Abstract
The morbidity and mortality rates of patients with non-small cell lung cancer (NSCLC) are increasing worldwide. Previous studies have demonstrated that long non-coding RNAs (lncRNAs) may serve critical roles in oncogenesis and cancer progression. The present study aimed to investigate the expression of lncRNA TCONS_00001798 and the clinicopathological factors and prognosis of patients with NSCLC. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was used to measure the expression of TCONS_00001798 in 118 paired NSCLC and adjacent non-tumor tissues. The association between TCONS_00001798 expression and patient clinicopathological factors and survival rates was subsequently analyzed. The results demonstrated that the expression of TCONS_00001798 was significantly downregulated in NSCLC tissues compared with adjacent non-tumor tissues (P<0.001). Additionally, the expression of TCONS_00001798 was negatively associated with lymph node metastasis (P<0.001) and an advanced pathological stage (P=0.003). A Kaplan-Meier analysis demonstrated that decreased TCONS_00001798 expression was significantly associated with shorter overall survival (OS) and disease-free survival (DFS) rates (each P<0.001). Furthermore, the results of multivariate analyses revealed that TCONS_00001798 expression may serve as an independent predictor for OS and DFS rates (each P=0.001). Therefore, the present study demonstrated that TCONS_00001798 may be involved in the oncogenesis and progression of NSCLC and that TCONS_00001798 may be a potential diagnostic and therapeutic target in patients with NSCLC.
Keywords: long non-coding RNA, TCONS_00001798, non-small cell lung cancer, survival
Introduction
Non-small cell lung cancer (NSCLC) accounts for 80–85% of all lung cancer cases and is the leading cause of cancer-associated mortality worldwide (1). Despite advancements in the understanding of the molecular mechanisms and clinical treatment of NSCLC, its prognosis remains poor and the 5-year overall survival rate of patients with NSCLC is ~15% (2). The primary cause of treatment failure and mortality in patients with NSCLC is the high metastatic potential of the tumor, combined with a lack of an effective early diagnostic method. Thus, it is important to identify novel biomarkers for the early diagnosis and treatment of patients with NSCLC.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs comprised of >200 nucleotides (3,4). Although lncRNAs were once classified as transcriptional noise (5), subsequent studies have demonstrated that lncRNAs participate in various biological processes, including epigenetic control, the regulation of gene expression, RNA maturation (including splicing and editing) and the maintenance of chromatin structure (3,4,6). The abnormal expression of lncRNA has been identified in various types of cancer, including prostate, breast, colorectal, bladder, lung and gastric cancer, as well as hepatocellular carcinoma and melanoma (7–14). Previous studies have demonstrated that certain lncRNAs may be potential prognostic biomarkers for patients with cancer. For example, HOTAIR, which is associated with human HOX loci and cancer metastasis, has been identified as a negative prognostic biomarker in certain types of cancer (15–17). Prensner et al (18) revealed that the expression of prostate cancer-associated transcript 1 (PCAT-1) was significantly upregulated in prostate tumors compared with normal prostate tissues, thus promoting cancer cell proliferation. It was also demonstrated that the knockdown of PCAT-1 expression inhibited the proliferation of prostate cancer cells, suggesting that PCAT-1 is involved in tumor progression and may be a potential prognostic biomarker of prostate cancer (18).
However, the function of lncRNAs in lung cancer remains largely unknown. Based on microarray analysis, the present study focused on a lncRNA termed TCONS_00001798 that demonstrated aberrant downregulation in NSCLC tissues compared with normal lung tissues. It was subsequently determined whether TCONS_00001798 is a novel prognostic marker for NSCLC. The expression of TCONS_00001798 in NSCLC and adjacent non-tumor tissues was examined and the association between the expression of lncRNATCONS_00001798 and the clinicopathological features and survival rates of patients with NSCLC was determined.
Materials and methods
Collection of patient tissue and clinical data
A total of 118 patients with NSCLC (76 males and 42 females) with a mean age of 57.9 years (range, 31–79 years), who underwent surgical resection between July 2008 and July 2010 at Tianjin Medical University Cancer Institute and Hospital (Tianjin, China), were enrolled in the present study. No patient received adjuvant therapy prior to surgery. Clinical and pathological features of NSCLC were obtained from patient reports. All resected NSCLC and adjacent non-tumor tissues (>5 cm from the tumor) were immediately frozen in liquid nitrogen and stored at −80°C prior to RNA extraction and reverse-transcription quantitative polymerase chain reaction (RT-qPCR). Tumors were staged according to the 7th edition of the tumor node metastasis classification (19). Following surgery, all patients were followed up every 3 months for the first year, every 6 months for the second year, and then every year until mortality or the last follow up. Patients were followed up until September 2016. Routine follow-up assessments included physical examinations, chest computed tomography, abdominal and cervical ultrasound, and laboratory tests. Further investigations included radionuclide bone scan and magnetic resonance imaging scans of the brain when patients had the symptoms of bone metastasis or brain metastasis. Overall survival (OS) was calculated from the date of surgery to the date of mortality or the last follow-up. Disease-free survival (DFS) was calculated from the date of surgery to the date of first recurrence or the last follow-up. The primary adjuvant treatment that patients underwent following surgery was chemotherapy or radiotherapy, either alone or in combination. The chemotherapywas a routine program for NSCLC, consisting of paclitaxel (135 mg/m2), vinorelbine (25 mg/m2 on days 1 and 8), pemetrexed (500 mg/m2) or gemcitabine (1 g/m2 on days 1 and 8) plus cisplatin (75 mg/m2) or carboplatin [target area under the curve, 5 (AUC5)] every 3 weeks for 4–6 cycles. All chemotherapy drugs were administered intravenously. The present study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (Tianjin, China) and written informed consent was obtained from each patient prior to enrolment.
RNA extraction and RT-qPCR
Total RNA from frozen tissue specimens was extracted using the TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol and reverse transcribed into cDNA using a Reverse Transcription kit (Takara Biotechnology Co., Ltd., Dalian, China). The reverse transcription reaction was performed under the following conditions: 37°C for 30 min; then 85°C for 5 sec; and holding at 4°C. Subsequently, qPCR was performed using a SYBR Premix Ex Taq II kit (Takara Biotechnology Co., Ltd.) on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA) to quantify the expression of TCONS_00001798. The PCR amplification was performed under the following conditions: 95°C for 30 sec, then 40 cycles of denaturation at 95°C for 5 sec, annealing at 60°C for 60 sec and elongation at 72°C for 30 sec. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. All experiments were repeated three times for accuracy. The primer sequences utilized were as follows: TCONS_00001798 forward, 5′CAGAAGGCTGCCCTGTAGAA3′ and reverse, 5′GCTTCTTCCACTCCTGCGT3′; GAPDH forward, 5′GGGAAACTGTGGCGTGAT3′ and reverse, 5′GAGTGGGTGTCGCTGTTGA3′.
Statistical analysis
SPSS 18.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) software were utilized for statistical analyses. χ2 and t-tests were performed to evaluate the association between TCONS_00001798 expression and patient clinicopathological features. Overall survival (OS) and disease-free survival (DFS) were estimated using the Kaplan-Meier method and the statistical differences between survival curves were calculated using the log-rank test. Univariate and multivariate analyses using the Cox proportional hazards regression model were performed to assess survival. All data are presented as the mean ± standard deviation and P<0.05 was considered to indicate a statistically significant difference.
Results
TCONS_00001798 expression is downregulated in NSCLC tumor tissues
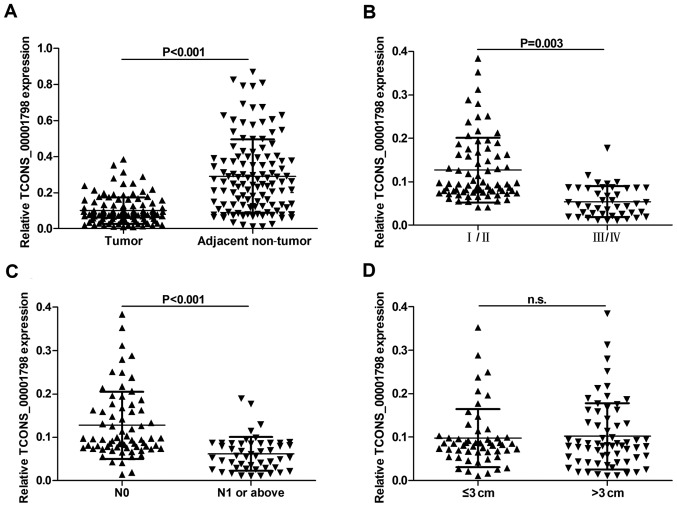
To assess whether TCONS_00001798 was present in NSCLC tissues, the expression of TCONS_00001798 in NSCLC and adjacent non-tumor tissues was determined using RT-qPCR. The results demonstrated that the expression of TCONS_00001798 was significantly lower in NSCLC tissues compared with adjacent non-tumor tissues (P<0.001; Fig. 1A).
Figure 1.
Association between the expression of TCONS_00001798 in NSCLC tissues and clinicopathological factors. (A) Relative expression of TCONS_00001798 in NSCLC tissues (n=118) compared with adjacent non-tumor tissues (n=118). TCONS_00001798 expression was detected by reverse transcription quantitative polymerase chain reaction and normalized to GAPDH. TCONS_00001798 expression was significantly lower in patients with (B) advanced pathological stage and (C) lymph node metastasis (N1 or above). (D) TCONS_00001798 expression was not associated with tumor size, measure by diameter. All data are presented as the mean ± standard deviation. NSCLC, non-small cell lung cancer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; n.s., not significant.
The association between TCONS_00001798 expression and patient clinicopathological factors
The association between TCONS_00001798 expression and the clinicopathological characteristics of patients with NSCLC was assessed and is presented in Fig. 1. According to the median value of TCONS_00001798 expression (0.084), patients with NSCLC were divided into high (≥median; n=59) and low (<median; n=59) expression groups. No significant differences between TCONS_00001798 expression in tumors >3 cm in diameter and tumors <3 cm in diameter were identified (Fig. 1D). However, a significant negative association between TCONS_00001798 expression and lymph node metastasis was identified. Tumors without lymph node metastasis (N0) were associated with higher TCONS_00001798 expression, whereas tumors with lymph node metastasis (≥N1) were associated with significantly lower TCONS_00001798 expression (P<0.001; Fig. 1C). Furthermore, statistical analysis revealed that low TCONS_00001798 expression was significantly associated with an advanced pathological stage (P=0.003; Fig. 1B). However, no associations were identified between TCONS_00001798 expression and other clinicopathological features, including patient age, sex, smoking status and histological subtype (Table I).
Table I.
Association between the expression of TCONS_00001798 and clinicopathological characteristics in 118 patients with NSCLC.
| Number of patients | ||||
|---|---|---|---|---|
| Patient characteristics | Number of patients | Low expression | High expression | P-value |
| Age (years) | 0.261 | |||
| ≤60 | 70 | 32 | 38 | |
| >60 | 48 | 27 | 21 | |
| Sex | 0.249 | |||
| Male | 76 | 41 | 35 | |
| Female | 42 | 18 | 24 | |
| Smoking | 0.431 | |||
| Yes | 80 | 42 | 38 | |
| No | 38 | 17 | 21 | |
| Histological subtype | 0.514 | |||
| AD | 41 | 21 | 20 | |
| SCC | 52 | 28 | 24 | |
| Others | 25 | 10 | 15 | |
| Pathological stage | 0.003a | |||
| I and II | 72 | 28 | 44 | |
| III and IV | 46 | 31 | 15 | |
| Lymph node metastasis (N) | <0.001a | |||
| N1 or above | 49 | 34 | 15 | |
| N0 | 69 | 25 | 44 | |
| Tumor size (cm) | 0.712 | |||
| ≤3 | 54 | 26 | 28 | |
| >3 | 64 | 33 | 31 | |
All data are presented as the mean ± standard deviation.
P<0.05. NSCLC, non-small cell lung cancer; AD, adenocarcinoma; SCC, squamous cell carcinoma.
Low TCONS_00001798 expression predicts unfavorable survival in patients with NSCLC
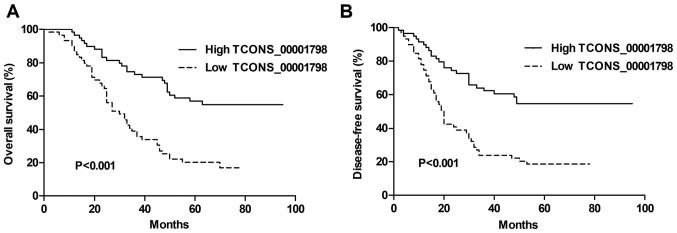
A Kaplan-Meier analysis and a log-rank test were used to further assess the association between TCONS_00001798 expression and the survival of patients with NSCLC. The results revealed that decreased TCONS_00001798 levels were significantly associated with shorter OS and DFS rates (each P<0.001; Fig. 2A and B). Univariate analysis using a Cox proportional hazards model demonstrated that TCONS_00001798 expression was a significant factor for the prediction of OS and DFS rates in patients with NSCLC, with a hazard ratio (HR) of 0.345 [P<0.001; 95% confidence interval (CI) 0.213–0.559] and 0.364 (P<0.001; 95% CI, 0.225–0.589), respectively (Tables II and III). In addition, other clinicopathological factors including lymph node metastasis and pathological stage were considered to be significant for OS and DFS (each P<0.001). Multivariate analysis revealed that TCONS_00001798 expression was significantly associated with OS (HR, 0.424; 95% CI, 0.257–0.699; P=0.001) and DFS (HR, 0.436; 95% CI,0.264–0.720; P=0.001), as was pathological stage (Tables II and III).
Figure 2.
Prognostic significance of TCONS_00001798 in patients with NSCLC. Kaplan-Meier curves for the (A) overall and (B) disease-free survival rates of patients with NSCLC, according to TCONS_00001798 expression. Patients with low levels of TCONS_00001798 expression exhibited a decreased overall and disease-free survival rate compared with patients that exhibited high expression. All data are presented as the mean ± standard deviation. NSCLC, non-small cell lung cancer.
Table II.
Univariate and multivariate analysis of clincopathological variables for OS rates in 118 patients with NSCLC.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | HR | P-value | 95% CI | HR | P-value | 95% CI |
| TCONS_00001798 expression (low, high) | 0.345 | <0.001a | 0.213–0.559 | 0.424 | 0.001a | 0.257–0.699 |
| Pathological stage (I/II, II/IV) | 2.894 | <0.001a | 1.813–4.620 | 2.017 | 0.047a | 1.010–4.029 |
| Lymph node metastasis (N0, N1 or above) | 0.376 | <0.001a | 0.235–0.600 | 0.798 | 0.526 | 0.396–1.605 |
| Sex (male, female) | 0.693 | 0.145 | 0.423–1.134 | |||
| Age (years) ≤60, <60) | 0.953 | 0.840 | 0.599–1.517 | |||
| Smoking (yes, no) | 0.913 | 0.715 | 0.561–1.486 | |||
| Histological subtype (AD, SCC, others) | 1.180 | 0.278 | 0.875–1.592 | |||
| Tumor size (≤3 cm, >3 cm) | 0.966 | 0.881 | 0.611–1.526 | |||
All data are presented as the mean ± standard deviation.
P<0.05. OS, overall survival; NSCLC, non-small cell lung cancer; HR, hazard ratio; CI, confidence interval; AD, adenocarcinoma; SCC, squamous cell carcinoma.
Table III.
Univariate and multivariate analysis of clinicopathological variables for DFS rates in 118 patients with NSCLC.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | HR | P-value | 95% CI | HR | P-value | 95% CI |
| TCONS_00001798 expression (low, high) | 0.364 | <0.001a | 0.225–0.589 | 0.436 | 0.001a | 0.264–0.720 |
| Pathological stage (I/II, III/IV) | 2.782 | <0.001a | 1.750–4.423 | 2.403 | 0.044a | 1.025–5.632 |
| Lymph node metastasis (N0, N1 or above) | 0.405 | <0.001a | 0.255–0.644 | 1.056 | 0.901 | 0.446–2.498 |
| Sex (male, female) | 0.720 | 0.192 | 0.440–1.179 | |||
| Age (years) (≤60, >60) | 1.045 | 0.854 | 0.657–1.662 | |||
| Smoking (yes, no) | 0.968 | 0.897 | 0.595–1.576 | |||
| Histological subtype (AD, SCC, others) | 1.174 | 0.287 | 0.874–1.577 | |||
| Tumor size (≤3 cm, >3 cm) | 1.031 | 0.894 | 0.653–1.630 | |||
All data are presented as the mean ± standard deviation.
P<0.05. DFS, disease-free survival; NSCLC, non-small cell lung cancer; HR, hazard ratio; CI, confidence interval; AD, adenocarcinoma; SCC, squamous cell carcinoma.
Discussion
With the rapid development of genome technology, it has been demonstrated that mammalian genomes encode thousands of lncRNAs with numerous mechanisms of action, in addition to protein-coding RNA (20,21). Although lncRNAs were previously considered to represent transcriptional noise, with no observable function in cells, it has since been demonstrated that lncRNAs serve a variety of important roles in biological processes, including epigenetic control, the regulation of gene expression, RNA maturation (including splicing and editing) and the maintenance of chromatin structure (3,4,6). Previous studies have revealed that lncRNAs are abnormally expressed in certain tumors, suggesting that they may serve as potential biomarkers predicting the prognosis of patients with cancer (15–18,22–25).
Several lncRNAs have been implicated in the initiation and progression of NSCLC and their expression may be regulated by transcription factors (26), interactions with microRNA (27) and Wnt/β-catenin signaling (28). Han et al (26) demonstrated that the decreased expression of the promoter of CDKN1A antisense DNA damage-activated RNA is negatively associated with NSCLC development and indicates the unfavorable prognosis of patients as it affects and cellular apoptosis via Bcl-2. Schmidt et al (29) and Tano et al (30) demonstrated that metastasis associated lung adenocarcinoma transcript 1 (MALAT-1) expression was significantly upregulated in NSCLC and is associated with poor patient prognosis. Additionally, MALAT-1 knockdown inhibited the migration of NSCLC cells by influencing the expression of motility-related genes (29,30). Weber et al (25) also revealed that MALAT-1 maybe used as a complementary blood biomarker in patients with NSCLC. Despite this, few lncRNAs have been associated with NSCLC oncogenesis.
In the present study, the association between TCONS_ 00001798 expression and the prognosis and clinicopathological features of patients with NSCLC was assessed. To the best of our knowledge, the present study was the first to demonstrate that lncRNA TCONS_00001798 is significantly downregulated in NSCLC tumor tissues compared with adjacent non-tumor tissues. In addition, the results of the present study revealed that the decreased expression of TCONS_00001798 is negatively associated with lymph node metastasis and advanced pathological stage. It was determined that TCONS_00001798 expression may be a potential independent prognostic factor for OS and DFS rates, and NSCLC pathological stage. The present study therefore indicates that lncRNA TCONS_00001798 may serve an essential role in the development and progression of NSCLC via invasion and metastasis.
One major limitation of the present study was that only a small number of patients were enrolled from the same hospital, potentially leading to inaccurate conclusions. Therefore, further studies involving cell models, animal models and a greater number of patients, are required to elucidate the precise molecular mechanisms that underlie the altered expression of TCONS_00001798.
In conclusion, to the best of our knowledge, the present study was the first to demonstrate that the expression of lncRNA TCONS_00001798 is significantly downregulated in NSCLC tumor tissues and indicate that it is negatively associated with lymph node metastasis and an advanced pathological stage. Furthermore, it was demonstrated that TCONS_00001798 expression may serve as an independent prognostic factor and potential therapeutic target in patients with NSCLC. However, further studies are required to clarify the molecular mechanisms that underlie this.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- lncRNAs
long non-coding RNAs
- NSCLC
non-small cell lung cancer
- OS
overall survival
- DFS
disease-free survival
- HR
hazard ratio
- CI
confidence interval
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81470137) and the Key Program for Anti-Cancer Research of Tianjin Municipal Science and Technology Commission (grant no. 12ZCDZSY15400).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LG performed the experiment and wrote the manuscript. HZ and BZ were responsible for the design of experiment and collection of the tumor samples. HZ and BZ analyzed the experiment data. CW was accountable for designing the experiment, performing data interpretation and revising the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (Tianjin, China) and written informed consent was obtained from each patient prior to enrolment.
Consent for publication
Written informed consent was obtained from each patient for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 3.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol. 2015;11:151–160. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB, Xu J. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis. 2011;32:1655–1659. doi: 10.1093/carcin/bgr187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuo YL, Li XM, Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur Rev Med Pharmacol Sci. 2015;19:3403–3411. [PubMed] [Google Scholar]
- 9.Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, Sun Y. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8:869–875. doi: 10.3892/ol.2014.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 11.Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L, Sha N. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One. 2014;9:e100893. doi: 10.1371/journal.pone.0100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Chen W, Yang C, Wu W, Wu S, Qin X, Li X. Long non-coding RNA UCA1a(CUDR) promotes proliferation and tumorigenesis of bladder cancer. Int J Oncol. 2012;41:276–284. doi: 10.3892/ijo.2012.1443. [DOI] [PubMed] [Google Scholar]
- 13.Zhao W, An Y, Liang Y, Xie XW. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:1930–1936. [PubMed] [Google Scholar]
- 14.Zhao JH, Sun JX, Song YX, Chen XW, Yang YC, Ma B, Wang J, Gao P, Wang ZN. A novel long noncoding RNA-LOWEG is low expressed in gastric cancer and acts as a tumor suppressor by inhibiting cell invasion. J Cancer Res Clin Oncol. 2016;142:601–609. doi: 10.1007/s00432-015-2071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 17.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobin LH, Gospodarowicz MK, Wittekind C. International Union Against Cancer (UICC): TNM classification of malignant tumours. 7th. John Willkey and Sons, Ltd.; UK: 2010. [Google Scholar]
- 20.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 23.Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM, Li CW, Wang Y, Hsu JL, Hung MC. Long non-coding RNAs: Versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4:127–150. [PMC free article] [PubMed] [Google Scholar]
- 24.Salagierski M, Schalken JA. Molecular diagnosis of prostate cancer: PCA3 and TMPRSS2:ERG gene fusion. J Urol. 2012;187:795–801. doi: 10.1016/j.juro.2011.10.133. [DOI] [PubMed] [Google Scholar]
- 25.Weber DG, Johnen G, Casjens S, Bryk O, Pesch B, Jöckel KH, Kollmeier J, Brüning T. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6:518. doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han L, Zhang EB, Yin DD, Kong R, Xu TP, Chen WM, Xia R, Shu YQ, De W. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 28.Fu X, Li H, Liu C, Hu B, Li T, Wang Y. Long noncoding RNA AK126698 inhibits proliferation and migration of non-small cell lung cancer cells by targeting Frizzled-8 and suppressing Wnt/β-catenin signaling pathway. Onco Targets Ther. 2016;9:3815–3827. doi: 10.2147/OTT.S100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, et al. The long noncoding MALAT-1RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 30.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.