Abstract
Background
Inflammatory bowel disease (IBD) is commonly divided into 2 entities: Crohn’s disease (CD) and ulcerative colitis (UC). Differentiating between these entities when dealing with IBD confined to the colon is important, especially when planning surgical treatment. Due to ambiguous histological or endoscopic findings, accurate diagnosis is not possible in up to 15% of cases. The aim of this study was to determine whether plasma microRNAs (miRNAs) can help differentiate Crohn’s colitis (CC) from ulcerative colitis.
Methods
Patients with isolated CC and with UC were enrolled in our study from January 2010 to May 2016. Peripheral blood was collected, and total RNA was isolated from plasma. Screening was performed for 380 common miRNAs. miRNAs that were differentially expressed between these 2 groups were chosen, and their differential expression was confirmed using single miRNA assays in a larger sample size. A predictive model was generated using these data. Significantly differentially expressed miRNAs were then validated utilizing the predictive model to assess blinded data from the single assays.
Results
Screening was performed on 8 patients from each group. Seven differentially expressed miRNAs were chosen for single assay confirmation. Two miRNAs (miR-598, miR-642) were consistently different between the patient groups (P = 0.013, P = 0.005). Using blinded data, these 2 miRNAs were validated using the predictive model, achieving an overall accuracy of 75% (95% confidence interval, 40.7–92.9).
Conclusions
We identified 2 plasma miRNAs that differentiated CC from UC. Our data indicate the promise and feasibility of a plasma miRNA–based assay to distinguish between these 2 conditions.
Keywords: microRNA, Crohn’s colitis, ulcerative colitis
Inflammatory bowel disease (IBD) refers to a chronic immune-mediated inflammatory state of the gastrointestinal system. IBD is commonly divided into 2 entities: Crohn’s disease (CD) and ulcerative colitis (UC). IBD is most common in westernized countries, with an incidence of up to 20 per 100,000 person-years for each (UC, CD) and with an estimated 1.5 million individuals currently affected in North America.1 Currently, the diagnosis is based upon clinical, endoscopic, radiographic, and laboratory findings. In most cases, the diagnosis of CD or UC is clear. However, when the disease is confined to the colon, distinguishing between the 2 can be problematic in up to 15% of cases due to ambiguous findings on histology or endoscopy.2–4 When planning surgical treatment, differentiating between these 2 conditions is important. Providing the wrong treatment can lead to serious morbidity (eg, performing a total proctocolectomy with ileo-anal pouch anastomosis for presumed UC, when in reality fistulizing CD is present).5, 6
There has been increasing interest in biomarkers to more precisely diagnose IBD and differentiate between its subtypes.7 Plasma microRNA (miRNA) represent 1 such group of potential biomarkers. miRNAs are a group of small (~18–24 nucleotides) noncoding RNAs that have an important role in the post-transcriptional regulation of gene expression. They bind to the 3′ untranslated region of messenger RNA (mRNA) and can inhibit translation directly or by promoting mRNA degradation. miRNAs have been shown to be associated with specific disease processes, such as cancer and inflammation.8, 9 Due to their stable nature in circulating plasma, miRNA panels have also been used for the diagnosis of various disease processes.10–13 There are few publications describing the use of specific plasma-based miRNAs in the diagnosis of IBD,14 and specifically regarding the differentiation between UC and Crohn’s colitis (CC).15, 16 We hypothesized that a plasma miRNA profile is different in CC as compared with UC. Our objective was to develop a panel of miRNAs capable of differentiating CC from UC.
MATERIALS AND METHODS
Patient selection
The study was approved by the University of Louisville Institutional Review Board (IRB), and written informed consent was obtained from all subjects. Patient data were managed in accordance with IRB guidelines. No intervention occurred during the study apart from blood and data collection. Patients were accrued from a large university digestive disease practice during the period January 2010 to May 2016. We included patients for whom we had access to medical records encompassing the diagnosis and treatment of IBD, and who at the time of blood sampling had at least part of the inflamed large bowel still in vivo. Within the CD patient group, we included only patients with isolated nonfistulizing CC without peri-anal CD, because this was the population in whom differentiation from UC was most difficult. Diagnostic features of CD included the presence of skip areas, fat wrapping and thickening of the bowel wall, and the microscopic features of CD (ie, non-necrotizing granulomas, transmural lymphoid aggregates without an overlying ulcer, and transmural inflammation). The diagnosis of UC was made in the presence of continuous disease from the dentate line to the proximal extent of disease, the presence of inflammation limited to the mucosa, and the presence of crypt abscesses, among others.3, 17 Patients without a clear diagnosis of either CC or UC were excluded, as were patients with a concomitant malignancy. Demographic and clinical data were collected from the patients’ medical records. Following consent, peripheral blood was collected from subjects in 8-mL EDTA-vacutainers (BD, Franklin Lakes, NJ). Plasma was immediately isolated from whole blood by centrifugation at 3500 rpm for 15 minutes as previously described and then frozen at –80°C for later use.10, 18 Total RNA was extracted using a 200-µL aliquot from each plasma sample using the miRNeasy Serum/Plasma Isolation Kit (Qiagen, Valencia, CA).19 Total RNA concentration and purity of each sample were determined using a Nanodrop 2000 spectrophotometer (ThermoFisher Scientific, Middlesex, MA) (Supplemental Table 1).20
Study design
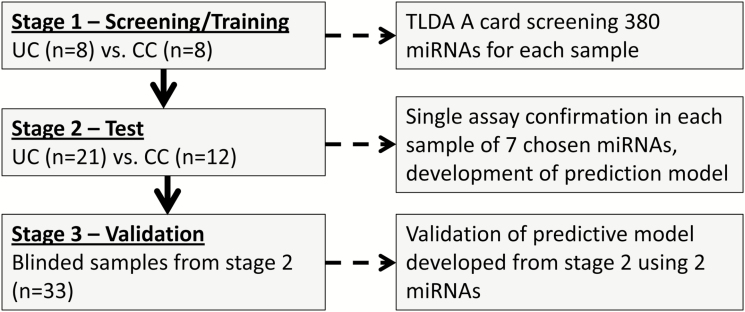
This study was performed in 3 stages (Fig. 1):
FIGURE 1.
Study design. See text for details.
Stage 1: Screening. Patients with isolated CC and UC were screened for expression of 380 human miRNAs. Plasma miRNAs with the potential to differentiate between the UC and CC groups (differentially expressed miRNA) were chosen for further investigation.
Stage 2: Test. Selected plasma miRNAs were confirmed using specific single assays in new patient samples, and a prediction model was developed.
Stage 3: Validation. Patient diagnosis was predicted using the prediction model as applied to blinded data from patient samples analyzed using single assays in stage 2.
Stage 1: screening phase
Total RNA was extracted as previously described. Complementary DNA was produced by reverse transcription using Megaplex RT (Applied Biosystems, Foster City, CA)21 without pre-amplification protocol using human pool primers A (Applied Biosystems). Each patient sample was screened for miRNA expression using Taqman low-density array (TLDA) pool A cards (Applied Biosystems), screening for 380 common human miRNAs. Quantitative real-time polymerase chain reaction (qRT-PCR) was completed using the ViiA7 real-time PCR system (Applied Biosystems), according to the manufacturer’s protocol, as previously described.18
Reference gene selection
NORMFINDER software was used on our screening data, together with published data on accepted reference genes (Supplemental Table 2). Based on this analysis, miR-16 was chosen as our endogenous reference gene both due to its stability and expression within our samples.
Stage 2: test phase (confirmation and sample enlargement)
The original samples used for the screening phase (8 UC and 8 CC) and additional samples (13 UC and 4 CC) were used. Total RNA was extracted as previously described. Reverse transcription and qRT-PCR was performed using TaqMan (Applied Biosystems, Foster City, CA) universal master mix II protocol with the relevant TaqMan microRNA assays (Applied Biosystems). qRT-PCR reactions were performed using a Step-One Plus RT-PCR System (Life Technologies, Carlsbad CA) using default thermal cycling conditions. All reactions were run in duplicate. A prediction model was then developed.
Stage 3: validation phase
Prediction of sample identity (UC or CC) was performed using blinded data from stage 2 using the predictive model developed during the test phase.
Statistical analysis
Stage 1: screening phase
miRNA expression was compared between the UC and the CC groups using the comparative delta cycle threshold method (ΔCT). In cases when the miRNA expression was undetermined, CT values were replaced with a numerical value of 40. miRNAs that were significantly dysregulated between UC and CC were identified from a potential list of 380 miRNAs.
Based on a literature search, about 70 unique miRNAs previously linked to IBD were identified. We therefore expected to identify at least 7 miRNAs that could differentiate these 2 groups (CC and UC). Using the method of Jung et al,22 in order to identify approximately 7 miRNAs (ca. 10% of 70 potentially significant miRNAs among 380 miRNAs) to be significant at a false discovery rate of 5%, the adjusted alpha was set at 0.0011. With an alpha of 0.0011 and a power of 90%, we needed a sample size of n1 = n2 = 8 to detect the effect size of 2.6 SD units’ effects, assuming a common SD in the 2 groups for comparing means using a 1-sided 2-sample t test. The effect size of 2.6 SDs is considered to be a very large effect size. For example, if the SD of ΔCT values for any miRNA is 4, we can then detect the difference of mean ΔCT of at least 10 units apart.
Stage 2: test phase
miRNAs identified in the screening stage were tested using single assays. CT values were obtained as previously described and using miR-16 as the reference gene. ΔCT values were calculated and compared between the UC and CC groups. As previously described, miRNAs with an undetermined CT expression were calculated using a numerical value of 40.
We used an adjusted alpha level of 0.007 (=0.05/7) based on 7 miRNAs that were examined. We enrolled additional subjects from each group (13 UC and 4 CC patients) with balanced age, race, and sex factors. Using the same approach for comparing UC and CD samples (n1 = 21 and n2 = 12), we had at least 90% power to detect the same effect size. We expected 1–2 miRNAs to remain highly significant, and therefore, we used a significance level of 0.025, which should have an 87% ability to detect the same effect size.
We used the test data of 12 subjects from each of the 2 disease groups to build a predictive model similar to others and that we had previously used in our collaborative group.10, 18 With the combined sample size of 18 of each of the sample types, we can differentiate sensitivity/specificity from 65% to 90% at an alpha of 0.05 and a power of 75%.
Stage 3: validation phase
Utilizing the sensitivity and specificity values calculated from stage 2, we validated our results using blinded patient data from stage 2, which consisted of the miRNAs that were differentially expressed between groups. The predictive model was applied to the combined sample size of 33 (n = 21 & n = 12) to predict disease diagnosis.
RESULTS
Patients
A total of 16 patients (8 from each group, UC and CC) were enrolled for screening. Thirty-three patients were included in the test and validation phases (stage 2 & stage 3) (21 UC and 12 CC patients). There was no difference between patient groups with regards to sex, race, and age (Table 1).
Table 1.
Demographic Parameters of the Patient Population
| Crohn’s Colitis | Ulcerative Colitis | P * | |
|---|---|---|---|
| Stage 1 (screening), No. pts | 8 | 8 | — |
| Female sex, No. | 6 | 5 | 0.58 |
| Caucasian race, No. | 8 | 8 | 1.0 |
| Age, average (±SD), y | 50.2 (±16.5) | 55 (±14.45) | 0.55 |
| Stage 2 (test), No. pts | 12 | 21 | — |
| Female sex, No. | 7 | 12 | 0.95 |
| Caucasian race, No. | 11 | 20 | 0.68 |
| Age, average (±SD), y | 47.7 (±15.1) | 46.6 (±18.1) | 0.86 |
*Chi-square test for categorical data and Student t test for continuous data.
Stage 1: screening
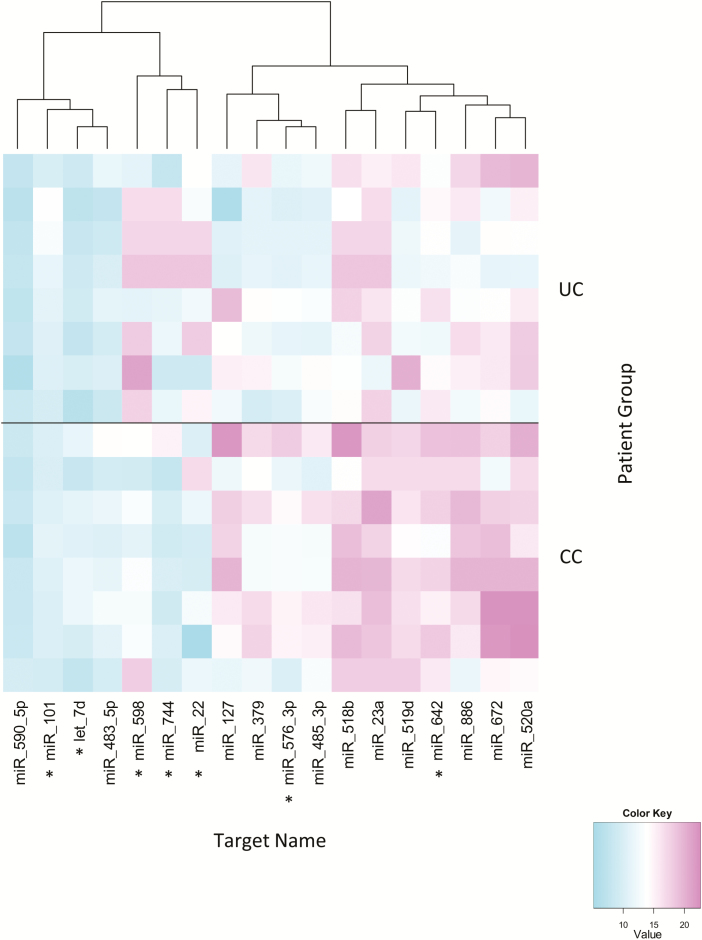
Total RNA isolated from plasma was screened for the presence of 380 common miRNAs using TLDA A cards (stage 1). Following exclusion of miRNAs that were more than 50% unexpressed in both groups, 13 miRNAs were found to be significantly differentially expressed between groups, with another 5 miRNAs approaching statistical significance. Using the screening data, miRNAs were chosen for further study based on P value and biological relevance (Fig. 2).16, 23 The following 7 miRNAs were chosen for the test phase: miR-484, miR-598, miR-576-3p, let-7d, miR-22, miR-101, miR-642, and miR-744.
FIGURE 2.
Results of screening data identifying differentially expressed miRNAs between ulcerative colitis (n = 8) and Crohn’s colitis (n = 8). The heat map shows delta cycle threshold values for 18 miRNAs differentially expressed between UC and CC. Seven miRNAs were chosen based on P value and biological relevance for single assay validation. *miRNA selected for further study.
Stage 2: test
The 7 miRNAs were tested using single assays. Two miRNAs were found to be significantly upregulated in UC as compared with CC: miR-598 (P = 0.013) and miR-642 (P = 0.005). The results of the single assay comparisons between the UC and CC groups are shown in Table 2.
Table 2.
Test Phase Using Single Assays for UC (n = 21) vs CC (n = 12)
| miRNA | P * | FDR† | Fold Change‡ | AUC (95% CI) | Sensitivity (95% CI), % | Specificity (95% CI), % | Accuracy (95% CI), % |
|---|---|---|---|---|---|---|---|
| let-7d | 0.361 | 0.615 | 1.654 | 0.67 (0.53–0.81) | 61.9 (46.8–75) | 75.0 (54.8–88.3) | 66.7 (54.6–76.9) |
| miR-101 | 0.541 | 0.659 | 1.220 | 0.63 (0.50–0.76) | 52.4 (37.7–66.6) | 79.2 (59.1–91.2) | 62.1 (50.0–72.9) |
| miR-22 | 0.659 | 0.659 | 0.856 | 0.44 (0.30–0.58) | 73.8 (58.8–84.8) | 100 (83.7–100) | 47.0 (35.4–58.8) |
| miR-576-3p | 0.596 | 0.659 | 1.351 | 0.66 (0.52–0.80) | 64.3 (49.1–77.1) | 79.2 (59.1–91.2) | 69.7 (57.7–79.5) |
| miR-598 | 0.013 | 0.054 | 1.831 | 0.67 (0.54–0.80) | 69.0 (53.9–81) | 62.5 (42.6–78.9) | 66.7 (54.6–76.9) |
| miR-642 | 0.005 | 0.040 | 4.327 | 0.69 (0.56–0.82) | 73.8 (58.8–84.8) | 66.7 (46.6–82.2) | 71.2 (59.3–80.8) |
| miR-744 | 0.172 | 0.458 | 1.598 | 0.61 (0.48–0.75) | 35.7 (22.9–50.9) | 95.8 (78.1–100) | 57.6 (45.6–68.8) |
Each reaction was performed in duplicate. miRNAs that were found to be significantly different are depicted in boldface.
*Student t test.
†False discovery rate, α = 0.05.
‡Comparative ΔCT method (2-ΔΔCT).
Using stage 2 data, a prediction model was developed: log (p/1-p) = 8.583-0.290*ΔmiR-598-0.324* ΔmiR-642. The model randomly selected 75% of the data for a fitting model and 25% for validation. This was performed 1000 times.
Stage 3: validation
The blinded data from the test phase were analyzed using the prediction model that was developed using the 2 significant miRNAs: miR-598 and miR-642. The prediction model achieved an average sensitivity of 71.7% (95% confidence interval [CI], 31.3–92%), a specificity of 85.8% (95% CI, 30.8–97.1%), area under the curve (AUC) of 73.2% (95% CI, 35.1–99.4%), and an accuracy of 75% (95% CI, 40.7–92.9%) at an alpha of 0.05 and a power of at least 90%.
DISCUSSION
We identified a differentially expressed plasma miRNA profile in CC as compared with UC. Thirteen of 380 miRNAs screened were significantly differentially expressed. Seven miRNAs chosen from the screening data were tested using single miRNA assays on a larger population, demonstrating 2 miRNAs (miR-598 and miR-642) to be consistently differentially expressed in the plasma of CC as compared with UC patients. Using blinded data from these single assays, these findings were validated using a prediction model, achieving an overall prediction accuracy of 75% (95% CI, 40.7%–92.9%).
Both miR-598 and miR-642 appear to have a biological basis for their differential expression. miR-598, found on chromosome 8, has been shown previously to be dysregulated in esophageal cancer, bile duct cancer, and breast cancer.24–26 A predicted repressional target of miR-598 is fibroblast growth factor receptor substrate 2 (FRS2), a docking protein involved in fibroblast growth factor receptor function.27 Fibroblast growth factors are believed to stimulate collagen production and are elevated in CD intestinal strictures.28 This target gene is linked to mitogen-activated protein kinase (MAPK) pathway activation, important in cell growth and differentiation.29 Therefore, the increased levels of miR-598 could potentially be influencing the regulation of fibrosis and stricturing CD. miR-642, found on chromosome 19, has been reported to be dysregulated in prostate cancer cells and in pancreatic cancer tissue.30, 31 Toll-like receptor (TLR)-4, a validated target of miR-642, is an important cell surface receptor for responding to stimuli from the gastrointestinal tract such as lipopolysaccharide (LPS).32 Excessive or uncontrolled stimulation of the TLR-4 pathways due to such epigenetic change could contribute to persisting inflammation such as that of CD. Consequently, differences in this miRNA could play a role in modulating mucosal immunity in IBD patients.
Distinguishing between CD and UC is extremely clinically important,5, 6 and yet difficult in many cases. Differentiation may be problematic, especially in patients with isolated colonic Crohn’s disease. To address this clinical question, our study has included specifically nonfistulizing CC patients without peri-anal disease in comparison with UC. Several groups have studied circulating miRNAs in Crohn’s patients vs controls, UC patients vs controls, and active vs inactive disease.23, 33, 34 As part of our screening process, we performed an initial comparison between UC (n = 8) and healthy controls (n = 10) and between CC (n = 8) and healthy controls (n = 10). Among 380 evaluated miRNAs, differential expression of 92 miRNAs was significantly different between UC patient as compared with controls and differential expression of 86 miRNAs between CD patients as compared with controls (Supplemental Figure 1a and b, respectively). Interestingly, miR-598 and miR-642 were among the significantly differentially expressed miRNAs compared with controls in both UC and CC patients, demonstrating that they likely change in inflammatory conditions compared with controls, but to a different magnitude in the different conditions. Only a small number of studies prior to ours have compared CC directly with UC. Wu et al.15 compared active UC and active CD patients and found 3 miRNAs that were significantly different between the 2 groups. Those miRNAs are different from our results. It is important to note, however, that their Crohn’s population also included patients with extracolonic disease, which could influence the miRNA signature. Schaefer et al.16 also examined the miRNA signatures in the whole blood, tissue, and saliva of patients diagnosed with UC and CD, as well as healthy controls, and found 4 miRNAs that were different in blood between the 2 disease states. Significantly, they pooled the blood from the different specimens from each group together before running the assays, and also included patients with extracolonic CD.
Reference genes are used in order to allow comparisons between different samples and different reaction efficiencies. The choice of the most suitable reference gene as well as reproducibility is an evolving science.35, 36 Several algorithms and software programs have been developed in order to assist in the choice of the most suitable reference gene. One of the most commonly used programs is the NORMFINDER software37 based on a referenced algorithm.38 Different miRNAs have been used as reference genes in different diseases and samples sets.39–45 RNU6B is one of the most commonly used; however, skepticism has emerged regarding its reliability, especially in plasma samples with repeated freeze/thaw cycles.46 Specifically, within our data, it was also found to be unstable (stability position 193 utilizing the NORMFINDER software). Considering the frequency of its usage in other manuscripts, together with its good stability value in our data set, we believe that miR-16 was the best reference gene for use in our study.
A large proportion of our patients received drugs such as steroids, biologics, and immunomodulators that may affect miRNA expression.47 Practically, it would be very difficult to accrue patients who had not received these drugs, and the clinical relevance for this study would mainly be for patients already receiving the medications. The vast majority of our participating patients were Caucasian due to our local IBD patient population. Different races may have different miRNA profiles. Our study demonstrated a significant difference in a fairly small number of patients. Future studies should focus on examining the miRNA profile on a larger scale and population in order to verify these results and perhaps identify further miRNAs that can help in the diagnosis.
In the current study, we used well-characterized patients with either nonfistulizing CC without peri-anal disease or UC. Future studies may be warranted in a different population of patients with indeterminate colitis, another subset of IBD, who, by definition, have an uncertain diagnosis. Follow-up and long-term correlative miRNA profile findings could have additional important clinical significance.
CONCLUSION
Differentiating between CC and UC is of great clinical importance. In an attempt to differentiate between these 2 conditions, we have identified 2 plasma-based miRNAs that have the potential to assist in this conundrum. Our results have shown the potential and feasibility for development of a plasma-miRNA-based assay to distinguish these conditions based on these and perhaps other biomarkers.
Supplementary Material
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.ibdjournal.org)
Supported by the John W. and Barbara Thruston Atwood Price Trust and the Mary K. Oxley Foundation
Address correspondence to: All authors declare no conflict of interest or relevant financial disclosure.
REFERENCES
- 1. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–217. [DOI] [PubMed] [Google Scholar]
- 2. Geboes K, Colombel JF, Greenstein A et al. . Indeterminate colitis: a review of the concept--what’s in a name?Inflamm Bowel Dis. 2008;14:850–857. [DOI] [PubMed] [Google Scholar]
- 3. Farmer M, Petras RE, Hunt LE et al. . The importance of diagnostic accuracy in colonic inflammatory bowel disease. Am J Gastroenterol. 2000;95:3184–3188. [DOI] [PubMed] [Google Scholar]
- 4. Tremaine WJ. Is indeterminate colitis determinable?Curr Gastroenterol Rep. 2012;14:162–165. [DOI] [PubMed] [Google Scholar]
- 5. Braveman JM, Schoetz DJ Jr, Marcello PW et al. . The fate of the ileal pouch in patients developing Crohn’s disease. Dis Colon Rectum. 2004;47:1613–1619. [DOI] [PubMed] [Google Scholar]
- 6. Brown CJ, Maclean AR, Cohen Z et al. . Crohn’s disease and indeterminate colitis and the ileal pouch-anal anastomosis: outcomes and patterns of failure. Dis Colon Rectum. 2005;48:1542–1549. [DOI] [PubMed] [Google Scholar]
- 7. Soubieres AA, Poullis A. Emerging role of novel biomarkers in the diagnosis of inflammatory bowel disease. World J Gastrointest Pharmacol Ther. 2016;7:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mi S, Zhang J, Zhang W et al. . Circulating microRNAs as biomarkers for inflammatory diseases. Microrna. 2013;2:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakraborty C, Das S. Profiling cell-free and circulating miRNA: a clinical diagnostic tool for different cancers. Tumour Biol. 2016;37(5):5705–5714. [DOI] [PubMed] [Google Scholar]
- 10. Kanaan Z, Roberts H, Eichenberger MR et al. . A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg. 2013;258:400–408. [DOI] [PubMed] [Google Scholar]
- 11. Zheng G, Du L, Yang X et al. . Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014;111:1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao Z, Liu C, Xu J et al. . Plasma microRNA panels to diagnose pancreatic cancer: results from a multicenter study. Oncotarget. 2016;7(27):41575–41583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan Y, Ge G, Pan T et al. . A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9:e107986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Ther Adv Gastroenterol. 2015;8:4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu F, Guo NJ, Tian H et al. . Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schaefer JS, Attumi T, Opekun AR et al. . MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 2015;16:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar V, Abbas A, Aster JC.. Robbin’s Basic Pathology. 9th ed Philadelphia: Elsevier; 2013:587–592. [Google Scholar]
- 18. Carter JV, Roberts HL, Pan J et al. . A highly predictive model for diagnosis of colorectal neoplasms using plasma microRNA: improving specificity and sensitivity. Ann Surg. 2016;264:575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qiagen. miRNeasy plasma total RNA extraction protocol from miRNEASY Serum/Plasma Handbook. https://www.qiagen.com/us/resources/resourcedetail?id=710c0168-e408-408b-95af-91df5b5b1dd6&lang=en (9 December 2017, date last accessed).
- 20. Wilfinger WW, Mackey K, Chomczynski P. Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques. 1997;22:474–476, 478–481. [DOI] [PubMed] [Google Scholar]
- 21. Applied Biosystems. MegaplexTM RT without pre-amplification protocol. http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_053965.pdf (9 December 2017, date last accessed). [Google Scholar]
- 22. Jung SH. Sample size for FDR-control in microarray data analysis. Bioinformatics. 2005;21:3097–3104. [DOI] [PubMed] [Google Scholar]
- 23. Iborra M, Bernuzzi F, Correale C et al. . Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin Exp Immunol. 2013;173:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao BS, Liu SG, Wang TY et al. . Screening of microRNA in patients with esophageal cancer at same tumor node metastasis stage with different prognoses. Asian Pac J Cancer Prev. 2013;14:139–143. [DOI] [PubMed] [Google Scholar]
- 25. Wang M, Wen TF, He LH et al. . A six-microRNA set as prognostic indicators for bile duct cancer. Int J Clin Exp Med. 2015;8:17261–17270. [PMC free article] [PubMed] [Google Scholar]
- 26. Fu L, Li Z, Zhu J et al. . Serum expression levels of microRNA-382-3p, -598-3p, -1246 and -184 in breast cancer patients. Oncol Lett. 2016;12:269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lawrance IC, Rogler G, Bamias G et al. . Cellular and molecular mediators of intestinal fibrosis. J Crohns Colitis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katoh M, Katoh M. FGF signaling network in the gastrointestinal tract (review). Int J Oncol. 2006;29:163–168. [PubMed] [Google Scholar]
- 30. Thorns C, Schurmann C, Gebauer N et al. . Global microRNA profiling of pancreatic neuroendocrine neoplasias. Anticancer Res. 2014;34:2249–2254. [PubMed] [Google Scholar]
- 31. Epis MR, Giles KM, Kalinowski FC et al. . Regulation of expression of deoxyhypusine hydroxylase (DOHH), the enzyme that catalyzes the activation of eIF5A, by miR-331-3p and miR-642-5p in prostate cancer cells. J Biol Chem. 2012;287:35251–35259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sato K, Yoshimura A, Kaneko T et al. . A single nucleotide polymorphism in 3’-untranslated region contributes to the regulation of Toll-like receptor 4 translation. J Biol Chem. 2012;287:25163–25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krissansen GW, Yang Y, McQueen FM et al. . Overexpression of miR-595 and miR-1246 in the sera of patients with active forms of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:520–530. [DOI] [PubMed] [Google Scholar]
- 34. Paraskevi A, Theodoropoulos G, Papaconstantinou I et al. . Circulating microRNA in inflammatory bowel disease. J Crohns Colitis. 2012;6:900–904. [DOI] [PubMed] [Google Scholar]
- 35. Mitchell PS, Parkin RK, Kroh EM et al. . Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rice J, Roberts H, Burton J et al. . Assay reproducibility in clinical studies of plasma miRNA. PLoS One. 2015;10:e0121948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Department of Molecular Medicine, Aarhus University Hospital, Denmark. Normfinder Software http://momadk/normfinder-software (9 December 2017, date last accessed).
- 38. Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. [DOI] [PubMed] [Google Scholar]
- 39. Song J, Bai Z, Han W et al. . Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012;57:897–904. [DOI] [PubMed] [Google Scholar]
- 40. Chen X, Liang H, Guan D et al. . A combination of Let-7d, Let-7g and Let-7i serves as a stable reference for normalization of serum microRNAs. PLoS One. 2013;8:e79652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y, Tang N, Hui T et al. . Identification of endogenous reference genes for RT-qPCR analysis of plasma microRNAs levels in rats with acetaminophen-induced hepatotoxicity. J Appl Toxicol. 2013;33:1330–1336. [DOI] [PubMed] [Google Scholar]
- 42. Zheng G, Wang H, Zhang X et al. . Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS One. 2013;8:e83025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szabo DR, Luconi M, Szabo PM et al. . Analysis of circulating microRNAs in adrenocortical tumors. Lab Invest. 2014;94:331–339. [DOI] [PubMed] [Google Scholar]
- 44. Barry SE, Chan B, Ellis M et al. . Identification of miR-93 as a suitable miR for normalizing miRNA in plasma of tuberculosis patients. J Cell Mol Med. 2015;19:1606–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gharbi S, Shamsara M, Khateri S et al. . Identification of reliable reference genes for quantification of microRNAs in serum samples of sulfur mustard-exposed veterans. Cell. 2015;17:494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiang M, Zeng Y, Yang R et al. . U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454:210–214. [DOI] [PubMed] [Google Scholar]
- 47. Fujioka S, Nakamichi I, Esaki M et al. . Serum microRNA levels in patients with Crohn’s disease during induction therapy by infliximab. J Gastroenterol Hepatol. 2014;29:1207–1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.