Abstract
Receptor recognition is a key step in the initiation of phage infection. Previously, we found that VP3, the T7 family phage of the Vibrio cholerae serogroup O1 biotype El Tor, can adsorb the core oligosaccharide (OS) of lipopolysaccharides of V. cholerae. However, some wildtype strains of V. cholerae possessing the intact OS gene cluster still have VP3 binding but are resistant to VP3 infection. Moreover, an OS gene–deletion mutant still exhibits weak VP3 binding, suggesting multiple factors are possibly involved in VP3 binding to V. cholerae. Here, we report that the outer-membrane protein TolC of V. cholerae is involved in the host adsorption of VP3. We observed that TolC directly interacts with the VP3 tail fiber protein gp44 and its C-terminal domains, and we also found that three amino acid residues in the outside loops of TolC, at positions 78, 290, and 291, are critical for binding to gp44. Among the VP3-resistant wildtype V. cholerae strains, frequent amino acid residue mutations were observed in the loops around the sites 78, 290, and 291, which were predicted to be exposed to the cell surface. These findings reveal a co-receptor–binding mechanism for VP3 infection of V. cholerae and that both outer-membrane TolC and OS are necessary for successful VP3 infection of V. cholerae. We conclude that mutations on the outside loops of the receptor may confer V. cholerae strains with VP3 phage resistance, enabling these strains to survive in environments containing VP3 or related phages.
Keywords: infection, lipopolysaccharide (LPS), membrane protein, receptor, Vibrio cholerae, TolC, phage
Introduction
Cholera is an acute diarrheal disease caused by Vibrio cholerae. Although 210 serogroups in V. cholerae have been identified according to its O antigen variability, only serogroups O1 and O139 are associated with known cholera epidemics and pandemics. Serogroup O1 includes two biological types, classical and El Tor (1). Subtyping protocols of bacterial pathogens, including serotyping, phage typing, and molecular typing, are widely used in microbiological and epidemiological studies. A phage-biotyping scheme for El Tor strains of V. cholerae serogroup O1 was established in the 1970s to divide the epidemic strains into subtypes and distinguish them from environmental non-toxigenic strains (2). Five lytic phages (VP1 to VP5) are included in the phage-typing scheme, and four other biotypes are included in the biotyping scheme. The phage-biotyping scheme has played an important role in cholera surveillance in China since the 1970s (3).
As the largest viral group in nature (4), phages are notable not only for their abundance but also for their specific interaction with specific bacterial hosts (5, 6). The first step of the phage infection process is its recognition of and adsorption onto one or more cell-surface constituents (7), which is followed by ejection of phage DNA into the host cell. In the course of phage–host interactions, phages tend to use easily accessible structures located on the outer membrane of the host and exposed to the environment as receptors (8, 9). As surface structures facing the surrounding environment, lipopolysaccharides (LPS)2 and outer-membrane proteins (OMPs) are the most common components acting as receptors for tailed phages that infect Gram-negative bacteria (10, 11). LPS is the receptor for many phages of the T7 family (12–14). Some phages infect hosts by interacting with the O antigen (13, 15), and others bind to the host through a core oligosaccharide (OS) (14, 16). OMPs forming trans-membrane channels (17–19) are important components involved in phage infection, and they include examples such as OmpA, OmpW, LamB, and OmpC (20–25). For some phages, LPS and OMPs are both necessary for binding to the host cell surface (26–31).
In phage–host interactions, host receptor mutations can confer phage resistance to the bacteria and improve survival in environments with phage. This bacteria–phage competition is relevant to ecological niches, food industries, and even phage therapy. Identification of host receptors allows us to understand the host specificity of a phage and the genetic basis of phage typing. Among the typing phages in the phage-biotyping scheme for the V. cholerae El Tor strains, our previous studies have identified the O antigen portion of LPS as receptor for VP4 (15) and OmpW as receptor for VP5 (24). The typing phage VP3 is a lytic V. cholerae phage belonging to the T7 superfamily (32). VP3 employs the OS component of LPS as a receptor (16), but a core oligosaccharide mutant strain was still bound by VP3 at lower efficiency under high VP3 concentrations. In addition, some wild El Tor V. cholerae strains have intact OS components but are resistant to VP3. These findings raise the question whether additional components on the V. cholerae cell surface play a role in VP3 adsorption. In this study, our data show that the successful infection of VP3 requires the outer-membrane protein TolC in addition to the OS of LPS, TolC may interact directly with the tail fiber protein of VP3. Our study reveals a complicated two-receptor adsorption process with the phage of T7 family to V. cholerae.
Results
Outer membrane protein TolC is needed for phage VP3 infection
VP3 is used as a typing phage for the subtype of V. cholerae O1 El Tor strains. Its adsorption on the surface of its host cell was observed by electron microscopy (Fig. 1). The head of VP3 has an icosahedral structure. In our previous study (16), the V. cholerae wav genes VC0229 and VC0231, involved in the synthesis of core oligosaccharides (OS) of lipopolysaccharide, were identified to be related to the resistance to VP3 infection, and OS was recognized as the receptor of VP3. However, we found that some wildtype El Tor strains possessing intact OS gene clusters can bind the tail fiber protein gp44 of VP3 but are still resistant to VP3 by the double-layer plaque assay; additionally, using a phage-binding assay, it was observed that VP3 still bound to the OS gene mutant strain C29 at lower efficiency.3 We thus suspected that other bacterial surface components of V. cholerae might be involved in VP3 infection.
Figure 1.
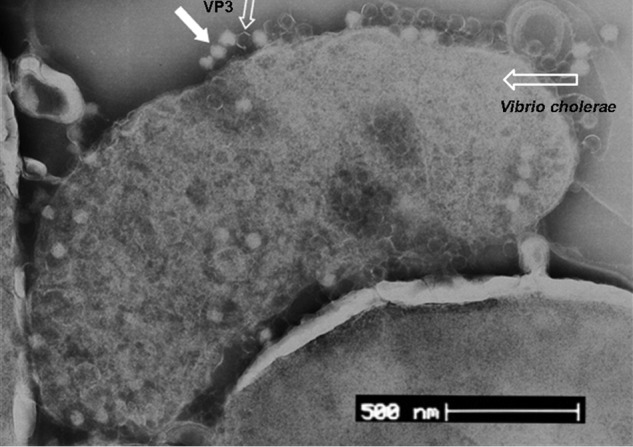
Adsorption of phage VP3 to its host cells observed with electron microscopy. VP3 particles were indicated with the solid arrow and the open arrow (empty particles).
In this study, we newly constructed a larger transposon mutant library to screen the mutants generating resistance to VP3 infection. More than 7,000 transposon insertion mutants of the El Tor strain N16961 were generated with the plasmid pSC123. During the co-incubation of the individual mutant strain with phage VP3, five mutants (24-H5, 61-A10, 17-B3, 26-A5, and 53-D9, see Table 1) showed growth rates comparable with that of the control (strain N16961 with no VP3 added). The sensitivity of these mutants to VP3 was further verified by double-layer plaque assays. Then, the transposon insertion sites in the genomes of these five strains were identified by arbitrary PCR. Five inserted genes other than the previously reported genes (16) were found (Table 1). One mutant (24-H5) had a transposon insertion within gene VC2436, which encodes the outer-membrane protein TolC. In Gram-negative bacteria, TolC forms a conserved, negatively charged entrance for virulence proteins and is also involved in antibiotic efflux to the external environment during infection (33–36). TolC confers Escherichia coli resistance to detergent, bile salts, and organic solvents (37–40) but sensitivity to colicin and phage (31, 41, 42). In V. cholerae, TolC has multiple functions in bile salt resistance, intestinal colonization, and RTX toxin protein secretion (43) and serves as the receptor for T7 and TLS phages (31). Therefore, in this study we focused on the validation of the possible role of TolC in VP3 infection.
Table 1.
Transposon mutants from V. cholerae strain N16961 with VP3 resistance phenotype
| Strains | Description | Gene function |
|---|---|---|
| 24-H5 | N16961 (Smr), VC2436 (tolC)::Tn, Smr Kanr VP3r | Outer membrane protein TolC |
| 61-A10 | N16961 (Smr), VCA0781::Tn, Smr Kanr VP3r | VCA0781 encodes a protein of unknown function with one MacB-PCD and two FtsX domains that were highly homologous to inner membrane protein MacB (ATP-binding-cassette-type efflux transporter) |
| 17-B3 | N16961 (Smr), VC0050::Tn, Smr Kanr VP3r | DNA topoisomerase I-related protein |
| 26-A5 | N16961 (Smr), VC0034::Tn, Smr Kanr VP3r | Disulfide interchange protein |
| 53-D9 | N16961 (Smr), VC0177::Tn, Smr Kanr VP3r | Hypothetical protein |
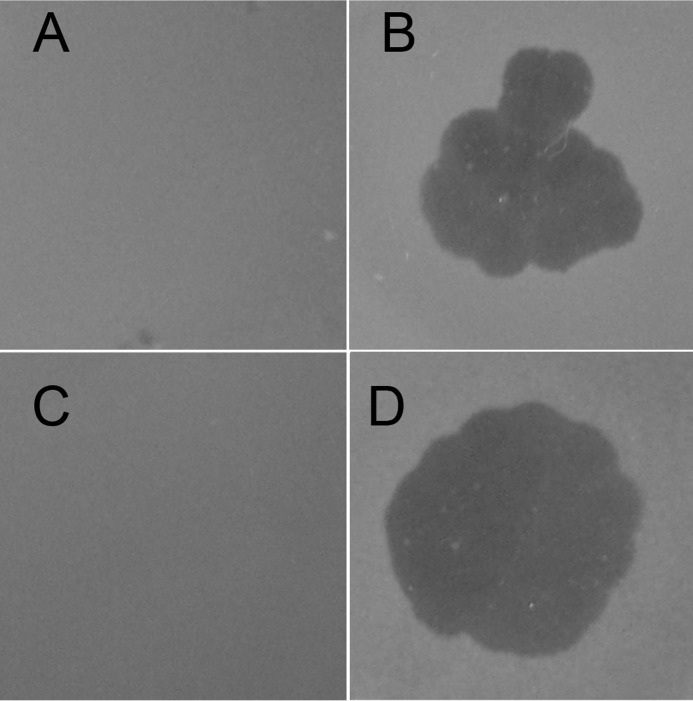
To exclude polar effects exerted by transposon insertion or a spontaneous mutation as a simultaneity and to confirm further the role of tolC during VP3 infection, the mutant strain NΔtolC with an in-frame deletion of tolC was constructed from the wild strain N16961 (Table 2). NΔtolC was resistant to VP3 when detected with the double-layer plaque assay, consistent with the transposon mutant strain 24-H5. When the plasmid pSRKTc-TolC was transformed into NΔtolC, the complementary strain NΔtolC-C re-acquired VP3 sensitivity (Fig. 2), thereby confirming its indispensable role in VP3 infection.
Table 2.
Strains and plasmids used in this study
| Strains or plasmids | Relevant property | Refs. |
|---|---|---|
| V. cholerae | ||
| 2477c | O1 El Tor, Ogawa | Lab collections stock |
| N16961 | Spontaneous mutant of N16961, Inaba, Smr | Lab collections stock |
| ICDC-VC5029 | O1 El Tor, Ogawa, with natural resistance phenotype resistant phenotype control | Lab collections |
| NΔOS | Core OS mutant strain, VC0231 deletion of N16961 (Smr) | 1 |
| NΔtolC | tolC (VC2346) deletion of N16961 (Smr) | This study |
| NΔtolC-OS | tolC and OS deletion of N16961 (Smr) | This study |
| NΔtolC-C | NΔtolC strains were complemented with pSRKTc-TolC | This study |
| NΔOS-C | NΔOS strains were complemented with pBAD33-VC0231 | This study |
| NΔtolC-OS-CtolC | NΔtolC-OS strains were complemented with pSRKTc-TolC | This study |
| NΔtolC-OS-COS ΔΔVC2436ΔVC0231C0231 | NΔtolC-OS strains were complemented with pBAD33-VC0231 | This study |
| NΔtolC-OS-C | NΔtolC-OS strains were complemented with pSRKTc-TolC and pBAD33-VC0231 | This study |
| NΔtolC-CA78D | NΔtolC strains were complemented with pSRKTc-TolCA78D | This study |
| NΔtolC-CΔ290–291 | NΔtolC strains were complemented with pSRKTc-TolCΔ290–291 | This study |
| E. coli | ||
| SM10 λpir | Km, thi thr leu tonA lacY supE recA:: RP4-2-TC::Mu λpir | 2 |
| DH5α λpir | sup E44, ΔlacU169 (ΦlacZΔM15), recA1, endA1, hsdR17, thi-1, gyrA96, relA1, λpir gyrA96, relA1, λpir | Lab collections stock |
| XL1blue | endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac | Lab collections |
| BTH101 | F−, cya-99, araD139, galE15, galK16, rpsL1 (Strr), hsdR2, mcrA1, mcrB1 | 3 |
| Plasmids | ||
| pSC123 | Suicide plasmid carrying transposon; Kanr Cmr | 4 |
| pWM91 | Suicide plasmid; oriR oriT lacZ tetAR sacB | 5 |
| pWM91-ΔtolC | pWM91 carrying upstream and downstream fragments flanking tolC | This study |
| pWM91-ΔVC0231 | pWM91 carrying upstream and downstream fragments flanking VC0231 | This study |
| pUT18C (abbreviated as pT18C) | pUT18C-derived vector, designed to create C-terminal heterologous protein fusion, Ampr | 3 |
| pKT25 (abbreviated as pT25) | lac promoter and the T25 fragment for C-terminal heterologous protein fusion. Kmr | 3 |
| pSRKTc | lac promoter and lacIq, Tetr | 6 |
| pSRKTc-TolC | pSRKTc-derived, VC2436 (TolC), Tetr | This study |
| pSRKTc-TolCA78D | pSRKTc-derived, TolCA78D, Tetr | This study |
| pSRKTc-TolCΔ290–291 | pSRKTc-derived, TolCΔ290–291, Tetr | This study |
| pBAD33-VC0231 | pBAD33-derived, VC0231, Cmr | This study |
| pT18C-TolC | pUT18C-derived, TolC23-end, Ampr | This study |
| pT25-TolC | pKT25-derived, TolC23-end, Kanr | This study |
| pT25-TolCA78D | pKT25-derived, TolCA78D, Kanr | This study |
| pT25-TolC Δ290–291 | pKT25-derived, TolCΔ290–291, Kanr | This study |
| pT25-gp44 | pKT25-derived, gp44, Kanr | This study |
| pT25-gp44(245–753) | pKT25-derived, gp44 (245–753), Kanr | This study |
| pT25-gp44(451–753) | pKT25-derived, gp44 (451–753), Kanr | This study |
| pT25-gp44(1–450) | pKT25-derived, gp44 (1–450), Kanr | This study |
| pT18C-TolCA78D | pUT18C-derived, TolCA78D, Ampr | This study |
| pT18C-TolCΔ290–291 | pUT18C-derived, TolCΔ290–291, Ampr | This study |
| pGEX-6p-1-TolC | pGEX-6p-1-derived, TolC23-end, Ampr | This study |
| pGEX-6p-1-TolCA78D | pGEX-6p-1-derived, TolCA78D, Ampr | This study |
| pGEX-6p-1-TolCΔ290–291 | pGEX-6p-1-derived, TolCΔ290–291, Ampr | This study |
| pET30a-gp44 | pET30a-derived, gp44 (245–753), Kanr | This study |
| pET30a-gp44(451–753) | pET30a-derived, gp44 (451–753), Kanr | This study |
| pET30a-gp44(1–450) | pET30a-derived, gp44 (1–450), Kanr | This study |
Figure 2.
Detection of VP3 infection to the V. cholerae mutants by double-layer plaque assay. A, wildtype V. cholerae El Tor strain ICDC-VC5029, which has natural VP3 resistance, was used as the resistance control (no plaque formation). B, VP3-sensitive wildtype strain N16961 was used as VP3-sensitive control with plaque formation. C, tolC mutant strain NΔtolC showed VP3 resistance. D, strain NΔtolC-C, carrying tolC expression plasmid cloned into strain NΔtolC, was sensitive to VP3.
Both TolC and OS of V. cholerae are necessary for host cell adsorption of VP3
In our previous study, deletion of the OS gene of V. cholerae strain N16961 may effectively block the binding and infection by VP3 (16). However, when the OS gene VC0231 deletion mutant (renamed as NΔOS in this study) was mixed with a high concentration of VP3 (109 cfu/ml, 10 times higher than in the previous study (16), with a multiplicity of infection >10), VP3 could still bind to NΔOS to some extent (Fig. 3). To validate the possible joint roles of TolC and OS in VP3 infection, a double-mutant strain NΔtolC-OS and complementary plasmids carrying gene tolC, VC0231, and both genes, respectively, were constructed (Table 2). The transcription of plasmid-carrying genes tolC and VC0231 in all the complementary V. cholerae strains was confirmed by reverse transcription-PCR. The sensitivity of each strain to VP3 was detected through growth rate and double-layer plaque assays (Table 3). All gene mutants, including NΔtolC, NΔOS, and NΔtolC-OS, had VP3-resistant phenotypes with no plaque formation and normal growth, whereas strains NΔtolC-C, NΔOS-C, and NΔtolC-OS-C supplemented with the corresponding recombinant plasmids were all VP3-sensitive (Table 3). When either single gene, tolC or VC0231, was replenished into the double mutant NΔtolC-OS, the resulting strains NΔtolC-OS-CtolC and NΔtolC-OS-COS were still resistant to VP3 (Table 3), showing that deletion of either gene of VC0231 or tolC conferred VP3 resistance to N16961, and both the VC0231 product and TolC would be necessary for VP3 infection.
Figure 3.
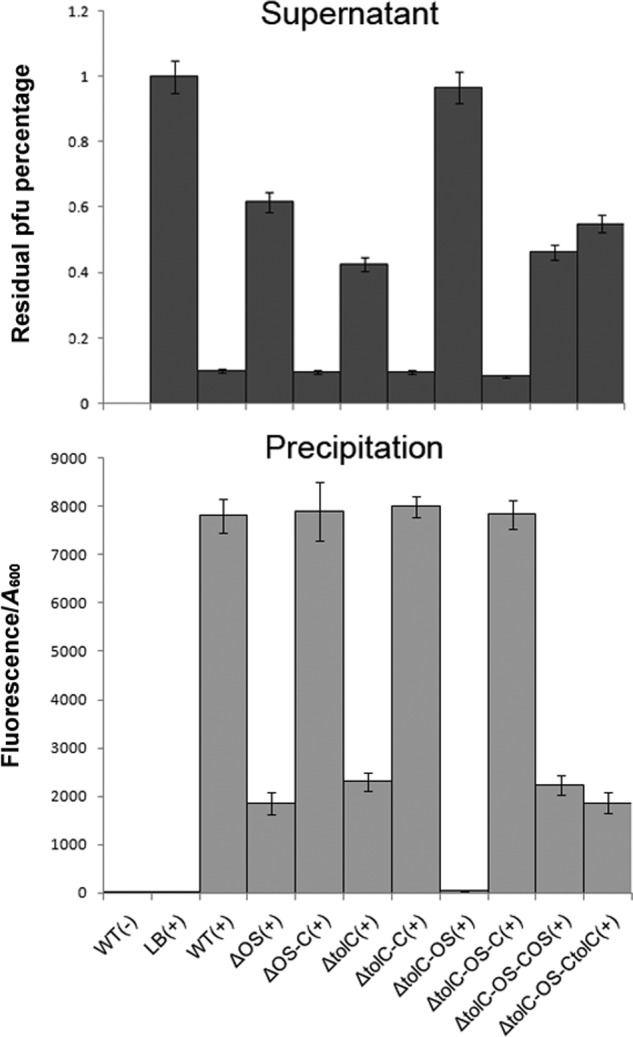
Analysis of the binding capacity between isolates and VP3 phage. The VP3 phage (109 cfu/ml) was labeled with SYBR Gold and mixed with fresh N16961 culture (A600 = 0.1) in a 1:1 (v/v) ratio for 5 min, and each sample was centrifuged at 4,000 rpm for 5 min. The remaining bacterial precipitation was resuspended in 200 μl of SM, and the fluorescence value was determined at 490 nm excitation/537 nm emission. Binding capacity between the strain and VP3 is measured via total fluorescence value/A600. The remaining phage titer in the supernatant of each sample was determined using a double-layer plaque assay. LB culture medium containing only wildtype strain or VP3 phage was used as a negative control, and the phage titer in the control supernatant was set to 100%. Error bars indicate ranges. +, VP3 present; −, VP3 not present, the detailed information of strains in Table 2.
Table 3.
Sensitivity of strains to VP3 detected by double-layer plaque assay
The following symbols and abbreviation were used: +, indicates strain with no plaque formation confirmed as VP3-resistant phenotype; −, indicates strain with plaque formation confirmed as VP3-sensitive phenotype; PC, N16961 (wildtype) without VP3 used as positive control (PC); NC, N16961 (wildtype) with VP3 used as negative control (NC).
| Strains | Growth rate (A600) | Double-layer plaque assay | Strains | Growth rate (A600) | Double-layer plaque assay |
|---|---|---|---|---|---|
| PC | 0.880 | + | NΔtolC-OS | 0.921 | + |
| NC | 0.021 | − | NΔtolC-OS-C | 0.017 | − |
| NΔOS | 0.889 | + | NΔtolC-OS-CtolC | 0.888 | + |
| NΔOS-C | 0.016 | − | NΔtolC-OS-COS | 0.904 | + |
| NΔtolC | 0.865 | + | NΔtolC-C | 0.021 | − |
Furthermore, phage-binding assays were performed to detect the binding ability of VP3 to the cell surface of various mutants and their corresponding complementary strains. After a short incubation of bacterial cells with SYBR Gold-labeled VP3, fluorescence values were measured in precipitated cells resuspended in SM buffer, and the remaining phage titer in the supernatant of each sample was subsequently determined by the plaque formation unit assay. Inverse correlations were observed with the phage titer in the supernatant of each sample and the fluorescence in the corresponding cell precipitate (Fig. 3). VP3 exhibited high adsorption to wildtype strain N16961, and most phages were removed from the suspension by binding to N16961. The complementary strains NΔtolC-C and NΔOS-C showed VP3 binding comparable with N16961, The double mutant NΔtolC-OS had no binding to VP3, whereas partial binding ability to VP3 was found for the single-gene mutants NΔtolC and NΔOS, Single gene complementation for NΔtolC-OS, referred to as NΔtolC-OS-CtolC or NΔtolC-OS-COS, only partially rescued adsorption to VP3. The double-gene complementary strain NΔtolC-OS-C behaved similarly to N16961 (Fig. 3). All these data indicated that both TolC and VC0231 are required for full adsorption of VP3.
TolC interacts directly with the C-terminal domains of tail protein gp44 of VP3
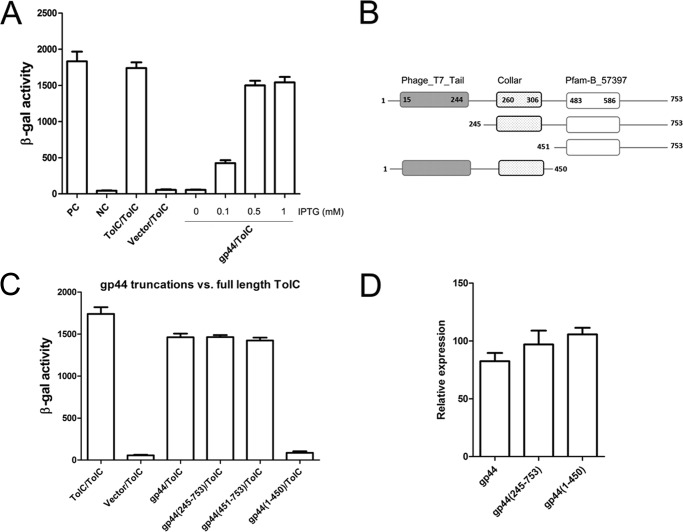
The phage VP3 of V. cholerae belongs to the T7 family. The T7 tail protein anchors phage onto its host cell surface (44, 45), and VP3 adsorbs to the OS of LPS through its tail fiber protein gp44 (16). We suspected that gp44 may also interact with TolC, and as such utilized the bacterial adenylate cyclase two-hybrid (BACTH) system (46) to detect the possible interaction between TolC and gp44. Previously, BACTH has been used to determine the interactions of TcpP–ToxR (47) and TcpP–TcpP in response to bile salt signals (48). In this study, we first fused the complementary fragments, T25 and T18, of the catalytic domain of adenylate cyclase (CyaA) from Bordetella pertussis (46, 49), with TolC lacking the signal peptide (22 amino acid residues in length) and with gp44, respectively, to generate the recombinant plasmids pT25-gp44 and pT18C-TolC (Table 2). Both plasmids were co-transformed into an E. coli Δcya mutant strain BTH101, and the β-galactosidase activities were measured in the presence of different concentrations of isopropyl β-d-thiogalactopyranoside (IPTG) as the inducer. The interaction between TolC and gp44 will bring the two Cya fragments together, generate cAMP, and increase β-galactosidase activity. It has been demonstrated that TolC can interact with itself to form polymers in V. cholerae (35). In this study, we used pT25-TolC and pT18C-TolC-transformed BTH101 as a positive control in addition to the leucine zipper of GCN4 (46) as a second positive control (Fig. 4A). Our results showed that gp44 could interact with TolC and that the interaction was IPTG concentration-dependent: 0.5 mm IPTG induced the maximum β-galactosidase activity, similar to both positive controls (Fig. 4A).
Figure 4.
Detection of the interaction between TolC and gp44 or its derivatives by BACTH. A, analysis of gp44–TolC interaction by BACTH. PC, positive control (leucine zipper of GCN4); NC, negative control (vector plasmid only); TolC/TolC, positive control (trimeric protein from V. cholerae); vector/TolC, negative control (vector pT18C and pT25-TolC). B, motif illustration of gp44 protein. Boxes represent fragments, and the numbers on the fragment denote the amino acid positions. C, C-terminal domains of gp44 have significant interactions with TolC. The plasmids pT25-TolC and pT18C-gp44 or their derivatives with truncations of gp44 were co-transformed into BTH101 separately and incubated to log phase at 37 °C, and β-galactosidase activity was measured. D, qRT-PCR assays of transcriptions of gp44 and its truncations in BTH101. Strains BTH101 (pT25-TolC and pT18C-gp44), BTH101 (pT25-TolC and pT18C-gp44(245–753)), and BTH101 (pT25-TolC and pT18C-gp44(1–450)) were grown in LB medium in the presence of 1 mm IPTG and incubated to log phase at 37 °C, and total RNA was extracted and determined by qRT-PCR. Each value is the average of three independent cultures. Error bars, standard deviations.
Three domains for gp44 can be predicted by searching the Pfam database, including N-terminal phage_T7_tail domain (15–244 aa), Collar domain (260–306 aa), and Pfam-B_57397 (483–586 aa) following the nomenclature in Pfam (Fig. 4B). (16) The N-terminal domain of gp44 of phage VP3 shows high similarity to those of the T7 tail fiber protein gp17 and the other tail fiber proteins of T7-like phages such as T3, YeO3012, and gh-1 (16), whereas the homology of their C-terminal domains is notably low (16, 50). To evaluate the gp44 domain(s) interacting with TolC, three fragments spanning different regions of gp44 were constructed in pT25 and co-transformed with pT18C-TolC into BTH101, and the interaction between TolC and each gp44 derivative was measured by the β-galactosidase activity (Fig. 4C). The gp44(245–753) and gp44(451–753) fragments showed significant interactions with TolC, whereas gp44(1–450), which includes two domains corresponding to the N-terminal phage_T7_tail and Collar domains (Fig. 4B), did not interact with TolC. To determine whether the low β-galactosidase activity in the gp44(1–450)/TolC pair was due to lower expression of gp44(1–450), we compared the transcript levels of gp44(1–450), gp44, and gp44(245–753) in BTH101 using qRT-PCR, and no obvious transcription differences were observed for these three genes (Fig. 4D). Thus, it could be deduced that the interaction domains of gp44 with TolC are located in the C-terminal domain of gp44 and that deletion of N-terminal domain does not affect the interaction of gp44 with TolC.
To further confirm the BACTH assay results, GST pulldown experiments were performed with GST-tagged TolC immobilized on glutathione-Sepharose resin and His-tagged truncations of gp44. The final samples from pulldown assays were separated on two SDS-polyacrylamide gels and transferred onto PVDF membranes, where anti-GST and anti-His monoclonal antibodies were used for protein detection. The results showed that both full-length gp44 and gp44(451–753) interacted with TolC, whereas no interaction was observed between gp44(1–450) and TolC (Fig. 5), which were consistent with the corresponding BACTH results, and indicated that gp44 interacts with TolC directly, and the C-terminal fragment gp44 (451–753) is necessary for interaction with TolC.
Figure 5.
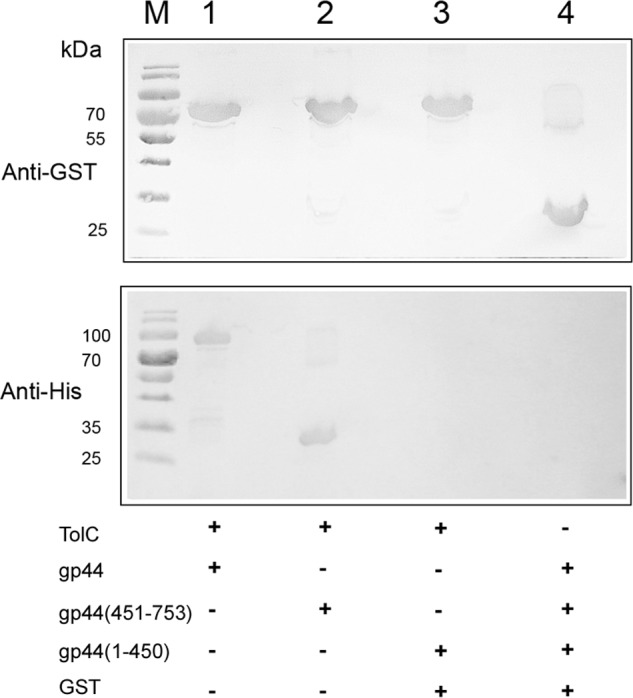
Analysis of the interactions of TolC with gp44 in vitro. Western blot analyses of GST pulldown experiments were performed with GST-TolC immobilized on glutathione-Sepharose resin and His-tagged protein extracts of full-length gp44 (lane 1), gp44(451–753) (lane 2), and gp44(1–450) (lane 3); the protein mixture (full-length gp44, gp44(451–753) and gp44(1–450)) described above was incubated with GST for pulldown analysis as a negative control (lane 4). Lane M indicates mass.
Ala-78 and Gly-290–Glu-291 sites of TolC are required for the interaction with gp44
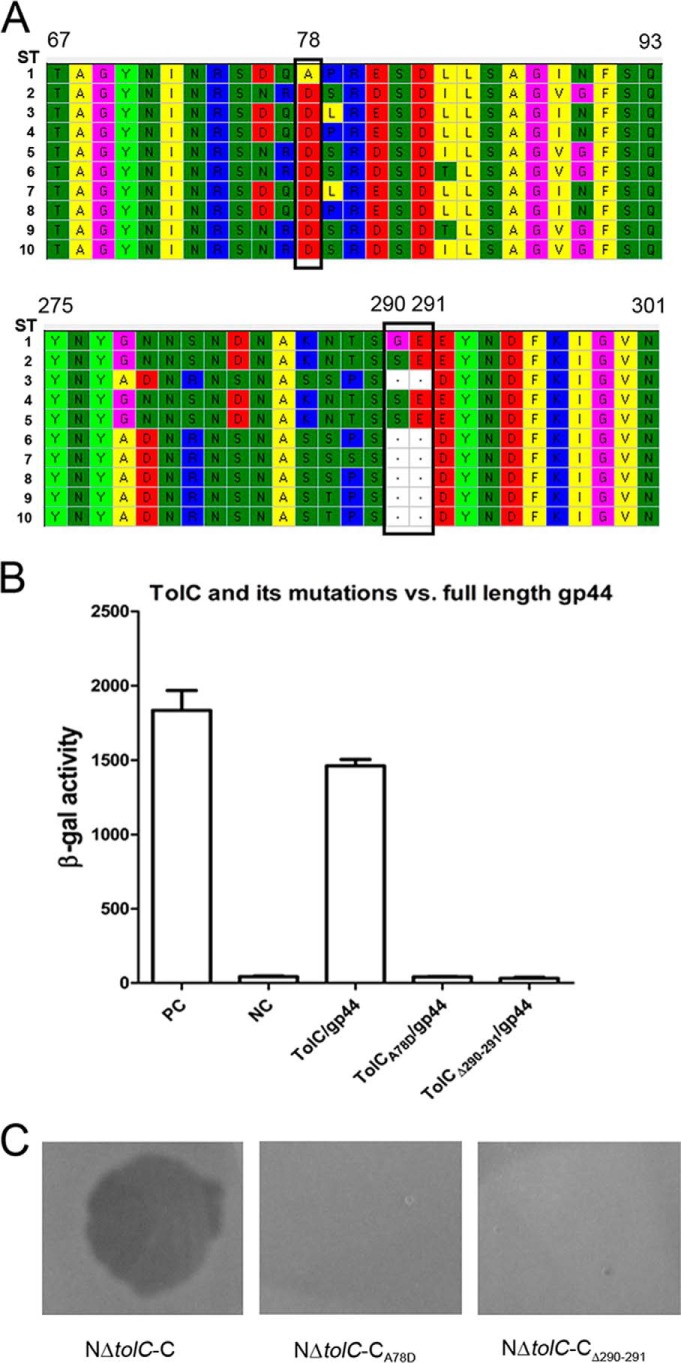
To determine the possible amino acid residues of TolC, which may have roles in its interaction to gp44, we first sequenced the tolC genes of the O1 El Tor VP3-sensitive and VP3-resistant strains to search the commonly different amino acid residues between both groups of strains. Here, 192 strains were selected, including 108 VP3-sensitive and 84 VP3-resistant strains. Of these, the genomes of 71 strains were previously sequenced (51), and the tolC sequences were retrieved directly from their genome sequence data, and the tolC sequences of remaining strains were amplified by PCR and sequenced. Ten TolC amino acid sequence types of these strains were obtained by alignment (Fig. 6A). Within them, all VP3-sensitive strains had the identical TolC sequences, as shown in sequence type (ST) no. 1. However, 96% (81/84) of the VP3-resistant strains had TolC amino acid variations compared with the VP3-sensitive strains. These variations clustered in the 76–90 and 278–291 regions (Fig. 6A). Among them, the common differences between the sensitive and resistant strains were the amino acid alterations A78D, G290S, or a Gly-290–Glu-291 deletion located in these two hypervariable regions (Fig. 6A).
Figure 6.
Sequence alignment of TolC of the VP3-sensitive and -resistant strains and the interaction analysis between gp44 and TolC or its amino acid site mutants. A, all the TolC amino acid sequence fragments of the VP3-sensitive and VP3-resistant strains containing the mutation sites were aligned using MEGA, 10 sequence types (ST1 to ST10) were found. All the VP3-sensitive strains have one sequence type, ST1. All the VP3-resistant strains belong to ST2–ST9 with the amino acid residue mutations. The common mutant residues in the VP3-resistant strains compared with the VP3-sensitive strains are marked with black boxes. The numbers of the amino acid residue sites are shown based on the sequence of ST1. B, BACTH assays of the interactions between gp44 and TolC or its mutants, including the mutation on A78D and the deletion of Gly-290–Glu-291. C, double-layer plaque assays were performed to detect the roles of the TolC protein mutants for the VP3 infection. The lysis plaque was observed when VP3 was added in the culture layer of strain NΔtolC-C but absent in the culture layers of the strains NΔtolC-CA78D or NΔtolC-CΔ290–291.
Then two plasmids, pT18C-TolCA78D and pT18C-TolCΔ290–291 (Table 2), were constructed to change the 78th amino acid Ala to Asp and to delete the Gly-290–Glu-291 of TolC from a VP3-sensitive strain. Each plasmid was co-transformed into BTH101 with pT25-gp44. Very weak β-galactosidase activity was detected for both mutations (Fig. 6B), suggesting that alteration of either site disabled the interaction between TolC and gp44. The recombinant plasmids pSRKTc-TolCA78D and pSRKTc-TolCΔ290–291 (Table 2) were constructed and transformed into strain NΔtolC, and the sensitivities of the resulting strains NΔtolC-CA78D and NΔtolC-CΔ290–291to VP3 were not restored when observed with the double-layer plaque assay (Fig. 6C).
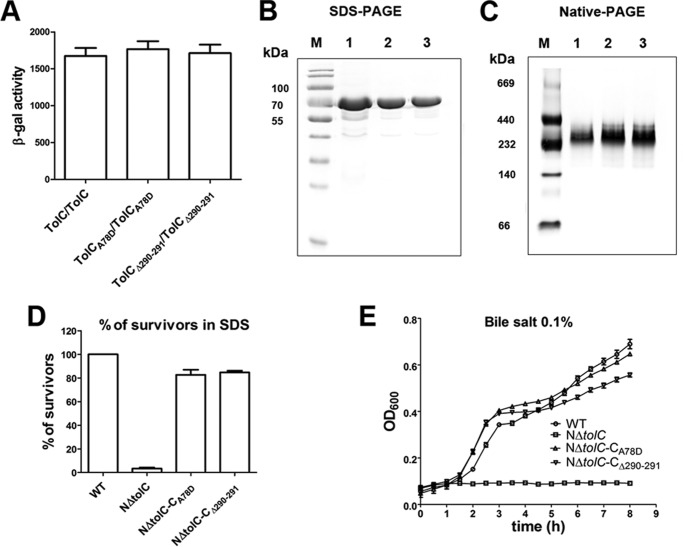
To detect whether the A78D mutation or Gly-290–Glu-291 deletion of TolC possibly impedes the TolC trimerization, we first evaluated the homotypic oligomerization of TolCA78D and TolCΔ290–291 with BACTH. The constructed plasmid pair pT25-TolCA78D/pT18C-TolCA78D or pT25-TolCΔ290–291/pT18C-TolCΔ290–291 was transformed, respectively, into BTH101, and their β-galactosidase activities were as high as the positive control pT25-TolC/pT18C-TolC (Fig. 7A), suggesting that TolCA78D and TolCΔ290–291 could still form oligomers. Second, SDS-PAGE and blue native-PAGE (52) were performed to detect the mass and oligomeric states of the proteins. The plasmids pEGX-6p-1-TolCA78D and pEGX-6p-1-TolCΔ290–291 were constructed and overexpressed in the E. coli strain BL21(DE3). TolCA78D-GST and TolCΔ290–291-GST were purified and analyzed by SDS-PAGE and native-PAGE. Both TolCA78D-GST and TolCΔ290–291-GST showed the same molecular weight as TolC-GST by SDS-PAGE (∼74 kDa, Fig. 7B). On the native-PAGE, all proteins migrated close to 240 kDa (Fig. 7C), showing that TolCA78D or TolCΔ290–291 could form polymers similar to the TolC control.
Figure 7.
Analysis of the mass, oligomeric states, and the physiological function of TolCA78D or TolCΔ290–291. A, BACTH assays of the trimerization of each TolC mutant protein with A78D and Gly-290–Glu-291 deletion, respectively. The resulting recombinant plasmid pair pT25-TolCA78D/pT18C-TolCA78D or pT25-TolCΔ290–291/pT18C-TolCΔ290–291 was co-transformed into BTH101, and the β-galactosidase activity was measured. TolC/TolC was used as the positive control. B, TolC-GST, TolCA78D-GST, and TolCΔ290–291-GST protein were separated by SDS-PAGE. A 74-kDa band representing the single subunit of TolC is shown. C, protein samples were separated by native-PAGE (4–15%, Solarbio). A 240-kDa band showing the trimer of TolC is shown. Lane M indicates mass. D, analysis of the SDS sensitivity of strains expressing TolCA78D or TolCΔ290–291 proteins. The cfu of each sample was normalized by cfu counting without SDS. E, growth kinetics of wildtype strain N16961 (WT), NΔtolC, NΔtolC-CA78D, and NΔtolC-CΔ290–291 in the presence of 0.1% bile salt.
Third, the sensitivity of the mutant V. cholerae strains encoding TolCA78D and TolCΔ290–291 against bile acids and SDS was examined as well. Growth of the strains NΔtolC-CA78D and NΔtolC-CΔ290–291 was assessed in LB media supplemented with 0.1% SDS or bile salt. Compared with the wildtype strain, NΔtolC exhibited hardly any growth in LB with SDS, whereas the growth of both strains NΔtolC-CA78D and NΔtolC-CΔ290–291was ∼80% survivors (Fig. 7D). In LB supplemented with bile salt, growth of NΔtolC was inhibited, but NΔtolC-CA78D and NΔtolC-CΔ290–291 grew vigorously, similar to the wildtype strain (Fig. 7E). These data showed that the mutations of Ala-78 or Gly-290–Glu-291 do not influence the oligomeric state of TolC. Both sites are necessary for the interaction of TolC with gp44 and VP3 infection.
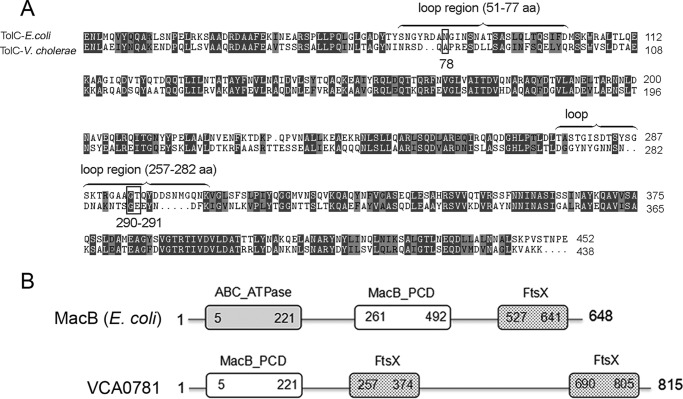
Sequence alignment showed that TolC of V. cholerae has 46% identity and 77% similarity with the TolC of E. coli (Fig. 8A) (53). The result showed that the corresponding sites of Ala-78 and Gly-290–Glu-291 all locate to the loop regions of TolC that are exposed to the cell surface.
Figure 8.
Sequence alignments of TolC of E. coli and V. cholerae and the predicted motifs of VCA0781. A, comparison of TolC sequence of V. cholerae and E. coli using program Clustal Omega. Identical amino acids are labeled in dark gray; similar amino acids are colored in light gray. The locations of the two loops of TolC (E. coli) are labeled with a brace. Sites Ala-78 and Gly-290–Glu-291 of TolC (V. cholerae) are marked with black box. B, predicted domains of VCA0781 based on the comparison with MacB of E. coli. The domains of VCA0781 are illustrated with the square frames, and the start and end amino acid positions are marked.
Discussion
Specific adsorption mediated by a receptor–ligand pair is the first step in phage infection. Bacterial envelope components such as LPS and OMP are the commonly recognized receptors for phages (54, 55). Many phages in the T7 family adsorb LPS for their infection (12–14), but for the Vibrio phage VP3, OS deficiency did not cause complete loss of VP3 adsorption. In this study, we identified TolC as another receptor for VP3 that mediates its nearly complete adsorption to V. cholerae O1 El Tor strains with OS, revealing a co-receptor–binding mechanism for Vibrio phage.
Our quantitative adsorption tests showed that in addition to the previously reported receptor OS (16), TolC is required for the complete binding of VP3 with V. cholerae. Mutation of either gene reduced binding capacity with VP3, and a double mutant completely abrogates VP3 binding. Co-receptors required for infection are also found in some members of the T7 family and other phages (26–29). In Yersinia pestis, LPS and OMPs (Ail and OmpF) are necessary for the infection of Yep-φ, which is used in the typing and identification of Y. pestis strains (30). Binding to LPS and TolC of E. coli is two separate but necessary events that function independently during TLS phage infection (31). The binding of the tail fiber of T7 and LPS is weak and reversible, perhaps allowing the phage to travel over the cell surface without dissociation (56). For some phages, binding is followed by conformational changes in the tail fiber (57, 58); interaction between phage and host initiates the injection of DNA into the host (59) through a channel formed by phage proteins (56, 57, 60, 61) or host-derived proteins (62, 63). We suspect that the binding of tail fiber protein gp44 to TolC uses the same strategy, forming a channel composed of host and phage proteins to transport phage DNA into the host cell.
We showed that VP3 bound to the surface of V. cholerae through the C-terminal region of gp44. The tail protein N terminus anchors the entire tail protein to the head of the phage particle (44), and the C-terminal domain determines the host range for T7 phage (45). The low homology of the tail protein C terminus with the T7 family may be explained by the diversity of receptors on different host cells. We suspect that the sequential binding to OS and TolC by different domains of gp44 mediates the successful infection of VP3. When either OS or TolC is deficient, the fibers of VP3 can bind only one site, and this weakens host cell surface adsorption efficiency. However, the precise structure and the dynamic process of gp44 binding to OS and TolC should be further studied.
Sequence alignment results showed that two highly variable regions of V. cholerae TolC, 76–90 and 278–291 aa, are composed of β-sheets located at the trans-membrane domain and at a neighbor-loop region that is cell-surface–exposed. Ala-78 and Gly-290–Glu-291 are located at these two exposed loop regions that are cell-surface–exposed. The surface distribution of these sites suggests a role in binding with gp44 of VP3.
Ala-78 and Gly-290–Glu-291 of TolC are critical for its interaction with the tail protein gp44 of VP3. The alanine at position 78 is a non-polar hydrophobic amino acid located in the 76–90 loop region of TolC, and it is changed to polar, negatively charged aspartic acid in most VP3-resistant strains. Another common mutation site is the 278–291 region of TolC, where the simplest amino acid Gly-290 is mutated to serine, or Gly-290–Glu-291 is deleted in the resistant strains. These mutations prevent TolC from binding to gp44 and confer strains with resistance to VP3 infection. The surface-exposed loop sequences of many outer-membrane proteins involved in phage infection, including TolC (31), OmpF (27), and FhuA (64), show little conservation. Bacteria may thus undergo conversion from phage-sensitive to phage-resistant via mutation of the loop region of the receptor protein, and this may help them to survive in the environment where the phage is present.
In addition to tolC, in this study, we identified four additional genes that are possibly related to VP3 resistance (Table 1). The hypothetical protein VCA0781 has two conserved domains (MacB-PCD and FtsX) that are similar to those of the E. coli inner membrane protein MacB (Fig. 8B). Many channels on bacterial membranes depend on TolC to form ABC-type transporters (38, 65, 66), and the macrolide transporter MacA–MacB–TolC was identified as the first ABC-type transporter that actively extrudes substrates, including macrolide antibiotics and polypeptide virulence factors (67, 68). Therefore VCA0781 probably localizes to the inner membrane, and together with TolC and a periplasmic protein they play a role in VP3 phage DNA injection or in the release of new phage particles from the cytoplasm. VC0050 encodes a DNA topoisomerase I-related protein, which is probably involved in the proper resolution of certain DNA structures (69, 70). We predicted that VC0050 may affect the assembly and formation of VP3 phage particles by destabilizing DNA. The product of VC0034 is predicted to be a disulfide isomerase involved in the formation of intra-protein disulfide bonds, and it may influence tertiary protein structure. The function of VC0177 is unknown. All of these four genes were preliminarily identified by transposon mutagenesis and were selected for resistance to VP3 infection. Further research is warranted to confirm the gene's roles in VP3 resistance.
In summary, we have identified a second receptor for phage VP3 and revealed the co-receptor manner for VP3 infection to V. cholerae O1 El Tor strains. Some wild VP3-resistant V. cholerae strains possessing mutations in the phage-binding regions of TolC cause resistance to VP3 infection. Our findings may help to understand the infection and anti-infection mechanisms of V. cholerae to its phage and the receptor recognition of the T7 phage family.
Materials and methods
Bacterial strains, phage, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are summarized in Table 2. VP3 phage was propagated on host strain 2477c as described in a previous study (16). The phage titers were determined by double-layer plaque assay (71). N16961 is resistant to streptomycin (Sm) and sensitive to VP3 (16). These experiments were used in conjugation tests and distinguished from E. coli SM10 λpir by its resistance to Sm. Unless otherwise stated, all strains were grown at 37 °C in Luria broth (LB) medium or on LB medium plates with 15 g/liter agar. Antibiotics were used at the following concentrations: ampicillin (Amp), 100 μg/ml; Sm, 100 μg/ml; kanamycin (Kan), 50 μg/ml. For E. coli, we used the following: chloramphenicol (Cm), 30 μg/ml; tetracycline, 10 μg/ml. For V. cholerae, we used the following: chloramphenicol, 2 μg/ml; tetracycline, 2 μg/ml.
Transmission electron microscopy
To observe the interaction of VP3 and V. cholerae, the phage suspension was mixed with an equal volume culture of its host strain 2477c and incubated for 5 min at 37 °C. Samples were adsorbed for 1 min onto a Formvar film on a carbon-coated 200 mesh copper grid. The adsorbed samples were washed three times in distilled water and negatively contrasted with 2% uranyl acetate (EMS, Hatfield, PA). Imaging was performed using a Tecnai 12 transmission electron microscope (FEI Co., Hillsboro, OR).
Double-layer plaque assay
The assay was performed as described previously (71). Briefly, 4 ml of 50 °C melted 0.7% LB agar was mixed with 100 μl of cell cultures and poured onto an LB agar plate, and 10 μl of VP3 was dropped onto the plate when the upper layer solidified. After overnight incubation at 37 °C, plaque formation indicates that the strain is sensitive to VP3.
Construction of a transposon insertion library and selection of VP3-resistant mutants
Plasmid pSC123 (72) was transformed into E. coli SM10 λpir (73) to obtain SM10-123 (Cmr, Kanr). Conjugation was performed between the recipient strain N16961 and the donor strain SM10-123 according to previously published protocols (15). Trans-conjugants were selected from LB agar plates (Kan, 50 μg/ml, and Sm, 100 μg/ml) and incubated in 96-well plates (Corning Costar 3599) until the absorbance at 600 nm (A600) reached 0.5 to 0.6. Then, 5 μl of culture was inoculated into 140 μl of LB with phage VP3 (1 × 109 pfu/ml) in new 96-well plates and incubated for 5 h. Cultures of strain N16961 (Smr) with and without VP3 were used as negative and positive controls, respectively. Wells with A600 significantly higher than the negative control and nearly as high as the positive control were selected as candidates for phage-resistant mutants. These candidates were subsequently tested using a double-layer plaque assay.
Arbitrary PCR (74, 75) was performed with two rounds of amplification to identify the transposon insertion site. Primers ARB-1, ARB-6, and 123-1 (Table 4) were used in the first round, and chromosomal DNA extracted from each mutant was used as a template. The PCR was performed as follows: 95 °C for 5 min; six cycles of 94 °C for 30 s, 30 °C for 30 s, and 72 °C for 1 min; 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; and finally 72 °C for 5 min. In the second round of arbitrary PCR, the PCR product from the first round was used as the template, and 123-2 and ARB-2 primers (Table 4) were used. The PCR assay was performed under the following conditions: 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, followed by 72 °C for 5 min. Amplicons were sequenced using the 123-2 primer.
Table 4.
Oligonucleotide primers used in this study
| Oligonucleotides | Sequence (5′–3′)a,b | Used in plasmid or reaction |
|---|---|---|
| VC2436-UP-Spe1-5′ | GG ACTAGTCCTCTCCGACCTCCGGCTGC | pWM91-ΔtolC |
| VC2436-UP-3′ | AAGCAGTTTTTTCATCGGTCC | |
| VC2436-DOWN-5′ | CCGATGAAAAAACTGCTTGTCGCGAAGAAGTAATCCATC | |
| VC2436-DOWN-XhoI-3′ | CCGCTCGAGACGCTATCGTGGTGCATCGCTG | |
| 123-1 | TCACCAACTGGTCCACCTAC | Primers for arbitrary PCR to identify transposon insertion sites |
| 123-2 | CGCTCTTGAAGGGAACTATG | |
| ARB1 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT | |
| ARB6 | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC | |
| ARB2 | GGCCACGCGTCGACTAGTAC | |
| TolC-NdeI-R | GCC CATATGAAAAAACTGCTTCCATTATTTG | pSRKTc-TolC |
| TolC-SpeI-F | CCG ACTAGTTTACTTCTTCGCGACTTTTAG | |
| VC0231-XbaI-F | GC TCTAGAAGGAGGAAAATC ATGTTCCTAATTATGTCT | pBAD33-VC0231 |
| VC0231-PstI-R | AA CTGCAGTGATCAAGCCTCAGTAAAATTTG | |
| VC0231-UP-BamHI-5′ | CGG GGATCCTTCTCGGCTGTCTGAATC | pWM91-ΔVC0231 |
| VC0231-UP-3′ | GATGATCGTCGCCGCCAA | |
| VC0231-DOWN-5′ | TTGGCGGCGACGATCATCAGCGACTTGATGCTGAAA | |
| VC0231- DOWN-XbaI-3′ | TCC TCTAGATGGTTTTCTGATGCTCAA | |
| TolC-KpnI-R | CGG GGTACCTTACTTCTTCGCGACTTTTAG | pT18C-TolC or pT25-TolC |
| TolC-XbaI-F | CGC TCTAGAGATGAAAAAACTGCTTCC | |
| gp44-XbaI-F | CGC TCTAGAGATGTCAGGCACTCGTGCTCC ATGTCAGGCACTCGTGCTCC | pT25-gp44 |
| gp44-BamHI-R | CCG GGATCCTTAATTTAAAGGGATAGTCC | |
| gp44245-XbaI-F | CGC TCTAGAG GTTAAAGACCTTGGTGCTG | pT25-gp44 (245–753) |
| gp44451-XbaI-F | CGC TCTAGAG CACATGACGGTCGTCTATG | pT25-gp44 (451–753) |
| gp44(1–450)-BamHI-R | CCG GGATCCGTTTAACTTCAAGAGCACTAC | pT25-gp44 (1–450) |
| TolCΔ290–291-F | CAATGCGAAAAACACTTCA GAGTACAACGATTTCAAAATC | pSRKTc-TolCΔ290–291 |
| TolCΔ290–291-R | GATTTTGAAATCGTTGTACTC TGAAGTGTTTTTCGCATTG | pT18C-TolCΔ290–291 |
| pT25-TolCΔ290–291 | ||
| TolCA78D-F | CCGCAGTGATCAA[b]GACCCACGCGAAAGTGATC | pSRKTc-TolCA78D |
| TolCA78D-R | GATCACTTTCGCGTGG[b]GTCTTGATCACTGCGG | pT18C-TolCA78D |
| pT25-TolCA78D | ||
| TolC-BamHI-F | CGCGGATCCGAAAACCTGGCAGAGATTTATAACC | pGEX-6p-1-TolC |
| TolC-XhoHI-3T-R | CCGCTCGAGTTACTTCTTCGCGACTTTTAGG | |
| gp44-NdeI-F | GCTCATATGATGTCAGGCACTCGTGCTCC | pET-30a-gp44 |
| gp44-XhoI-R | CCGCTCGAGATTTAAAGGGATAGTCCAGTTG | |
| gp44451-NdeI-F | GCTCATATGCACATGACGGTCGTCTATGCG | pET-30a-gp44 (451–753) |
| gp44(1–450)-XhoI-R | CCGCTCGAGTTTAACTTCAAGAGCACTAC | pET-30a-gp44 (1–450) |
| Amp-F | ATGAGTATTCAACATTTCCG | RT-PCR |
| Amp-R | CAAGTCATTCTGAGAATAGTG | |
| gp(44–350)-F | ATGGAAATCGGTGGTGAGTTC | RT-PCR |
| gp(44–450)-R | CTTCAAGAGCACTACCAATC |
a Restriction sites are underlined.
b Bold letters indicate amino acid change.
Construction of mutants and complementation experiments
An in-frame tolC (VC2436) mutant of N16961 was constructed by homologous recombination using the suicide plasmid pWM91 (76). The 1-kb flanking regions upstream and downstream of tolC gene were amplified by PCR from N16961 chromosomal DNA using the primers pairs VC2436-UP-SpeI-5′/VC2436-UP-3′ and VC2436-DOWN-5′/VC2436-DOWN-XhoI-3′ (Table 4), respectively. The two amplicons overlapped and were used as templates to generate the full fragment using the primers VC2436-UP-SpeI-5′/VC2436-DOWN-XhoI-3′. The fragment was digested with SpeI/XhoI and cloned into pWM91, generating the plasmid pWM91-ΔtolC, which was conjugally transferred into N16961 from the donor strain E. coli SM10 λpir. Trans-conjugants were selected on LB agar (Amp, 100 μg/ml, and Sm, 100 μg/ml) and re-streaked onto LB agar with 10% sucrose and without NaCl at 22 °C. Colonies from the sucrose selection medium that failed to grow on LB agar plates with Amp (100 μg/ml) indicated that the suicide plasmid was absent and a double crossover had occurred. Clones were amplified with the primers VC2436-UP-SpeI-5′/VC2436-DOWN-XhoI-3′ (Table 4), producing amplicons of 2 kb, ∼1.3 kb shorter than the wildtype strain. The resulting mutants were confirmed by sequencing.
The primers used for the construction of plasmid pWM91-ΔVC0231 are listed in Table 4. The tolC-OS double mutant of N16961 was constructed based on tolC mutant in a similar fashion.
Genetic complementation analysis
Plasmid pSRKTc was used to construct complementary plasmids with inserted wildtype TolC or its derivatives, including TolCA78D and TolCΔ290–291. When the complementary plasmid pSRKTc-TolC was constructed, the fragment containing tolC was PCR-amplified from chromosomal DNA of N16961 with the primers TolC-NdeI-R/TolC-SpeI-F, digested by restriction enzymes NdeI/SpeI and inserted into pSRKTc. Using the same method, the plasmids pSRKTc-TolCA78D and pSRKTc-TolCΔ290–291 were constructed. The primers TolCΔ290–291-F/TolCΔ290–291-R for pSRKTc-TolCΔ290–291 and the primers TolCA78D-F/TolCA78D-R for pSRKTc-TolCA78Dare listed in Table 4. The complementary plasmid pBAD33-VC0231 was constructed with the primers VC0231-XbaI-F/VC0231-PstI-R (Table 4). The complementary plasmids were transformed into different mutants and induced with 0.01% arabinose for pBAD33 or 1 μm IPTG for pSRKTc as mentioned under the “Results.” The empty plasmid pSRKTc or pBAD33 was transformed into mutants as a control. Phage VP3 sensitivity analysis for the complemented mutants was performed by double-layer plaque assay.
Phage-binding assays
Pure VP3 phage with a titer of at least 1010 pfu/ml was mixed at 100,000:1 (v/v) with a SYBR Gold nucleic acid gel stain stock solution (S11494; Invitrogen) and incubated for 30 min in the dark at room temperature. The mixture was filtered through 0.02-μm pore filters (6809-5002; Waterman, Germany) and washed once using an equal volume of SM buffer (100 mm NaCl, 8 mm MgSO4·7H2O, 50 mm Tris-HCl, pH 7.5). The filters were washed with 1 ml of SM three times to gather labeled phage. Next, 700 μl of bacterial culture (A600 = 0.2) was mixed with phage (1010 pfu/ml) at 1:1 (v/v), incubated for 5 min, and centrifuged at 5,000 rpm for 5 min. The cell precipitate was resuspended with 200 μl of SM, and the adsorption of each strain with VP3 phage was measured at 537 nm (490 nm blue-light excitation) using a Multiscan Spectrum TECAN infinite M200 Pro and reported in fluorescence units/A600. The residual phage titers of the supernatant were tested by double-layer plaque assay (3).
SDS and bile salt sensitivity assays
The SDS sensitivity assay was performed as described previously (77). Cells of the strains were grown to the A600 of 1.0, diluted with LB medium supplemented with IPTG (0.5 mm) and SDS (0.1%) at about 106 cells/ml, and then cultured at 37 °C for 17 h. The viable cell amounts were counted by cell plating. The culture media were supplemented with tetracycline for strains NΔtolC-CA78D and NΔtolC-CΔ290–291. Percentages of survivors of strains NΔtolC, NΔtolC-CA78D, and NΔtolC-CΔ290–291 were calculated by their colony formation units to the wildtype strain N16961.
The bile salt sensitivity assay was performed by culture of the strains in the presence of 0.1% bile salt in LB media. Growth kinetics were measured in the 100-well plates (Bioscreen C, Finland). The A600 was monitored every 30 min in Bioscreen C at 37 °C with shaking for 8 h.
Bacterial two-hybrid system for analysis of the TolC and gp44 interaction
Overnight cultures of cyaA mutant E. coli BTH101 containing each plasmid pair pT25-gp44/pT18C-TolC, TolC derivatives including pT18C-TolCA78D and pT18C-TolCΔ290–291, plasmid pairs pT18C-TolC/pT25-gp44, or derivatives and various truncations of gp44 were subcultured in LB containing different amounts of IPTG and grown with shaking at 220 rpm at 37 °C until the A600 reached ∼0.4. The β-galactosidase activity was measured and recorded in Miller units as described previously (78).
RT-PCR
Total RNA was isolated from the culture of BTH101 strains by using the RNeasy kit (Qiagen). The RNA samples were analyzed by qRT-PCR using One-Step SYBR Primer script RT-PCR kit II (TaKaRa, Japan). Relative expression values (R) were calculated as 2−(ΔCt target − ΔCt reference), where Ct is the fractional threshold cycle. The mRNA of ampicillin-resistant gene in pUT18C was used as a reference. The following primer combinations were used: Amp-F and Amp-R for bla mRNA, gp44–350-F, and GP44–450-R for mRNA of gp44 and its truncated derivatives. A control mixture lacking reverse transcriptase was performed in each reaction to exclude chromosomal DNA contamination.
Protein expression and purification
TolC and gp44 genes, either wildtype or truncated, were subcloned into prokaryotic expression vectors for protein production. All primers and restriction enzymes used are included in Table 4. Genes of TolC (encoding 22–438 aa), TolCA78D, and TolCΔ290–291 were cloned, respectively, into pGEX-6p-1, overexpressed in E. coli strain BL21(DE3), and purified following detergent extraction as described (35). Full-length gp44 and its different truncations were cloned into pET-30a and expressed in E. coli strain BL21 (DE3). Cells containing the expression plasmids were grown in LB culture supplemented with corresponding antibiotics to a density of 0.5–0.6 A600/ml and induced with 0.5 mm IPTG for 4 h at 37 °C. Cells were collected, pelleted, and resuspended in buffer A (20 mm Tris-HCl, pH 9.0, and 300 mm NaCl, supplemented with protease inhibitors). The cells were lysed by sonication and centrifuged at 12,000 rpm for 1 h. The His6-tagged proteins, including soluble gp44 and various truncations, were first purified using Ni2+ resin (Invitrogen), and the elution samples were dialyzed using buffer B (20 mm Tris-HCl, pH 9.0, and 300 mm NaCl) and used in the interaction analysis with GST-tagged TolC protein. Protein concentration was measured using the BCA protein assay reagent kit (Pierce).
Protein binding assays in vitro
The proteins of His6-tagged full-length gp44 or its truncation variant gp44(451–753), 0.3 mg in each, was mixed, respectively, with 0.1 mg of GST-tagged TolC protein affixed to GS4B resin and incubated for 1 h at 4 °C. GST proteins loaded onto glutathione-Sepharose resin were added to a mixture of all gp44 His6-tagged proteins and used as a negative control. After an extensive washing step to remove unbound protein, the bound proteins were eluted with an elution buffer containing 10 mm reduced glutathione, and 1% of each sample was subjected to SDS-PAGE and Western blot analysis. Unless otherwise noted, all samples were boiled for 5 min in SDS loading buffer before separation on 12% SDS-polyacrylamide gels. After electrophoresis, proteins were transferred to PVDF membranes (Immobilon-P, Millipore). Western blottings were probed with anti-GST and anti-His monoclonal antibodies from mouse (Tiangen Biotech, Beijing, China). Anti-mouse peroxidase-conjugated AffiniPure IgG (H+L) secondary antibody (Zhong Shan Jin Qiao, Beijing, China) was used for protein detection.
Author contributions
B. K. and Fx. F. designed the study and wrote the paper. Fx. F., X. L., Z. L., and Jy. Z. purified protein and performed the experiments. B. P. and C. Z. contributed DNA and protein-sequence analysis. My. Y. and Wl. L. provided technical assistance and contributed to the preparation of the figures. J. L. and Lj. Z. cryopreserved and saved the strains. All authors analyzed the results and approved the final version of the manuscript.
This work was supported by National Science Foundation of China Youth Fund 81501724, National Basic Research Priorities Program Grant 2015CB554201, and Science Priority Grant 2014SKLID101 from the State Key Laboratory of Infections Disease Prevention and Control. The authors declare that they have no conflicts of interest with the contents of this article.
F. Fan, J. Zhang, and B. Kan, unpublished data.
- LPS
- lipopolysaccharide
- OS
- oligosaccharide
- OMP
- outer-membrane protein
- IPTG
- isopropyl β-d-thiogalactopyranoside
- aa
- amino acid
- qRT-PCR
- quantitative real-time reverse transcription-PCR
- pfu
- plaque-forming unit
- GST
- glutathione S-transferase
- cfu
- colony-forming unit
- ST
- sequence type
- BACTH
- bacterial adenylate cyclase two-hybrid
- Sm
- streptomycin
- Amp
- ampicillin
- Kan
- kanamycin
- Cmm
- chloramphenicol.
References
- 1. Feeley J. C. (1965) Classification of Vibrio cholerae (Vibrio comma), including El Tor vibrios, by infrasubspecific characteristics. J. Bacteriol. 89, 665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gao S., S. W., Liu B. (1984) Characteristics of typing phages of Vibrio cholerae biotype El Tor. Fu Huo Luan Zi Liao Hui Bian. 4, 237–245 [Google Scholar]
- 3. Disease Control Bureau of the Ministry of Health of China (2013) 2013 Manual of Cholera Prevention (6th Ed.) People's Medical Publishing House, Beijing, China [Google Scholar]
- 4. Brüssow H., and Hendrix R. W. (2002) Phage genomics: small is beautiful. Cell 108, 13–16 10.1016/S0092-8674(01)00637-7 [DOI] [PubMed] [Google Scholar]
- 5. Hyman P., and Abedon S. T. (2010) Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70, 217–248 10.1016/S0065-2164(10)70007-1 [DOI] [PubMed] [Google Scholar]
- 6. Duplessis M., and Moineau S. (2001) Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41, 325–336 10.1046/j.1365-2958.2001.02521.x [DOI] [PubMed] [Google Scholar]
- 7. Krüger D. H., and Schroeder C. (1981) Bacteriophage T3 and bacteriophage T7 virus-host cell interactions. Microbiol. Rev. 45, 9–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shin H., Lee J. H., Kim H., Choi Y., Heu S., and Ryu S. (2012) Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PLoS One 7, e43392 10.1371/journal.pone.0043392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. São-José C., Baptista C., and Santos M. A. (2004) Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J. Bacteriol. 186, 8337–8346 10.1128/JB.186.24.8337-8346.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindberg A. A. (1973) Bacteriophage receptors. Annu. Rev. Microbiol. 27, 205–241 10.1146/annurev.mi.27.100173.001225 [DOI] [PubMed] [Google Scholar]
- 11. São-José C., Lhuillier S., Lurz R., Melki R., Lepault J., Santos M. A., and Tavares P. (2006) The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J. Biol. Chem. 281, 11464–11470 10.1074/jbc.M513625200 [DOI] [PubMed] [Google Scholar]
- 12. al-Hendy A., Toivanen P., and Skurnik M. (1991) Expression cloning of Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb. Pathog. 10, 47–59 10.1016/0882-4010(91)90065-I [DOI] [PubMed] [Google Scholar]
- 13. al-Hendy A., Toivanen P., and Skurnik M. (1992) Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect. Immun. 60, 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kiljunen S., Datta N., Dentovskaya S. V., Anisimov A. P., Knirel Y. A., Bengoechea J. A., Holst O., and Skurnik M. (2011) Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage phiA1122. J. Bacteriol. 193, 4963–4972 10.1128/JB.00339-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu J., Zhang J., Lu X., Liang W., Zhang L., and Kan B. (2013) O antigen is the receptor of Vibrio cholerae serogroup O1 El Tor typing phage VP4. J. Bacteriol. 195, 798–806 10.1128/JB.01770-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J., Li W., Zhang Q., Wang H., Xu X., Diao B., Zhang L., and Kan B. (2009) The core oligosaccharide and thioredoxin of Vibrio cholerae are necessary for binding and propagation of its typing phage VP3. J. Bacteriol. 191, 2622–2629 10.1128/JB.01370-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakae T. (1976) Identification of the outer-membrane protein of E. coli that produces transmembrane channels in reconstituted vesicle membranes. Biochem. Biophys. Res. Commun. 71, 877–884 10.1016/0006-291X(76)90913-X [DOI] [PubMed] [Google Scholar]
- 18. Nikaido H. (1994) Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem. 269, 3905–3908 [PubMed] [Google Scholar]
- 19. Schirmer T., Keller T. A., Wang Y. F., and Rosenbusch J. P. (1995) Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science 267, 512–514 10.1126/science.7824948 [DOI] [PubMed] [Google Scholar]
- 20. Porcek N. B., and Parent K. N. (2015) Key residues of S. flexneri OmpA mediate infection by bacteriophage Sf6. J. Mol. Biol. 427, 1964–1976 10.1016/j.jmb.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parent K. N., Erb M. L., Cardone G., Nguyen K., Gilcrease E. B., Porcek N. B., Pogliano J., Baker T. S., and Casjens S. R. (2014) OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Mol. Microbiol. 92, 47–60 10.1111/mmi.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morona R., Klose M., and Henning U. (1984) Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J. Bacteriol. 159, 570–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morona R., Krämer C., and Henning U. (1985) Bacteriophage receptor area of outer membrane protein OmpA of Escherichia coli K-12. J. Bacteriol. 164, 539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu D., Zhang J., Liu J., Xu J., Zhou H., Zhang L., Zhu J., and Kan B. (2014) Outer membrane protein OmpW is the receptor for typing phage VP5 in the Vibrio cholerae O1 El Tor biotype. J. Virol. 88, 7109–7111 10.1128/JVI.03186-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gehring K., Charbit A., Brissaud E., and Hofnung M. (1987) Bacteriophage λ receptor site on the Escherichia coli K-12 LamB protein. J. Bacteriol. 169, 2103–2106 10.1128/jb.169.5.2103-2106.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Datta D. B., Arden B., and Henning U. (1977) Major proteins of the Escherichia coli outer cell envelope membrane as bacteriophage receptors. J. Bacteriol. 131, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Traurig M., and Misra R. (1999) Identification of bacteriophage K20 binding regions of OmpF and lipopolysaccharide in Escherichia coli K-12. FEMS Microbiol. Lett. 181, 101–108 10.1111/j.1574-6968.1999.tb08831.x [DOI] [PubMed] [Google Scholar]
- 28. Vakharia H., and Misra R. (1996) A genetic approach for analysing surface-exposed regions of the OmpC protein of Escherichia coli K-12. Mol. Microbiol. 19, 881–889 10.1046/j.1365-2958.1996.430957.x [DOI] [PubMed] [Google Scholar]
- 29. Yu F., and Mizushima S. (1982) Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J. Bacteriol. 151, 718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao X., Cui Y., Yan Y., Du Z., Tan Y., Yang H., Bi Y., Zhang P., Zhou L., Zhou D., Han Y., Song Y., Wang X., and Yang R. (2013) Outer membrane proteins ail and OmpF of Yersinia pestis are involved in the adsorption of T7-related bacteriophage Yep-φ J. Virol. 87, 12260–12269 10.1128/JVI.01948-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. German G. J., and Misra R. (2001) The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. J. Mol. Biol. 308, 579–585 10.1006/jmbi.2001.4578 [DOI] [PubMed] [Google Scholar]
- 32. Li W., Zhang J., Chen Z., Zhang Q., Zhang L., Du P., Chen C., and Kan B. (2013) The genome of VP3, a T7-like phage used for the typing of Vibrio cholerae. Arch. Virol. 158, 1865–1876 10.1007/s00705-013-1676-9 [DOI] [PubMed] [Google Scholar]
- 33. Wandersman C., and Delepelaire P. (1990) TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. U.S.A. 87, 4776–4780 10.1073/pnas.87.12.4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Glaser P., Sakamoto H., Bellalou J., Ullmann A., and Danchin A. (1988) Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 7, 3997–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koronakis V., Li J., Koronakis E., and Stauffer K. (1997) Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol. Microbiol. 23, 617–626 10.1046/j.1365-2958.1997.d01-1880.x [DOI] [PubMed] [Google Scholar]
- 36. Thanassi D. G., and Hultgren S. J. (2000) Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12, 420–430 10.1016/S0955-0674(00)00111-3 [DOI] [PubMed] [Google Scholar]
- 37. Fralick J. A., and Burns-Keliher L. L. (1994) Additive effect of tolC and rfa mutations on the hydrophobic barrier of the outer membrane of Escherichia coli K-12. J. Bacteriol. 176, 6404–6406 10.1128/jb.176.20.6404-6406.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nikaido H. (1994) Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264, 382–388 10.1126/science.8153625 [DOI] [PubMed] [Google Scholar]
- 39. Aono R., Tsukagoshi N., and Yamamoto M. (1998) Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180, 938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosenberg E. Y., Bertenthal D., Nilles M. L., Bertrand K. P., and Nikaido H. (2003) Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48, 1609–1619 10.1046/j.1365-2958.2003.03531.x [DOI] [PubMed] [Google Scholar]
- 41. Nagel de Zwaig R., and Luria S. E. (1967) Genetics and physiology of colicin-tolerant mutants of Escherichia coli. J. Bacteriol. 94, 1112–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davies J. K., and Reeves P. (1975) Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J. Bacteriol. 123, 102–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boardman B. K., and Satchell K. J. (2004) Vibrio cholerae strains with mutations in an atypical type I secretion system accumulate RTX toxin intracellularly. J. Bacteriol. 186, 8137–8143 10.1128/JB.186.23.8137-8143.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steven A. C., Trus B. L., Maizel J. V., Unser M., Parry D. A., Wall J. S., Hainfeld J. F., and Studier F. W. (1988) Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200, 351–365 10.1016/0022-2836(88)90246-X [DOI] [PubMed] [Google Scholar]
- 45. Garcia-Doval C., and van Raaij M. J. (2012) Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc. Natl. Acad. Sci. U.S.A. 109, 9390–9395 10.1073/pnas.1119719109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karimova G., Pidoux J., Ullmann A., and Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fan F., Liu Z., Jabeen N., Birdwell L. D., Zhu J., and Kan B. (2014) Enhanced interaction of Vibrio cholerae virulence regulators TcpP and ToxR under oxygen-limiting conditions. Infect. Immun. 82, 1676–1682 10.1128/IAI.01377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang M., Liu Z., Hughes C., Stern A. M., Wang H., Zhong Z., Kan B., Fenical W., and Zhu J. (2013) Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc. Natl. Acad. Sci. U.S.A. 110, 2348–2353 10.1073/pnas.1218039110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ladant D. (1988) Interaction of Bordetella pertussis adenylate cyclase with calmodulin. Identification of two separated calmodulin-binding domains. J. Biol. Chem. 263, 2612–2618 [PubMed] [Google Scholar]
- 50. Veesler D., and Cambillau C. (2011) A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 75, 423–433 10.1128/MMBR.00014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Didelot X., Pang B., Zhou Z., McCann A., Ni P., Li D., Achtman M., and Kan B. (2015) The role of China in the global spread of the current cholera pandemic. PLoS Genet. 11, e1005072 10.1371/journal.pgen.1005072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wittig I., Braun H. P., and Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 10.1038/nprot.2006.62 [DOI] [PubMed] [Google Scholar]
- 53. Koronakis V., Sharff A., Koronakis E., Luisi B., and Hughes C. (2000) Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919 10.1038/35016007 [DOI] [PubMed] [Google Scholar]
- 54. Heller K. J. (1992) Molecular interaction between bacteriophage and the Gram-negative cell envelope. Arch. Microbiol. 158, 235–248 10.1007/BF00245239 [DOI] [PubMed] [Google Scholar]
- 55. Nikaido H., and Vaara M. (1985) Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49, 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Molineux I. J. (2001) No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40, 1–8 10.1046/j.1365-2958.2001.02357.x [DOI] [PubMed] [Google Scholar]
- 57. Feucht A., Schmid A., Benz R., Schwarz H., and Heller K. J. (1990) Pore formation associated with the tail-tip protein pb2 of bacteriophage T5. J. Biol. Chem. 265, 18561–18567 [PubMed] [Google Scholar]
- 58. Eckert B., and Schmid F. X. (2007) A conformational unfolding reaction activates phage fd for the infection of Escherichia coli. J. Mol. Biol. 373, 452–461 10.1016/j.jmb.2007.07.060 [DOI] [PubMed] [Google Scholar]
- 59. Riechmann L., and Holliger P. (1997) The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell 90, 351–360 10.1016/S0092-8674(00)80342-6 [DOI] [PubMed] [Google Scholar]
- 60. Letellier L., Plançon L., Bonhivers M., and Boulanger P. (1999) Phage DNA transport across membranes. Res. Microbiol. 150, 499–505 10.1016/S0923-2508(99)00107-2 [DOI] [PubMed] [Google Scholar]
- 61. Moffatt B. A., and Studier F. W. (1988) Entry of bacteriophage T7 DNA into the cell and escape from host restriction. J. Bacteriol. 170, 2095–2105 10.1128/jb.170.5.2095-2105.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Berrier C., Bonhivers M., Letellier L., and Ghazi A. (2000) High-conductance channel induced by the interaction of phage λwith its receptor maltoporin. FEBS Lett. 476, 129–133 10.1016/S0014-5793(00)01705-1 [DOI] [PubMed] [Google Scholar]
- 63. Roessner C. A., and Ihler G. M. (1986) Formation of transmembrane channels in liposomes during injection of λ DNA. J. Biol. Chem. 261, 386–390 [PubMed] [Google Scholar]
- 64. Killmann H., Videnov G., Jung G., Schwarz H., and Braun V. (1995) Identification of receptor–binding sites by competitive peptide mapping: phages T1, T5, and φ0 and colicin M bind to the gating loop of FhuA. J. Bacteriol. 177, 694–698 10.1128/jb.177.3.694-698.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Putman M., van Veen H. W., and Konings W. N. (2000) Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64, 672–693 10.1128/MMBR.64.4.672-693.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Paulsen I. T., Park J. H., Choi P. S., and Saier M. H. Jr. (1997) A family of Gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from Gram-negative bacteria. FEMS Microbiol. Lett. 156, 1–8 10.1016/S0378-1097(97)00379-0 [DOI] [PubMed] [Google Scholar]
- 67. Kobayashi N., Nishino K., and Yamaguchi A. (2001) Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183, 5639–5644 10.1128/JB.183.19.5639-5644.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tikhonova E. B., Devroy V. K., Lau S. Y., and Zgurskaya H. I. (2007) Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol. Microbiol. 63, 895–910 [DOI] [PubMed] [Google Scholar]
- 69. Wang J. C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 10.1038/nrm831 [DOI] [PubMed] [Google Scholar]
- 70. Kwan K. Y., Moens P. B., and Wang J. C. (2003) Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase III β. Proc. Natl. Acad. Sci. U.S.A. 100, 2526–2531 10.1073/pnas.0437998100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Frost J. A., Kramer J. M., and Gillanders S. A. (1999) Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol. Infect. 123, 47–55 10.1017/S095026889900254X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chiang S. L., and Mekalanos J. J. (2000) Construction of a Vibrio cholerae vaccine candidate using transposon delivery and FLP recombinase-mediated excision. Infect. Immun. 68, 6391–6397 10.1128/IAI.68.11.6391-6397.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Simon R., Priefer U., and Pühler A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1, 784–791 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 74. Judson N., and Mekalanos J. J. (2000) TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 18, 740–745 10.1038/77305 [DOI] [PubMed] [Google Scholar]
- 75. Tsou A. M., Liu Z., Cai T., and Zhu J. (2011) The VarS/VarA two-component system modulates the activity of the Vibrio cholerae quorum-sensing transcriptional regulator HapR. Microbiology 157, 1620–1628 10.1099/mic.0.046235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Metcalf W. W., Jiang W., Daniels L. L., Kim S. K., Haldimann A., and Wanner B. L. (1996) Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35, 1–13 10.1006/plas.1996.0001 [DOI] [PubMed] [Google Scholar]
- 77. Cescau S., Debarbieux L., and Wandersman C. (2007) Probing the in vivo dynamics of type I protein secretion complex association through sensitivity to detergents. J. Bacteriol. 189, 1496–1504 10.1128/JB.01480-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Miller J. H. (1972) Experiments in Molecular Genetics, pp. 352–355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]