Heparin-mediated inhibition of Chlamydia psittaci adherence to HeLa cells. Microb. Pathog. 1997;22:47–57. doi: 10.1006/mpat.1996.0090. [DOI] [PubMed] [Google Scholar]
- 49.DeBoer EC, Bevers RFM, Kurth K-H, Schamhart DHJ. Double fluorescent flow cytometric assessment of bacterial internalization and binding by epithelial cells. Cytometry. 1996;25:381–387. doi: 10.1002/(SICI)1097-0320(19961201)25:4<381::AID-CYTO10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 50.Okagaki L, Strain A, Nielsen K, Chartier C, Baltes NJ, Chretien F, Heitman J, Dromer F, Nielsen K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010;6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alanio A, Vernel-Pauillac F, Sturny-Leclere A, Dromer F. Cryptococcus neoformans host adaptation: toward biological evidence of dormancy. MBio. 6:e02580–14. doi: 10.1128/mBio.02580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson-Corley KN, Bockenstedt MM, Li H, Boggiato PM, Phanse Y, Petersen CA, Bellaire BH, Jones DE. An in vitro model of antibody-enhanced killing of intracellular parasite Leishmania amazonensis. PLoS ONE. 2014;9:e106426. doi: 10.1371/journal.pone.0106426. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koshi AA, Dietrich HK, Christian DA, Melehani JH, Shastri AJ, Hunter CA, Boothroyd JC. Toxoplasma co-opts host cells it does not invade. PLoS Pathogens. 2012;8:e1002825. doi: 10.1371/journal.ppat.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Der Heijden J, Bosman ES, Reynolds LA, Finlay BB. Direct measurement of oxidative and nitrosative stress dynamics in Salmonella inside macrophages. Proc. Natl. Acad. Sci. USA. 2015;112:560–565. doi: 10.1073/pnas.1414569112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Souwer Y, Griekspoor A, de Wit J, Martinoli C, Zagato E, Jorritsima T, et al. Selective infection of antigen-specific B-lymphocytes by Salmonella mediates bacterial survival and systemic spreading of infection. PLoS ONE. 2012;7:e50667. doi: 10.1371/journal.pone.0050667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee YQ, Goh AS, Ch’ng JH, Nosten FH, Preiser PR, Pervaiz S, Yadav SK, Tan KS. A high-content phenotypic screen reveals the disruptive potency of quinacrine and 3',4'-dichlorobenzamil on the digestive vacuole of Plasmodium falciparum. Antimicrob. Agents Chemother. 2014;58:550–558. doi: 10.1128/AAC.01441-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Safekui I, Buffet PA, Perrot S, Sauvanet A, Aussilhou B, Dokmak S, et al. Surface area loss and increased sphericity account for the splenic entrapment of subpopulations of Plasmodium falciparum ring-infected erythrocytes. PLoS ONE. 2013;8:e60150. doi: 10.1371/journal.pone.0060150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mantel PY, Hoang AN, Goldowitz I, Potashnikova D, Hamza B, Vorobjev I, Ghiran I, Toner M, Irimia D, Ivanov AR, Barteneva N, Marti M. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe. 2013;13:521–534. doi: 10.1016/j.chom.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yason JA, Tan KS. Seeing the whole elephant: imaging flow cytometry reveals extensive morphological diversity with Blastocystis isolates. PLoS ONE. 2015;10:e0143974. doi: 10.1371/journal.pone.0143974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Havixbeck JJ, Wong ME, Bayona JAM, Barreda DR. Multi-parametric analysis of phagocyte antimicrobial responses using imaging flow cytometry. J. Immunol. Methods. 2015;423:85–92. doi: 10.1016/j.jim.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 61.Zenaro E, Donini M, Dusi S. Induction of Th1/Th17 immune response by Mycobacterium tuberculosis: role of dectin-1, mannose receptor, and DC-SIGN. J. Leukocyte Biol. 2009;86:1393–1401. doi: 10.1189/jlb.0409242. [DOI] [PubMed] [Google Scholar]
- 62.Bonay M, Roux A-L, Floquet J, Retory Y, Herrmann J-L, Lofaso F, Deramaudt TB. Caspase-independent apoptosis in infected macrophages, triggered by sulfloraphane via Nrf2/p38 signaling pathways. Cell Death Discov. 2015;1:15022. doi: 10.1038/cddiscovery.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buss SN, Hamano S, Vidrich A, Evans C, Zhang Y, Crasta OR, Sobral BW, Gilchrist CA, Petri WA. Members of the Entamoeba histolytica transmembrane kinase family play non-redundant roles in growth and phagocytosis. Int. J. Parasitol. 2010;40:833–843. doi: 10.1016/j.ijpara.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilchrist CA, Moore ES, Zhang Y, Bousquet CB, Lannigan JA, Mann BJ, Petri WA. Regulation of virulence of Entamoeba histolytica by the URE3-BP transcription factor. MBio. 2010;1:e00057–10. doi: 10.1128/mBio.00057-10. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ralston KS, Solga MD, Mackey-Lawrence NM, Somlata, Bhattacharya A, Petri WA. Trogocytosis by Entamoeba histolytica contributes to cell killing and tissue invasion. Nature. 2014;508:526–530. doi: 10.1038/nature13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, Gigliotti F, Bhagwat SP, George TC, Wright TW. Immune modulation with sulfasalazine attenuates immunopathogenesis but enhances macrophage-mediated fungal clearance during Pneumocystis pneumonia. PLoS Pathog. 2010;6:e1001058. doi: 10.1371/journal.ppat.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Copenhaver AM, Casson CN, Nguen HT, Fung TC, Duda MM, Roy CR, Shin S. Alveolar macrophages and neutrophils are the primary reservoirs for Legionella pneumophila and mediate cytosolic surveillance of Type 4 secretion. Inf. Immun. 2014;82:4325–4336. doi: 10.1128/IAI.01891-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shakerley NL, Chandrasekaran A, Trebak M, Miller BA, Melendez JA. Francisella tularensis catalase restricts immune function by impairing TRPM2 channel activity. J. Biol. Chem. 2016;291:3871–3881. doi: 10.1074/jbc.M115.706879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vitry M-A, Hanot MD, Deghelt M, Hack K, Machelart A, Lhomme F, Vanderwinden JM, Vermeersch M, De Trez C, Perez-Morga D, Lefesson JJ, Muraille E. Brucella melitensis invades murine erythrocytes during infection. Infect. Immun. 2014;82:3927–3938. doi: 10.1128/IAI.01779-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Evani SJ, Ramasubramanian AK. Biophysical regulation of Chlamydia pneumoniae-infected monocyte recruitment to atherosclerotic foci. Scientific Rep. 2016;6:19058. doi: 10.1038/srep19058.D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kenzaka T, Nalahara M, Higuchi S, Maeda K, Tani K. Draft genome sequences of amoeba-resistant Aeromonas spp. isolated from aquatic environments. Genome Announc. 2014;2:e01115–14. doi: 10.1128/genomeA.01115-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindguist RA, Moffat J, Golland P, Sabatini DM. Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hwang RD, Chen CC, Knecht DA. ReAsH: another viable option for in vivo protein labelling in Dictyostelium. J. Microsc. 2009;234:9–15. doi: 10.1111/j.1365-2818.2009.03149.x. [DOI] [PubMed] [Google Scholar]
- 75.Lyons AAB. Analysing cell division in vivo and in vitro using flow cytometric measurement of VFSE dye dilution. J. Immunol. Methods. 2000;243:147–154. doi: 10.1016/s0022-1759(00)00231-3. [DOI] [PubMed] [Google Scholar]
- 76.Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt WD. The cost of virulence: retarded growth of Salmonella typhimurium cells expressing type III secretion system 1. PLoS Pathog. 2011;7:e1002143. doi: 10.1371/journal.ppat.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gego A, Silvie O, Franetich JF, Farhati K, Hannoun L, Luty AJ, Sauerwein RW, Boucheix C, Rubinstein E, Mazier D. New approach for high-throughput screening of drug activity on Plasmodium liver stages. Antimicrob. Agents Chemother. 2006;50:1586–1589. doi: 10.1128/AAC.50.4.1586-1589.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Franzen C, Mueller A. Molecular techniques for detection, species differentiation, and phylogenetic analysis of microsporidia. Clin. Microbiol. Rev. 1999;12:243–285. doi: 10.1128/cmr.12.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bruna-Romero O, Hafalla JC, Gonzalez-Aseguinolaza G, Sano G, Tsuji M, Zavala F. Detection of malaria liver-stages in mice infected through the bite of a single Anopheles mosquito using a highly sensitive real-time PCR. Int. J. Parasitol. 2001;31:1499–1502. doi: 10.1016/s0020-7519(01)00265-x. [DOI] [PubMed] [Google Scholar]
- 80.Evans RT, Woodland RM. Detection of Chlamydia by isolation and direct examination. Br. Med. Bull. 1983;39:181–186. doi: 10.1093/oxfordjournals.bmb.a071813. [DOI] [PubMed] [Google Scholar]
- 81.Endoh T, Yagita K. Toxoplasma gondii, a simple method of titration of infectivity with monolayer cells. Japan J. Med. Sci. Biol. 1989;42:13–23. doi: 10.7883/yoken1952.42.13. [DOI] [PubMed] [Google Scholar]
- 82.Nakao M, Konishi E. Proliferation of Toxoplasma gondii in human neutrophils in vitro. Parasitology. 1991;103:23–27. doi: 10.1017/s0031182000059242. [DOI] [PubMed] [Google Scholar]
- 83.Alvarez-Barrientos A, Arroyo J, Canton R, Nombela C, Sanchez-Perez M. Applications of flow cytometry to clinical microbiology. Clin. Microb. Rev. 2000;13:167–195. doi: 10.1128/cmr.13.2.167-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lapinsky SE, Glenross D, Car NG, Kallenbach JM, Zwi S. Quantification and assessment of viability of Pneumocystis carinii organisms by flow cytometry. J. Clin. Microbiol. 1991;29:911–915. doi: 10.1128/jcm.29.5.911-915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gay-Andrieu F, Cozon GIN, Ferrandiz J, Kahl S, Peyron F. Flow cytometric quantification of Toxoplasma gondii cellular infection and replication. J. Parasitol. 1999;85:545–549. [PubMed] [Google Scholar]
- 86.Franzen C, Mueller A, Hartmann P, Salzberger B. Quantitation of microsporidia in cultured cells by flow cytometry. Cytometry. 2004;60A:107–114. doi: 10.1002/cyto.a.20028. [DOI] [PubMed] [Google Scholar]
- 87.Prudencio M, Rodrigues CD, Ataide R, Mota MM. Dissecting in vitro host cell infection by Plasmodium sporozoites using flow cytometry. Cell Microbiol. 2008;10:218–224. doi: 10.1111/j.1462-5822.2007.01032.x. [DOI] [PubMed] [Google Scholar]
- 88.Kentner D, Sourjik V. Use of fluorescence microscopy to study intracellular signaling in bacteria. Annu. Rev. Microbiol. 2010;64:373–390. doi: 10.1146/annurev.micro.112408.134205. [DOI] [PubMed] [Google Scholar]
- 89.Maguire O, Collins C, O’Loughlin K, Miecznikowski J, Minderman H. Quantifying nuclear p65 as a parameter for NF-kB activation: correlation between Imagestream cytometry, microscopy and western blot. Cytometry. 2011;79a:461–469. doi: 10.1002/cyto.a.21068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ranjbar S, Haridas V, Jasenosky LD, Falvo JV, Goldfeld AE. A Role for IFITM proteins in restriction of Mycobacterium tuberculosis infection. Cell Rep. 2015;13:874–83. doi: 10.1016/j.celrep.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Engel JC, Ang KKH, Chen S, Arkin MR, McKerrow JH, Doyle PS. Image-based high-throughput drug screening targeting the intracellular stage of Trypanosoma cruzi, the agent of Chagas disease. Antimicrob. Agents Chemother. 2010;54:3326–3334. doi: 10.1128/AAC.01777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maudet C, Mano M, Sunkavalli U, Sharan M, Giacca M, Foerstner KU, Eulalio A. Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction for Salmonella infection. Nat. Commun. 2014;5:4718. doi: 10.1038/ncomms5718. [DOI] [PubMed] [Google Scholar]
- 93.Ryning FW, Remington JS. Effect of cytochalasin D on Toxoplasma gondii cell entry. Infect. Immun. 1978;20:739–743. doi: 10.1128/iai.20.3.739-743.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dobrowolski JM, Sibley LD. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell. 1996;84:933–939. doi: 10.1016/s0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- 95.Drewry LL, Sibley LD. Toxoplasma actin is required for efficient host cell invasion. MBio. 2015;6:e00557–15. doi: 10.1128/mBio.00557-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bhatt K, Salgame P. Host innate immune response to Mycobacterium tuberculosis. J. Clin. Immunol. 2007;27:347–362. doi: 10.1007/s10875-007-9084-0. [DOI] [PubMed] [Google Scholar]
- 97.Baena A, Porcelli SA. Evasion and subversion of antigen presentation by Mycobacterium tuberculosis. Tissue Antigens. 2009;74:189–204. doi: 10.1111/j.1399-0039.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stein MP, Mueller MP, Wandinger-Ness A. Bacterial pathogens commandeer Rab GTPases to establish intracellular niches. Traffic. 2012;13:1565–1588. doi: 10.1111/tra.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Via LE, Deretic D, Ulmer RJ, Hibler NS, Hibler LA, Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 1997;272:13326–13331. doi: 10.1074/jbc.272.20.13326. [DOI] [PubMed] [Google Scholar]
- 100.Seto S, Matsumoto S, Ohta I, Tsujimura K, Koide Y. Dissection of Rab 7 localization on Mycobacterium tuberculosis phagosome. Biochem. Biophys. Res. Commun. 2009;387:272–277. doi: 10.1016/j.bbrc.2009.06.152. [DOI] [PubMed] [Google Scholar]
- 101.Seto S, Matsumoto S, Tsujimura K, Koide Y. Differential recruitment of CD63 and Rab7-interacting lysosomal protein to phagosomes containing Mycobacterium tuberculosis in macrophages. Microbiol. Immunol. 2010;54:170–174. doi: 10.1111/j.1348-0421.2010.00199.x. [DOI] [PubMed] [Google Scholar]
- 102.Seto S, Tsujimura K, Koide Y. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic. 2011;12:407–420. doi: 10.1111/j.1600-0854.2011.01165.x. [DOI] [PubMed] [Google Scholar]
- 103.Xu S, Cooper A, Sturgill-Koszycki S, van Heyningen T, Chatterjee D, Orne I, Allen P, Russel DG. Intracellular trafficking in Mycobacterium tuberculosis and Mycobacterium avium-infected macrophages. J. Immunol. 1994;153:2568–2578. [PubMed] [Google Scholar]
- 104.Welin A, Winberg ME, Abdalla H, Sarandahl E, et al. Incorporation of Mycobacterium tuberculosis lipoarabinomannan into macrophage membrane rafts is a prerequisite for the phagosomal maturation block. Infect. Immun. 2008;76:2882–2887. doi: 10.1128/IAI.01549-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O’Leary S, O’Sullivan MP, Keane J. IL-10 blocks phagosome maturation in Mycobacterium tuberculosis-infected human macrophages. Am. J. Respic. Cell Mol. Biol. 2011;45:172–180. doi: 10.1165/rcmb.2010-0319OC. [DOI] [PubMed] [Google Scholar]
- 106.Erdbrugger U, Rudy CK, Etter ME, Dryden KA, Yeager M, Klibanov AL, Lannigan J. Imaging flow cytometry elucidates limitations of microparticle analysis by conventional flow cytometry. Cytometry A. 2014;85:756–770. doi: 10.1002/cyto.a.22494. [DOI] [PubMed] [Google Scholar]
- 107.Headland SE, Jones HR, D’Sa AS, Perretti M, Norling LV. Cutting-edge analysis of extracellular microparticles using ImageStream (X) imaging flow cytometry. Sci. Rep. 2014;4:5237. doi: 10.1038/srep05237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barteneva NS, Maltsev N, Vorobjev IA. Microvesicles and intercellular communication in the context of parasitism. Front. Cell Infect. Microbiol. 2013;3:49. doi: 10.3389/fcimb.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Evans-Osses I, Reichembach LH, Ramirez MI. Exosomes or microvesicles? Two kinds of extracellular vesicles with different routes to modify protozoan-host cell interaction. Parasitol. Rev. 2015;114:3567–3575. doi: 10.1007/s00436-015-4659-9. [DOI] [PubMed] [Google Scholar]
- 110.Schorey JS, Harding CV. Extracellular vesicles and infectious diseases: new complexity to an old story. J. Clin. Invest. 2016;126:1181–1189. doi: 10.1172/JCI81132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archea: a conserved yet underappreciated aspects of microbial life. Infect. Immun. 2012;80:1948–1957. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barteneva NS, Fasler-Kan E, Bernimoulin M, Stern JN, Ponomarev ED, Duckett L, Vorobjev IA. Circulating microparticles: square the circle. BMC Cell Biol. 2013;14:23. doi: 10.1186/1471-2121-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Satory D, Gordon AJ, Halliday JA, Herman C. Epigenetic switches: can infidelity govern fate in microbes? Curr. Opin. Microbiol. 2011;14:212–217. doi: 10.1016/j.mib.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 114.Helaine S, Holden DW. Heterogeneity of intracellular replication of bacterial pathogens. Curr. Opin. Microbiol. 2013;16:184–191. doi: 10.1016/j.mib.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 115.Maser P, Wittlin S, Rottmann M, Wenzler T, Kaiser M, Brun R. Antiparasitic agents: new drugs on the horizon. Curr. Opin. Pharmacol. 2012;12:562–566. doi: 10.1016/j.coph.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 116.Mukhopadhyay S, Nair S, Ghosh S. Pathogenesis in tuberculosis: transcriptomic approaches to unraveling virulence mechanisms and finding new drug targets. FEMS Microbiol. Rev. 2012;36:463–485. doi: 10.1111/j.1574-6976.2011.00302.x. [DOI] [PubMed] [Google Scholar]
- 117.Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 2012;28:504–514. doi: 10.1016/j.pt.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aulner N, Danckaert A, Rouault-Hardoin E, Desrivot J, Helynck O, et al. High-content of primary macrophages hosting proliferating Leishmania amastigotes: application to anti-leishmanial drug discovery. PLoS Negl. Trop. Dis. 2013;7:e2154. doi: 10.1371/journal.pntd.0002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hoagland D, Liu J, Lee RB, Lee RE. New agents for the treatment of drug-resistant Mycobacterium tuberculosis. Adv. Drug Deliv. Rev. 2016;102:55–72. doi: 10.1016/j.addr.2016.04.026. pii: S0169-409X(16)30136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sharlow ER, Close D, Shun T, Leimgruber S, Reed R, Mustafa G, Wipf P, Johnson J, MO’Neil, Groegl M, Magill AJ, Lazo JS. Identification of potent chemotypes targeting Leishmania major using a high throughput, low-stringency, computationally enhanced, small molecule screen. PLoS Negl. Trop. Dis. 2009;3:e540. doi: 10.1371/journal.pntd.0000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Siqueira-Neto JL, Song OR, Oh H, Sohn JH, Yang G, et al. Antileishmanial high-throughput drug screening reveals drug candidates with new scaffolds. PLoS Negl. Trop. Dis. 2010;4:e675. doi: 10.1371/journal.pntd.0000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coombs GH, Hart DT, Capaldo J. Leishmania mexicana: drug sensitivities of promastigotes and transforming amastigotes. J. Antimicrob. Chemother. 1983;11:151–162. doi: 10.1093/jac/11.2.151. [DOI] [PubMed] [Google Scholar]
- 123.De Muylder G, Ang KK, Chen S, Arkin MR, Engel JC, Mckerrow JH. A screen against Leishmania intracellular amastigotes: comparison to a promastigote screen and identification of a host cell-specific hit. PLoS Negl. Trop. Dis. 2011;5:e1253. doi: 10.1371/journal.pntd.0001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vermeersch M, da Luz RI, Tote K, Timmermans JP, Cos P, Maes L. In vitro susceptibilities of Leishmania donovani promastigote and amastigote stages to antileishmanial reference drugs: practical relevance of stage-specific differences. Antimicrob. Agents Chemother. 2009;53:3855–3859. doi: 10.1128/AAC.00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lira R, Sundar S, Makharia A, Kenney R, Gam A, Saraiva E, Sacks D. Evidence that the high incidence of treatment failures in Indian kala-azar is due to the emergence of antimony-resistant strains of Leishmania donovani. J. Infect. Dis. 1999;180:564–567. doi: 10.1086/314896. [DOI] [PubMed] [Google Scholar]
- 126.Jasek E, Mirecka J, Litwin JA. Effect of differentiating agents (all-trans retinoic acid and phorbol 12-myristate 13-acetate) on drug sensitivity of HL60 and NB4 cells in vitro. Folia Histochem. Cytobiol. 2008;46:323–330. doi: 10.2478/v10042-008-0080-x. [DOI] [PubMed] [Google Scholar]
- 127.Matikainen S, Hurme M. Comparison of retinoic acid and phorbol myristate acetate as inducers of monocytic differentiation. Int. J. Cancer. 1994;57:98–103. doi: 10.1002/ijc.2910570118. [DOI] [PubMed] [Google Scholar]
- 128.Lee YQ, Hall BE, Tan KSW. Screening for drugs against the Plasmodium falciparum digestive vacuole by imaging flow cytometry. In: Barteneva Natasha S, Vorobjev IA., editors. Imaging Flow Cytometry: Methods and Protocols. Humana Press; New York: 2016. pp. 195–205. [DOI] [PubMed] [Google Scholar]