Abstract
CoA is the major acyl carrier in mammals and a key cofactor in energy metabolism. Dynamic regulation of CoA in different tissues and organs supports metabolic flexibility. Two mammalian Nudix hydrolases, Nudt19 and Nudt7, degrade CoA in vitro. Nudt19 and Nudt7 possess conserved Nudix and CoA signature sequences and specifically hydrolyze the diphosphate bond of free CoA and acyl-CoAs to form 3′,5′-ADP and 4′-(acyl)phosphopantetheine. Limited information is available on these enzymes, but the relatively high abundance of Nudt19 and Nudt7 mRNA in the kidney and liver, respectively, suggests that they play specific roles in the regulation of CoA levels in these organs. Here, we analyzed Nudt19−/− mice and found that deletion of Nudt19 elevates kidney CoA levels in mice fed ad libitum, indicating that Nudt19 contributes to the regulation of CoA in vivo. Unlike what was observed for the regulation of Nudt7 in the liver, Nudt19 transcript and protein levels in the kidney did not differ between fed and fasted states. Instead, we identified chenodeoxycholic acid as a specific Nudt19 inhibitor that competed with CoA for Nudt19 binding but did not bind to Nudt7. Exchange of the Nudix and CoA signature motifs between the two isoforms dramatically decreased their kcat. Furthermore, substitutions of conserved residues within these motifs identified amino acids playing different roles in CoA binding and hydrolysis in Nudt19 and Nudt7. Our results reveal that the kidney and liver each possesses a distinct peroxisomal CoA diphosphohydrolase.
Keywords: coenzyme A (CoA), metabolic regulation, peroxisome, kidney, hydrolase, Nudix hydrolase
Introduction
CoA is an obligate cofactor utilized as an acyl carrier in hundreds of metabolic reactions. Numerous CoA thioesters are also involved in the post-translational modification of histones and thousands of other proteins (1–5). Three major subcellular CoA pools are found in the cytosol, mitochondria, and peroxisomes to support specific metabolic pathways (6, 7). These include, among others, fatty acid synthesis in the cytosol, the TCA cycle and oxidation of long-chain and medium-chain fatty acids in the mitochondria, and bile acid conjugation and oxidation of very long-chain and branched-chain fatty acids in the peroxisomes. A dedicated pool of acetyl-CoA is also found in the endoplasmic reticulum, where it is involved in protein quality control and autophagy (8), whereas nuclear acyl-CoAs, which freely equilibrate with the cytosolic pool across the nuclear pores, contribute to the regulation of gene expression (5).
Tight control over the concentration of CoA in different organs is essential to maintain normal metabolism and organ function. Indeed, elevated CoA levels in the liver of diabetic mice promote excessive gluconeogenesis and hyperglycemia (9), whereas a supraphysiological concentration of CoA in skeletal muscle leads to decreased ATP levels and exercise performance (10). Adverse consequences are also associated with lower-than-normal CoA levels. Decreased synthesis of this cofactor in the nervous system is associated with a rare neurological disorder in humans (11, 12), whereas decreased synthesis of CoA in the liver leads to fasting hypoglycemia and hepatic triglyceride accumulation (13). Dynamic regulation of CoA within the normal physiological range is important to support the metabolic reprogramming that underlies the capacity to respond to changes in the metabolic state. For example, the increase in CoA that characterizes the fed-to-fasted transition in the liver is driven by the activation of the biosynthetic pathway and is required to sustain the high rates of hepatic fatty acid oxidation and gluconeogenesis under fasting conditions (13, 14). On the other hand, the net decrease in the concentration of CoA observed upon refeeding after a fast requires both inhibition of de novo CoA synthesis and active degradation of the cofactor accumulated in the fasted state.
The existence of a CoA-degradation pathway in the liver and kidneys is supported by the fact that mice treated with an inhibitor of the CoA biosynthetic pathway exhibit a dramatic reduction in hepatic and renal CoA, indicating rapid CoA turnover in these tissues (15). Liver and kidneys contain high transcript levels of two Nudix hydrolases, Nudt7 and Nudt19, respectively, which exhibit CoA diphosphohydrolase activity in vitro (16, 17). Nudix hydrolases hydrolyze the diphosphate bond of a variety of substrates consisting of a nucleoside diphosphate linked to another moiety, x (18, 19). All members of the Nudix superfamily contain the conserved Nudix box motif G1N[5X]E7N[7X]R15NE16NXXE19NE20NXG22NU (where U is a hydrophobic residue, X is any residue, and superscript N denotes the position of a residue of the signature motif within a specific protein sequence) (20, 21). Additionally, Nudix hydrolases that specifically hydrolyze CoA also contain a putative CoA binding motif, (L/M)(L/F)TXR(S/A)[3X](R/K)[3X]G[3X]FPGG (PROSITE accession number PS01293, formerly UPF0035), upstream of the Nudix box motif (22). The crystal structure of human NUDT7 has recently been released (PDB3 code 5T3P). Similar to the structure of the bacterial homolog from Deinococcus radiodurans (24), the crystal structure of the human enzyme does not contain CoA bound. Thus, the contribution of the CoA motif to CoA binding and the organization of the CoA-binding site in these enzymes are currently unknown.
Independent studies and approaches have demonstrated the localization of Nudt7 in the peroxisomes (17, 25). Nudt7 mRNA and protein levels in the liver are regulated by the nutritional state and are inversely correlated with the tissue concentration of CoA (9, 26). Additionally, overexpression of a catalytically active, cytosolic form of Nudt7 has also recently been shown to cause a decrease in CoA levels in vivo (25).
Significantly less is known about Nudt19. Nudt19 exhibits modest sequence similarity to Nudt7 (∼30%), is larger than a typical Nudix hydrolase, including Nudt7, and contains a unique 45–49-amino acid insertion of unknown function within its Nudix box (16). This enzyme, also known as RP2p, was originally reported to be a kidney protein whose transcript levels were robustly up-regulated by androgens (27). Later, recombinant Nudt19 was shown to specifically hydrolyze CoA species (16), although modest activity against capped RNA has also been detected (28). Based on proteomics studies, Nudt19 localization has been reported in both peroxisomes and mitochondria (16, 29); however, it is currently unclear whether the enzyme has dual localization or exclusively resides in one of the two subcellular compartments, and which one. Given the presence of dedicated CoA pools and complementary metabolic processes in peroxisomes and mitochondria, this distinction is important to gain insight into the physiological role of Nudt19, even in relation to the other mammalian isoform, Nudt7. Indeed, it is presently unclear whether Nudt7 and Nudt19, which seem to be differentially expressed in liver and kidneys, differ in key aspects of their function.
We have investigated the effect of Nudt19 deletion on CoA levels, the localization of this enzyme in intact cells, and its regulation by selected metabolites. Additionally, we used a combination of computational modeling, substrate docking, and site-directed mutagenesis to identify residues involved in CoA binding and hydrolysis in Nudt19 and analyzed the role of equivalent residues in Nudt7. We show that Nudt19 is a peroxisomal enzyme that contributes to the regulation of kidney CoA levels in vivo and provide evidence supporting the conclusion that the CoA-binding sites of Nudt19 and Nudt7 are different.
Results
Nudt19 regulates kidney CoA levels in vivo
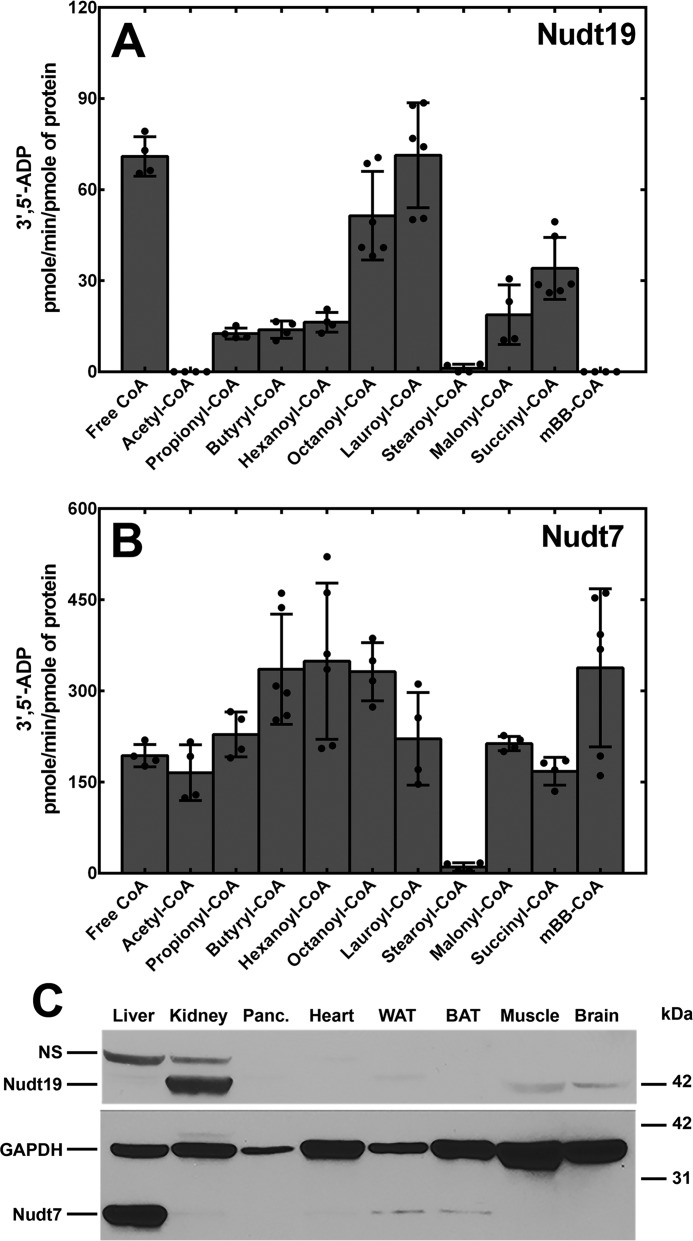
Previous work has shown that Nudt19 hydrolyzes free CoA and acyl-CoAs to 3′,5′-ADP and 4′-(acyl)phosphopantetheine in vitro (16). We confirmed these findings by measuring the activity of recombinant Nudt19 against several acyl-CoAs and comparing it to the better characterized Nudt7 (17, 25) (Fig. 1, A and B). We determined that the optimal concentration of MgCl2 for Nudt7 and Nudt19 was 4 and 10 mm, respectively, when free CoA was used as the substrate (data not shown). However, in experiments where we tested multiple acyl-CoAs, it was necessary to keep the concentration of MgCl2 at 4 mm for both Nudt7 and Nudt19 to prevent the precipitation of lauroyl- and stearoyl-CoA. Under these conditions, the specific activity per pmol of Nudt19 was lower than that of Nudt7, but the enzyme readily hydrolyzed short and medium chain acyl-CoAs, malonyl- and succinyl-CoA, in addition to free CoA (Fig. 1A). Exceptions were acetyl-CoA and the synthetic monobromobimane (mBB)-CoA, which were excellent substrates for Nudt7, but not for Nudt19. Nudt19 also had no detectable hydrolytic activity against ATP, ADP, NAD+ NADH, NADP+, and NADPH (data not shown), confirming the specificity of this enzyme for CoA (16).
Figure 1.
In vitro activity and tissue distribution of Nudt19 and Nudt7. A and B, recombinant mouse Nudt19 (A) and Nudt7 (B) were incubated with 250 μm of each CoA substrate, and the 3′,5′-ADP produced was quantified as described under “Experimental procedures.” The data are plotted as the mean (bars) of duplicate measurements from two or three independent experiments (circles) ± S.D. C, immunoblot analysis to detect Nudt19 and Nudt7 proteins in mouse tissues. Tissue lysate proteins (150 μg) were fractionated by SDS-PAGE, transferred to a PVDF membrane, and blotted with the Nudt19 antibody. The same membrane was then stripped and blotted with antibodies against GAPDH (loading control) and Nudt7. Note that although the same amount of protein was loaded, different tissues contain different amounts of GAPDH. Panc., pancreas; WAT, white adipose tissue; BAT, brown adipose tissue; NS, nonspecific band.
The mRNA levels of Nudt19 are highly abundant in mouse kidneys (16, 30). To examine the distribution of this protein in mouse tissues, we generated a polyclonal antibody against the full-length mouse Nudt19. Consistent with the mRNA expression pattern, Nudt19 protein showed the highest abundance in the kidneys, with lower but still detectable levels in skeletal muscle and brain (Fig. 1C). This expression pattern was strikingly different from Nudt7, which was expressed at the highest levels in the liver and, to a significantly lower extent, in white and brown adipose tissue (Fig. 1C). The high expression level of Nudt19, combined with the minimally detectable Nudt7 protein in the kidneys (Fig. 1C), suggested that Nudt19 was the major of the two isoforms in these organs.
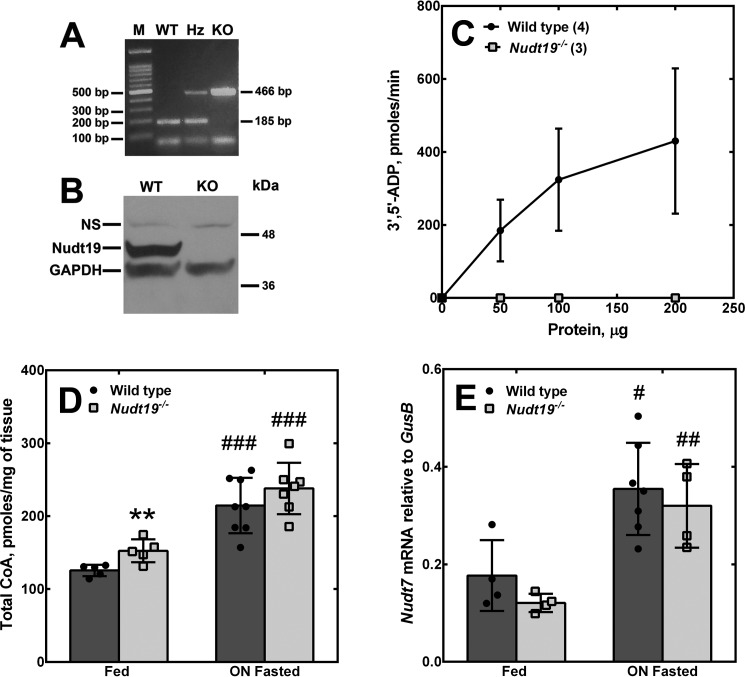
To determine whether Nudt19 regulated CoA levels in vivo, we obtained Nudt19+/− mice from the KOMP repository and bred them to generate Nudt19−/− mice and wildtype littermate controls. The Nudt19−/− mice were viable, fertile, and outwardly normal. Successful deletion of the gene was verified by PCR analysis of tail biopsies (Fig. 2A), whereas Western blotting analysis confirmed the lack of Nudt19 protein expression (Fig. 2B). Additionally, enzymatic assays confirmed the lack of any residual formation of 3′,5′-ADP in kidney homogenates obtained from Nudt19−/− mice (Fig. 2C). The concentration of CoA was measured in the kidneys of Nudt19−/− and control mice in the fed state and following an overnight fast. In the fed state, the kidneys of the Nudt19−/− mice exhibited a significant 20% increase in the concentration of CoA compared with the wildtype mice (Fig. 2D). Fasting induced an increase in kidney CoA levels in both wildtype and Nudt19−/− mice. Under these conditions, the concentration of CoA in the Nudt19−/− kidneys tended to be higher than control mice, but the difference did not reach statistical significance. No compensatory increase in Nudt7 transcript levels was detected in the kidneys of the Nudt19−/− mice in either the fed or fasted states (Fig. 2E). Combined, these data showed that Nudt19 was the major CoA diphosphohydrolase in the kidneys and that this enzyme contributed to the regulation of kidney CoA levels in vivo.
Figure 2.
Effect of Nudt19 deletion on kidney CoA levels. A, mouse genotyping. Multiplex PCRs were set up to detect the wildtype and/or the knockout allele, as described under “Experimental procedures.” A PCR product of 185 bp indicated a wildtype allele; a PCR product of 466 bp indicated a knockout allele. Heterozygous mice exhibited both bands. M, marker; WT, wildtype; Hz, heterozygous; KO, knockout. B, Western blotting analysis of kidney homogenates (150 μg) from wildtype and Nudt19−/− mice using the Nudt19 antibody. GAPDH was used as a loading control. NS, nonspecific band. C, production of 3′,5′-ADP in reaction mixtures containing free CoA and kidney extracts from wildtype and Nudt19−/− mice. Numbers in parentheses represent the number of animals analyzed; data are reported as the means ± S.D. D, total CoA measured in the kidneys of wildtype and Nudt19−/− mice fed ad libitum or fasted overnight (ON). E, Nudt7 mRNA abundance in the kidneys of fed and fasted wildtype and Nudt19−/− mice. D and E, data are reported as the mean (bars) of measurements on individual mice (circles and squares) ± S.D. **, p < 0.01 relative to wildtype mice; #, p < 0.05; ##, p < 0.01; ###, p < 0.001 relative to the fed state.
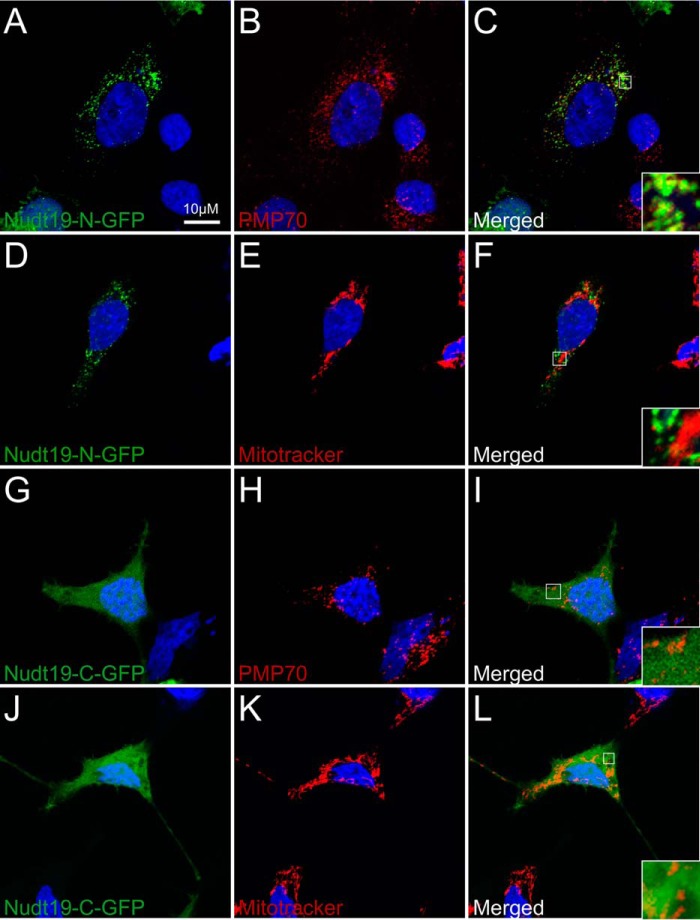
Mouse Nudt7 and other CoA diphosphohydrolases from Saccharomyces cerevisiae and Caenorhabditis elegans localize to the peroxisomes (17, 22, 31), which suggests that these enzymes might specifically regulate the peroxisomal pool of CoA (32, 33). Nudt19 contains a C-terminal peroxisome targeting signal type 1 (PTS1), Ala-His-Leu, and was originally identified as a peroxisomal enzyme by the proteomic analysis of kidney peroxisomes (16). Recently, proteomic studies conducted on isolated mitochondria have also led to the annotation of Nudt19 as a mitochondrial enzyme in the Mitocarta2.0 inventory (29). Given the common contamination of isolated peroxisomes and mitochondria with variable amounts of other organelles, we used immunofluorescence and confocal microscopy to determine the intracellular localization of Nudt19 in whole HEK 293 cells (Fig. 3). HEK 293 cells were transiently transfected with constructs expressing Nudt19 with N- or C-terminal GFP tags. Nuclei were stained with DAPI, mitochondria were stained with MitoTracker® Orange CMTMRos, and peroxisomes were visualized with an antibody against the endogenous peroxisomal protein PMP70. When expressed with an N-terminal GFP tag that left the PTS1 exposed, Nudt19 colocalized with PMP70, as shown by the yellow pixels resulting from the overlap of the green and red contributions (Fig. 3, A–C). Masking the PTS1 of Nudt19 with a C-terminal GFP tag prevented the peroxisomal localization (Fig. 3, G–I) but did not direct the protein to the mitochondria (Fig. 3, J–L), leading instead to a diffuse cytoplasmic localization. Overall, these results supported the conclusion that Nudt19 was a peroxisomal enzyme. As such, the 20% increase measured in the whole kidney homogenates from fed Nudt19−/− mice (Fig. 2D) could be an underestimation of the local accumulation of CoA in the kidney peroxisomes of these animals.
Figure 3.
Localization of Nudt19 to the peroxisomes. HEK 293 cells were transfected with expression plasmids encoding for GFP fused to the N terminus (Nudt19-N-GFP) (A–F) or the C terminus of Nudt19 (Nudt19-C-GFP) (G–L). Fixed cells were visualized using confocal microscopy. B, C, H, and I, the endogenous PMP70 protein was used as a marker for the peroxisomes, shown in red. E, F, K, and L, mitochondria were visualized using MitoTracker Orange CMTMRos and shown in red. A–L, cell nuclei were stained with DAPI, in blue. C, F, I, and L, insets in merged images show details at higher magnification. The results are representative of at least two independent experiments.
Nudt19, but not Nudt7, is competitively inhibited by specific bile acids
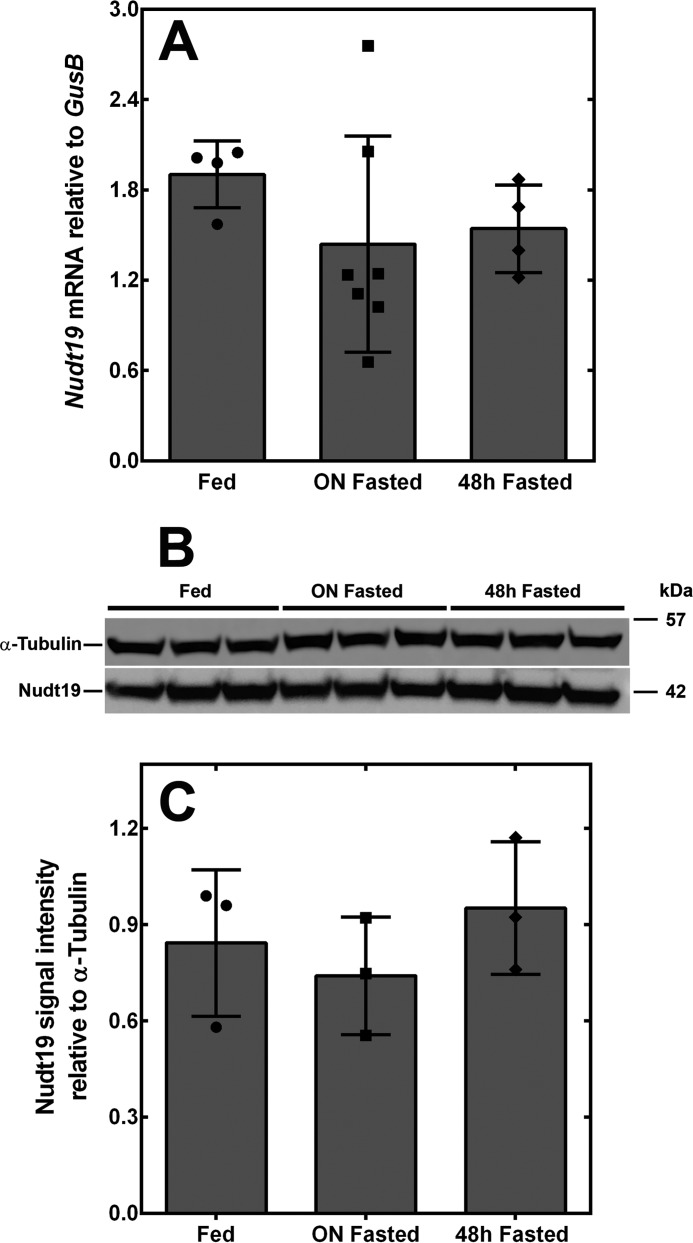
Deletion of Nudt19 led to a significant increase in kidney CoA levels in fed but not fasted mice (Fig. 2D). This suggested that, in a wildtype mouse, the activity of Nudt19 might be higher in the fed state compared with the fasted state. We analyzed Nudt19 mRNA and protein expression levels in the kidneys of mice fed ad libitum and mice fasted for up to 48 h, but we did not detect any difference (Fig. 4, A–C). We then focused our attention on metabolite regulation.
Figure 4.
Nudt19 expression in the kidneys of fed and fasted mice. A, Nudt19 transcript levels in the kidneys of fed, overnight (ON) fasted, and 48 h fasted mice. B, Western blotting analysis showing that Nudt19 protein levels in the kidneys did not change between fed, overnight fasted, and 48 h fasted mice. α-Tubulin was used as a loading control. C, quantification of the Western blot in B. The data are reported as the mean (bars) of measurements on individual mice (circles, squares, and diamonds) ± S.D.
We screened a panel of 30 compounds, including a large number of substrates and products of peroxisomal metabolism, for their effect on the activity of recombinant Nudt19 (Table 1). None of the compounds tested significantly stimulated Nudt19 activity. Instead, we found that a select group of bile acids inhibited the enzyme by >50%. More specifically, chenodeoxycholic acid (CDCA) and its conjugated derivatives, taurochenodeoxycholic acid and glycochenodeoxycholic acid, decreased Nudt19 activity by 60–75% with respect to vehicle control (Table 1). More hydrophobic bile acids, such as lithocholic acid and ursocholanic acid, were also potent inhibitors of Nudt19, and α-muricholic acid, which derives from CDCA by the addition of a 6β-hydroxyl group, still retained significant potency against the enzyme. In contrast, cholic acid and its glycine and taurine conjugates exhibited decreased potency against Nudt19 compared with CDCA. Furthermore, other steroid compounds, such as progesterone, pregnenolone, and corticosterone, had no significant effect on the activity of the enzyme, confirming the specificity of the inhibitory effect of CDCA and derived bile acids on Nudt19 (Table 1).
Table 1.
Effect of selected metabolites on the activity of Nudt19 and Nudt7
Nudt19 and Nudt7 were assayed in the presence of 250 μm of each compound, unless otherwise indicated, and free CoA as substrate. A minus sign indicates a decrease in activity with respect to the vehicle control. A plus sign indicates an increase in activity with respect to the vehicle control. NMN, nicotinamide mononucleotide.
| Metabolite | Activity relative to vehicle |
|
|---|---|---|
| Nudt19 | Nudt7 | |
| % | ||
| Hydrogen peroxide (10 μm) | +16 | −11 |
| Hydrogen peroxide (100 μm) | +3 | +5 |
| ATP | +6 | +2 |
| NAD+ | −5 | +1 |
| NADH | −13 | +20 |
| NADP+ | −16 | +4 |
| NADPH | −5 | +7 |
| NMN | −8 | −1 |
| Pyruvate | +13 | −17 |
| α-Ketoglutarate | 0 | −8 |
| Isocitrate | −9 | −5 |
| Sarcosine | +8 | +4 |
| Carnitine | −3 | −1 |
| Glycine | −4 | +16 |
| Taurine | +5 | +24 |
| Lauric acid | −16 | −4 |
| Palmitic acid (50 μm) | −2 | −32 |
| Palmitoylcarnitine (50 μm) | +23 | +11 |
| Cholic acid | −24 | +8 |
| Glycocholic acid | −30 | +10 |
| Taurocholic acid | −17 | +3 |
| Chenodeoxycholic acid | −62a | +1 |
| Glycochenodeoxycholic acid | −67a | +3 |
| Taurochenodeoxycholic acid | −75a | +11 |
| α-Muricholic acid | −50a | −8 |
| Lithocholic acid (50 μm) | −83a | −15 |
| Ursocholanic acid (50 μm) | −70a | +12 |
| Corticosterone | −26 | −7 |
| Progesterone | −37 | 0 |
| Pregnenolone | −15 | +14 |
a Metabolites that decreased or increased the activity ≥50%.
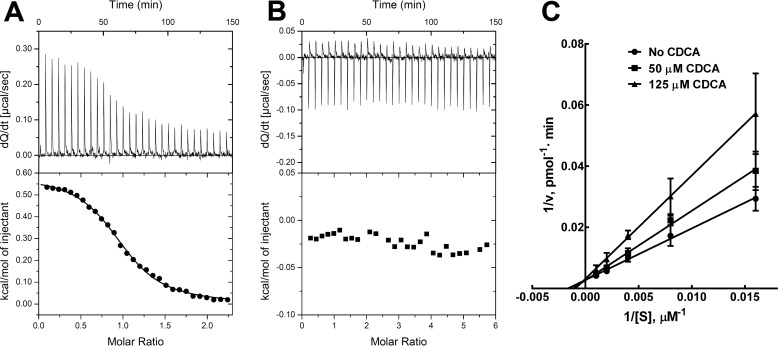
Interestingly, none of the compounds tested, including CDCA, significantly affected the activity of Nudt7 (Table 1). In addition, isothermal titration calorimetry (ITC) studies showed no detectable binding of CDCA to the liver enzyme, whereas the Kd of the Nudt19-CDCA complex was determined to be 8.3 μm (ΔG = −6.9 kcal/mol) (Fig. 5, A and B). Further kinetic analysis of the mechanism of Nudt19 inhibition revealed that CDCA was a competitive inhibitor with respect to free CoA (Fig. 5C). Combined, these results identified CDCA as a specific competitive inhibitor of Nudt19 in vitro.
Figure 5.
CDCA binds to Nudt19 but not to Nudt7. A and B, ITC binding curves of Nudt19 (A) and Nudt7 (B). The binding curves are representative of two or three independent experiments. C, graphical analysis of CDCA inhibition of Nudt19 using fixed concentrations of the inhibitor and variable concentrations of free CoA revealed that CDCA was competitive with free CoA. The data are reported as the mean of two independent experiments in duplicate ± S.D.
The CoA boxes and the Nudix signature sequences of Nudt19 and Nudt7 are not interchangeable
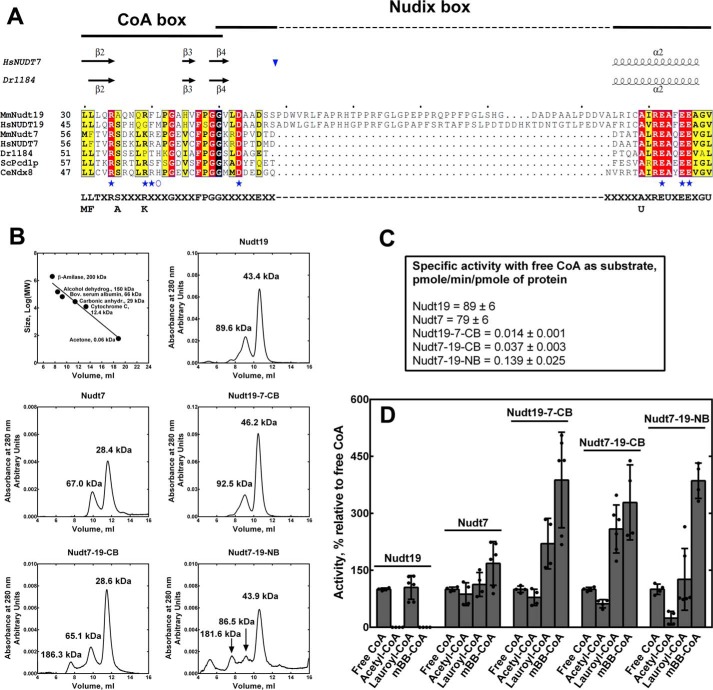
Bile acids are primarily synthesized and conjugated in the liver. Although the potential relevance of Nudt19 inhibition by a select group of these compounds in vivo is currently not known, the lack of CDCA binding to Nudt7 and the competitive nature of the CDCA inhibition of Nudt19 strongly suggested that the CoA-binding sites of the two enzymes might be different. Nudix hydrolases that degrade CoA contain a conserved motif, (L/M)(L/F)TXR(S/A)[3X](R/K)[3X]G[3X]FPGG, here referred to as the CoA box, which is proposed to play a role in CoA binding (24) (Fig. 6A). Based on the structures of both human NUDT7 and the D. radiodurans homolog, the CoA box comprises the three β strands β2, β3, and β4, whereas the Nudix signature sequence, G1N[5X]E7N[7X]R15NE16NXXE19NE20NXG22NU, here referred to as the Nudix box, forms a β-strand-loop/helix/loop structural motif (Fig. 6A) (21, 34, 35). In several validated CoA diphosphohydrolases, the Nudix box exhibits a variable number of X residues between E7N and R15N. The most extreme case is represented by mouse and human Nudt19, which contain a 45–49-amino acid insertion (Fig. 6A). Given its position within the Nudix box, this insertion would occur between strand β4 and helix α2, i.e. in the first loop of the Nudix β-strand-loop/helix/loop structural motif (Fig. 6A).
Figure 6.
Effect of exchanging the CoA and Nudix boxes of Nudt19 and Nudt7. A, alignment of the CoA and Nudix boxes of characterized CoA diphosphohydrolases from different organisms. Invariant residues are highlighted in red, conserved residues are highlighted in yellow, and groups of conserved residues are boxed. The Nudix and the CoA boxes overlap at a glycine highlighted in black. Residues mutated in both Nudt19 and Nudt7 are marked with blue stars. Residues mutated just in Nudt19 are marked with a blue open circle. Secondary structure elements of the D. radiodurans CoAse (Dr1184; PDB code 1NQZ) and human NUDT7 (HsNUDT7; PDB code 5T3P) are shown on top of the alignment. The unique insertion sequence found in mouse and human Nudt19 would be inserted (blue inverted triangle) in the loop region between the β strand and the α helix of the Nudix box structural motif. Mm, Mus musculus; Hs, Homo sapiens; Dr, D. radiodurans; Sc, S. cerevisiae; Ce, C. elegans. GenBankTM accession numbers for the aligned proteins are as follows: MmNudt7, 165972342; HsNUDT77, 157785656; MmNudt19, 187954021; HsNUDT19, 157739940; Dr1184, 6458920; ScPcd1p, 6323180; and CeNdx8, 68565843. B, calibration curve of the Superdex 75 10/300 GL column and elution profile of wildtype Nudt19, Nudt7, and chimeras. The native molecular weights of the protein peaks, as estimated from the calibration curve, are reported for each enzyme. The calculated molecular weights of the recombinant His-tagged proteins, based on the amino acid composition, are: wildtype Nudt19, 42.6 kDa; Nudt19-7-CB, 42.6 kDa; wildtype Nudt7, 29.2 kDa; Nudt7-19-NB, 34.0 kDa; and Nudt7-19-CB, 29.1 kDa. C, specific activity of the recombinant Nudt19, Nudt7, and chimeric proteins. Specific activities ± standard error were calculated from the linear regression of five-point protein curves obtained for each enzyme from at least two independent experiments in duplicate. D, substrate specificity of Nudt19, Nudt7, and chimeric proteins. Activity with each CoA species is expressed as percentage relative to the activity with free CoA. The data are reported as the mean (bars) of duplicate measurements from two or three independent experiments (circles) ± S.D.
We swapped the CoA box in Nudt7 and Nudt19, generating a chimeric Nudt7 with the Nudt19 CoA box, Nudt7-19-CB, and a chimeric Nudt19 with the Nudt7 CoA box, Nudt19-7-CB. We also substituted the Nudix box of Nudt7 with that of Nudt19 producing a Nudt7-19-NB chimera. All three chimeras were expressed and purified in amounts similar or higher compared with the wildtype Nudt7 and Nudt19. Conversely, exchanging the Nudix box in Nudt19 with the shorter Nudt7 Nudix box produced a protein that could be expressed but not purified as a soluble enzyme. Interestingly, a similar result was obtained when we expressed a Nudt19(Δ58–103) mutant lacking the unique insertion sequence in the Nudix box (data not shown), suggesting that this amino acid stretch could be important for the folding and/or stability of Nudt19. The three soluble chimeras were analyzed by analytical gel filtration to assess their folding. Based on their calculated molecular weight, ∼70% of Nudt19-7-CB, Nudt7-19-CB, and Nudt7-19-NB eluted as globular monomers, similar to wildtype Nudt7 and Nudt19 (Fig. 6B). Smaller peaks, indicating the formation of multimers, were also detected in both the chimeras and the wildtype enzymes (Fig. 6B). Despite evidence of protein folding, the specific activities of Nudt19-7-CB, Nudt7-19-CB, and Nudt7-19-NB were decreased 2–3 orders of magnitude compared with the respective wildtype enzymes (Fig. 6C). In particular, the substitution of the CoA box of Nudt19 with that of Nudt7 caused >5,000-fold decrease in the kcat and a 3-fold increase in the Km for free CoA compared with Nudt19 (Table 2). Similarly, swapping the Nudt7 CoA box with the CoA box of Nudt19 in Nudt7-19-CB led to a 500-fold decrease in kcat and a 5-fold increase in Km. Finally, exchanging the Nudix box in Nudt7-19-NB caused a 200-fold decrease in kcat and a 3-fold increase in Km compared with Nudt7 (Table 2).
Table 2.
Kinetic parameters of Nudt19 and Nudt7 mutants and chimeras
The experiments were conducted in duplicate as described under “Experimental procedures.” Kinetic constants were determined by nonlinear regression analysis of the data and expressed ± S.E.
| Mutation | Km | kcat |
|---|---|---|
| μm | 1/min | |
| Nudt19 | ||
| WT | 300 ± 40 | 284 ± 12 |
| Nudt19-7-CB | 1134 ± 284 | 0.05 ± 0.01 |
| E112A | N.D.a | N.D. |
| E115A | 1019 ± 226 | 0.16 ± 0.02 |
| E116A | N.D. | N.D. |
| R34M | 680 ± 186 | 0.12 ± 0.01 |
| R39A | 511 ± 64 | 91 ± 4 |
| F40A | 1053 ± 153 | 210 ± 12 |
| L41A | 1250 ± 125 | 101 ± 4 |
| D53A | 434 ± 59 | 323 ± 16 |
| R189A | 1193 ± 184 | 118 ± 11 |
| R189M | 455 ± 80 | 177 ± 10 |
| Nudt7 | ||
| WT | 283 ± 94 | 125 ± 12 |
| Nudt7-19-CB | 1362 ± 328 | 0.28 ± 0.03 |
| Nudt7-19-NB | 836 ± 173 | 0.65 ± 0.04 |
| E92A | N.D. | N.D. |
| E95A | 444 ± 87 | 10 ± 0.7 |
| E96A | N.D. | N.D. |
| R60M | 1010 ± 155 | 0.55 ± 0.03 |
| K65A | 958 ± 277 | 463 ± 56 |
| R66A | 3069 ± 407 | 220 ± 10 |
| R66M | 496 ± 79 | 167 ± 7 |
| D79A | 334 ± 29 | 159 ± 5 |
a N.D. indicates that the parameters were not determined because of undetectable enzymatic activity.
Wildtype Nudt19 and Nudt7 readily degrade free CoA and lauroyl-CoA but exhibit different activities against acetyl-CoA and mBB-CoA, which are good substrates for Nudt7 but not Nudt19 (Fig. 1, A and B). We next determined the effect of swapping the Nudix and CoA boxes on the substrate specificity of the three soluble chimeras Nudt19-7-CB, Nudt7-19-CB, and Nudt7-19-NB (Fig. 6D). For each enzyme, the activity data with the different substrates were plotted relative to free CoA to allow for comparison across enzymes with dramatically different specific activities. All three proteins, including the Nudt19 chimera Nudt19-7-CB, degraded acetyl-CoA and mBB-CoA, with a significantly higher relative preference for the latter substrate compared with wildtype Nudt7 and Nudt19. Combined with the dramatic reduction in kcat and robust decrease in Km (Table 2), the altered substrate specificity of the chimeras suggested that swapping the Nudix and CoA boxes between Nudt19 and Nudt7 might have caused a substantial deformation of the respective active sites.
Mutagenesis-guided modeling of Nudt19
The recently released crystal structure of human NUDT7 is the only currently available structure of a mammalian CoA diphosphohydrolase. This structure does not contain any divalent cation, essential to correctly orient the diphosphate bond of the substrate for hydrolysis, or CoA bound (PDB code 5T3P). To identify Nudt19 residues involved in CoA and possibly CDCA binding, we decided to build a model of the putative structure of Nudt19 in complex with Mg2+ and free CoA using a computational approach guided by mutational analysis.
Within the Nudix box, the carbonyl group of G1N and two of the three glutamates in the E16N, E19N, and E20N series, participate in the coordination of the essential divalent cation (24, 34–39). E16N, E19N, and E20N correspond to Glu-112, Glu-115, and Glu-116 in Nudt19, respectively (Figs. 6A and 7C). Mutation of these residues to alanine abolished the enzymatic activity of Nudt19 E112A and Nudt19 E116A (Table 2). The activity of Nudt19 E115A was detectable; however, the enzyme exhibited a ∼2000-fold decrease in kcat and a 3-fold increase in Km (Table 2). Interestingly, the effect of this mutation on the Km suggested that this residue of the Nudix box contributed to CoA binding. We also mutated the equivalent Nudt7 residues E1692, E1995, and E2096. Similar to the Nudt19 mutants, Nudt7 E92A and Nudt7 E96A did not show any detectable activity. Mutation of Nudt7 Glu-95 to alanine decreased the kcat >10-fold, but unlike the equivalent mutant in Nudt19, it had no significant effect on the Km for CoA. Overall, these results were consistent with mutagenesis studies conducted on other Nudix hydrolases (36, 40, 41) and confirmed the importance of E16N, E19N, and E20N for the activity of both Nudt19 and Nudt7. The crystal structure of the CoA diphosphohydrolase from D. radiodurans, CoAse, contains Mg2+ bound (24). Although unable to fit completely, docking of free CoA onto the “closed” structure of the bacterial CoAse suggested the existence of a potential contact between the side chain of Arg-55, the first arginine in the CoA box (Fig. 6A), and the diphosphate bond of CoA (24). Additionally, mutation of the equivalent residue to a glutamine in the Arabidopsis thaliana homolog AtCoAse led to a dramatic decrease in activity (42). This arginine residue is highly conserved across CoA diphosphohydrolases, and corresponds to Arg-34 in Nudt19 and Arg-60 in Nudt7 (Fig. 6A). Mutation of these residues to a methionine, an amino acid with a comparable side chain but no charge, dramatically decreased the kcat of the Nudt19 R34M and Nudt7 R60M mutants by 2000- and 200-fold, respectively, and caused a 2–3-fold increase in the Km. Combined, these data supported the importance of this residue for catalysis.
Figure 7.
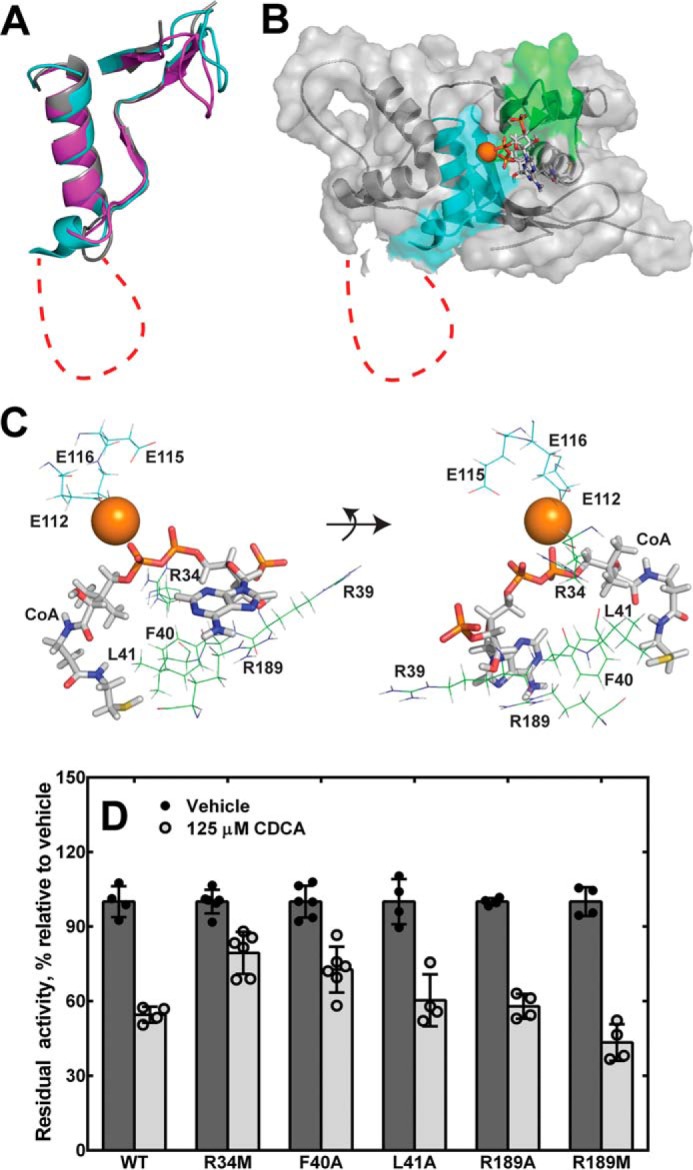
Homology model of Nudt19 with magnesium ion and free CoA bound. A, alignment of the CoA and Nudix box regions of the Nudt19 model (cyan) with the crystal structures of human NUDT7 (purple; PDB code 5T3P) and D. radiodurans CoAse (gray; PDB code 1NQZ). B, the homology model for Nudt19 with Mg2+ bound (orange sphere) was obtained using the YASARA modeling software suite. Free CoA (sticks) was then docked using MOE, and the resultant complex was evaluated and selected as described in the text. Structural elements in the CoA and Nudix boxes are shown in green and cyan, respectively. The unique amino acid insertion in the Nudix box of Nudt19 is shown as a red dotted line in both A and B. C, mode of CoA binding to Nudt19, with potentially interacting residues shown. D, activity of Nudt19 mutants incubated with or without the competitive inhibitor CDCA. The data are reported as the mean (bars) of duplicate measurements from two or three independent experiments (circles) ± S.D.
We used this information as a guide to evaluate computational models of Nudt19 generated by using the YASARA modeling software suite (43). Specifically, the final model that we selected exhibited 1) the correct positioning of E16112, E19115, and E20116 with respect to Mg2+, 2) the appropriate orientation of the diphosphate moiety of free CoA with respect to Mg2+ to allow for catalysis, and 3) interaction of Arg-34 with the diphosphate group of the substrate CoA (Fig. 7, B and C). In this model, the CoA and Nudix boxes of Nudt19 folded into the expected structural motifs with very good superposition with the CoA and Nudix boxes of the D. radiodurans CoAse and human NUDT7 (Fig. 7A). Within the β-strand-loop/helix/loop structural motif of the Nudix box, the 45-amino acid insertion in Nudt19 was modeled as an expansion in the loop connecting the strand β4 to the helix α2 and was mostly unstructured.
Identification of additional residues involved in CoA binding and hydrolysis
Based on the model, we selected and mutated a set of Nudt19 residues predicted to participate in the binding of CoA (Table 2). In particular, we mutated Arg-39, Phe-40, Leu-41, which are part of the CoA box, and Arg-189 to alanine (Figs. 6A and 7C). In the Nudt19 model, the side chains of Phe-40 and Leu-41 extended toward the free thiol end of the CoA molecule, whereas the side chain of Arg-39 extended toward the 3′-phosphate of the CoA adenine ring (Fig. 7C). Mutation of Phe-40 and Leu-41 to alanine caused a robust 3–4-fold increase in the Km, confirming that these residues played a role in CoA binding. In addition, the Nudt19 L41A mutant also showed a ∼3-fold decrease in kcat. Mutation of Arg-39 to an alanine did not have a significant effect (<2-fold) on the Km; however, removal of the positive charge caused a 3-fold decrease in kcat, indicating that this residue was nevertheless important for CoA hydrolysis. Arg-189 was located in a loop region outside the CoA box and stretched toward the adenosine moiety of CoA (Fig. 7C). The Nudt19 R189A mutant exhibited a 4-fold increase in Km and a 2-fold decrease in kcat, confirming that Arg-189 was important for both CoA binding and hydrolysis. We further investigated the role of this residue by mutating Arg-189 to a methionine. Unlike alanine, the side chain of methionine was able to functionally replace Arg-189, thus suggesting the existence of potential non-polar side chain interaction(s) between Arg-189 and CoA (Table 2). Because Nudt19 R34M, F40A, L41A, and R189A mutants exhibited a reduced affinity for CoA (Table 2), we tested the sensitivity of these mutants to inhibition by the competitive inhibitor CDCA (Fig. 5C). At a concentration of 250 μm of free CoA, the IC50 of CDCA was determined to be 125 μm against wildtype Nudt19. Among the above mutants, Nudt19 R34M retained ∼80% of activity in the presence of CDCA (Fig. 7D), suggesting that Arg-34 might be involved in both CoA and CDCA binding.
Although one of the variable X residues in the Nudix box, Nudt19 Asp-53 is highly conserved among the CoA diphosphohydrolases (Fig. 6A). In the Nudt19 model, this residue showed no interaction with CoA, but we nevertheless mutated it to alanine to determine the effect of the mutation on Km and kcat. Consistent with the model, the Nudt19 D53A mutant exhibited kinetic parameters similar to the wildtype enzyme, and we obtained similar results with the equivalent Nudt7 D79A mutant (Table 2). The sequence alignment of the CoA diphosphohydrolases also showed that it was common to find a positively charged residue in the same position as Nudt19 Arg-39, a residue involved in catalysis but not CoA binding (Table 2). We mutated the equivalent residue in Nudt7, Lys-65, to alanine. The Nudt7 K65A mutant exhibited a ∼3-fold increase in both Km and kcat, showing a differential effect on both kinetic parameters with respect to Nudt19. Finally, because Phe-40 in Nudt19 was involved in substrate binding, we mutated the equivalent Nudt7 Arg-66 to both alanine and methionine. Nudt7 R66A showed a >10-fold increase in Km, whereas the Nudt7 R66M mutant exhibited kinetic parameters similar to the wildtype enzyme (Table 2). These results supported the conclusion that the side chain of Arg-66 participated in CoA binding through hydrophobic interactions and suggested that Phe-40 in Nudt19 and Arg-66 in Nudt7 might be functionally similar.
Discussion
To gain insight into the physiological role of Nudt19, a Nudix enzyme with CoA diphosphohydrolase activity, one the major goals of this study was to determine the ability of the enzyme to regulate CoA levels in vivo. The present work demonstrates that Nudt19 degrades CoA and contributes to the regulation of this cofactor in the kidneys. Indeed, Nudt19−/− mice exhibited a significant 20% increase in total CoA levels in whole kidney homogenates obtained from fed mice (Fig. 2D). This result is consistent with the very narrow tissue distribution of the protein, which showed the highest expression levels in the kidneys, with brain and skeletal muscle containing barely detectable levels of Nudt19 (Fig. 1C).
The second goal of this study was to address conflicting reports on the peroxisomal and mitochondrial localization of Nudt19, because peroxisomes and mitochondria contain separate pools of CoA and are the sites of distinct CoA-dependent metabolic processes (7, 44). Using immunofluorescence and constructs expressing Nudt19 with either a C-terminal or an N-terminal GFP tag, we showed that the enzyme resides in the peroxisomes and not in the mitochondria (Fig. 3), thus suggesting a specific role for Nudt19 in peroxisomal metabolism. In the peroxisomes, free CoA and acyl-CoAs are imported through dedicated transporters (45, 46) and participate in several processes including the α- and β-oxidation of fatty acids that are not substrates for the mitochondrial oxidative machinery, the synthesis of ether phospholipids and, in the liver, the final steps in bile acid synthesis and conjugation. Additionally, because imported CoA species and other molecules >400 Da cannot freely exit the peroxisomes, the Nudix-mediated hydrolysis of CoA species to 3′,5′-ADP and 4′-(acyl)phosphopantetheine likely prevents CoA buildup in this organelle (32, 33). Indeed, it has been proposed that the function of peroxisomal CoA-degrading enzymes, including Nudt7 and now Nudt19, is to regulate the peroxisomal CoA pool and, in turn, peroxisomal lipid metabolism (32). Consistent with this proposed role, Nudt19 activity was highest with free CoA, which is required in multiple peroxisomal pathways, and with succinyl-CoA and C8-C12 acyl-CoAs, which are the oxidation products of dicarboxylic and very long-chain fatty acids, respectively (Fig. 1A). Nudt19 was previously reported to possess mRNA decapping activity in vitro (28). Although detectable, the hydrolytic activity of Nudt19 against capped RNA was minimal when compared with that of established mRNA decapping enzymes such as Dcp2 and Nudt16 (28). Additionally, mRNA decapping occurs in cytosolic foci (47). Thus, the peroxisomal localization of Nudt19, combined with its effect on CoA levels in vivo, argues against a role for this enzyme in mRNA decapping.
The peroxisomal localization of Nudt19 and fact that the enzyme could affect just one of the subcellular CoA pools could also explain the relatively small increase in total CoA levels observed in whole kidney homogenates obtained from Nudt19−/− mice in the fed state. In the fasted state, the concentration of kidney CoA was higher than in the fed state and similar between wildtype control and Nudt19−/− mice. These results suggest that the increase in kidney CoA levels that occurs upon fasting may be promoted more by activation of the CoA biosynthetic pathway than by a decrease in CoA degradation. The flux through the CoA biosynthetic pathway is controlled by the pantothenate kinase (PanK) isoforms, and PanK1, which is the isoform that drives the increase in CoA levels observed in the liver of fasted mice, is also the major PanK isoform in the kidneys (13, 48). Upon refeeding, a net decrease in CoA levels requires not only inhibition of CoA synthesis, but also active degradation of the cofactor accumulated in the fasted state. This might explain why the effect of deleting Nudt19 on kidney CoA levels was mostly detected in fed mice. Interestingly, although the Nudt19−/− mice exhibited higher kidney CoA levels in the fed state compared with control mice, this concentration of the cofactor was still significantly lower than in the fasted state, suggesting the existence of residual CoA-degrading activity in the absence of Nudt19. Nudt7 and Nudt19 are the only currently confirmed CoA-degrading enzymes, and in the kidneys, Nudt7 is a minor isoform (Fig. 1C, and compare Fig. 2E to Fig. 4A). Under our experimental conditions, the production of 3′,5′-ADP in kidney extracts from Nudt19−/− mice was undetectable (Fig. 2C), and we did not observe any compensatory up-regulation of Nudt7 (Fig. 2E). Despite this, an accurate estimate of the contribution of Nudt7, if any, to the regulation of kidney CoA levels, will require the generation of mice with a deletion in both Nudt7 and Nudt19. Alternatively, CoA might be hydrolyzed to products other than 3′,5′-ADP and 4′-(acyl)phosphopantetheine by a novel class of CoA-degrading enzymes that may or may not reside in the peroxisomes.
The narrow and non-overlapping tissue distribution of Nudt19 and Nudt7 in kidneys and liver, respectively, is intriguing. Another major goal of this study was to investigate the distinctive biochemical and regulatory properties of these enzymes to gain insight into the physiological roles that Nudt19 and Nudt7 may play in their respective organs. Nudt19 (42 kDa) is significantly larger than Nudt7 (27 kDa) and contains unique sequence regions, the most notable of which is the 45-amino acid insertion in the Nudix box, which are not present in Nudt7 (Fig. 6A). Nudt7 expression in the liver decreases in the fasted state and increases in the fed state, showing an inverse correlated with the hepatic concentration of CoA (9, 26). Conversely, Nudt19 mRNA and protein levels in the kidneys did not change between the fed and fasted states, despite the fact that this enzyme contributes to the regulation of kidney CoA levels.
While searching for potential small molecule regulators, we identified CDCA as a competitive inhibitor of Nudt19 that did not bind to Nudt7, strongly suggesting that these two enzymes possess distinct CoA-binding sites (Fig. 5). Interestingly, binding of CDCA to Nudt19 depended, at least in part, on Arg-34 (Arg-60 in Nudt7), a highly conserved residue involved in CoA binding and hydrolysis in both Nudt19 and Nudt7 (Figs. 6A and 7D and Table 2). Additional evidence in support of a different organization of the CoA-binding sites between the two isoforms is provided by the different activities that Nudt19 and Nudt7 displayed against substrates such as acetyl-CoA and mBB-CoA and by the dramatic decrease in kcat observed upon exchange of the CoA and Nudix boxes between the two enzymes. Furthermore, the invariant E19N in the Nudix box was found to participate in CoA binding in Nudt19 but not Nudt7, whereas the conserved (Arg/Lys) residue in the CoA box (corresponding to Lys-65 in Nudt7 and Arg-39 in Nudt19) was found to be involved in CoA binding in Nudt7 but not Nudt19.
No structural information on CoA diphosphohydrolases with CoA bound is currently available, and this represents a limitation to the accurate prediction of the mode of CoA binding through homology models. Despite this, mutational analysis based on the Nudt19 model that we generated allowed us to identify residues that are important for CoA binding and hydrolysis (Table 2). These residues were found both within and outside the CoA box. The Nudt19 model also shows that the 45-amino acid insertion within the Nudix box may be mostly disordered. Within a protein, disordered regions can exert different functions, including providing display sites for post-translational modification and/or mediating protein–protein interactions (49). There are only a few known examples of peroxisomal proteins regulated through reversible post-translational modifications (50, 51), and the role of this potentially disordered sequence, if any, in the regulation Nudt19 activity remains to be elucidated. We also docked Mg2+ and free CoA to the NUDT7 crystal structure to determine whether we could find a pose that would agree with the mutational analysis on the closely related mouse Nudt7 and provide us with a model of CoA binding to NUDT7. As for Nudt19, we selected the most likely models using the position of E1692, E1995, E2096, Arg-60, CoA, and Mg2+. In these models, Arg-66 (equivalent to Arg-66 in mouse Nudt7) would form a salt bridge with the diphosphate moiety of CoA (data not shown). Although the 10-fold increase in Km of the Nudt7 R66A mutant supports the importance of Arg-66 for substrate binding, the lack of effect of the R66M mutation on the kinetic parameters of the enzyme indicates that the requirement for Arg-66 is likely spatial and geometric and not about the charge (Table 2). Thus, in this case, the docking of CoA to the structure of NUDT7 does not seem to provide a close representation of how CoA binds to this enzyme. Crystallization of NUDT7 and possibly Nudt19 with a substrate bound will be required to yield further details of CoA binding.
Finally, bile acids are synthesized de novo exclusively in the liver, with the last steps in the synthesis and conjugation to glycine and taurine occurring in the peroxisomes (52). Although it may be unlikely for Nudt19 to encounter high concentrations of CDCA in kidney peroxisomes, Nudt7 is presumably exposed to high concentrations of CDCA, cholic acid, and conjugated derivatives in liver peroxisomes. Thus, it is tempting to speculate that the ability of Nudt7 to remain active even at high micromolar concentrations of bile acids is one possible reason for the selection of this enzyme as the major CoA diphosphohydrolase in the liver. As for Nudt19, further studies, including an in depth characterization of the Nudt19−/− mice, will be required to determine what selective advantage(s) this isoform may confer to the kidneys.
Experimental procedures
Materials
Reagents were purchased from the following suppliers: oligonucleotides, Lipofectamine 2000, cell culture reagents, Alexa Fluor 555- and horseradish peroxidase-conjugated goat anti-rabbit IgG from Thermo Fisher Scientific; restriction enzymes, DNA polymerases, and the Q5® site-directed mutagenesis kit from New England Biolabs; HEK 293 cells from the American Type Culture Collection; and the GAPDH antibody from Cell Signaling. The PMP70 antibody was purchased from Sigma–Aldrich. The antibody against the full-length mouse Nudt19 was raised in rabbit and purified by antigen affinity chromatography. The Nudt7 antibody was generated as previously described (9). mBB-CoA was synthesized from free CoA and monobromobimane (Echelon Biosciences) as described by Shimada and Mitamura (53). The conversion of free CoA to mBB-CoA was >99.0%, as estimated by HPLC. All other reagents were of analytical grade or better and were purchased from Sigma–Aldrich or Fisher Scientific, unless otherwise stated.
Animal studies
Nudt19tm1(KOMP)Vlcg embryos, obtained by replacing the Nudt19 gene with a ZEN-UB1 cassette harboring the lacZ reporter and neo resistance gene, were purchased from the Knock-out Mouse Project (KOMP) repository and implanted into pseudopregnant foster mothers by the Transgenic Animal Core Facility at West Virginia University. The progeny was screened for the deletion of the Nudt19 gene by multiplex PCR analysis using the Accustart II PCR genotyping kit (QuantaBio) and the following KOMP primers: Reg-NeoF (GCAGCCTCTGTTCCACATACACTTCA), Reg-Nudt19-R (TTCACAACCTGCCAAAATGGCTACC), Reg-Nudt19-wtF2 (GGAAACGGGAACAGAGAAACAGG), and Reg-Nudt19-wtR (AAGGACTCAACTCTCACCCTAAGCG). The Nudt19 knockout allele yielded a product of 466 bp. Although smaller than the size of 584 bp predicted by the KOMP repository, this 466-bp amplicon contained the expected 3′ end of the ZEN-UB1 cassette (124 bases) and a sequence corresponding to the genomic region downstream of the Nudt19 gene (342 bp), as confirmed by sequencing. The wildtype allele yielded a product of the expected size of 185 bp. Homozygous Nudt19−/− mice and wildtype C57BL6/N littermate controls were obtained through heterozygous × heterozygous breeding. The mice were fed a standard chow diet (Tekland 2018S) and maintained at a room temperature of 72.0 ± 0.3 °F, room humidity of 40% ± 2%, and a 12-h light/12-h dark cycle, with the dark cycle starting at 18:00 h. For experiments that required fasting, the mice were transferred to clean cages containing floor grids, and food was removed for the indicated time. All studies were conducted on 10–14-week-old male mice and were approved by the Institutional Animal Care and Use Committees of West Virginia University.
CoA analysis, immunoblotting, and RT-PCR
The concentration of total CoA (free CoA plus CoA thioesters) in kidney homogenates was determined after conversion of the cofactor to the monobromobimane derivative, as previously described (25). For Western blotting analysis, flash frozen tissues were homogenized in ice-cold radioimmunoprecipitation assay buffer supplemented with protease inhibitors (Biotool) and centrifuged at 10,000 × g for 10 min at 4 °C. Proteins were fractionated on 4–12% bis-Tris polyacrylamide gels and transferred onto PVDF membranes. The GAPDH antibody was used at a 1:3000 dilution; the Nudt7 and Nudt19 antibodies were used at a 1:10,000 dilution. Bound primary antibodies were detected by chemiluminescence with horseradish peroxidase-conjugated goat anti-rabbit IgG at a 1:45,000 dilution. RNA was isolated from flash frozen tissue as previously described (54). Following the removal of genomic DNA with Turbo DNase-free kit (Thermo Fisher Scientific), Nudt19 mRNA levels were quantified in triplicate by RT-PCR using the Quantitect SYBR Green RT-PCR kit (Qiagen), and the primers were previously reported (54). The relative abundance of the Nudt19 mRNA was calculated using the CT method, and the amount (2−ΔCT) was reported relative to the mRNA of β-glucuronidase (GusB) (25).
Plasmid construction and mutagenesis
Wildtype mouse Nudt7 and Nudt19 were expressed as N-terminal hexahistine-tagged proteins from plasmids pKM204 (25) and pKM249, respectively. Plasmid pKM249 was a kind gift from Dr. Suzanne Jackowski (St. Jude Children's Research Hospital). The above plasmids were mutagenized with the Q5® site-directed mutagenesis kit to generate single-point Nudt7 and Nudt19 mutants and a Nudt19 deletion mutant lacking amino acids 58–103. Insertion of the correct mutations was confirmed by sequencing analysis by the Genomics Core Facility at West Virginia University. Sequences encoding Nudt7 and Nudt19 chimeras were custom synthesized (Thermo Fisher Scientific) to contain NdeI and NotI restriction sites at the 5′ and 3′ ends, respectively, codon-optimized for expression in Escherichia coli and subcloned into pET-28a(+) (Millipore). For the expression and localization of mouse Nudt19 in HEK 293 cells, custom pcDNA3.1(+)-derived constructs (GenScript) were assembled to express Nudt19 proteins, codon-optimized for expression in human cells, with N- or C-terminal GFP tags.
Protein purification, analytical gel filtration chromatography, and ITC
All Nudt7 and Nudt19 proteins with N-terminal hexahistidine tags were expressed in BL21 CodonPlus (DE3)-RIPL (Agilent Technologies) cells, following induction with 1 mm isopropyl β-d-1-thiogalactopyranoside and growth for 18–20 h at 18–25 °C. For enzymatic assay, the enzymes were purified as previously described (25). Fractions containing ∼90% pure enzymes were combined and dialyzed overnight at 4 °C in buffer containing 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol. Glycerol was added to a final concentration of 50%, and proteins were stored at −20 °C. Protein samples analyzed by analytical gel filtration chromatography were loaded (50 μl) onto a Superdex 75 10/300 GL column and eluted at 0.3 ml/min with 50 mm Tris-HCl, 150 mm NaCl, pH 8.0. Protein elution was monitored by measuring the absorbance at 280 nm. For ITC studies, the purification procedure included three chromatographic steps to increase the purity of the proteins to >98%, and the hexahistidine tag was removed. Briefly, cell pellets were lysed in 50 mm Na2HPO4/NaH2PO4, pH 8.0, 300 mm NaCl, 10 mm imidazole and centrifuged at 13,000 rpm for 45 min at 4 °C. The clarified supernatant was allowed to bind to the nickel-nitrilotriacetic acid resin for 2 h with stirring, before the suspension was transferred into a column. The resin-bound proteins were washed with 50 mm Na2HPO4/NaH2PO4, pH 8.0, 300 mm NaCl, 20 mm imidazole, followed by elution with wash buffer containing 50–100 mm of imidazole. Protein-containing fractions were combined, dialyzed in buffer A (50 mm Na2HPO4/NaH2PO4, pH 8.0, 200 mm NaCl), and incubated with 0.04 unit/ml thrombin (Sigma–Aldrich) overnight at room temperature to remove the hexahistine tag. The salt concentration of the protein samples was then lowered with an additional dialysis step in buffer B (50 mm Na2HPO4/NaH2PO4, 50 mm NaCl), before applying the proteins onto a Resource15Q anion exchange column equilibrated in buffer B. Proteins were eluted with a 0–100% gradient of buffer C (50 mm Na2HPO4/NaH2PO4, pH 8.0, 500 mm NaCl), and the fractions containing the proteins of interested were pooled, dialyzed in buffer A, and concentrated to 5 ml. The concentrated protein samples were subsequently loaded onto a HiLoad 26/60 Superdex 75-pg size exclusion column equilibrated in buffer A, eluted in the same buffer, concentrated, and stored at −80 °C.
Binding affinity of wildtype Nudt7 and Nudt19 for CDCA was measured by ITC at 25 °C using a VP-ITC instrument (Microcal Inc., Northhampton, MA) and 1.4 ml of purified Nudt7 and Nudt19 dialyzed in ITC buffer (30 mm PIPES, pH 7.5, 50 mm NaCl, 2 mm MgCl2) and diluted to 124 and 55.5 μm, respectively. Protein solutions were degassed for at least 20 min. CDCA stock solutions were prepared in the same ITC buffer. After an initial 2-μl injection of CDCA solution with a 240-s equilibration time, the compound was titrated in 30 injections of 10 μl each, added at 300-s intervals. The duration of each injection was 12 s, and the filter period was 2 s. The stirring speed and Feedback Mode/gain option were set up to 300 rpm and “High” respectively. The heat evolved at 25 °C after each ligand injection was obtained by integration of the calorimetric signal after subtracting the average heat of dilution. Binding parameters were determined from titration binding curves using the MicroCal Data Analysis software (Malvern, Westborough, MA).
Enzymatic assays
The CoA diphosphohydrolase activity of the recombinant enzymes was assayed, in duplicate, as previously described (25), with minor modifications. The standard assay contained 10 ng of recombinant Nudt7 or 30 ng of Nudt19, 100 mm Tris-HCl, pH 8.0, 0.1 mg/ml γ-globulin, an optimized concentration of MgCl2, which was 4 mm for Nudt7 or 10 mm for Nudt19, and 250 μm of free CoA or acyl-CoA substrates, in a total volume of 40 μl. The reactions were incubated at 37 °C for 10 min and stopped by the addition of perchloric acid. Following neutralization with potassium carbonate, the reaction mixtures were diluted 5-fold with water, and the UV-visible 3′,5′-ADP product was separated and quantified by HPLC coupled with UV detection on a 4.6 × 150 mm, 3-μm C-18 column (Acclaim 120) kept at 35 °C. Elution was performed at 0.7 ml/min as follows: 0–2.5 min, 100% buffer A (50 mm KH2PO4, pH 4.6); 2.5–9.0 min linear gradient to 50% B (100% acetonitrile); 9.0–10.0 min isocratic at 50% B; 10.0–15.0 min linear gradient from 50% B to 75% B; 15.0–19.0 min isocratic at 75% B; 19.0–32.0 min return to 100% A, and column equilibration. A standard curve of 3′,5′-ADP (Sigma–Aldrich) subjected to the same reaction conditions was used to quantify the amount of product formed. For each mutant and chimeric enzyme, protein curves were generated to select concentrations within the activity linear range. The length of the assay was also extended to 2 h for mutants exhibiting marginal activity. The kinetic parameters Km, Vmax, and kcat were determined using free CoA as the substrate, with concentrations ranging from 31.25 μm to 10 mm, and obtained from curves fitted to a Michaelis–Menten equation using GraphPad Prism 6 (GraphPad Software).
To monitor the production of 3′,5′-ADP in kidney extracts, flash-frozen tissue (∼50 mg) was homogenized in 750 μl of 20 mm Tris-HCl, pH 8.0. The homogenate was incubated on ice for 10 min and centrifuged at 20,000 × g for 10 min. The supernatant was loaded on to a PD10 column equilibrated in 20 mm Tris-HCl, pH 8.0, to remove the small molecules. The eluted proteins (50–200 μg) were incubated with 250 μm free CoA for 5 min at 37 °C. The reactions were then stopped and analyzed as described above.
Immunofluorescence
HEK 293 cells were cultured at 37 °C and 5% CO2 in phenol red-free Opti-MEM I medium supplemented with 4% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B. To localize Nudt19, the cells were seeded on 12-mm coverslips at low density and grown in the same medium to ∼80% confluence. Transfections were performed with 1.25 μg of plasmid DNA using Lipofectamine 2000, as per the manufacturer's instructions. Where indicated, the cells were incubated with MitoTracker® Orange CMTMRos at a concentration of 25 nm for 30 min at 37 °C. The cells were fixed with 4% paraformaldehyde and blocked in PBS containing 5% FBS and 0.3% Triton X-100 for 1 h at room temperature. The PMP70 antibody was used at a 1:1000 dilution in 1% BSA, 0.3% Triton X-100 for 1 h at room temperature. Following 10 washes with PBS, the cells were incubated for 1 h at room temperature with Alexa Flour 555 donkey anti-mouse IgG, used at a dilution of 1:1000 in 1% BSA, 0.3% Triton X-100. Slides were mounted using Prolong Gold Antifade with DAPI (Thermo Fisher). Imaging experiments were performed in the West Virginia University Imaging Facilities using a Nikon A1R/N SIM-E confocal microscope. Image analysis was conducted using ImageJ.
Molecular modeling
Homology models with bound Mg2+ were built using the software suite YASARA (43) (www.yasara.org).4 The models showing the expected orientation of E16N, E19N, and E20N with respect to the Mg2+ ion were selected and prepared for docking studies with free CoA by adding hydrogens at pH 7.4 in the modeling program MOE 2016 (www.chemcomp.com; Ref. 57).4 After an induced fit docking step, the top five poses were evaluated for the correct relative orientation of the diphosphate moiety of free CoA, the bound Mg2+, and Nudt19 Arg-34 or NUDT7 Arg-60. Sequence alignments were obtained using Clustal W (55) and combined with secondary structure elements using ESPript 3 (56). The Nudt19 model and the crystal structures of human NUDT7 and the bacterial CoAse were aligned using Phenix Ensembler (23).
Statistical analysis
Unless otherwise stated, all data are reported as the means with ± S.D. Statistical significance was calculated by unpaired two-tailed Student's t test using GraphPad Prism 6 (GraphPad Software).
Author contributions
S. A. S., W. J. G., S. B. G., and R. L. conceptualization; S. A. S., E. W. K., W. J. G., P. F., P. T., S. B. G., and R. L. data curation; S. A. S., E. W. K., W. J. G., G. E. M., P. F., P. T., H. S., S. B. G., and R. L. formal analysis; S. A. S., W. J. G., G. E. M., S. B. G., and R. L. funding acquisition; S. A. S., E. W. K., W. J. G., P. T., H. S., S. B. G., and R. L. visualization; S. A. S., E. W. K., W. J. G., P. F., S. B. G., and R. L. writing-original draft; P. F. and P. T. validation.
Acknowledgments
We are grateful to Dr. Aaron Robart (West Virginia University) for assistance with the structure alignment. We thank Deborah Corbin and Claire Smathers, and the staff of the Genomics Core and the Imaging Facilities (West Virginia University) for their expert technical assistance.
This work was supported by West Virginia University's School of Medicine startup foundation funding (RL), the Allegheny Health Network-Johns Hopkins Cancer Research Fund (SBG), and National Institutes of Health (NIH) Grants GM119528 (RL), F31 GM126838 (SAS), P20 GM103434 awarded to the West Virginia IDeA Network for Biomedical Research Excellence (GEM), WVU Stroke CoBRE grant P20 GM109098 (WJG), and CA062924 (SBG). The West Virginia University Imaging Facilities were supported by the WVU Cancer Institute and NIH grants P20RR016440, P30RR032138/P30GM103488 and U54GM104942. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PDB
- Protein Data Bank
- CDCA
- chenodeoxycholic acid
- mBB
- monobromobimane
- PTS1
- peroxisome targeting signal type 1
- ITC
- isothermal titration calorimetry
- KOMP
- Knock-out Mouse Project.
References
- 1. Choudhary C., Weinert B. T., Nishida Y., Verdin E., and Mann M. (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell. Biol. 15, 536–550 10.1038/nrn3747,10.1038/nrm3841 [DOI] [PubMed] [Google Scholar]
- 2. Hirschey M. D., and Zhao Y. (2015) Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell. Proteomics 14, 2308–2315 10.1074/mcp.R114.046664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Resh M. D. (2016) Fatty acylation of proteins: The long and the short of it. Prog. Lipid Res. 63, 120–131 10.1016/j.plipres.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daniotti J. L., Pedro M. P., and Valdez Taubas J. (2017) The role of S-acylation in protein trafficking. Traffic 18, 699–710 10.1111/tra.12510 [DOI] [PubMed] [Google Scholar]
- 5. Sabari B. R., Zhang D., Allis C. D., and Zhao Y. (2017) Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell. Biol. 18, 90–101 10.1038/nrm.2016.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horie S., Ishii H., and Suga T. (1981) Changes in peroxisomal fatty acid oxidation in the diabetic rat liver. J. Biochem. 90, 1691–1696 10.1093/oxfordjournals.jbchem.a133645 [DOI] [PubMed] [Google Scholar]
- 7. Van Broekhoven A., Peeters M. C., Debeer L. J., and Mannaerts G. P. (1981) Subcellular distribution of coenzyme A: evidence for a separate coenzyme A pool in peroxisomes. Biochem. Biophys. Res. Commun. 100, 305–312 10.1016/S0006-291X(81)80097-6 [DOI] [PubMed] [Google Scholar]
- 8. Peng Y., and Puglielli L. (2016) N-lysine acetylation in the lumen of the endoplasmic reticulum: a way to regulate autophagy and maintain protein homeostasis in the secretory pathway. Autophagy 12, 1051–1052 10.1080/15548627.2016.1164369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leonardi R., Rock C. O., and Jackowski S. (2014) Pank1 deletion in leptin-deficient mice reduces hyperglycaemia and hyperinsulinaemia and modifies global metabolism without affecting insulin resistance. Diabetologia 57, 1466–1475 10.1007/s00125-014-3245-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corbin D. R., Rehg J. E., Shepherd D. L., Stoilov P., Percifield R. J., Horner L., Frase S., Zhang Y. M., Rock C. O., Hollander J. M., Jackowski S., and Leonardi R. (2017) Excess coenzyme A reduces skeletal muscle performance and strength in mice overexpressing human PANK2. Mol. Genet. Metab. 120, 350–362 10.1016/j.ymgme.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dusi S., Valletta L., Haack T. B., Tsuchiya Y., Venco P., Pasqualato S., Goffrini P., Tigano M., Demchenko N., Wieland T., Schwarzmayr T., Strom T. M., Invernizzi F., Garavaglia B., Gregory A., et al. (2014) Exome sequence reveals mutations in CoA synthase as a cause of neurodegeneration with brain iron accumulation. Am. J. Hum. Genet. 94, 11–22 10.1016/j.ajhg.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou B., Westaway S. K., Levinson B., Johnson M. A., Gitschier J., and Hayflick S. J. (2001) A novel pantothenate kinase gene (PANK2) is defective in Hallervorden–Spatz syndrome. Nat. Genet. 28, 345–349 10.1038/ng572 [DOI] [PubMed] [Google Scholar]
- 13. Leonardi R., Rehg J. E., Rock C. O., and Jackowski S. (2010) Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state. PLoS One 5, e11107 10.1371/journal.pone.0011107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jackowski S., and Leonardi R. (2014) Deregulated coenzyme A, loss of metabolic flexibility and diabetes. Biochem. Soc. Trans. 42, 1118–1122 10.1042/BST20140156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y. M., Chohnan S., Virga K. G., Stevens R. D., Ilkayeva O. R., Wenner B. R., Bain J. R., Newgard C. B., Lee R. E., Rock C. O., and Jackowski S. (2007) Chemical knockout of pantothenate kinase reveals the metabolic and genetic program responsible for hepatic coenzyme A homeostasis. Chem. Biol. 14, 291–302 10.1016/j.chembiol.2007.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ofman R., Speijer D., Leen R., and Wanders R. J. (2006) Proteomic analysis of mouse kidney peroxisomes: identification of RP2p as a peroxisomal nudix hydrolase with acyl-CoA diphosphatase activity. Biochem. J. 393, 537–543 10.1042/BJ20050893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gasmi L., and McLennan A. G. (2001) The mouse Nudt7 gene encodes a peroxisomal nudix hydrolase specific for coenzyme A and its derivatives. Biochem. J. 357, 33–38 10.1042/0264-6021:3570033,10.1042/bj3570033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bessman M. J., Frick D. N., and O'Handley S. F. (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271, 25059–25062 10.1074/jbc.271.41.25059 [DOI] [PubMed] [Google Scholar]
- 19. Carreras-Puigvert J., Zitnik M., Jemth A. S., Carter M., Unterlass J. E., Hallström B., Loseva O., Karem Z., Calderón-Montaño J. M., Lindskog C., Edqvist P. H., Matuszewski D. J., Ait Blal H., Berntsson R. P. A., Häggblad M., et al. (2017) A comprehensive structural, biochemical and biological profiling of the human NUDIX hydrolase family. Nat. Commun. 8, 1541 10.1038/s41467-017-01642-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McLennan A. G. (2006) The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63, 123–143 10.1007/s00018-005-5386-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de la Peña A. H., Suarez A., Duong-Ly K. C., Schoeffield A. J., Pizarro-Dupuy M. A., Zarr M., Pineiro S. A., Amzel L. M., and Gabelli S. B. (2015) Structural and enzymatic characterization of a nucleoside diphosphate sugar hydrolase from Bdellovibrio bacteriovorus. PLoS One 10, e0141716 10.1371/journal.pone.0141716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cartwright J. L., Gasmi L., Spiller D. G., and McLennan A. G. (2000) The Saccharomyces cerevisiae PCD1 gene encodes a peroxisomal nudix hydrolase active toward coenzyme A and its derivatives. J. Biol. Chem. 275, 32925–32930 10.1074/jbc.M005015200 [DOI] [PubMed] [Google Scholar]
- 23. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang L. W., Gabelli S. B., Bianchet M. A., Xu W. L., Bessman M. J., and Amzel L. M. (2003) Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family. J. Bacteriol. 185, 4110–4118 10.1128/JB.185.14.4110-4118.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shumar S. A., Fagone P., Alfonso-Pecchio A., Gray J. T., Rehg J. E., Jackowski S., and Leonardi R. (2015) Induction of neuron-specific degradation of coenzyme A models pantothenate kinase-associated neurodegeneration by reducing motor coordination in mice. PLoS One 10, e0130013 10.1371/journal.pone.0130013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reilly S. J., Tillander V., Ofman R., Alexson S. E., and Hunt M. C. (2008) The nudix hydrolase 7 is an Acyl-CoA diphosphatase involved in regulating peroxisomal coenzyme A homeostasis. J. Biochem. 144, 655–663 10.1093/jb/mvn114 [DOI] [PubMed] [Google Scholar]
- 27. Rheaume C., Barbour K. W., Tseng-Crank J., and Berger F. G. (1989) Molecular genetics of androgen-inducible RP2 gene transcription in the mouse kidney. Mol. Cell. Biol. 9, 477–483 10.1128/MCB.9.2.477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song M. G., Bail S., and Kiledjian M. (2013) Multiple Nudix family proteins possess mRNA decapping activity. RNA 19, 390–399 10.1261/rna.037309.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Calvo S. E., Clauser K. R., and Mootha V. K. (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 10.1093/nar/gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Snider L. D., King D., and Lingrel J. B. (1985) Androgen regulation of MAK mRNAs in mouse kidney. J. Biol. Chem. 260, 9884–9893 [PubMed] [Google Scholar]
- 31. AbdelRaheim S. R., and McLennan A. G. (2002) The Caenorhabditis elegans Y87G2A.14 Nudix hydrolase is a peroxisomal coenzyme A diphosphatase. BMC Biochem. 3, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hunt M. C., Tillander V., and Alexson S. E. (2014) Regulation of peroxisomal lipid metabolism: the role of acyl-CoA and coenzyme A metabolizing enzymes. Biochimie 98, 45–55 10.1016/j.biochi.2013.12.018 [DOI] [PubMed] [Google Scholar]
- 33. Antonenkov V. D., and Hiltunen J. K. (2006) Peroxisomal membrane permeability and solute transfer. Biochim. Biophys. Acta 1763, 1697–1706 10.1016/j.bbamcr.2006.08.044 [DOI] [PubMed] [Google Scholar]
- 34. Gabelli S. B., Bianchet M. A., Bessman M. J., and Amzel L. M. (2001) The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat. Struct. Biol. 8, 467–472 10.1038/87647 [DOI] [PubMed] [Google Scholar]
- 35. Gabelli S. B., Bianchet M. A., Xu W., Dunn C. A., Niu Z. D., Amzel L. M., and Bessman M. J. (2007) Structure and function of the E. coli dihydroneopterin triphosphate pyrophosphatase: a Nudix enzyme involved in folate biosynthesis. Structure 15, 1014–1022 10.1016/j.str.2007.06.018 [DOI] [PubMed] [Google Scholar]
- 36. Boto A. N., Xu W., Jakoncic J., Pannuri A., Romeo T., Bessman M. J., Gabelli S. B., and Amzel L. M. (2011) Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition. Proteins 79, 2455–2466 10.1002/prot.23069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Swarbrick J. D., Bashtannyk T., Maksel D., Zhang X. R., Blackburn G. M., Gayler K. R., and Gooley P. R. (2000) The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L. J. Mol. Biol. 302, 1165–1177 10.1006/jmbi.2000.4085 [DOI] [PubMed] [Google Scholar]
- 38. Lin J., Abeygunawardana C., Frick D. N., Bessman M. J., and Mildvan A. S. (1996) The role of Glu 57 in the mechanism of the Escherichia coli MutT enzyme by mutagenesis and heteronuclear NMR. Biochemistry 35, 6715–6726 10.1021/bi953071x [DOI] [PubMed] [Google Scholar]
- 39. O'Handley S. F., Thirawatananond P., Kang L. W., Cunningham J. E., Leyva J. A., Amzel L. M., and Gabelli S. B. (2016) Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis. J. Bioenerg. Biomembr. 48, 557–567 10.1007/s10863-016-9681-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abdelghany H. M., Bailey S., Blackburn G. M., Rafferty J. B., and McLennan A. G. (2003) Analysis of the catalytic and binding residues of the diadenosine tetraphosphate pyrophosphohydrolase from Caenorhabditis elegans by site-directed mutagenesis. J. Biol. Chem. 278, 4435–4439 10.1074/jbc.M211983200 [DOI] [PubMed] [Google Scholar]
- 41. Iwai T., Kuramitsu S., and Masui R. (2004) The Nudix hydrolase Ndx1 from Thermus thermophilus HB8 is a diadenosine hexaphosphate hydrolase with a novel activity. J. Biol. Chem. 279, 21732–21739 10.1074/jbc.M312018200 [DOI] [PubMed] [Google Scholar]
- 42. Kupke T., Caparrós-Martín J. A., Malquichagua Salazar K. J., and Culiáñez-Macià F. A. (2009) Biochemical and physiological characterization of Arabidopsis thaliana AtCoAse: a Nudix CoA hydrolyzing protein that improves plant development. Physiol. Plant. 135, 365–378 10.1111/j.1399-3054.2009.01205.x [DOI] [PubMed] [Google Scholar]
- 43. Krieger E., and Vriend G. (2014) YASARA View: molecular graphics for all devices: from smartphones to workstations. Bioinformatics 30, 2981–2982 10.1093/bioinformatics/btu426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Horie S., Isobe M., and Suga T. (1986) Changes in CoA pools in hepatic peroxisomes of the rat under various conditions. J. Biochem. 99, 1345–1352 10.1093/oxfordjournals.jbchem.a135602 [DOI] [PubMed] [Google Scholar]
- 45. Agrimi G., Russo A., Scarcia P., and Palmieri F. (2012) The human gene SLC25A17 encodes a peroxisomal transporter of coenzyme A, FAD and NAD+. Biochem. J. 443, 241–247 10.1042/BJ20111420 [DOI] [PubMed] [Google Scholar]
- 46. Kemp S., Theodoulou F. L., and Wanders R. J. (2011) Mammalian peroxisomal ABC transporters: from endogenous substrates to pathology and clinical significance. Br. J. Pharmacol. 164, 1753–1766 10.1111/j.1476-5381.2011.01435.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sheth U., and Parker R. (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dansie L. E., Reeves S., Miller K., Zano S. P., Frank M., Pate C., Wang J., and Jackowski S. (2014) Physiological roles of the pantothenate kinases. Biochem. Soc. Trans. 42, 1033–1036 10.1042/BST20140096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van der Lee R., Buljan M., Lang B., Weatheritt R. J., Daughdrill G. W., Dunker A. K., Fuxreiter M., Gough J., Gsponer J., Jones D. T., Kim P. M., Kriwacki R. W., Oldfield C. J., Pappu R. V., Tompa P., et al. (2014) Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ortega-Galisteo A. P., Rodríguez-Serrano M., Pazmiño D. M., Gupta D. K., Sandalio L. M., and Romero-Puertas M. C. (2012) S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J. Exp. Bot. 63, 2089–2103 10.1093/jxb/err414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Knoblach B., and Rachubinski R. A. (2010) Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J. Biol. Chem. 285, 6670–6680 10.1074/jbc.M109.094805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ferdinandusse S., and Houten S. M. (2006) Peroxisomes and bile acid biosynthesis. Biochim. Biophys. Acta 1763, 1427–1440 10.1016/j.bbamcr.2006.09.001 [DOI] [PubMed] [Google Scholar]
- 53. Shimada K., and Mitamura K. (1994) Derivatization of thiol-containing compounds. J. Chromatogr. B Biomed. Appl. 659, 227–241 10.1016/0378-4347(93)E0444-U [DOI] [PubMed] [Google Scholar]
- 54. Garcia M., Leonardi R., Zhang Y. M., Rehg J. E., and Jackowski S. (2012) Germline deletion of pantothenate kinases 1 and 2 reveals the key roles for CoA in postnatal metabolism. PLoS One 7, e40871 10.1371/journal.pone.0040871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thompson J. D., Higgins D. G., and Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chemical Computing Group ULC (2017) Molecular Operating Environment (MOE) 2013.08, Montreal, Quebec, Canada [Google Scholar]