Abstract
March-I is a membrane-bound E3 ubiquitin ligase belonging to the membrane-associated RING-CH (March) family. March-I ubiquitinates and down-regulates the expression of major histocompatibility complex (MHC) class II and cluster of differentiation 86 (CD86) in antigen-presenting cells. March-I expression is regulated both transcriptionally and posttranslationally, and it has been reported that March-I is ubiquitinated and that this ubiquitination contributes to March-I turnover. However, the molecular mechanism regulating March-I ubiquitination and the importance of March-I's E3 ligase activity for March-I ubiquitination are not fully understood. Here we confirmed that, although March-I is ubiquitinated, it is not ubiquitinated on a lysine residue, as a lysine-less March-I variant was ubiquitinated similarly as wildtype March-I. We found that March-I E3 ligase activity is not required for its ubiquitination and does not regulate March-I protein expression, suggesting that March-I does not undergo autoubiquitination. Knocking down ubiquitin-conjugating enzyme E2 D1 (Ube2D1) impaired March-I ubiquitination, increased March-I expression, and enhanced March-I–dependent down-regulation of MHC-II proteins. Taken together, our results suggest that March-I undergoes lysine-independent ubiquitination by an as yet unidentified E3 ubiquitin ligase that, together with Ube2D1, regulates March-I expression.
Keywords: ubiquitin ligase, major histocompatibility complex (MHC), antigen presentation, ubiquitin, dendritic cell, ubiquitin-conjugating enzyme (E2 enzyme), lysine-independent ubiquitination
Introduction
Ubiquitination is a posttranslational modification process in which the 8-kDa protein ubiquitin (Ub)2 is covalently attached to a substrate protein (1). Ubiquitination is a multistep process involving three classes of enzymes termed E1, E2, and E3 (2). E1s are ubiquitin-activating enzymes that activate Ub in an ATP-dependent manner, resulting in the formation of a thioester linkage between Ub and the E1 protein. Thereafter, ubiquitin-conjugating enzymes, termed E2s, transfer the Ub from the E1 to the E2 itself. Unlike HECT-type E3 ubiquitin ligases, RING-type E3 ligases confer substrate specificity for ubiquitination; however, it is the E2-conjugating enzyme itself that catalyzes the formation of an isopeptide bond between the C-terminal Gly of Ub and an acceptor residue (generally a Lys) on a substrate protein (3, 4). Ub conjugation to Lys residues in a substrate-attached Ub leads to the formation of polymeric Ub chains. There are seven Lys residues on ubiquitin, and each of them can be used for chain formation, resulting in Ub chains of different topologies (5). It has been shown that the different E2/E3 combination confers the specificity of chain formation and results in different outcomes for substrates (6). For example, Lys-48–linked Ub chains with a length of four or more Ubs serve as the predominant signal for proteasome-dependent substrate degradation, whereas Lys-63–linked Ub chains are generally thought to function in proteasome-independent processes such as DNA repair, signal transduction, and receptor endocytosis.
The E3 ubiquitin ligase March-I regulates the ubiquitination, expression, intracellular transport, and stability of MHC class II (MHC-II) and CD86 proteins in antigen-presenting cells (APCs) (7–11). March-I is a member of the family of membrane-associated RING-CH (March) domain–containing E3 ubiquitin ligases, a family of proteins that were identified by their relation to viral immune evasion molecules (12, 13). Unlike other members of the March family, March-I is almost exclusively expressed on hematopoietic APCs in lymphoid tissues (7, 12). March-I is expressed in resting dendritic cells and B cells and ubiquitinates a single Lys residue on the MHC-II β-chain cytosolic domain (7) and a cluster of Lys residues on CD86 (10). The expression of March-I is regulated transcriptionally and posttranslationally. When resting dendritic cells are activated by treatment with lipopolysaccharide, March-I mRNA expression is terminated (14). By contrast, treatment of monocytes with interferon-γ and the anti-inflammatory cytokine IL-10 actually stimulates March-I transcription and leads to down-regulation of MHC-II and CD86 (15, 16). Pulse-chase studies have shown that March-I is a very unstable protein with an estimated half-life of 30 min (17). Unidentified sequences present in the first 66 amino acids of the March-I N terminus regulate March-I stability (17), as March-I variants with N-terminal truncations have significantly higher expression and reduced degradation rates compared with wildtype March-I.
Ubiquitination has been reported to contribute to March-I turnover. Bourgeois-Daigneault and Thibodeau (18) reported that March-I was ubiquitinated when overexpressed in HEK293 and HeLa cells; however, others have failed to detect significant March-I ubiquitination when the protein was overexpressed in various cell types (17). An N-terminal deletion mutant of March-I that also contained mutations of every Lys residue in the C-terminal cytosolic domain of March-I was not ubiquitinated and was reported to have a prolonged half-life relative to wildtype March-I (18); however, this mutant contained a deletion of the first 30 amino acids of the March-I N terminus, and the precise Lys sites for ubiquitination were not identified. Thus, it is unclear whether the prolonged half-life of this mutant was due to deletion of the March-I N terminus or due to mutation of March-I ubiquitination site(s) in the March-I C terminus.
In this report, we confirm that March-I is ubiquitinated in HeLa cells and in mouse B cells. Contrary to our expectation, we found that March-I undergoes Lys-independent ubiquitination, as a Lys-less mutant of March-I remained fully functional and was still ubiquitinated. We found that knocking down expression of the E2 ubiquitin-conjugating enzyme Ube2D1 reduces March-I ubiquitination, enhances March-I expression, and enhances the ability of March-I to down-regulate its substrate MHC-II. Thus, our data show that Ube2D1 mediates Lys-independent ubiquitination of March-I and that ubiquitination regulates the expression of March-I.
Results and discussion
March-I is ubiquitinated and degraded in the endocytic pathway
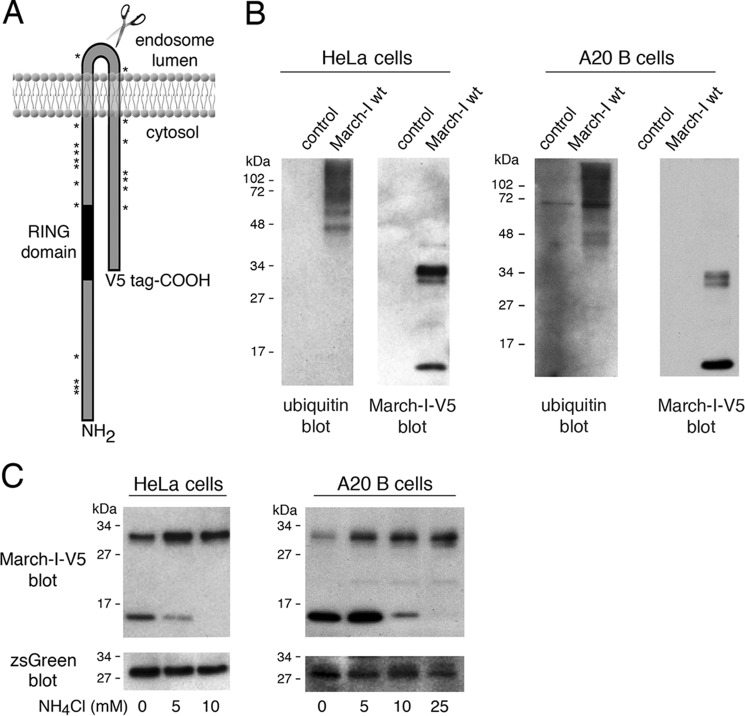
The regulation of March-I protein expression is essential for immunity, as March-I regulates the expression of CD86 and MHC-II complexes on APCs that are critical for stimulation of naïve CD4 T cells. APCs express very small amounts of endogenous March-I protein (7, 14, 17), and every commercial March-I antibody we have tested shows immunoreactivity even to cell lysates obtained from March-I–deficient mice. For this reason, we and others (14, 17, 18) have analyzed the regulation of epitope-tagged March-I protein expression in both HeLa cells and B cells (Fig. 1A). When expressed as a C-terminal V5 epitope–tagged fusion protein, we found that March-I is oligo-ubiquitinated in both HeLa cells and B cells, and the 46-kDa size of the smallest observed ubiquitinated species suggests that it consists of two ubiquitin moieties (16 kDa) attached to the 32-kDa March-I protein. We noted in our immunoprecipitates that March-I exists as a 32-kDa full-length form and also as a smaller, 17-kDa C-terminal fragment in both HeLa cells and B cells (Fig. 1B). The size of this fragment suggests that it is generated in a region of March-I that is located in the lumen of endosomes. Perturbing endosomal proteinase activity with the lysosomotropic amine NH4Cl abrogates the appearance of this March-I fragment (Fig. 1C), demonstrating that this fragment is a product of proteolytic degradation of March-I. March-I turnover has been shown previously to be suppressed by endo/lysosomal proteinase inhibitors (17); however, to our knowledge, this is the first observation of March-I ubiquitination and identification of a March-I degradation intermediate in a professional antigen-presenting cell.
Figure 1.
March-I is ubiquitinated and undergoes proteolytic degradation. A, schematic of the structure of March-I, showing the location of the C-terminal V5 epitope tag, the catalytic RING domain, and the location of each of the 19 Lys residues in the full-length 272 amino acid human March-I (variant 2) protein. B, HeLa cells (left panel) or A20 B cells (right panel) were transfected with an expression vector encoding V5 epitope–tagged March-I containing an IRES and a zsGreen1 protein reporter or the same IRES/zsGreen1 protein reporter alone as a control. After 18 h, cells were harvested and lysed, and each sample was incubated with anti-V5-agarose beads. The samples were analyzed by immunoblotting using anti-ubiquitin or anti-V5 antibodies. C, HeLa cells (left panel) or A20 B cells (right panel) were transfected with an expression vector encoding V5 epitope–tagged March-I containing an IRES and a zsGreen1 protein reporter. After 4 h, the cells were washed and incubated in complete medium containing the indicated concentration of NH4Cl for an additional 16 h before harvesting. Cell lysates were prepared, and aliquots of each lysate were analyzed for expression of V5-March-I as well as the zsGreen1 reporter. Representative gels from more than three independent experiments are shown.
March-I is not autoubiquitinated
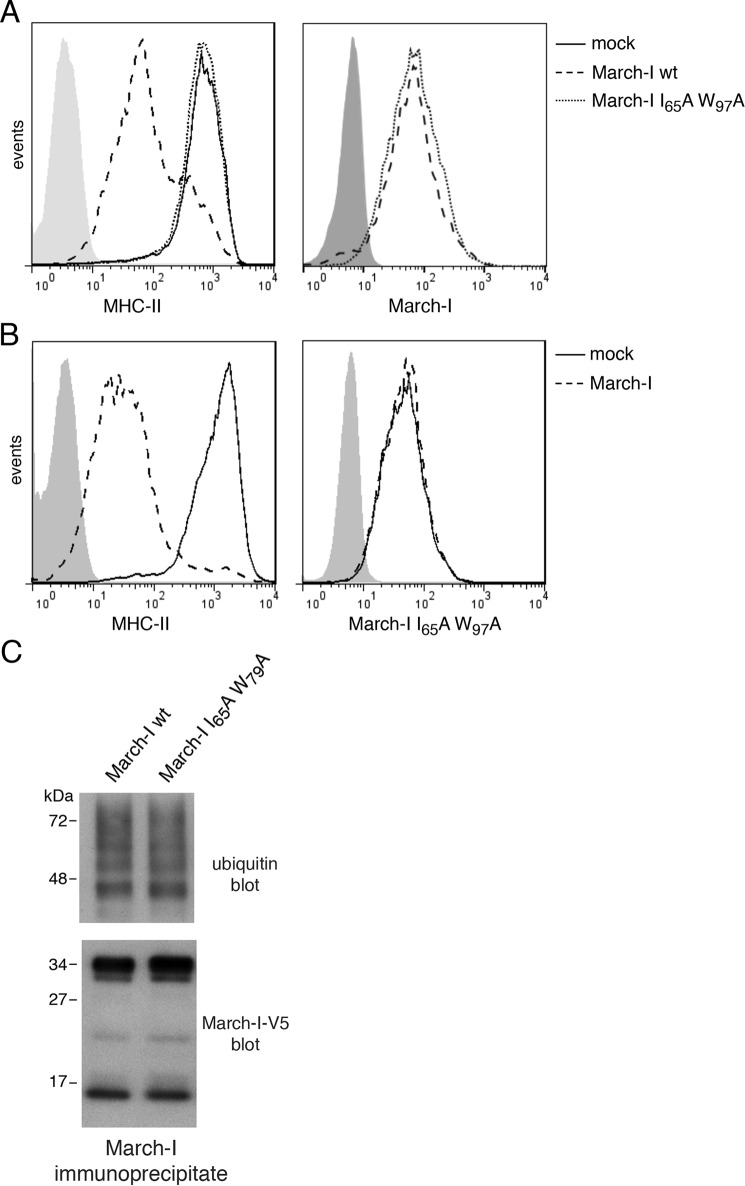
The expression of many E3 ligases, including the March family member March-VII, is regulated by autoubiquitination (19–21). To find out whether the catalytic activity of March-I regulates March-I expression, we have generated a catalytically inactive form of V5-tagged March-I by replacing an essential Ile and Trp residue in the RING domain with Ala residues. This March-I I65A/W97A mutant does not possess any ubiquitin ligase activity and is unable to down-regulate expression of the March-I substrate MHC-II (Fig. 2A). The March-I expression vector used in these studies contains an internal ribosome entry site allowing expression of the zsGreen1 reporter protein, and, therefore, FACS gating on cells possessing identical levels of zsGreen1 (as a transfection control) allows us to accurately monitor the expression of March-I in transfected cells. The catalytically inactive March-I mutant and wildtype March-I were both expressed at essentially identical levels in HeLa cells (Fig. 2A), demonstrating that March-I E3 ligase activity does not regulate March-I expression. Furthermore, although overexpression of FLAG-tagged wildtype March-I dramatically down-regulated the expression of MHC-II, it had no effect on the expression of the V5 epitope–tagged catalytically inactive form of March-I (Fig. 2B). Most importantly, both wildtype and catalytically inactive March-I were ubiquitinated identically in HeLa cells (Fig. 2C), demonstrating that March-I E3 ligase activity is not required for March-I ubiquitination and that another E3 ubiquitin ligase, and not March-I itself, is responsible for March-I ubiquitination.
Figure 2.
March-I E3 ligase activity is not required for its ubiquitination. HeLa-CIITA cells were transfected with an expression vector encoding V5-tagged wildtype March-I/IRES/zsGreen1 protein, a V5-tagged I65A/W97A RING domain mutant in the IRES/zsGreen1 vector, or the IRES/zsGreen1 protein reporter alone as a control. After 18 h, cells were harvested and analyzed. A, expression of MHC-II on the surface of live cells (left panel) or V5-March-I present in fixed/permeabilized cells (right panel) was analyzed by FACS. B, HeLa-CIITA cells were co-transfected with vectors expressing a V5-tagged March-I I65A/W97A RING domain mutant together with FLAG-tagged March-I or an empty vector control. After 18 h, cells were harvested, and the expression of MHC-II on live cells (left panel) or V5-tagged I65A/W97A RING domain mutant March-I present in fixed/permeabilized cells (right panel) was analyzed by FACS. Representative FACS profiles from three independent experiments are shown. C, lysates of cells transfected with the V5 epitope–tagged wildtype or I65A/W97A RING domain March-I mutant were incubated with anti-V5-agarose beads to immunoprecipitate March-I, and the extent of March-I ubiquitination in each sample was determined by anti-ubiquitin and anti-V5 immunoblotting. A representative blot of three independent experiments is shown.
March-I is the target of non-canonical oligo-ubiquitination
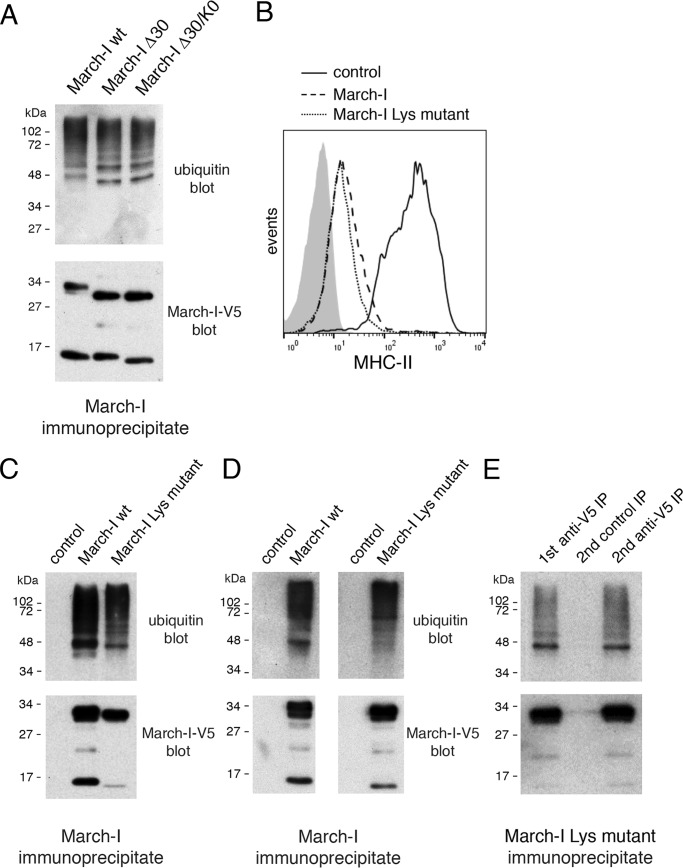
To unambiguously demonstrate that ubiquitination regulates the expression of March-I, one must block March-I ubiquitination by either mutating/eliminating March-I ubiquitination sites or silencing the enzyme(s) responsible for March-I ubiquitination. It has been reported that the extreme N terminus and/or the C-terminal cytosolic domain of March-I harbor ubiquitination site(s) (18). In an attempt to examine March-I ubiquitination in greater detail, we analyzed the ubiquitination of V5 epitope–tagged forms of wildtype March-I, March-I lacking only the N-terminal 30 amino acids (March-I Δ30), and March-I Δ30 containing C-terminal Lys mutations (March-I Δ30/K0) reported previously (18). We clearly see the effect of the deletion of the N-terminal 30 amino acids on the mobility of March-I; however, wildtype March-I, March-I Δ30, and March-I Δ30/K0 show nearly identical ubiquitination patterns (Fig. 3A). We therefore generated a March-I mutant in which all 19 Lys residues in March-I were mutated to either Ala or Arg residues (the relative location of each Lys is shown in Fig. 1A). This Lys-less March-I mutant was functional in its ability to down-regulate expression of MHC-II (Fig. 3B); however, simultaneous mutation of every Lys in March-I did not affect March-I expression and has little effect on the pattern of March-I ubiquitination (Fig. 3C). Curiously, we routinely observed variations in the intensity of the 47-kDa ubiquitination species of March-I in different experiments. These variations were not due to differences in the relative amount of the 17-kDa March-I degradation intermediate because blocking the appearance of this fragment by ammonium chloride treatment had little effect on the March-I ubiquitination pattern.3 It is possible that these variations reflect differences in the activity of deubiquitinating enzyme activity during cell lysis/immunoprecipitation; however, additional studies will be needed to address this issue.
Figure 3.
Ubiquitination of March-I is independent of Lys. A, HeLa cells were transfected with expression vectors encoding IRES/zsGreen1 containing either wildtype March-I, March-I with a 30-amino acid N-terminal truncation (March-I Δ30), or March-I Δ30 containing Lys mutations at the March-I C terminus as described under “Experimental procedures.” After 18 h, cells were harvested, and March-I in cell lysates was immunoprecipitated with a V5-specific antibody. March-I ubiquitination in each sample was revealed by blotting with anti-ubiquitin and anti-V5 antibodies. A, a representative blot of three independent experiments. B–E, every Lys in March-I was mutated by site-directed mutagenesis. HeLa-CIITA cells were transfected with expression vectors encoding IRES/zsGreen1 alone (as a control), wildtype V5-tagged March-I, or V5-tagged March-I lacking every Lys residue (March-I Lys mutant). After 18 h, the cells were harvested for analysis. B, the expression of MHC-II on the surface of each sample was analyzed by FACS analysis. C and D, HeLa-CIITA cells were transfected with control plasmid, wildtype March-I, or March-I Lys mutant either in the wildtype V5 epitope tag vector (C) or the V5-Lys>Arg tag vector (D). After 18 h, cell lysates were prepared, March-I was immunoprecipitated using a V5-specific antibody, and March-I ubiquitination in each sample was revealed by blotting with anti-ubiquitin and anti-V5 antibodies. A representative blot of multiple independent experiments is shown. E, V5-tagged March-I Lys mutant was isolated by immunoprecipitation (IP) as described above. The sample was subjected to boiling in SDS and to a second round of immunoprecipitation using control IgG-agarose beads or anti-V5-agarose beads. The samples were analyzed by SDS-PAGE and immunoblotting as indicated. The blot shown is representative of results obtained in two independent experiments.
The V5 epitope tag used in this study contains a single Lys residue, so to rule out the possibility that the Lys in the V5 epitope tag was itself ubiquitinated, this residue was mutated to Arg, and both the wildtype and the Lys-less March-I mutant were expressed with the V5-Lys>Arg epitope tag. Robust ubiquitination of the Lys-less March-I mutant was observed using the V5-Lys>Arg epitope tag (Fig. 3D). The hemagglutinin epitope tag does not contain any Lys residues, and as was observed with the V5-Lys>Arg–tagged proteins, robust March-I ubiquitination was observed using hemagglutinin epitope–tagged wildtype or Lys-less March-I.3
To confirm that the ubiquitinated protein identified in March-I immunoprecipitates was March-I (and not simply a ubiquitinated March-I–binding protein), immunoprecipitated V5 epitope–tagged March-I was denatured by boiling in SDS and then reimmunoprecipitated with anti-V5 mAb for immunoblot analysis (Fig. 3E). The results of this experiment confirmed that the isolated ubiquitinated protein was indeed March-I and, taken together with the data presented above, demonstrate that March-I is ubiquitinated on a non-canonical (i.e. non-Lys) amino acid residue.
The E2 ubiquitin-conjugating enzyme Ube2D1 regulates March-I ubiquitination and expression
The E2 ubiquitin-conjugating enzyme Ube2D1 has been shown to facilitate the autoubiquitination of a March-I RING domain–glutathione S-transferase fusion protein (12). Despite the fact that our data argue that March-I is not autoubiquitinated in vivo, we nevertheless asked whether Ube2D1 is an E2 ubiquitin-conjugating enzyme that functions, together with an unidentified E3 ubiquitin ligase, to ubiquitinate March-I. Treatment of HeLa cells with Ube2D1 shRNA dramatically increased March-I protein expression, as determined by quantitative immunoblot analysis (Fig. 4, A and B). Ube2D1 shRNA treatment resulted in a greater than 90% reduction in Ube2D1 protein and a 3-fold increase in March-I expression. The increase in March-I protein expression in Ube2D1 knockdown cells was correlated precisely with reduced ubiquitination of March-I (Fig. 4, C and D). It is important to note that the 17-kDa C-terminal March-I degradation intermediate is still observed in Ube2D1 knockdown cells, demonstrating that March-I ubiquitination is not required for March-I trafficking to endosomes. This finding is consistent with published data showing that Tyr-based sorting signals are required for March-I localization to the endosomes (17, 22).
Figure 4.
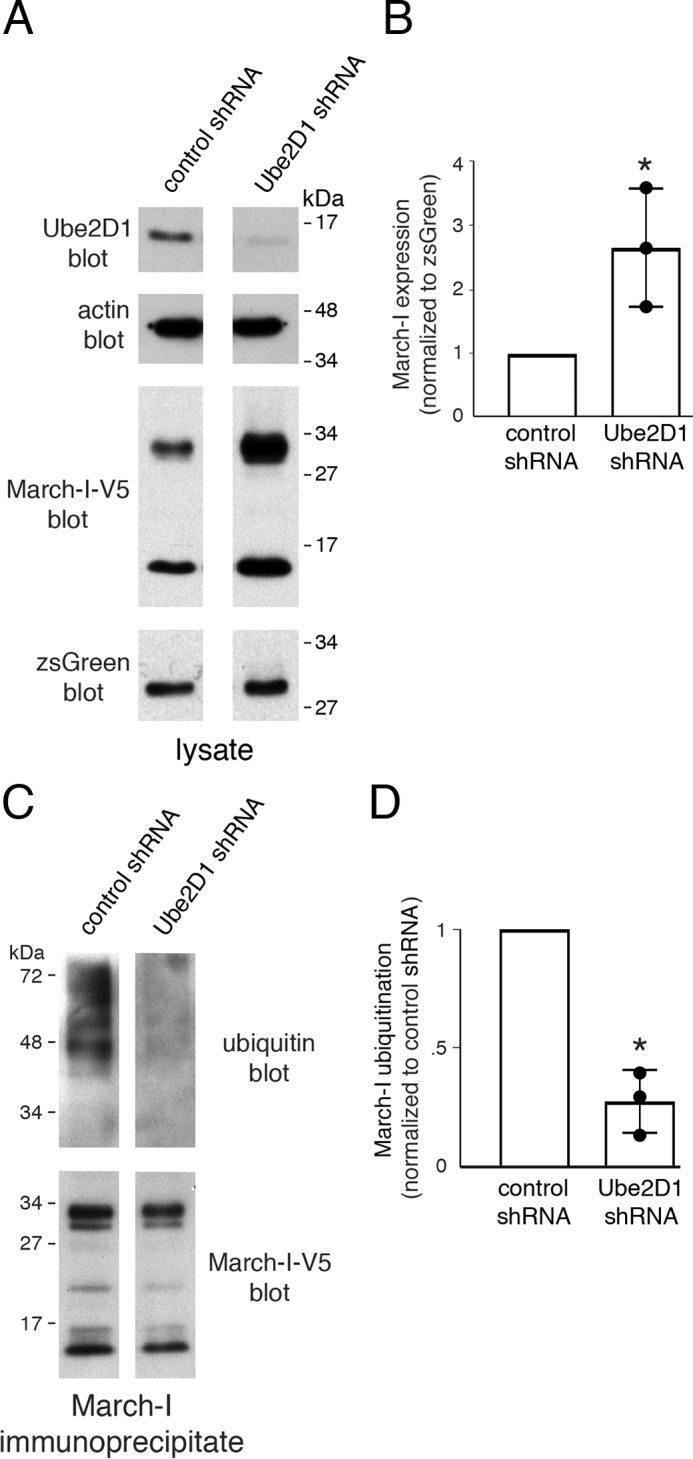
Knocking down Ube2D1 decreases March-I ubiquitination and increases March-I expression. HeLa cells were transfected with control shRNA or Ube2D1 shRNA for 2 days. Cells were then transfected with V5-tagged March-I, and 18 h later, cells were harvested. The expression of Ube2D1, actin, March-I, and zsGreen1 was analyzed by immunoblot of cell lysates. A, a representative blot of three independent experiments. B, the expression of March-I (normalized to zsGreen1) was determined by quantitative densitometry and was expressed relative to that in the control shRNA sample. Error bars represent ± S.D. obtained from three different experiments (each experimental value is indicated with a filled circle). *, p < 0.05. C, lysates of cells transfected as described above were incubated with anti-V5-agarose beads to immunoprecipitate March-I, and the extent of March-I ubiquitination was determined by anti-ubiquitin immunoblotting. The amount of sample loaded was corrected to allow visualization of the extent of March-I ubiquitination for identical amounts of total March-I (determined by anti-V5 immunoblot). A representative blot of three independent experiments is shown. D, the extent of March-I ubiquitination present in each sample was determined by quantitative densitometry and was expressed relative to that in the control shRNA sample. Error bars represent ± S.D. obtained from three different experiments (each experimental value is indicated with a filled circle). *, p < 0.05.
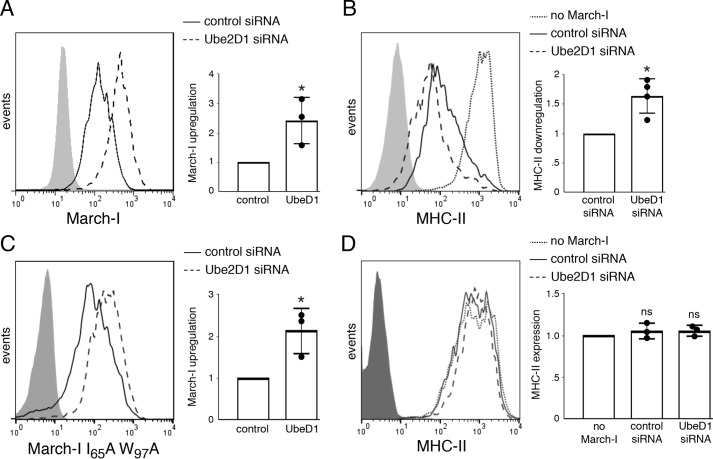
March-I is known to ubiquitinate and down-regulate the expression of MHC-II and CD86, two important mediators of CD4 T cell activation by APCs (7, 10, 12, 13). We therefore set out to determine whether Ube2D1 knockdown affected March-I–dependent down-regulation of MHC-II. In agreement with the immunoblot analysis, FACS analysis of fixed/permeabilized cells demonstrated that Ube2D1 siRNA increased March-I protein expression in HeLa-CIITA cells (Fig. 5A). Ube2D1 siRNA enhanced the ability of March-I–expressing cells to down-regulate MHC-II expression (Fig. 5B), confirming that enhanced expression of March-I in Ube2D1 knockdown cells was functionally significant. The enhanced down-regulation of MHC-II in Ube2D1 siRNA–treated cells was dependent on March-I activity, as the catalytically inactive March-I I65A/W97A mutant was still up-regulated in Ube2D1 siRNA–treated cells (Fig. 5C), but MHC-II expression was unaltered (Fig. 5D). Taken together, these data demonstrate that Ube2D1 is the E2-conjugating enzyme responsible for regulating the ubiquitination and expression of March-I in living cells.
Figure 5.
Knocking down Ube2D1 enhances March-I-dependent down-regulation of MHC-II. HeLa-CIITA cells were transfected with control siRNA or Ube2D1 siRNA for 2 days. Cells were then transfected with an expression vector encoding wildtype March-I (A and B) or catalytically inactive March-I I65A/W97A mutant (C and D) in the V5-tagged March-I/IRES/zsGreen1 vector. After 18 h, cells were harvested and examined by FACS analysis for expression of V5-tagged March-I, MHC-II, and zsGreen1. A and C, the extent of March-I up-regulation in the Ube2D1 siRNA sample was expressed relative to the expression of March-I present in the control siRNA sample (normalized to 1.0 for each independent experiment). A representative FACS plot is shown, and error bars represent ± S.D. obtained from three different experiments. *, p < 0.05. B and D, MHC-II expression on siRNA-treated HeLa-CIITA cells transfected with empty zsGreen1 vector (no March-I) or V5-tagged wildtype March-I (B) or V5-tagged March-I I65A/W97A mutant (D) was determined by FACS analysis. B, the extent of MHC-II down-regulation by wildtype March-I in the Ube2D1 siRNA sample was expressed relative to the expression of MHC-II present in the control siRNA sample (normalized to 1.0 for each independent experiment). D, the expression of MHC-II in cells expressing V5-tagged March-I I65A/W97A with either control siRNA or Ube2D1 siRNA was expressed relative to the expression of MHC-II in control-transfected cells (normalized to 1.0 for each independent experiment). A representative FACS plot is shown, and error bars represent ± S.D. obtained from at least three different experiments (each experimental value is indicated with a filled circle). *, p < 0.05; ns, not significant.
For reasons we do not understand, in our hands, a V5 epitope–tagged March-I mutant identical to the “ubiquitination-deficient” March-I mutant described previously (18) is ubiquitinated in a manner that is nearly identical to that of wildtype March-I. In fact, we found that simultaneous mutation of every Lys of March-I did not affect March-I expression, did not affect March-I E3 ligase activity, and did not alter the pattern of March-I ubiquitination. These data demonstrate that March-I is ubiquitinated on a non-Lys residue. Although Lys is the “canonical” Ub acceptor, there are examples in which Ub is attached to proteins at the extreme N-terminal amino group (23, 24), to cysteine through a thioester linkage (25, 26), or to serine, threonine, or tyrosine through a hydroxyester linkage (27, 28). Given the sheer number of serine, threonine, and tyrosine residues in March-I, identifying the March-I ubiquitination site(s) is beyond the scope of this study.
Our identification of the E2 ubiquitin-conjugating enzyme Ube2D1 as a regulator of March-I ubiquitination and March-I stability confirms that ubiquitination of March-I regulates its expression. Previous studies showed that sequences present in first 30 amino acids of the March-I N terminus conferred stability to March-I (17), and based on our Ube2D1 knockdown studies, it is possible that this region of the protein harbors non-canonical ubiquitination site(s) that affect March-I stability. Because we found that March-I expression is independent of March-I E3 ligase activity, our data argue that Ube2D1 is the E2 that functions together with an as yet unidentified E3 ligase to catalyze March-I ubiquitination. Our attempts to identify March-I ubiquitination sites by mass spectroscopy have, unfortunately, not been successful. Nevertheless, although we have not yet identified this E3 ligase, our data clearly show that ubiquitination regulates the expression of the March-I E3 ubiquitin ligase.
Experimental procedures
Cells and antibodies
HeLa cells (ATCC, CCL2, Manassas, VA) and HeLa cells expressing the class II transactivator CIITA (a gift from Peter Cresswell, Yale University Medical Center, New Haven, CT) were grown in 6-well plates or 10-cm dishes in Dulbecco's modified Eagle's medium (Gibco) containing 10% fetal bovine serum (Gibco) at 37 °C in an incubator supplied with 5% CO2. A20 cells (ATCC, TIB-208) were grown in RPMI 1640 medium containing 10% fetal bovine serum. Biotinylated anti-ubiquitin antibody (P4D1) was from eBiosciences (Waltham, MA). Anti-V5 epitope tag antibody was from Invitrogen. Phosphatidylethanolamine-conjugated anti-FLAG epitope antibody was from Biolegend (San Diego, CA). Rabbit anti-Ube2D1 antibody was from Santa Cruz Biotechnology (Dallas, TX). Mouse anti-β-actin antibody was from Sigma. Mouse anti-zsGreen1 antibody was from Origene (Rockville, MD).
Plasmids
Human March-I (variant 2) with a V5 epitope tag sequence was amplified using a plasmid encoding C-terminal V5-tagged March-I provided by Satoshi Ishido (Hyogo College of Medicine, Nishinomiya, Japan) as a template and the following primers: forward, 5′-CCGGAATTCGCCATGACCAGCAGCCACGTTTG-3′; backward, 5′-CGCGGATCCTCACGTAGAATCGAGACCGAGG-3′. Then the PCR product was cloned into the EcoRI and BamHI sites in the pLVX-EF1α-IRES-ZsGreen1 vector (Clontech). A mutant lacking the first 30 amino acids of March-I (March-I Δ30) was generated using the primers 5′-CCG-GAATTCGGCATGTTATCTAACTTGTTTCTCCAGG-3′ (forward) and 5′-CGC-GGATCCTCACGTAGAATCGAGACCGAGG-3′ (backward) and cloned into the pLVX-EF1α-IRES-ZsGreen1 vector as above.
Site-directed mutagenesis was conducted using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Lys residues in the March-I C-terminal domain were mutated to either Ala or Arg to generate a “March-I K0” construct (K203R, K213A, K229A, K230A, K233A, and K244R) containing identical substitutions as described previously (18). A March-I Lys-less mutant was made by mutating all 19 Lys to Arg or Ala as indicated: K15R, K16R, K18R, K30R, K99R, K110R, K118R, K120R, K124R, K127R, K137R, K168R, K183R, K203R, K213A, K229A, K230A, K233A, and K244R. Catalytically inactive March-I was made by mutating Ile-65 and Trp-97 to Ala. The Lys residue in the V5 epitope tag in the pLVX-EF1α-IRES-ZsGreen1 expression vector was changed to Arg by site-directed mutagenesis. All mutations were verified by DNA sequence analysis. Purified plasmid DNA was transfected into HeLa or HeLa-CIITA cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions, and cells were generally analyzed 18 h after transfection. Plasmid DNA was transfected into A20 cells by electroporation as described previously (29), and cells were analyzed 18 h after transfection.
shRNA and siRNA
shRNA vectors were used in all experiments requiring large amounts of cells, whereas siRNA was used in experiments requiring smaller numbers of cells. Control experiments showed that each system resulted in comparable knockdown efficiency. To construct shRNA vectors, 60-bp hairpin oligonucleotides were designed and subcloned into pSuper.basic vectors (Oligoengine, Seattle, WA) following the manufacturer's instructions. The Ube2D1 sequence targeted was 5′-GAGAATGGACTCAGAAATA-3′ (30). The nonsilencing control shRNA sequence was described previously, targeting 5′-AATTCTCCGAACGTGTCACGT-3′ (31). siRNAs were purchased from Origene. siRNA and shRNA were transfected into HeLa cells using Lipofectamine 2000, following the manufacturer's instructions.
Immunoprecipitation and immunoblotting
HeLa cells or A20 cells were lysed in lysis buffer (10 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100, 1 mg/ml BSA, 50 mm phenylmethylsulfonyl fluoride, 0.1 mm Nα-tosyl-l-lysine chloromethyl ketone hydrochloride, and 25 mm N-ethylmaleimide) for 1 h at 4 °C and analyzed by immunoprecipitation. Briefly, cell lysates were precleared by binding to mouse IgG–agarose beads (Sigma), and precleared lysates were then incubated with anti-V5–agarose beads (Sigma) at 4 °C. After washing, proteins were eluted by adding SDS-PAGE loading buffer and analyzed by SDS-PAGE and immunoblotting. In reimmunoprecipitation experiments, immunoprecipitated V5-tagged March-I was removed from anti-V5–agarose beads by boiling in 100 μl of Tris/NaCl/1% SDS for 3 min. 900 μl of 10 mm Tris-HCl (pH 7.4), 150 mm NaCl/1% Triton X-100 was added to quench the SDS. The sample was cooled to 4 °C and subjected to a second round of immunoprecipitation using control IgG– or V5–agarose beads. Immunoblots were quantitated and analyzed using a GS-900 calibrated densitometer and the software Image Lab (Bio-Rad) following the manufacturer's instructions.
Flow cytometry
For surface staining of MHC-II, 18 h after transfection, HeLa-CIITA cells were harvested and suspended in FACS buffer (PBS containing 2% fetal bovine serum). Mouse anti-HLA-DR mAb L243-APC (Biolegend) was used for surface staining of MHC-II on ice for 30 min. After washing with FACS buffer three times, cells were analyzed on a BD FACSCalibur flow cytometer (BD Biosciences). For intracellular staining of V5-tagged March-I or FLAG-tagged March-I, HeLa cells were harvested and fixed/permeabilized in BD Cytofix/Cytoperm solution on ice for 30 min. After washing with FACS buffer, Alexa Fluor 647–conjugated mouse anti-V5 mAb (Invitrogen) or phosphatidylethanolamine-conjugated mouse anti-FLAG mAb (Biolegend) was added to detect intracellular epitope-tagged March-I. In all experiments examining expression in siRNA/shRNA-treated cells, co-expressed zsGreen1 was used to specifically identify transfected cells. Identical zsGreen1 expression levels between different samples were identified by gating, and MHC-II or V5-March-I expression on zsGreen-gated cells was included in the FACS analyses.
Statistical analysis
A two-tailed Student's t test was used to compare the extent of ubiquitination in different samples (the ubiquitination level was calculated as ubiquitin blot intensity/V5 blot intensity) between samples. Quantitation of blot intensities was carried out by analysis of data obtained in the linear range of exposure. Statistical differences were considered to be significant only when p < 0.05.
Author contributions
L.L., J.B.-S., and P.A.R. formal analysis; L.L., J.B.-S., and P.A.R. supervision; L.L. investigation; L.L. writing-original draft; L.L., J.B.-S., and P.A.R. writing-review and editing; P.A.R. conceptualization; P.A.R. data curation; P.A.R. project administration.
Acknowledgments
We thank Peter Cresswell and Satoshi Ishido for the generous gifts of reagents used in this study.
This work was supported by the Intramural Research Program of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
L. Lei, unpublished observation.
- Ub
- ubiquitin
- MHC-II
- major histocompatibility complex class II
- APC
- antigen-presenting cell
- shRNA
- short hairpin RNA.
References
- 1. Callis J. (2014) The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 12, e0174 10.1199/tab.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- 3. Deshaies R. J., and Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 4. Metzger M. B., Pruneda J. N., Klevit R. E., and Weissman A. M. (2014) RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60 10.1016/j.bbamcr.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. d'Azzo A., Bongiovanni A., and Nastasi T. (2005) E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6, 429–441 10.1111/j.1600-0854.2005.00294.x [DOI] [PubMed] [Google Scholar]
- 6. Ye Y., and Rape M. (2009) Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 10.1038/nrm2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matsuki Y., Ohmura-Hoshino M., Goto E., Aoki M., Mito-Yoshida M., Uematsu M., Hasegawa T., Koseki H., Ohara O., Nakayama M., Toyooka K., Matsuoka K., Hotta H., Yamamoto A., and Ishido S. (2007) Novel regulation of MHC class II function in B cells. EMBO J. 26, 846–854 10.1038/sj.emboj.7601556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walseng E., Furuta K., Goldszmid R. S., Weih K. A., Sher A., and Roche P. A. (2010) Dendritic cell activation prevents MHC class II ubiquitination and promotes MHC class II survival regardless of the activation stimulus. J. Biol. Chem. 285, 41749–41754 10.1074/jbc.M110.157586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furuta K., Walseng E., and Roche P. A. (2013) Internalizing MHC class II-peptide complexes are ubiquitinated in early endosomes and targeted for lysosomal degradation. Proc. Natl. Acad. Sci. U.S.A. 110, 20188–20193 10.1073/pnas.1312994110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baravalle G., Park H., McSweeney M., Ohmura-Hoshino M., Matsuki Y., Ishido S., and Shin J. S. (2011) Ubiquitination of CD86 is a key mechanism in regulating antigen presentation by dendritic cells. J. Immunol. 187, 2966–2973 10.4049/jimmunol.1101643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oh J., Wu N., Baravalle G., Cohn B., Ma J., Lo B., Mellman I., Ishido S., Anderson M., and Shin J. S. (2013) MARCH1-mediated MHCII ubiquitination promotes dendritic cell selection of natural regulatory T cells. J. Exp. Med. 210, 1069–1077 10.1084/jem.20122695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartee E., Mansouri M., Hovey Nerenberg B. T., Gouveia K., and Früh K. (2004) Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 78, 1109–1120 10.1128/JVI.78.3.1109-1120.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ohmura-Hoshino M., Goto E., Matsuki Y., Aoki M., Mito M., Uematsu M., Hotta H., and Ishido S. (2006) A novel family of membrane-bound E3 ubiquitin ligases. J. Biochem. 140, 147–154 10.1093/jb/mvj160 [DOI] [PubMed] [Google Scholar]
- 14. De Gassart A., Camosseto V., Thibodeau J., Ceppi M., Catalan N., Pierre P., and Gatti E. (2008) MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc. Natl. Acad. Sci. U.S.A. 105, 3491–3496 10.1073/pnas.0708874105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thibodeau J., Bourgeois-Daigneault M. C., Huppé G., Tremblay J., Aumont A., Houde M., Bartee E., Brunet A., Gauvreau M. E., de Gassart A., Gatti E., Baril M., Cloutier M., Bontron S., Fruh K., et al. (2008) Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur. J. Immunol. 38, 1225–1230 10.1002/eji.200737902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mittal S. K., Cho K. J., Ishido S., and Roche P. A. (2015) Interleukin 10 (IL-10)-mediated immunosuppression: March-I induction regulates antigen presentation by macrophages but not dendritic cells. J. Biol. Chem. 290, 27158–27167 10.1074/jbc.M115.682708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jabbour M., Campbell E. M., Fares H., and Lybarger L. (2009) Discrete domains of MARCH1 mediate its localization, functional interactions, and posttranscriptional control of expression. J. Immunol. 183, 6500–6512 10.4049/jimmunol.0901521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bourgeois-Daigneault M. C., and Thibodeau J. (2012) Autoregulation of MARCH1 expression by dimerization and autoubiquitination. J. Immunol. 188, 4959–4970 10.4049/jimmunol.1102708 [DOI] [PubMed] [Google Scholar]
- 19. Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., and Weissman A. M. (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275, 8945–8951 10.1074/jbc.275.12.8945 [DOI] [PubMed] [Google Scholar]
- 20. Bruce M. C., Kanelis V., Fouladkou F., Debonneville A., Staub O., and Rotin D. (2008) Regulation of Nedd4–2 self-ubiquitination and stability by a PY motif located within its HECT-domain. Biochem. J. 415, 155–163 10.1042/BJ20071708 [DOI] [PubMed] [Google Scholar]
- 21. Nathan J. A., Sengupta S., Wood S. A., Admon A., Markson G., Sanderson C., and Lehner P. J. (2008) The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic 9, 1130–1145 10.1111/j.1600-0854.2008.00747.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bourgeois-Daigneault M. C., and Thibodeau J. (2013) Identification of a novel motif that affects the conformation and activity of the MARCH1 E3 ubiquitin ligase. J. Cell Sci. 126, 989–998 10.1242/jcs.117804 [DOI] [PubMed] [Google Scholar]
- 23. Ciechanover A., and Ben-Saadon R. (2004) N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14, 103–106 10.1016/j.tcb.2004.01.004 [DOI] [PubMed] [Google Scholar]
- 24. Breitschopf K., Bengal E., Ziv T., Admon A., and Ciechanover A. (1998) A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17, 5964–5973 10.1093/emboj/17.20.5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cadwell K., and Coscoy L. (2005) Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309, 127–130 10.1126/science.1110340 [DOI] [PubMed] [Google Scholar]
- 26. Williams C., van den Berg M., Sprenger R. R., and Distel B. (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534–22543 10.1074/jbc.M702038200 [DOI] [PubMed] [Google Scholar]
- 27. Wang X., Herr R. A., Chua W. J., Lybarger L., Wiertz E. J., and Hansen T. H. (2007) Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177, 613–624 10.1083/jcb.200611063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McDowell G. S., and Philpott A. (2013) Non-canonical ubiquitylation: mechanisms and consequences. Int. J. Biochem. Cell Biol. 45, 1833–1842 10.1016/j.biocel.2013.05.026 [DOI] [PubMed] [Google Scholar]
- 29. Khandelwal S., and Roche P. A. (2010) Distinct MHC class II molecules are associated on the dendritic cell surface in cholesterol-dependent membrane microdomains. J. Biol. Chem. 285, 35303–35310 10.1074/jbc.M110.147793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Popov N., Schülein C., Jaenicke L. A., and Eilers M. (2010) Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat. Cell Biol. 12, 973–981 10.1038/ncb2104 [DOI] [PubMed] [Google Scholar]
- 31. Yamashita M., Fatyol K., Jin C., Wang X., Liu Z., and Zhang Y. E. (2008) TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-β. Mol. Cell 31, 918–924 10.1016/j.molcel.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]