Abstract
Podocin is a key component of the slit diaphragm in the glomerular filtration barrier, and mutations in the podocin-encoding gene NPHS2 are a common cause of hereditary steroid-resistant nephrotic syndrome. A mutant allele encoding podocin with a p.R138Q amino acid substitution is the most frequent pathogenic variant in European and North American children, and the corresponding mutant protein is poorly expressed and retained in the endoplasmic reticulum both in vitro and in vivo. To better understand the defective trafficking and degradation of this mutant, we generated human podocyte cell lines stably expressing podocinwt or podocinR138Q. Although it has been proposed that podocin has a hairpin topology, we present evidence for podocinR138Q N-glycosylation, suggesting that most of the protein has a transmembrane topology. We find that N-glycosylated podocinR138Q has a longer half-life than non-glycosylated podocinR138Q and that the latter is far more rapidly degraded than podocinwt. Consistent with its rapid degradation, podocinR138Q is exclusively degraded by the proteasome, whereas podocinwt is degraded by both the proteasomal and the lysosomal proteolytic machineries. In addition, we demonstrate an enhanced interaction of podocinR138Q with calnexin as the mechanism of endoplasmic reticulum retention. Calnexin knockdown enriches the podocinR138Q non-glycosylated fraction, whereas preventing exit from the calnexin cycle increases the glycosylated fraction. Altogether, we propose a model in which hairpin podocinR138Q is rapidly degraded by the proteasome, whereas transmembrane podocinR138Q degradation is delayed due to entry into the calnexin cycle.
Keywords: genetic disease, glycosylation, intracellular trafficking, kidney, endoplasmic-reticulum-associated protein degradation (ERAD), proteasome, podocin, steroid-resistant nephrotic syndrome
Introduction
Nephrotic syndrome is clinically characterized by proteinuria, edema, hypoalbuminemia, and hyperlipidemia, and is a consequence of glomerular filtration barrier (GFB)2 dysfunction. The prognosis of steroid-resistant nephrotic syndrome (SRNS) is poor, with a high proportion of patients rapidly developing end-stage renal disease, requiring dialysis or transplantation (1, 2). The GFB is comprised principally of podocytes, specialized epithelial cells that interdigitate at junctions known as slit diaphragms (SDs). Mutations in the NPHS2 gene, encoding the slit diaphragm (SD) protein podocin, are the most frequent monogenic cause of SRNS in childhood (3, 4). Importantly, the missense mutation NPSH2: R138Q (p.R138Q) accounts for 20% of all SRNS-causing alleles in Europe and North America (4), and is associated with an early-onset and rapidly progressing form of the disease (5). In accordance with the severe clinical phenotype, podocin p.R138Q (PodR138Q) is retained in the ER of podocytes and does not reach the SD, thus impairing correct functioning of the GFB (6, 7).
Podocin belongs to the stomatin and prohibitin homology domain (PHB) protein family and is specifically expressed in podocytes. It has been proposed that podocin acts as a molecular scaffold for other SD proteins in lipid raft membrane subdomains. For example, podocin interacts with both nephrin and CD2AP through its carboxyl terminus, and participates in various signaling events at the SD (8–10). Podocin has a predicted hairpin-loop topology, with both the N and C termini facing the cytoplasm, and its hydrophobic domain anchored either to the inner leaflet of the plasma membrane (PM) or to the outer leaflet of the ER-membrane (7). However, an alternative transmembrane topology has been described for stomatin and podocin, particularly when a conserved proline residue, critical for the kink of the hairpin topology, found at position 118 in podocin, is mutated, and has been linked to N-glycosylated forms of these proteins (11, 12). In addition, although the stability and degradation of podocin has been associated with a short internalization motif located in its C terminus and to an interaction with the ubiquitin ligase Ubr4 (13, 14), the mechanisms of PodR138Q degradation are still unexplored. In this study, we report the degradation pathways followed by PodR138Q, which might aid to establish new therapeutic strategies.
Results
PodocinR138Q is predominantly N-glycosylated
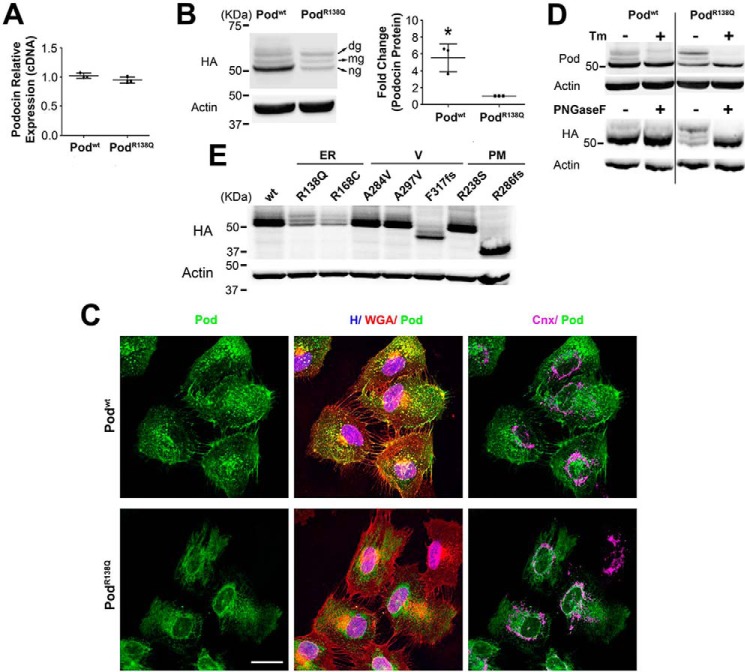
We generated two cell lines stably expressing two human influenza hemagglutinin (HA) tags at the N terminus of wildtype (WT) podocin (Podwt) and PodR138Q by lentiviral transduction of a human podocyte cell line. Although both cell lines expressed comparable levels of podocin mRNA (Fig. 1A), we found significantly higher levels of Podwt protein in comparison to PodR138Q (Fig. 1B). We then confirmed by immunofluorescence the expected subcellular localization at the PM for Podwt (co-localization with the wheat germ agglutinin, WGA) and at the ER for PodR138Q (a reticular pattern staining partially colocalizing with calnexin) (Fig. 1C) (7). We demonstrated that neither Podwt nor PodR138Q stable overexpression caused ER stress as evidenced by the lack of up-regulated levels of BiP protein (GRP78), an ER-chaperone widely used as an ER-stress indicator (Fig. S1) (15–17). As positive controls of ER stress induction caused by misfolded mutant proteins, we generated human podocyte cell lines stably expressing V5-tagged nephrinwt (Nephwt) or nephrinS366R (NephS366R), because this latter mutant has been previously shown to induce ER stress (Fig. S1A) (18).
Figure 1.
Stably over-expressed podocinR138Q is largely N-glycosylated, a feature shared only with other ER-retained podocin mutants. qRT-PCR (A), Western blot (B, D, and E), and immunofluorescence analysis (C) comparing podocin expression levels, protein band distribution, and subcellular localization, respectively, in 2HA-Podwt and 2HA-PodR138Q expressing podocyte cell lines. B, ng, non-glycosylated podocin; mg, mono-glycosylated podocin; dg, double-glycosylated podocin. Quantification of total podocin (ng, mg and dg) from three independent experiments are shown as mean ± S.D. *, p < 0.05. HA monoclonal antibody was used to identify podocin and β-actin served as loading control. C, polyclonal anti-podocin AP-P35 (Pod) and monoclonal calnexin AF18 (Cnx) were used as primary antibodies. Cell membrane and nuclei were labeled with WGA and Hoechst (H), respectively. Scale bar = 30 μm. D, cells were treated overnight with 10 μg/ml of tunicamycin (Tm) to impair N-glycosylation (upper immunoblot). Alternatively, cell lysates were treated with PNGase F, an enzyme that digests N-glycans from glycoproteins (lower immunoblot). Polyclonal AP-P35 or monoclonal anti-HA were used to immunoblot podocin. E, immunoblots of different podocin mutants known to possess different subcellular localizations. ER, endoplasmic reticulum–retained podocin mutants; V, vesicular podocin mutants; PM, plasma membrane localized podocin mutants.
Interestingly, we observed three specific bands for both Podwt and PodR138Q on podocin immunoblots. The fastest migrating band (lower band) was predominantly observed in Podwt protein extracts, whereas the slowest migrating band (upper band) was predominantly found in PodR138Q extracts (Fig. 1B). To determine whether the upper bands in the WT and mutant podocin triplets were N-glycosylated forms, we either inhibited N-glycosylation by treating cells with tunicamycin, or treated cell lysates with peptide:N-glycosidase F (PNGase F) to digest N-glycan groups. Upper and middle bands of the podocin triplets disappeared using both strategies (Fig. 1D) allowing us to define the upper, middle, and lower bands of the podocin triplets as double-glycosylated (dg), monoglycosylated (mg), and non-glycosylated (ng) podocin. To test whether the difference in protein levels of Podwt and PodR138Q was due to these differential glycosylation patterns, we also created podocyte cell lines stably expressing HA-tagged Podwt and PodR138Q bearing mutations in the amino acid residues required for N-glycosylation, Asn199 and Asn355 (Podwt(N199Q,N355S) and PodR138Q(N199Q,N355S)). Immunoblots of protein extracts from these cells revealed that although the podocin triplet disappears with mutation of residues Asn199 and Asn355 (Fig. S1B), glycosylation alone does not account for the differences in protein levels of Podwt and PodR138Q, as there remains increased amounts of Podwt(N199Q,N355S) compared with PodR138Q(N199Q,N355S).
Our results showed that the mutant PodR138Q was predominantly N-glycosylated, similarly to PodP118L (12), another ER-retained podocin mutant, suggesting that the majority of intracellular PodR138Q has a transmembrane topology (11, 12). However, a small part of the Podwt pool was also glycosylated, consistent with the observations of WT podocin and stomatin by other authors (11, 12). We tested by immunoblot the protein expression pattern of a series of podocin mutants that present different subcellular localizations, confirming that only ER-retained mutants, such as PodR168C, were enriched in N-glycosylated forms (Fig. 1E).
PodocinR138Q has a shorter half-life than Podwt and is rapidly degraded by the proteasome
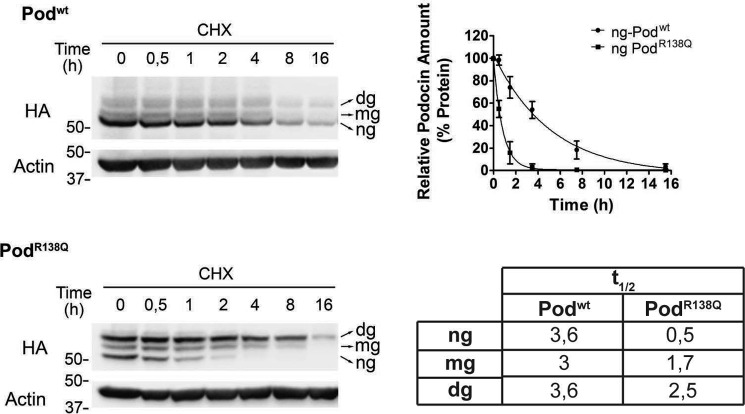
To investigate whether the difference in protein levels of Podwt and PodR138Q was due to a higher degradation rate of the mutant protein, we determined the half-life (t½) of both proteins using a cycloheximide time course experiment, quantifying separately the glycosylated and non-glycosylated forms (Fig. 2). No differences were observed between the t½ of the three Podwt forms, whereas a drastically reduced t½ was detected for ng-PodR138Q when compared with Podwt (7-fold). Consistent with this finding, we also demonstrated that the t½ of the glycosylation mutant PodR138Q(N199Q,N355S) was also reduced around 7-fold compared with Podwt(N199Q,N355S) (Fig. S2). Interestingly, ng-PodR138Q (i.e. hairpin-like topology) appeared to be degraded faster than the N-glycosylated forms of PodR138Q (i.e. those with transmembrane topology), implying that the hairpin-like structure is more exposed to the intracellular degradative machinery (Fig. 2 and Fig. S2).
Figure 2.
PodocinR138Q has a short half-life. Immunoblot analysis of the time course of podocin degradation after inhibition of protein synthesis with cycloheximide (CHX) (25 μm) in podocyte cell lines. Densitometry data from three independent experiments are represented as mean ± S.D. Half-lives were estimated by fitting a one-phase exponential decay curve to the data, as in the graph shown for ng-Podwt and ng-PodR138Q, and are summarized in the table.
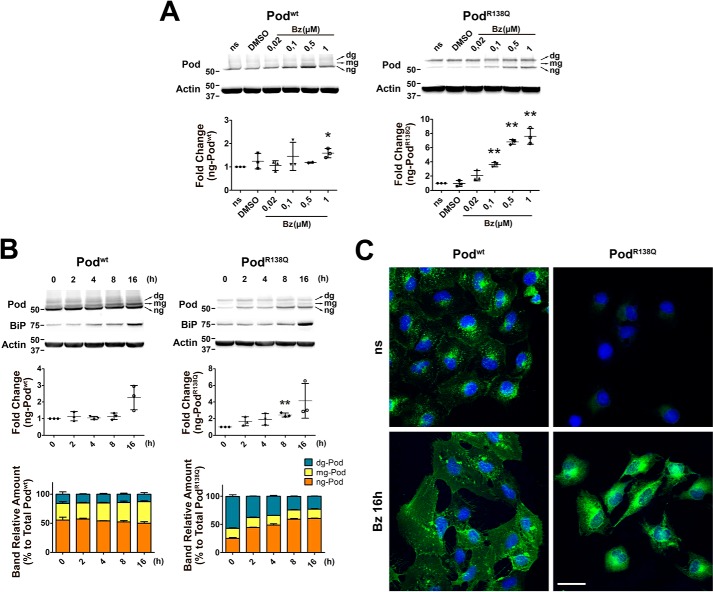
Because ng-PodR138Q is degraded very quickly, we hypothesized that the proteasome was mediating its clearance. We therefore used the reversible proteasome inhibitor bortezomib (Bz) to perform dose-response and time course experiments (Fig. 3). We observed that ng-PodR138Q accumulated in a dose-dependent manner, whereas levels of ng-Podwt increased significantly only at the highest Bz dose of 1 μm (Fig. 3A). Consistent with their longer t½, both glycosylated Podwt and glycosylated PodR138Q were not increased at any dose of Bz after 2 h (Fig. 3A). Thus, we next performed an overnight time course (16 h) using the lowest effective dose of Bz (0.1 μm) (Fig. 3B). We found that only ng-PodR138Q accumulated with time, and, most interestingly, the balance between ng- and dg-PodR138Q was inverted, thus after 16 h the proportions of each PodR138Q band resembled the proportions of each Podwt band in podocin immunoblots (Fig. 3B, colored graph). Levels of all forms of Podwt were increased only at the longest time points. Similar findings were observed when we performed an overnight Bz time course on cells expressing the glycosylation mutants Podwt(N199Q,N355S) and PodR138Q(N199Q,N355S), with protein levels of the WT protein only significantly increased at the longest time point of 16 h, and a trend for levels of PodR138Q(N199Q,N355S) to accumulate in the presence of 0.1 μm Bz (Fig. S3A). The increased amount of Podwt and PodR138Q after an overnight treatment with 0.1 μm Bz was confirmed by immunofluorescence analysis (Fig. 3C). As expected, a sustained exposure to Bz led to ER stress, as indicated by BiP induction (Fig. 3C), but to a higher extent in cells expressing PodR138Q (Fig. S3). Therefore, we concluded that the proteasome contributes to the degradation of both Podwt and PodR138Q, but more actively for ng-PodR138Q.
Figure 3.
PodocinR138Q is degraded by the proteasome. Podocyte cell lines were treated with the proteasome inhibitor Bz or vehicle (DMSO) and processed for immunoblot (A and B) or immunofluorescence analysis of podocin (C). A, Bz dose-response (0.02 to 1 μm) after 2 h of treatment. B, time course of podocin after addition of 0.1 μm Bz. A and B, podocin was identified using AP-P35 (Pod) primary antibody. BiP analysis was included to track ER stress (B). Quantification of ng bands is shown in the lower graphs in A and in the middle graphs in B. Color graphs in B represent the percentage amount of each podocin band relative to total podocin. Quantitative results are shown as mean ± S.D. (n = 3). Asterisks refer to non-stimulated cells (ns or 0 h). *, p < 0.05 and **, p < 0.01. C, podocin was detected by incubation with monoclonal HA primary antibody (in green). Hoechst nuclei labeling was included (in blue). All images were taken using the same confocal microscope settings to allow comparison of the intensity of fluorescence. Scale bar = 40 μm.
PodocinR138Q is not degraded by lysosomes
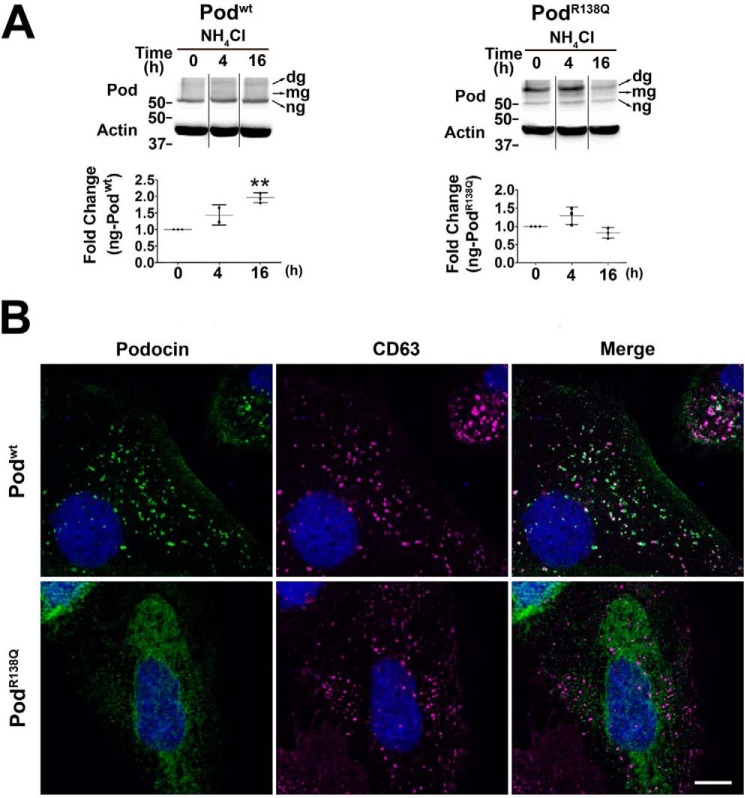
Because protein degradation proceeds via two major routes: the proteasomal and the autophagic-lysosomal pathways, we investigated whether lysosomal degradation contributes to Podwt and PodR138Q proteolysis. We therefore treated the cells with ammonium chloride (NH4Cl), a weak base known to inhibit lysosomal proteases. NH4Cl treatment significantly increased levels of the ng-Podwt protein fraction and levels of Podwt(N199Q,N355S), implying that the lysosomal machinery is involved in Podwt degradation but not in PodR138Q proteolysis; indeed, we observed levels of both PodR138Q and PodR138Q(N199S,N355S) to fall after a 16-h NH4Cl treatment (Fig. 4A and Fig. S4B). Then, by confocal microscopy, we confirmed the absence of PodR138Q in the late endosome/lysosome compartment labeled with CD63 (Fig. 4B). Conversely, Podwt is predominantly present in this compartment, as already described (13). Taken together, our data suggest that Podwt is mainly degraded in lysosomes, in contrast to PodR138Q, which is exclusively degraded by the proteasome.
Figure 4.
PodocinR138Q is not degraded in the lysosomal compartment. A, time course immunoblot analysis of podocin content after the impairment of lysosomal degradation with NH4Cl (50 mm). The polyclonal antibody AP-P35 (Pod) was used to immunolabel podocin. Graphs show densitometry quantification of ng-podocin in three independent experiments. Data are normalized to β-actin and then to non-treated cells (0 h). Asterisks refer to 0 h. **, p < 0.01. B, double immunofluorescence staining of podocin (AP-P35; green) and the lysosome/late endosome marker CD63 (cyan). Nuclei labeling by Hoechst is included (blue). Scale bar = 10 μm.
PodocinR138Q enhanced interaction with calnexin
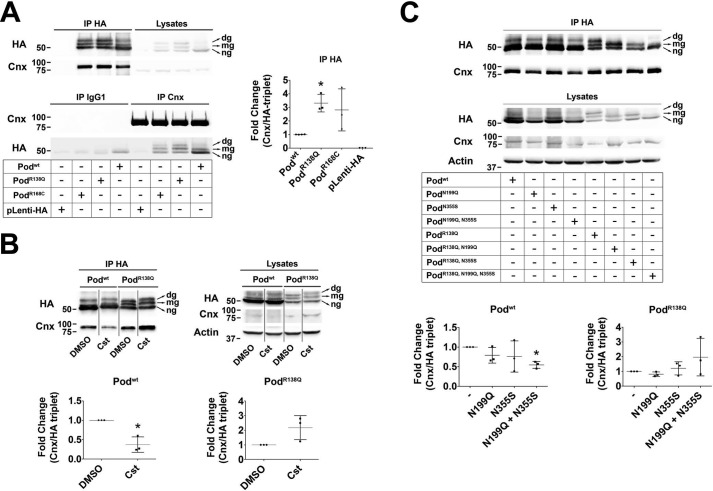
Misfolded N-glycosylated proteins located in the ER membrane are submitted to a strict quality control (QC) by the so-called calnexin (Cnx) cycle, being retained by the 90-kDa chaperone Cnx until they reach their native conformation or are otherwise sent for ER-associated degradation (ERAD). We hypothesized that if PodR138Q is mostly N-glycosylated and possesses a transmembrane topology, interaction with Cnx may be a mechanism of PodR138Q ER retention, as has already been described for some nephrin mutants (18). Immunoblots of HEK293T cells transfected with HA-tagged WT podocin and the two ER-retained mutants, PodR138Q and PodR168C (7), showed that similarly to our findings from podocyte cell lines stably expressing Podwt and PodR138Q podocin was present as three differentially glycosylated species (Fig. 5A). Co-immunoprecipitation studies revealed an interaction between Cnx and both WT and mutant podocin, with the mutant proteins interacting to a greater extent compared with the WT protein (Fig. 5A). That we found Cnx also interacted with Podwt was not surprising, because HA immunoprecipitation enriched a certain proportion of N-glycosylated Podwt (Fig. 5A, upper panel). Surprisingly, ng-podocin, WT, and mutants, also co-immunoprecipitated with Cnx, suggesting that hairpin podocin might interact with Cnx either indirectly, perhaps through oligomerization with the podocin transmembrane fraction via its N terminus (19), or directly, through membrane or cytosolic domains. To test whether impairing the interaction of PodR138Q with Cnx would allow its ER exit and promote membrane localization, we first treated the cells with castanospermine (Cst), a specific inhibitor of glucosidases I and II that prevents the sugar trimming necessary for the recognition of the substrate protein N-glycan groups by the lectin domain of Cnx. Cst decreased Cnx interaction with Podwt, but not with PodR138Q (Fig. 5B), suggesting that PodR138Q interaction with Cnx is N-glycan independent. Similar results were obtained when these experiments were performed using cells expressing glycosylation mutants Podwt(N199Q,N355S) and PodR138Q(N199Q,N355S) (Fig. 5C), supporting the idea that the interaction with Cnx is more lectin dependent for Podwt than for PodR138Q. Finally, we found that introduction of p.N199Q and p.N355S substitutions into the PodR138Q mutant in podocytes did not prevent its ER retention, implying that suppression of the N-glycan-dependent interaction with Cnx is not sufficient to bring PodR138Q to the PM (Fig. S4A), and supporting the idea that podocin is capable of lectin-dependent binding to Cnx.
Figure 5.
PodocinR138Q has an enhanced interaction with calnexin. A–C, co-immunoprecipitation analyses of podocin and calnexin in HEK293T cells. A, co-immunoprecipitation of HA-tagged Podwt, PodR138Q, PodR168C, or an empty HA lentiviral vector (pLenti-HA) with Cnx. B, cells were treated with the glucosidase I and II inhibitor castanospermine (Cst; 500 μm, 16 h) before performing HA-immunoprecipitation to study the lectin-dependent interaction of Cnx with Podwt and PodR138Q. C, co-immunoprecipitation of Cnx with podocin in cells overexpressing HA-tagged Podwt and PodR138Q with or without mutated N-glycosylation sites Asn199 and Asn355. A–C, monoclonal anti-HA and anti-Cnx AF18 were used to identify podocin and Cnx, respectively. Graphs represent the densitometry quantification of Cnx when podocin is immunoprecipitated (IP HA). Data are normalized to total immunoprecipitated podocin (HA triplet in IP HA) and represent at least three independent experiments. *, p < 0.05.
Glycosylated podocinR138Q enters the calnexin cycle
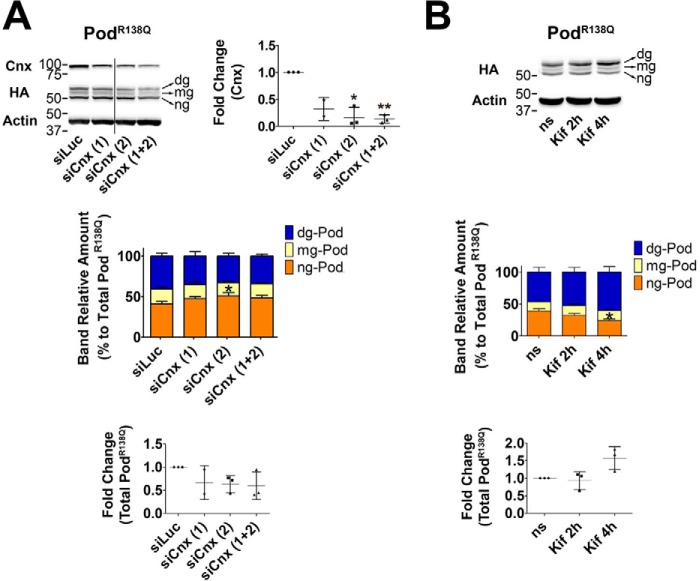
Because we observed PodR138Q(N199Q,N355S) is still able to interact with Cnx, we next knocked-down Cnx in the two stable podocyte cell lines using siRNA (Fig. 6A and Fig. S5A). Interestingly, we observed that levels of N-glycosylated forms of PodR138Q were decreased upon Cnx knockdown, in contrast to levels of ng-PodR138Q, which were significantly increased (Fig. 6A, colored graph), suggesting that Cnx may play a role in stabilization of the transmembrane form of N-glycosylated PodR138Q, or indeed, that interaction with Cnx promotes glycosylation of PodR138Q. However, in support of the former statement, the total amount of PodR138Q was slightly decreased after Cnx knockdown, suggesting that interaction with Cnx might delay PodR138Q degradation (Fig. 6A, lower graph). Next, we treated cells with kifunensine (Kif), a drug that inhibits the activity of α-mannosidase I, and thus the mannose trimming that tags Cnx substrates for ERAD (Fig. 6B). Here, our results were precisely the opposite to those obtained upon Cnx knockdown, that is, levels of ng-PodR138Q were significantly reduced, whereas levels of dg-PodR138Q were increased (Fig. 6B, colored graph). Furthermore, the total amount of PodR138Q was augmented after 4 h of treatment (Fig. 6B, lower graph), demonstrating that Kif treatment partially prevented PodR138Qdegradation. These last data support the idea that only N-glycosylated, and thus transmembrane PodR138Q, enters the Cnx cycle before being directed to ERAD. Finally, we observed by immunofluorescence that neither knockdown of Cnx nor blocking entry into the ERAD pathway with Kif were sufficient to target PodR138Q to the PM (data not shown). No significant changes on Podwt levels were observed upon Cnx knockdown, but 4 h of Kif treatment did increase the levels of dg-Podwt, suggesting that glycosylated Podwt may also enter the Cnx cycle (Fig. S5B).
Figure 6.
PodocinR138Q enters the calnexin cycle. A and B, immunoblot analysis of podocin (HA) after Cnx knockdown (siCnx) (A) or impairment of Cnx cycle exit through the inhibition of α-mannosidase I with kifunensine (Kif) (B) in HA WT and R138Q stably expressing podocyte cell lines. A, two siRNA oligonucleotides against Cnx were tested, alone (siCnx (1) and siCnx (2)) or in combination (siCnx (1 + 2)). Luciferase siRNA (siLuc) served as control. Cnx protein amount was quantified to confirm Cnx knockdown (upper left graph). B, effect of kifunensine on podocin protein content. ns, non-stimulated cells. A and B, podocin was quantified as the relative percentage amount of each band within the triplet (color graph) or as total protein (the three bands altogether; lower graph). *, p < 0.05 and **, p < 0.01.
Bortezomib partially re-addresses PodR138Q to the plasma membrane
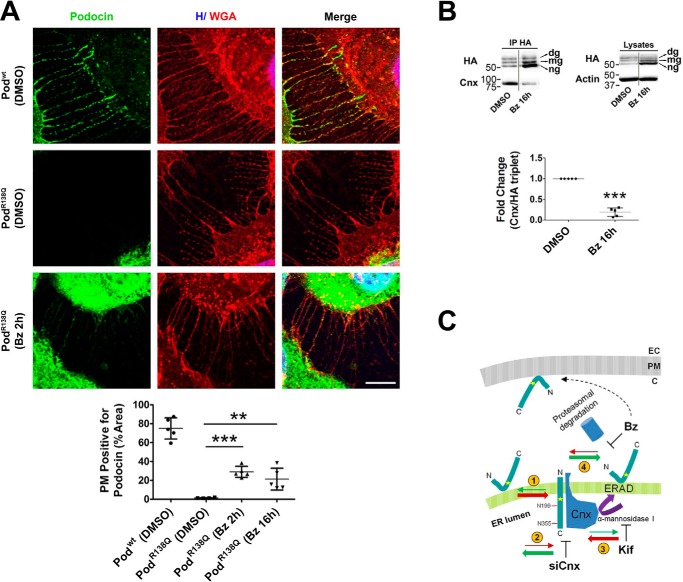
While studying the proteasomal degradation of PodR138Q, we observed by immunofluorescence that a fraction of PodR138Q was localized, after Bz exposure, to thin filopodia PM protrusions, similarly to Podwt in untreated cells, whereas PodR138Q was completely absent from these structures before treatment (Fig. 7A). Targeting of PodR138Q to filopodia, quantified as the percentage of WGA-positive filopodia that were also positive for podocin, revealed that Bz treatment increased plasma membrane targeting of PodR138Q, as soon as 2 h after Bz treatment, although levels did not reach those of Podwt (Fig. 7A, graph). We also observed a decreased interaction of PodR138Q with Cnx in HEK293T cells treated with Bz, which may serve as an indicator of PodR138Q accumulation outside the ER (Fig. 7B). Because we do not see PodR138Q at the PM (Fig. 7A) and the glycosylated forms of PodR138Q seem to be insensitive to Bz (Fig. 3), we hypothesize that only ng-PodR138Q is reaching the PM upon Bz treatment. Taking into account all our data we propose a scheme depicting a dynamic interchange between the hairpin and the transmembrane topology at the different steps of ER QC and subsequent ERAD (Fig. 7C). Briefly, hairpin PodR138Q is detected by the cytoplasmic QC machinery and rapidly degraded by the proteasome. In the meantime, N-glycosylated transmembrane PodR138Q enters the Cnx cycle to be eventually sent for ERAD. Because proteasomal degradation takes place in the cytosol, transmembrane PodR138Q would lose its N-glycan groups at some point between retrotranslocation and proteasomal degradation (20–22), possibly adopting a hairpin topology when exposed to the cytosol. Bz treatment would then inhibit the degradation of ng-PodR138Q, giving it the chance to follow the secretory pathway and reach the PM.
Figure 7.
Bortezomib partially re-addresses podocinR138Q to the plasma membrane. A, immunofluorescence analysis showing PodR138Q (green) in the presence of filopodia, through colocalization with the PM marker WGA (red), after a 2-h treatment with Bz (0.1 μm) or vehicle (DMSO). Podwt co-localization with WGA is included as a positive control. Hoechst nuclei labeling (H) is also shown (blue). Scale bar = 10 μm. Image analysis of the WGA co-localization with podocin only at filopodia was obtained through the quantification of regions of interest (ROIs) that carefully delimit cell perimeters. Graph corresponds to the quantification of one representative experiment. One-way analysis of variance followed by Dunnett's post-test was used as statistical analysis. **, p < 0.01 and ***, p < 0.001. B, co-immunoprecipitation (IP HA) of HA-podocin and Cnx after Bz addition (16 h at 0.1 μm). The graph represents the densitometry quantification of Cnx when podocin is immunoprecipitated. Data are normalized to total immunoprecipitated podocin (HA triplet) and represent to at least three independent experiments. ***, p < 0.001. C, schematic summarizing the influence of different treatments on PodR138Q topology and subcellular localization. Red and green arrows indicate a dynamic change to the transmembrane “wrong” or hairpin “right” topology, respectively. Numbers circled in yellow: (1) there is an imbalance of hairpin PodR138Q toward a transmembrane topology, (2) decreasing PodR138Q interaction with Cnx through siCnx transfection favors the hairpin topology, (3) stabilization of PodR138Q interaction with Cnx, through inhibition of Cnx cycle exit with Kif, enhances podocin transmembrane topology, and (4) inhibition of PodR138Q proteasomal degradation with Bz increases the proportion of hairpin PodR138Q and is the only treatment that allows partial relocalization to the PM. EC, extracellular matrix; PM, plasma membrane; C, cytosol.
Discussion
Despite p.R138Q being the most common podocin mutation causing SRNS in European and North American children (4), little is known about how this mutation affects podocin stability and degradation. In this study, we have found that human PodR138Q, when stably overexpressed in human podocyte cell lines, is clearly resolved as a triplet on immunoblots. The upper bands of these triplets correspond to N-glycosylated forms, which is particularly intriguing because it implies that the C terminus of most PodR138Q is inside the ER lumen. N-Glycosylation, and evidence of transmembrane topology, has been already described for stomatin, for the short isoform of podocin and for podocinP118L, another ER-retained podocin mutant (11, 12, 23). Whereas Pro118, an amino acid that is highly conserved throughout the stomatin family, is located within the hydrophobic intramembrane region and is responsible for the kink of the hairpin topology (12), Arg138 and Arg168, which are also very well conserved, are located within the PHB domain, far away from this region. Nevertheless, we hypothesize that missense mutations in the PHB region may also destabilize podocin hairpin topology, the folding of which is already intrinsically inefficient (11). Indeed, most of the missense mutations affecting the PHB domain typically result in ER-retention (7), and only podocin mutants that are known to be retained in the ER present N-glycosylation levels comparable with PodR138Q. The switch in topology has important implications for the stability and the degradation of PodR138Q. We found the glycosylated PodR138Q forms to be more stable than ng-PodR138Q. The latter, with a hairpin structure, can be rapidly degraded by the proteasome. In contrast, we found evidence that the transmembrane isoform of PodR138Q enters the calnexin cycle, which may explain its longer t½. Of course, it remains that glycosylation and deglycosylation events may be affecting the observed differences in stability of the different glycosylated podocin species, and undoubtedly these processes are also contributing to our results. That said, based on our experiments with Podwt(N199Q,N355S) and PodR138SQ(N199Q,N355S), which show these proteins behave in the same way as ng-Podwt and ng-PodR138Q, we believe that glycosylation of ng-podocin is not contributing to the observed increased stability of the glycosylated species, and in fact the longer t½ of mg- and dg-PodR138Q are due to decreased exposure to the intracellular degradative machinery. According to our results, transmembrane PodR138Q would be exclusively located in the ER as an intermediary of PodR138Q QC, and is there stabilized by interaction with Cnx. Indeed, in contrast to what has been shown for PodP118L (12), we did not find PodR138Q to be localized at the PM by immunofluorescence. This same model may apply for misfolded Podwt, because it also interacts with Cnx and its glycosylated forms are enriched after Kif treatment. Nevertheless, the increased levels of Podwt following NH4Cl addition, together with the lack of response to Bz at short times, suggest that Podwt is mainly degraded in the endosome/lysosome compartment, a finding in accordance with results from other authors (13).
Cnx recognizes a complex code of glucose and mannose trimming in the N-glycan groups of its substrate proteins (21, 24–26). The fact that PodR138Q topology and degradation is sensitive to mannose trimming implies that most probably there is an interaction with Cnx through its N-glycan groups. This result apparently contradicts the data obtained by the specific inhibition of the N-glycan-dependent interaction of Cnx with PodR138Q, because the interaction with Cnx was not decreased. Nevertheless, there is an increasing amount of literature suggesting alternative sites of interaction to the lectin domain of Cnx (i.e. the Cnx transmembrane domain), especially in the case of mutant proteins (27–29). An interesting proposal is that there are at least two different types of interaction occurring sequentially or simultaneously; one carbohydrate based, through the N-glycan groups, and one peptide based, outside the Cnx lectin domain (30–32). This would explain why inhibiting lectin binding is insufficient to impair PodR138Q interaction with Cnx. In contrast, Cnx interaction with Podwt seemed to be more dependent on the N-glycan groups, even though Podwt is predominantly non-glycosylated. However, we cannot rule out a second Cnx interaction site also for Podwt, because deletion of N-glycan sites did not completely impair the interaction with Cnx and ng-Podwt also co-immunoprecipitated with Cnx. Finally, it is tempting to speculate that N-glycosylation and interaction with Cnx may be part of normal podocin biosynthesis and initial folding, and not only a mechanism of podocin quality control. In fact, Cnx is part of the ribosome–translocon complex and its initial association to nascent glycoproteins is usually co-translational (33, 34).
The targeting of PodR138Q to the cell membrane using chemical chaperones as glycerol, trimethylamine-N-oxide, and DMSO has already been reported in transiently transfected human podocytes (35). Nevertheless, there is still no treatment available for SRNS patients carrying the p.R138Q mutation. Here, we report the therapeutic potential of Bz (Velcade®), a reversible inhibitor of the 26S proteasome that has been approved and successfully used in multiple myeloma therapy (36, 37). PodR140Q, the mouse equivalent to PodR138Q, is poorly expressed in the glomeruli of the constitutive Nphs2R140Q/R140Q knock-in mice (6), similarly to our findings in podocyte cell lines. By studying PodR138Q t½ and the degradation pathways it follows, we have found that low intracellular levels of PodR138Q are due to rapid proteasomal degradation. Bz not only increases PodR138Q protein levels, but allows a significant amount of PodR138Q to reach the PM, at least at filopodia. This could be simply caused by an overflow from the ER, because 2 h of Bz treatment already quadruples PodR138Q protein levels. Indeed, proteasomal inhibition has been found to inhibit the retrotranslocation of most ERAD substrates, thus giving them the chance to remain in the secretory pathway (21, 38, 39). Alternatively, because de-glycosylation by cytoplasmic peptide:N-glycanases (PNGases) is an additional step in glycoprotein ERAD (40), Bz would specifically rescue the ng-PodR138Q already targeted for proteasomal degradation. In support of this idea, a promising effect of Bz is that it enriches the ng-PodR138Q fraction corresponding to the potentially functional hairpin topology of PodR138Q at the SD. A successful therapy would need not only to bring PodR138Q to the PM, but also promote its hairpin topology. Interestingly, Bz has been reported to rescue other misfolded mutant proteins back to the PM in vitro and in vivo (42). Additionally, second generation proteasomal inhibitors are currently being tested in clinical trials with less severe side effects than those of Bz, such as neurotoxicity or lymphopenia (43). Altogether, although we lack in vivo studies, we propose that increasing ng-PodR138Q protein levels through the inhibition of the proteasomal degradation may be a reasonable strategy to treat patients with p.R138Q mutation and possibly with other ER-retained podocin mutations.
Experimental procedures
Plasmids, cell culture, and establishment of lentiviral cell lines
Human podocin-coding constructs were generated as described by Tory et al. (44). Human wildtype NPHS1 cDNA, encoding nephrin, was amplified from the construct described by Philippe et al. (6) and subcloned into NotI and SpeI sites of LentiORF pLEX-MCS (Open Biosystems). Thus, the encoded proteins consist of podocin with two hemagglutinin tags (2HA) fused to its N terminus, and nephrin with a V5 tag fused to its N terminus. Site-directed mutagenesis (QuikChange kit, Stratagene) was used to generate the missense mutations used in this study: p.R138Q, p.R168C, p.N199Q, p.N355S for podocin, and p.S366R for nephrin. All constructs were verified by Sanger sequencing. A human immortalized podocyte cell line (AB8/13), obtained by transfection of the temperature-sensitive mutant tsA58 of the SV40-T-antigen-encoding gene, was kindly provided by M. A. Saleem (45). Stable podocyte cell lines were obtained by transduction of the above cell line with lentiviral vectors expressing either podocin (wildtype or mutants) or nephrin (wildtype or mutants) at a multiplicity of infection of 1, and subsequently selected by puromycin (2 μg/ml). Podocytes were cultured at 33 °C with 7% CO2 in RPMI 1640 medium supplemented with 10% fetal bovine serum, insulin/transferrin/selenium, glutamine, and penicillin/streptomycin (all from Life Technologies). At this growth-permissive temperature, podocytes are proliferating and undifferentiated, and do not express either endogenous podocin or nephrin. HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, glutamine, and penicillin/ streptomycin (all from Life Technologies). Cell lines used in this study were tested mycoplasma-free.
Antibodies, enzymes, and chemical compounds
PNGase F was purchased from New England BioLabs. Tunicamycin, cycloheximide, bortezomib, NH4Cl, castanospermine, and kifunensine were purchased from Sigma. The commercial antibodies used were as follows: mouse anti-HA (HA.11 clone 16B12, Covance), mouse anti-calnexin (Cnx) (clone AF18, Enzo Life Sciences), mouse anti-CD63 (clone H5C6, DSHB), and mouse anti-calnexin (clone C5C9, Cell Signaling Technology). Rabbit anti-podocin AP-P35 (Pod) was described previously (46). Alexa Fluor 555-conjugated wheat germ agglutinin (WGA555) (W32464, Life Technologies) was used to stain the plasma membrane. Rat IgG2a (isotype control clone 2H3, MBL) for immunoprecipitation control experiments was purchased from MBL. Secondary antibodies for immunoblotting were sheep anti-mouse and donkey anti-rabbit HRP-conjugated antibodies (GE Healthcare UK). Secondary antibodies for immunofluorescence were donkey anti-rabbit and anti-mouse Alexa Fluor 488- and 647-conjugated antibodies (Life Sciences).
siRNA experiments
The following siRNAs were used to transfect podocyte cell lines stably expressing podocin to specifically knockdown the human CNX (calnexin) gene: siRNA-1, 5′-AAGACGAUACCGAUGAUGAAA-3′ and siRNA-2, 5′AAUGUGGUGGUGCCUAUGUGA-3′. siRNA against the luciferase gene (Luc, 5′-GCCAUUCUAUCCUCUAGAGGAUG-3′) was used as a siRNA control. siRNAs were transfected using Lipofectamine® RNAiMAX Reagent (Invitrogen) at a concentration of 20 nm and the efficacy of siRNA was tested 3 days after transfection.
Quantitative real-time PCR
Total RNA was extracted from podocyte cell lines using the Qiagen extraction RNeasy® kit (Qiagen) and treated with DNase I. One μg of total RNA was reverse-transcribed using SuperScript II according to the manufacturer's protocol (Invitrogen). The relative expression levels of the studied mRNAs were determined by quantitative real-time PCR using Absolute SYBR Green ROX Mix (ABgene) with the following specific primers for human NPHS2: forward (929F) 5′-GGCTGAAGCGCAAAGACAAG-3′ and reverse (988R) 5′-GCAGCCTTTTCCGCTTCTG-3′. Human hypoxanthine phosphoribosyltransferase (Hprt) was used as an internal standard. Data were analyzed with the 2−ΔΔCt method (47).
Immunofluorescence
Podocytes were cultured on type I collagen-coated coverslips and either transiently transfected using FuGENE® HD (Promega) or treated with different drugs. When appropriate, cells were washed once with cold PBS, incubated with Alexa Fluor 555-conjugated WGA (1:600) for 15 min at 4 °C, and then fixed with ice-cold ethanol for 5 min. Fixed cells were blocked with 1× PBS, 1% BSA for 30 min before incubation with mouse anti-HA primary antibodies (1:500) or rabbit anti-podocin AP-P35 (1:300) when combined with mouse anti-calnexin (1:500) or mouse anti-CD63 (1:200), followed by Alexa Fluor 488- or 647-conjugated secondary antibodies (1:200). Confocal images were captured using a ×40 oil objective attached to a Leica SP8 confocal microscope. At least five random fields, with the only condition that filopodia were well visualized in the WGA channel, were considered for the quantification of the percentage area of WGA colocalization with podocin using ImageJ 1.48i software. Masks were carefully created to specifically quantify the labeling of the filopodia. Confocal settings and Image J thresholds were kept the same.
Immunoblotting
Proteins from podocyte cell lines were extracted in lysis buffer containing 150 mm NaCl, 50 mm Tris-HCl, pH 7, 0.5% Triton X-100 with CompleteTM protease inhibitors (Roche Applied Science). Protein dosage was then performed using the BCA protein assay kit (Thermo Scientific). Fifty micrograms of protein were loaded on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. Membranes were blocked in 5% skimmed milk in 1× Tris-buffered saline, 0.1% Tween 20 (TBST) for 1 h and incubated with primary antibodies at a 1:1,000 dilution. After washing, membranes were incubated with HRP-conjugated secondary antibodies diluted 1:10,000 in TBST, 5% milk for 1 h at room temperature. Signals were detected using ECL reagents (Amersham Biosciences) and acquired in a Fusion Fx7 darkroom (Vilber Lourmat). Densitometry quantification was performed using Bio-1D software.
Immunoprecipitation
HEK293T cells were transiently transfected with HA-tagged WT podocin and mutants using calcium phosphate. Forty-eight h post-transfection, cells were lysed in 150 mm NaCl, 25 mm Tris-HCl, pH 8, 0.5% Triton X-100 with protease inhibitors, and HA-tagged podocin was immunoprecipitated using the μMACSTM Epitope Tag Protein Isolation Kit (Miltenyi Biotec). Briefly, fresh lysates (1–1.5 mg of protein) were incubated either with mouse anti-calnexin antibodies, followed by a 30-min incubation with magnetic beads-coupled to protein A, or directly with magnetic beads coupled to a HA antibody. A rat IgG2a isotype control was included to discard unspecific interactions. Immunoprecipitated proteins were isolated using μMACS® Separation Columns in a magnetic μMACS separator and subsequently eluted with 1× Laemmli buffer. Lysates and immunoprecipitated samples were subjected to immunoblot.
Statistical analyses
All immunoblots were normalized to β-actin and then to the corresponding control group or the immunoprecipitated protein in control conditions. Statistical analysis of at least three independent experiments was done using the one-sample two-tailed t test (41) or a one-way analysis of variance, p values: *, p < 0.05; **, p < 0.01; ***, p < 0.001. GraphPad Prism 5 software was used to perform all statistical analyses (mean ± S.D.).
Author contributions
M.-C. S.-P., F. C. T., C. Antignac, and G. Mollet conceptualization; M.-C. S.-P., F. C. T., F. N., and G. Mollet formal analysis; M.-C. S.-P., F. C. T., K. T., and G. Mollet validation; M.-C. S.-P., F. C. T., F. N., C. Arrondel, S. S., G. Martin, and K. T. investigation; M.-C. S.-P., F. C. T., F. N., C. Arrondel, S. S., and G. Martin methodology; M.-C. S.-P., F. C. T., C. Antignac, and G. Mollet writing-original draft; M.-C. S.-P., F. C. T., F. N., C. Arrondel, S. S., G. Martin, K. T., C. Antignac, and G. Mollet writing-review and editing; C. Antignac supervision; C. Antignac and G. Mollet project administration; G. Mollet funding acquisition.
Supplementary Material
Acknowledgments
We thank Ana Maria Cuervo (Albert Einstein College, NY) for helpful discussion and advice; Gisèle Froment, Didier Nègre, and Caroline Costa from the Lentivectors Production Facility/SFR BioSciences Gerland-Lyon Sud (UMS3444/US8) and the Confocal Platform at Imagine Institute.
This work was supported by European Union's Seventh Framework Programme Grant FP7/2007–2013/n°305608-EURenOmics (to C. Antignac), State funding from the Agence Nationale de la Recherche under “Investissements d'avenir” program Grant ANR-10-IAHU-01 (to C. Antignac), and Association des Malades d'un Syndrome Néphrotique (AMSN) (Programme Ambition Recherche 2008–2011 (to G. Mollet). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- GFB
- glomerular filtration barrier
- SRNS
- steroid-resistant nephrotic syndrome
- SD
- slit diaphragm
- PHB
- prohibitin homology domain
- ER
- endoplasmic reticulum
- HA
- human influenza hemagglutinin
- WGA
- wheat germ agglutinin
- PNGase F
- peptide:N-glycosidase F
- Bz
- bortezomib
- QC
- quality control
- ERAD
- endoplasmic reticulum–associated degradation
- Cnx
- calnexin
- PM
- plasma membrane
- Cst
- castanospermine
- Kif
- kifunensine
- dg
- double-glycosylated
- mg
- mono-glycosylated
- ng
- non-glycosylated.
References
- 1. Mekahli D., Liutkus A., Ranchin B., Yu A., Bessenay L., Girardin E., Van Damme-Lombaerts R., Palcoux J. B., Cachat F., Lavocat M. P., Bourdat-Michel G., Nobili F., and Cochat P. (2009) Long-term outcome of idiopathic steroid-resistant nephrotic syndrome: a multicenter study. Pediatr. Nephrol. 24, 1525–1532 10.1007/s00467-009-1138-5 [DOI] [PubMed] [Google Scholar]
- 2. Tune B. M., and Mendoza S. A. (1997) Treatment of the idiopathic nephrotic syndrome: regimens and outcomes in children and adults. J. Am. Soc. Nephrol. 8, 824–832 [DOI] [PubMed] [Google Scholar]
- 3. Boute N., Gribouval O., Roselli S., Benessy F., Lee H., Fuchshuber A., Dahan K., Gubler M. C., Niaudet P., and Antignac C. (2000) NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat. Genet. 24, 349–354 10.1038/74166 [DOI] [PubMed] [Google Scholar]
- 4. Sadowski C. E., Lovric S., Ashraf S., Pabst W. L., Gee H. Y., Kohl S., Engelmann S., Vega-Warner V., Fang H., Halbritter J., Somers M. J., Tan W., Shril S., Fessi I., Lifton R. P., et al. (2015) A single-gene cause in 29.5 % of cases of steroid-resistant nephrotic syndrome. J. Am. Soc. Nephrol. 26, 1279–1289 10.1681/ASN.2014050489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hinkes B. G., Mucha B., Vlangos C. N., Gbadegesin R., Liu J., Hasselbacher K., Hangan D., Ozaltin F., Zenker M., Hildebrandt F., and Arbeitsgemeinschaft für Paediatrische Nephrologie Study, G. (2007) Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2). Pediatrics 119, e907–919 10.1542/peds.2006-2164 [DOI] [PubMed] [Google Scholar]
- 6. Philippe A., Weber S., Esquivel E. L., Houbron C., Hamard G., Ratelade J., Kriz W., Schaefer F., Gubler M. C., and Antignac C. (2008) A missense mutation in podocin leads to early and severe renal disease in mice. Kidney Int. 73, 1038–1047 10.1038/ki.2008.27 [DOI] [PubMed] [Google Scholar]
- 7. Roselli S., Moutkine I., Gribouval O., Benmerah A., and Antignac C. (2004) Plasma membrane targeting of podocin through the classical exocytic pathway: effect of NPHS2 mutations. Traffic 5, 37–44 10.1046/j.1600-0854.2003.00148.x [DOI] [PubMed] [Google Scholar]
- 8. Huber T. B., Kottgen M., Schilling B., Walz G., and Benzing T. (2001) Interaction with podocin facilitates nephrin signaling. J. Biol. Chem. 276, 41543–41546 10.1074/jbc.C100452200 [DOI] [PubMed] [Google Scholar]
- 9. Huber T. B., Schermer B., Müller R. U., Höhne M., Bartram M., Calixto A., Hagmann H., Reinhardt C., Koos F., Kunzelmann K., Shirokova E., Krautwurst D., Harteneck C., Simons M., Pavenstädt H., et al. (2006) Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc. Natl. Acad. Sci. U.S.A. 103, 17079–17086 10.1073/pnas.0607465103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schwarz K., Simons M., Reiser J., Saleem M. A., Faul C., Kriz W., Shaw A. S., Holzman L. B., and Mundel P. (2001) Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J. Clin. Invest. 108, 1621–1629 10.1172/JCI200112849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kadurin I., Huber S., and Gründer S. (2009) A single conserved proline residue determines the membrane topology of stomatin. Biochem. J. 418, 587–594 10.1042/BJ20081662 [DOI] [PubMed] [Google Scholar]
- 12. Schurek E. M., Völker L. A., Tax J., Lamkemeyer T., Rinschen M. M., Ungrue D., Kratz J. E. 3rd, Sirianant L., Kunzelmann K., Chalfie M., Schermer B., Benzing T., and Höhne M. (2014) A disease-causing mutation illuminates the protein membrane topology of the kidney-expressed prohibitin homology (PHB) domain protein podocin. J. Biol. Chem. 289, 11262–11271 10.1074/jbc.M113.521773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gödel M., Ostendorf B. N., Baumer J., Weber K., and Huber T. B. (2013) A novel domain regulating degradation of the glomerular slit diaphragm protein podocin in cell culture systems. PLoS ONE 8, e57078 10.1371/journal.pone.0057078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rinschen M. M., Bharill P., Wu X., Kohli P., Reinert M. J., Kretz O., Saez I., Schermer B., Höhne M., Bartram M. P., Aravamudhan S., Brooks B. R., Vilchez D., Huber T. B., Müller R. U., Krüger M., and Benzing T. (2016) The ubiquitin ligase Ubr4 controls stability of podocin/MEC-2 supercomplexes. Hum. Mol. Genet. 25, 1328–1344 10.1093/hmg/ddw016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W. W., Sistonen L., Morimoto R. I., and Lee A. S. (1994) Stress induction of the mammalian GRP78/BiP protein gene: in vivo genomic footprinting and identification of p70CORE from human nuclear extract as a DNA-binding component specific to the stress regulatory element. Mol. Cell. Biol. 14, 5533–5546 10.1128/MCB.14.8.5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wooden S. K., Li L. J., Navarro D., Qadri I., Pereira L., and Lee A. S. (1991) Transactivation of the grp78 promoter by malfolded proteins, glycosylation block, and calcium ionophore is mediated through a proximal region containing a CCAAT motif which interacts with CTF/NF-I. Mol. Cell. Biol. 11, 5612–5623 10.1128/MCB.11.11.5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshida H., Haze K., Yanagi H., Yura T., and Mori K. (1998) Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273, 33741–33749 10.1074/jbc.273.50.33741 [DOI] [PubMed] [Google Scholar]
- 18. Drozdova T., Papillon J., and Cybulsky A. V. (2013) Nephrin missense mutations: induction of endoplasmic reticulum stress and cell surface rescue by reduction in chaperone interactions. Physiol. Rep. 1, e00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huber T. B., Simons M., Hartleben B., Sernetz L., Schmidts M., Gundlach E., Saleem M. A., Walz G., and Benzing T. (2003) Molecular basis of the functional podocin-nephrin complex: mutations in the NPHS2 gene disrupt nephrin targeting to lipid raft microdomains. Hum. Mol. Genet. 12, 3397–3405 10.1093/hmg/ddg360 [DOI] [PubMed] [Google Scholar]
- 20. Hirsch C., Blom D., and Ploegh H. L. (2003) A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22, 1036–1046 10.1093/emboj/cdg107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lederkremer G. Z., and Glickman M. H. (2005) A window of opportunity: timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem. Sci. 30, 297–303 10.1016/j.tibs.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 22. Olzmann J. A., Kopito R. R., and Christianson J. C. (2013) The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5, A013185 10.1101/cshperspect.a013185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Völker L. A., Schurek E. M., Rinschen M. M., Tax J., Schutte B. A., Lamkemeyer T., Ungrue D., Schermer B., Benzing T., and Höhne M. (2013) Characterization of a short isoform of the kidney protein podocin in human kidney. BMC Nephrol. 14, 102 10.1186/1471-2369-14-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frenkel Z., Gregory W., Kornfeld S., and Lederkremer G. Z. (2003) Endoplasmic reticulum-associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6–5GlcNAc2. J. Biol. Chem. 278, 34119–34124 10.1074/jbc.M305929200 [DOI] [PubMed] [Google Scholar]
- 25. Hammond C., Braakman I., and Helenius A. (1994) Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. U.S.A. 91, 913–917 10.1073/pnas.91.3.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ware F. E., Vassilakos A., Peterson P. A., Jackson M. R., Lehrman M. A., and Williams D. B. (1995) The molecular chaperone calnexin binds Glc1Man9GlcNAc2 oligosaccharide as an initial step in recognizing unfolded glycoproteins. J. Biol. Chem. 270, 4697–4704 10.1074/jbc.270.9.4697 [DOI] [PubMed] [Google Scholar]
- 27. Cannon K. S., Hebert D. N., and Helenius A. (1996) Glycan-dependent and -independent association of vesicular stomatitis virus G protein with calnexin. J. Biol. Chem. 271, 14280–14284 10.1074/jbc.271.24.14280 [DOI] [PubMed] [Google Scholar]
- 28. Fontanini A., Chies R., Snapp E. L., Ferrarini M., Fabrizi G. M., and Brancolini C. (2005) Glycan-independent role of calnexin in the intracellular retention of Charcot-Marie-tooth 1A Gas3/PMP22 mutants. J. Biol. Chem. 280, 2378–2387 10.1074/jbc.M405104200 [DOI] [PubMed] [Google Scholar]
- 29. Swanton E., High S., and Woodman P. (2003) Role of calnexin in the glycan-independent quality control of proteolipid protein. EMBO J. 22, 2948–2958 10.1093/emboj/cdg300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arunachalam B., and Cresswell P. (1995) Molecular requirements for the interaction of class II major histocompatibility complex molecules and invariant chain with calnexin. J. Biol. Chem. 270, 2784–2790 10.1074/jbc.270.6.2784 [DOI] [PubMed] [Google Scholar]
- 31. Lederkremer G. Z. (2009) Glycoprotein folding, quality control and ER-associated degradation. Curr. Opin. Struct. Biol. 19, 515–523 10.1016/j.sbi.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 32. Williams D. B. (2006) Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J. Cell Sci. 119, 615–623 10.1242/jcs.02856 [DOI] [PubMed] [Google Scholar]
- 33. Chen W., Helenius J., Braakman I., and Helenius A. (1995) Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc. Natl. Acad. Sci. U.S.A. 92, 6229–6233 10.1073/pnas.92.14.6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lakkaraju A. K., Abrami L., Lemmin T., Blaskovic S., Kunz B., Kihara A., Dal Peraro M., and van der Goot F. G. (2012) Palmitoylated calnexin is a key component of the ribosome-translocon complex. EMBO J. 31, 1823–1835 10.1038/emboj.2012.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohashi T., Uchida K., Uchida S., Sasaki S., and Nihei H. (2003) Intracellular mislocalization of mutant podocin and correction by chemical chaperones. Histochem. Cell Biol. 119, 257–264 [DOI] [PubMed] [Google Scholar]
- 36. Adams J. (2004) The proteasome: a suitable antineoplastic target. Nat. Rev. Cancer 4, 349–360 10.1038/nrc1361 [DOI] [PubMed] [Google Scholar]
- 37. Hideshima T., Richardson P. G., and Anderson K. C. (2003) Targeting proteasome inhibition in hematologic malignancies. Rev. Clin. Exp. Hematol. 7, 191–204 [PubMed] [Google Scholar]
- 38. Bubeck A., Reusch U., Wagner M., Ruppert T., Muranyi W., Kloetzel P. M., and Koszinowski U. H. (2002) The glycoprotein gp48 of murine cytomegalovirusL proteasome-dependent cytosolic dislocation and degradation. J. Biol. Chem. 277, 2216–2224 10.1074/jbc.M104178200 [DOI] [PubMed] [Google Scholar]
- 39. Kamhi-Nesher S., Shenkman M., Tolchinsky S., Fromm S. V., Ehrlich R., and Lederkremer G. Z. (2001) A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 12, 1711–1723 10.1091/mbc.12.6.1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suzuki T., Park H., and Lennarz W. J. (2002) Cytoplasmic peptide:N-glycanase (PNGase) in eukaryotic cells: occurrence, primary structure, and potential functions. FASEB J. 16, 635–641 10.1096/fj.01-0889rev [DOI] [PubMed] [Google Scholar]
- 41. Fay D. S., and Gerow K. (2013) A biologist's guide to statistical thinking and analysis. WormBook: the Online Review of C. elegans Biology, 1–54 10.1895/wormbook.1.159.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bonuccelli G., Sotgia F., Capozza F., Gazzerro E., Minetti C., and Lisanti M. P. (2007) Localized treatment with a novel FDA-approved proteasome inhibitor blocks the degradation of dystrophin and dystrophin-associated proteins in mdx mice. Cell Cycle 6, 1242–1248 10.4161/cc.6.10.4182 [DOI] [PubMed] [Google Scholar]
- 43. Buac D., Shen M., Schmitt S., Kona F. R., Deshmukh R., Zhang Z., Neslund-Dudas C., Mitra B., and Dou Q. P. (2013) From bortezomib to other inhibitors of the proteasome and beyond. Curr. Pharmaceut. Design 19, 4025–4038 10.2174/1381612811319220012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tory K., Menyhárd D. K., Woerner S., Nevo F., Gribouval O., Kerti A., Stráner P., Arrondel C., Huynh Cong E., Tulassay T., Mollet G., Perczel A., and Antignac C. (2014) Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat. Genet. 46, 299–304 10.1038/ng.2898 [DOI] [PubMed] [Google Scholar]
- 45. Saleem M. A., O'Hare M. J., Reiser J., Coward R. J., Inward C. D., Farren T., Xing C. Y., Ni L., Mathieson P. W., and Mundel P. (2002) A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol. 13, 630–638 [DOI] [PubMed] [Google Scholar]
- 46. Roselli S., Gribouval O., Boute N., Sich M., Benessy F., Attié T., Gubler M. C., and Antignac C. (2002) Podocin localizes in the kidney to the slit diaphragm area. Am. J. Pathol. 160, 131–139 10.1016/S0002-9440(10)64357-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.