Abstract
Spermatozoa are not mature until they transit the epididymis where they acquire motility and the ability to fertilize an egg through sequential modifications. The epididymis has three functional regions, caput, corpus, and cauda, and the luminal proteins of the epididymis play important roles in the above modifications. However, the proteins with differential enrichment between the caput and cauda are still largely unknown. To reveal the functions of the caput and cauda during sperm maturation, luminal proteins from caput and cauda of mice were analyzed by isobaric tag for relative and absolute quantitation (iTRAQ). Overall, 128 differentially enriched proteins were found, of which 46 were caput enriched and 82 were cauda enriched. Bioinformatic analysis showed that lipid metabolism was active in the caput; while anion- and cation-binding activity and phosphorus and organophosphate metabolism were active in the cauda. A new epididymal luminal protein, the caput-enriched PDZ domain containing 1 (Pdzk1), also named Na+/H+ exchange regulatory cofactor 3 (NHERF3), which plays a critical role in cholesterol metabolism and carnitine transport, was found in the lipid metabolism. Western blotting and immunofluorescence analyses showed that Pdzk1 was expressed in the epididymis but not in the testis, and localized at the middle piece of the sperm tail. Pdzk1 protein level was also reduced in the spermatozoa in case of asthenozoospermic patients compared with that in normozoospermic men, suggesting that Pdzk1 may participate in sperm maturation regulation and may be associated with male infertility. These results may provide new insights into the mechanisms of sperm maturation and male infertility.
Keywords: asthenozoospermia, epididymis, fertility, PDZ domain containing 1, sperm
INTRODUCTION
According to the report of World Health Organization (WHO), infertility affects 10%–15% of couples of reproductive age worldwide, and half of the cases are due to male factors.1 Many factors contribute to male infertility, such as impaired spermatogenesis in the testis2 and sperm maturational disorders in the epididymis.3,4 It has been reported that up to 40% of idiopathic male infertility cases are related to sperm maturational disorders. However, the mechanisms underlying the disorders are still unknown.5
Spermatozoa in the testis are morphologically complete, but not motile or fertile. They mature in the epididymis to acquire the abilities to move, become capacitated, bind to the zona, and fuse with the oocyte to form an embryo.6 Mature spermatozoa have a reduced cholesterol level, increased surface-negative charge and disulfide bonds, and modified surface proteins. All of these characteristics are acquired through sequential modifications occurring in the epididymis.7
The epididymis is divided into the caput, corpus, and cauda regions, each of which has distinctive functions.8 The proteins in epididymal fluid are essential for sperm maturation, since spermatozoa are synthetically inactive.9 Since the function of the epididymis is region dependent, identifying the differentially enriched proteins in the caput and cauda is critical to the understanding of the mechanism of sperm maturation.
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) is a proteomic method that is widely used for the study of luminal proteins. However, it has been reported that no more than twenty proteins account for 80%–90% of the total epididymal luminal proteins, and many proteins are beyond the detection limit of 2D-PAGE.9 Thus, the proteins that are differentially enriched between the caput and cauda fluid are still largely unknown. Moreover, very little research has been performed on this issue using mouse model.
In this study, iTRAQ was used to profile the different protein compositions of mouse caput and cauda epididymal fluids. We found that the caput-enriched PDZ domain containing 1 (Pdzk1), also called Na+/H+ exchange regulatory cofactor 3 (NHERF3), was associated with male infertility. Our research indicated that identification of the differentially enriched proteins in the caput and cauda could provide new clues to how sperm maturation and male infertility develop.
MATERIALS AND METHODS
Materials
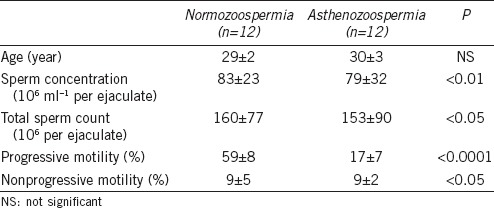
C57BL/6 mice were purchased from the Department of Laboratory Animal Science, School of Medicine, Shanghai Jiaotong University. The mice were kept at a temperature of 22°C with light cycles of 14 h light and 10 h dark; mice were provided food and water ad libitum. Human semen samples were collected at the International Peace Maternity and Child Health Hospital, School of Medicine, Shanghai Jiaotong University, from normozoospermic men (mean age: 29 years) and asthenozoospermic patients (mean age: 30 years) (Table 1). Informed consent was provided by all participants. Semen samples were obtained according to the fifth edition of the WHO Laboratory Manual for the Examination and Processing of Human Semen. This study was approved by the Ethics Committee of the International Peace Maternity and Child Health Hospital.
Table 1.
Ejaculated sperm characteristics of the 24 men included in the analysis (mean±standard deviation)
Protein extraction
Epididymal luminal proteins were extracted by a modified version of a previously described method.10 Briefly, five mice were sacrificed and both epididymides of each mouse were collected; thus, ten epididymides in total were harvested. Then, the caput and cauda were dissected from the epididymides, and five capita epididymidum were pooled together as were those from the cauda. The epididymal heads and tails were placed in PBS with protease inhibitor cocktail (Roche, Indianapolis, IN, USA), and several small slits were made in the tubules to promote the release of caput and cauda fluid and spermatozoa into the media. After being incubated at 4°C for 40 min with shaking, the medium was harvested and centrifuged at 800 g for 5 min at 4°C to remove the sperm cells. Then, the supernatant was collected and centrifuged again at 12 000 g for 10 min at 4°C. Finally, proteins of each sample were precipitated using chilled acetone, and the pellets were dried by a vacuum freeze dryer (Thermo Scientific Savant, San Jose, CA, USA). Then, they were dissolved with 50 μl of Dissolution Buffer supplied in the iTRAQ 8-plex Kit (AB SCIEX, Framingham, MA, USA).
iTRAQ proteome analysis
Proteins were digested by Trypsin Gold (Promega, Madison, WI, USA); the iTRAQ labeling procedure was performed in accordance with the manufacturer's instructions (AB SCIEX). The caput samples were labeled with iTRAQ tag 117 or 119, and the cauda samples were labeled with tags 118 or 121. The labeled peptide mixtures were purified by strong cation exchange chromatography on the Agilent 1200 System (Agilent, Santa Clara, CA, USA). They were analyzed on a TripleTOF 5600 System (AB SCIEX) coupled online to the nanoLC-Ultra 2D System (Eksigent Technologies, Dublin, CA, USA).
Data were processed with Protein Pilot Software version 5.0 (AB SCIEX) against the Mus musculus database (UniProt release 2015_12) using the Paragon algorithm.11 Epididymal luminal proteins that were differentially enriched between the caput and cauda were identified using the following criteria: 1.2-fold cutoff and P < 0.05. Differentially enriched proteins were analyzed by QuickGO for gene ontology (GO) annotation and enrichment to obtain the information of biological processes and molecular function.12
Western blotting
Western blotting was carried out as described previously.13 The mouse tissues were lysed in RIPA buffer (50 mmol l−1 Tris-HCl, pH 7.4, 150 mmol l−1 NaCl, 1% Triton X-100, 1% SDS, 1% sodium deoxycholate, 1 mmol l−1 EDTA) with protease inhibitor cocktail (Roche), 1 mmol l-1 PMSF, and 5 mmol l-1 sodium orthovanadate. In order to harvest enough luminal fluid, especially the corpus fluid, epididymides from twenty mice were used and processed as mentioned above. The remaining tissues and released spermatozoa were then collected for further analysis. Spermatozoa from the mouse epididymis and human semen were lysed in RIPA buffer with sonication. Proteins were separated by SDS-PAGE and stained by Coomassie Brilliant Blue R-250 (Sigma-Aldrich, St. Louis, MO, USA) or transferred onto nitrocellulose membranes (Amersham Biosciences, Piscataway, NJ, USA). The membranes were blocked by 5% fat-free milk in TBST (10 mmol l-1 Tris, pH 7.5, 200 mmol l-1 NaCl, and 0.2% Tween 20) followed by incubation with primary antibodies: Pdzk1 antibody (Abcam, Cambridge, UK), GAPDH antibody (Proteintech, Rosemont, IL, USA), α-Tubulin (Sigma-Aldrich), and protamine 1 (Briar Patch Biosciences, Livermore, CA, USA). The membranes were probed by goat anti-mouse or rabbit IgG conjugated to HRP (Proteintech), visualized by ECL substrate (Pierce, Rockford, IL, USA) and captured by ImageQuant LAS 4000 mini (GE Healthcare Bioscience, Carlsbad, CA, USA).
Immunofluorescence
To determine the localization of Pdzk1 in the epididymis and spermatozoa, immunofluorescence was performed. Briefly, 10 μm frozen sections of mouse epididymis, testis, and kidney, or slides spread with spermatozoa from the mouse caput and cauda, and from human semen, were fixed in 4% paraformaldehyde (PFA), permeabilized by 0.1% Triton X-100, and blocked in 5% BSA. The slides were incubated with the Pdzk1 antibodies at 4°C overnight, followed by incubation with Alexa Fluor-488 conjugated donkey anti-rabbit IgG (Molecular Probes, Eugene, OR, USA), and nuclei were stained with Hoechst 33342 (Sigma-Aldrich). Fluorescent signals were captured by confocal microscope LSM 510 (Carl Zeiss, Jena, Germany).
Statistical analysis
The signal intensity of bands was analyzed by ImageJ software (NIH, Bethesda, MD, USA), and data were analyzed using GraphPad Prism 5.01 (GraphPad Prism, La Jolla, CA, USA). The differences between the normozoospermia and asthenozoospermia group were analyzed by the two-tailed Mann–Whitney U-test, and P < 0.05 was considered statistically significant.
RESULTS
The differentially enriched proteins between the caput and cauda
In this study, luminal proteins were extracted from mouse caput and cauda, and the absence of nuclear protein Protamine 1 (Prm1) in all samples showed that they were not contaminated by sperm (Supplementary Figure 1 (97.9KB, tif) ). In the analysis by Protein Pilot 5.0 search engine, a total of 66 794 spectra and 20 799 distinct peptides were identified. These peptides could be to 2720 proteins with a confidence ≥ 95%, and a total of 2132 nonredundant proteins with a false discovery rate < 1% were detected (Supplementary Table 1 (45.6KB, tif) ). With a criterion of 1.2-fold cutoff and P < 0.05, 128 differentially enriched proteins were identified, of which 46 were caput fluid enriched and 82 were cauda fluid enriched (Supplementary Table 2 (3.8MB, tif) ). The proteins with no difference in enrichment between the two fluids are listed in Supplementary Table 3 (39.5MB, tif) .
Western blotting analysis of protamine 1 in the fluid of caput and cauda, and the cauda sperm. Gel is stained by Coomassie Brilliant Blue R-250.
The identifications by isobaric tag for relative and absolute quantitation
The differentially enriched proteins between the mouse caput and cauda luminal fluids
The unchanged proteins between the mouse caput and cauda luminal fluids
The different functions of the caput and cauda epididymidis
The 128 differentially enriched proteins were subjected to bioinformatic analysis by GO annotation and enrichment to analyze the different functions of the caput and cauda regions. Biological process analysis found that processes related to small-molecule metabolism, organic acid metabolism, oxidation–reduction, organonitrogen compound metabolism, and organic substance catabolism existed in both the caput and cauda. However, lipid metabolism was found in the caput, while those associated with phosphorus and organophosphate metabolism were found in the cauda (Figure 1a and 1b). The results of molecular function annotation indicated that binding activity was the main function of these proteins. Identical protein binding, protein domain-specific binding, protein complex binding, and coenzyme binding activities were all identified in the caput and cauda while anion and cation binding activities were identified only in cauda (Figure 1c and 1d). These results indicate that the caput and cauda play different roles in the process of sperm maturation.
Figure 1.
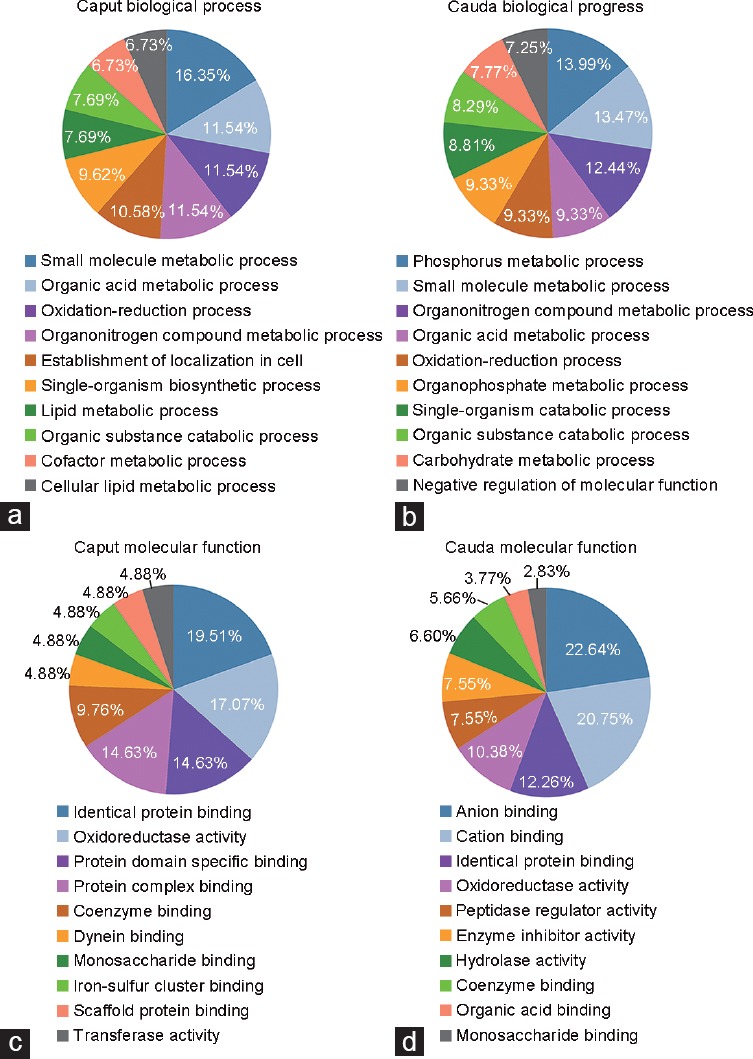
GO analysis of differentially enriched proteins in the fluids of mouse caput and cauda. A total of 128 differentially enriched proteins were identified by iTRAQ, 46 caput-enriched and 82 cauda-enriched proteins are annotated. The caput (a) and cauda (b) enriched luminal proteins are classified in different categories based on the biological processes, the caput (c) and cauda (d) enriched luminal proteins are also classified based on the molecular functions. The top ten terms of GO enrichment with P < 0.05 were listed. iTRAQ: isobaric tag for relative and absolute quantitation; GO: gene ontology.
Pdzk1 is a newly identified epididymal luminal protein
GO analysis showed that Pdzk1 was found in the lipid metabolism group, as an epididymal luminal protein newly identified by iTRAQ (Supplementary Table 2 (3.8MB, tif) ). The expression and localization of Pdzk1 were studied by western blotting and immunofluorescence analyses.
Western blot analysis showed that Pdzk1 was expressed in heart, liver, kidney, epididymis, and efferent ducts, but not in the testis or ovary14 (Figure 2a and Supplementary Figure 2a (467.2KB, tif) ). The expression of Pdzk1 in the caput, corpus, and cauda (Figure 2b) was confirmed, and the expression level of Pdzk1 in the spermatozoa and fluid from the caput, corpus, and cauda was also analyzed (Figure 2c and 2d). Immunoblotting result confirmed that Pdzk1 was caput fluid enriched (Figure 2b-2d).
Figure 2.
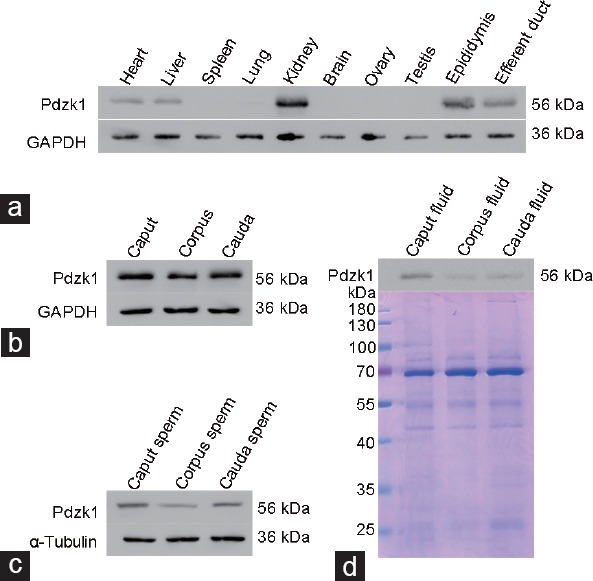
The expression of Pdzk1 in different organs of the mouse (a) and in the caput, corpus, and cauda (b) is analyzed by western blotting analysis, and also the expression of Pdzk1 in spermatozoa (c) and luminal fluid (d) from the caput, corpus, and cauda is analyzed. The gel is stained by Coomassie Brilliant Blue R-250. Pdzk1: PDZ domain containing 1.
Complete scans of the different blots presented in Figure 2 and Figure 6 indicating the specificity of Pdzk1 antibody. (a) Uncropped picture presented in Figure 2a showing the expression of Pdzk1 in different organs of mouse. (b) Uncropped picture presented in Figure 6b showing the expression of Pdzk1 in human spermatozoa. Pdzk1: PDZ domain containing 1.
The localization of Pdzk1 was shown by immunofluorescence to have a dynamic expression pattern in the epididymis (Figure 3). In the proximal and distal caput, Pdzk1 was expressed over a wide area and had a stronger signal in the distal caput. Then, an intense signal was observed at the surface of corpus and cauda epithelia. The expression pattern of Pdzk1 in the kidney has been reported and was used here as a positive control.15
Figure 3.
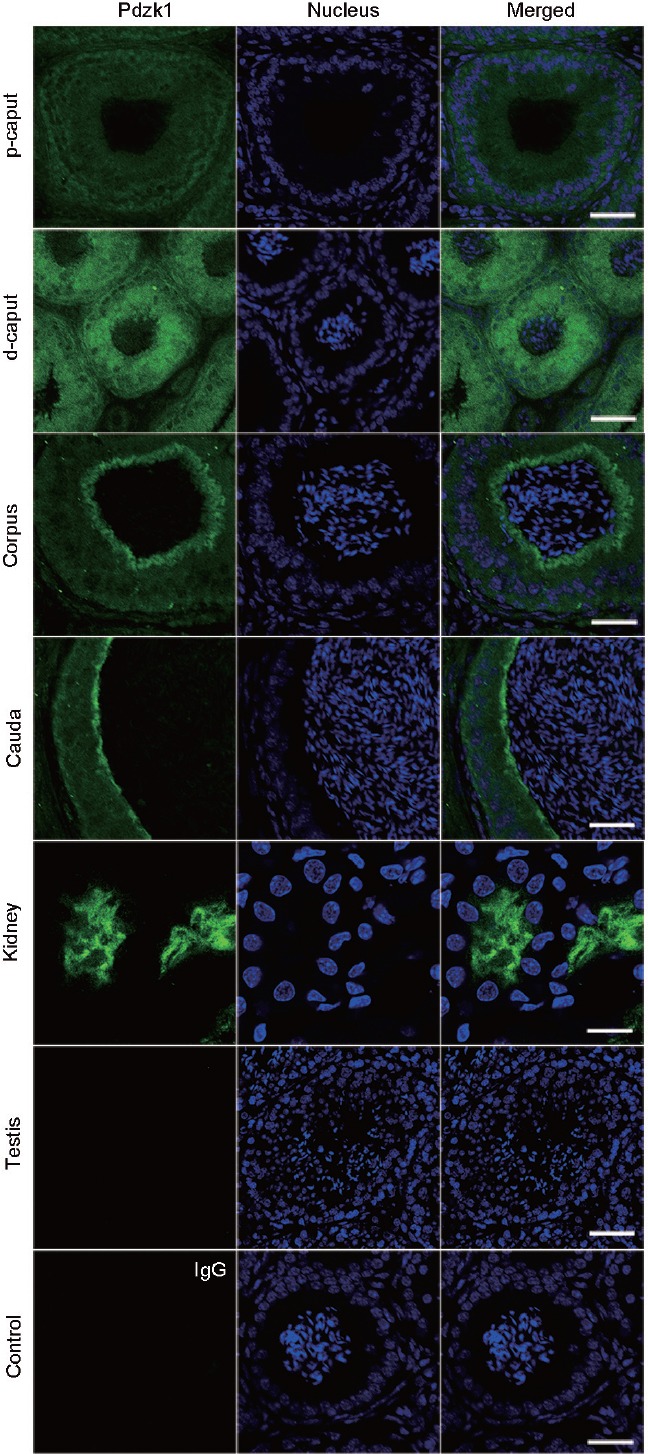
Immunofluorescence analysis of the localization of Pdzk1 in the epididymis. p-caput: proximal caput; d-caput: distal caput. IgG was used as control. Antibody dilution: 1:400. Scale bar: 12 μm in kidney and 50 μm in other panels. Pdzk1: PDZ domain containing 1.
In order to illuminate the sperm localization of Pdzk1, spermatozoa from the mouse caput, corpus, and cauda were analyzed by immunofluorescence (Figure 4). Pdzk1 is localized to the sperm tail, especially in the middle piece, where the mitochondrial sheath lies (Figure 4).
Figure 4.
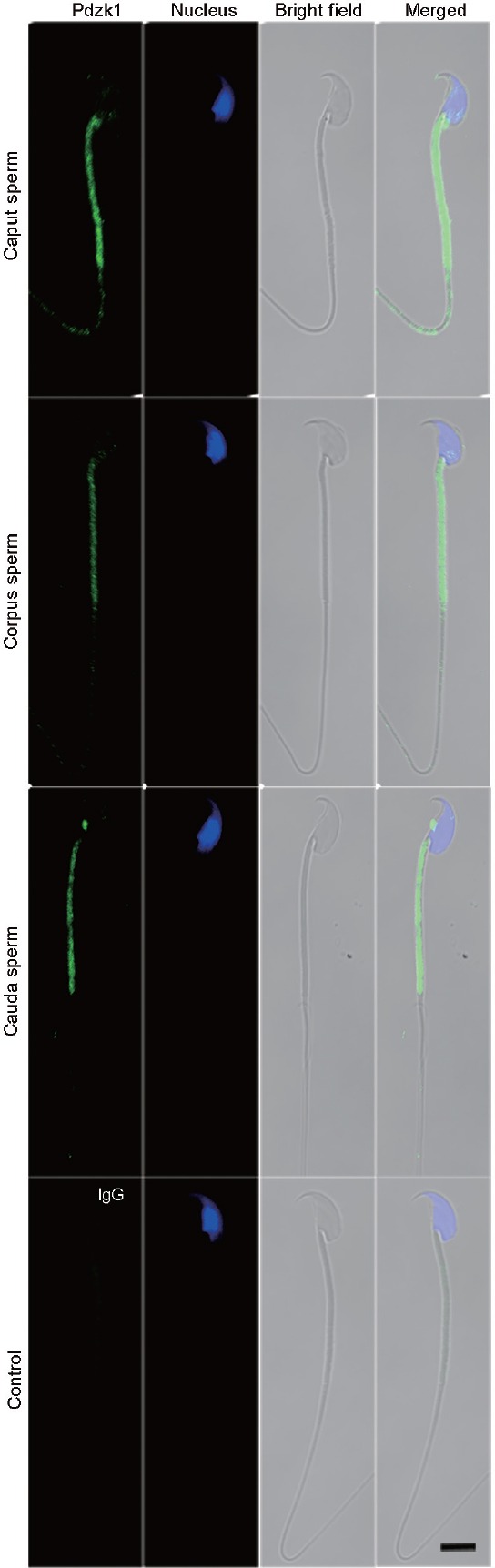
Immunofluorescence analysis of the localization of Pdzk1 in spermatozoa from the mouse. Kidney and testis were used as positive and negative controls, respectively. Antibody dilution: 1:100. IgG was used as control. Scale bar = 5 μm. Pdzk1: PDZ domain containing 1.
The expression of Pdzk1 in asthenozoospermia
Spermatozoa from normozoospermic men (mean age: 29 years) and asthenozoospermic patients (mean age: 30 years) were collected to investigate the role of Pdzk1 in male fertility (Table 1). The localization of Pdzk1 in human spermatozoa was similar to that in the mouse, and the localization pattern did not differ between normozoospermia and asthenozoospermia (Figure 5). The expression of Pdzk1 was analyzed by western blotting analysis, which showed that Pdzk1 was decreased in the spermatozoa in asthenozoospermia (Figure 6 and Supplementary Figure 2b (467.2KB, tif) ), suggesting that Pdzk1 may function in sperm maturation and may be associated with male infertility.
Figure 5.
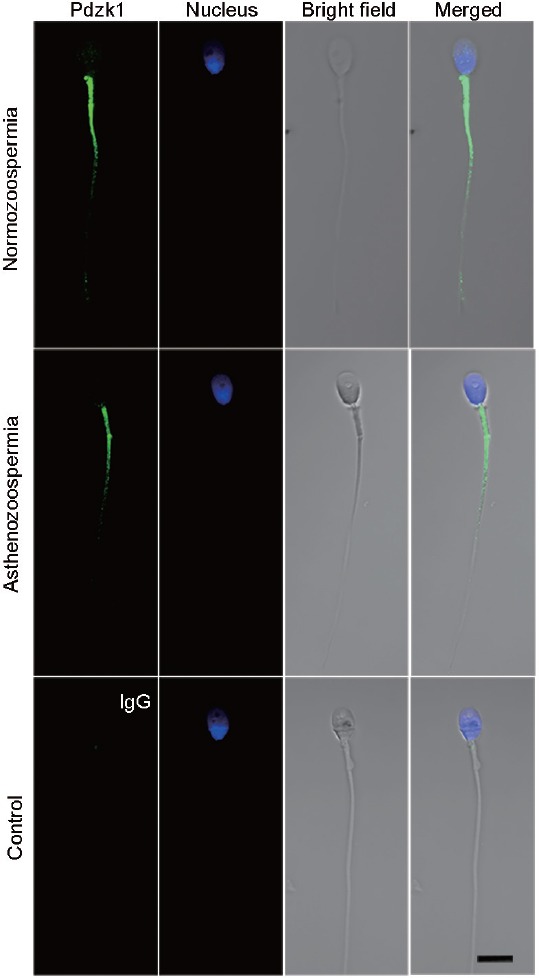
Immunofluorescence analysis of the localization of Pdzk1 in spermatozoa from normozoospermic and asthenozoospermic men. Spermatozoa from normozoospermic men were marked as N1 and asthenozoospermia as A1. Antibody dilution: 1:100. IgG was used as control. Scale bar = 10 μm. Pdzk1: PDZ domain containing 1.
Figure 6.
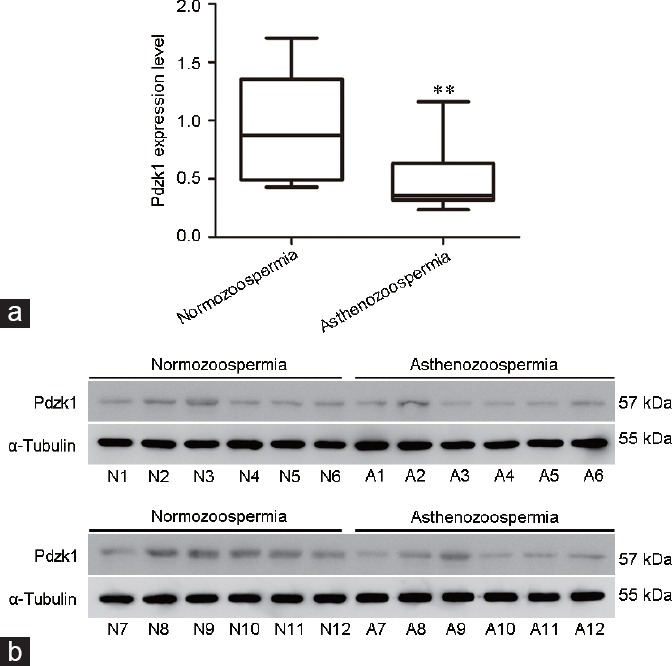
The sperm expression of Pdzk1 in normozoospermia and asthenozoospermia. (a) Box and whiskers plot of Pdzk1 expression in spermatozoa. Boxes indicate the 25th and 75th percentiles, the middle line indicates arithmetic mean. **P < 0.01, compared with normal control. (b) Western blot analysis of the expression of Pdzk1. α-Tubulin was used as normal control. N: normozoospermia; A: asthenozoospermia; Pdzk1: PDZ domain containing 1.
DISCUSSION
When spermatozoa are transiting through the convoluted epididymal tubule, they successively interact with the epididymal fluid to acquire progressive motility and fertilizing capacity, a process that is called epididymal sperm maturation. To reveal the mechanism of sperm maturation, transcriptomic and proteomic analyses of epididymis,16,17,18,19 proteomic analyses of sperm,20,21 and proteomic analyses of epididymal fluid6,9 have been performed by other groups.
Proteomic analyses of luminal proteins were mainly carried out by 2D-PAGE.6,9 However, this method has drawbacks, in that it is difficult to use it to analyze low-abundance proteins, hydrophobic proteins, and membrane proteins.22 The epididymal fluid contains many low-abundance proteins, and studies by 2D-PAGE can only identify hundreds of proteins or fewer in this fluid in various species, such as 525 proteins in human,23 146 proteins in boars,24 117 proteins in stallions,25 172 proteins in bulls,26 12 proteins in rams,27 198 proteins in monkeys,28 27 proteins in platypuses,29 19 proteins in rats,30,31 and 23 proteins in mice.32 Thus, the epididymal luminal proteins, especially the proteins differentially enriched between the caput and cauda, are still largely unknown.
Several methods for epididymal fluid collection have been developed. Mincing and pressing the epididymis was the earliest used method. However, samples processed by this method may be contaminated with serum, lymph, or epididymal cellular proteins. As an alternative, the microperfusion method can provide much purer epididymal fluid, but this method is mainly performed on large animals, and is not suitable for mice, especially for the collection of mouse caput fluid.25,33 Thus, the method of making small slits in the epididymis tubules with pressing was adopted to collect mouse caput and cauda luminal fluids in the study.
In the present study, the iTRAQ method was applied to reveal the proteins differentially enriched between the caput and cauda. The results of bioinformatic analysis showed that these proteins took part in different biological processes and played different molecular functions. These results indicate that the caput and cauda have distinct functions during sperm maturation.
Pdzk1 is a newly identified epididymal luminal protein by iTRAQ, which was found in lipid metabolism. Pdzk1, also named NHERF3, belongs to the Na+/H+ exchange regulatory cofactor (NHERF) family. It is a scaffold protein with four PDZ domains and participates in the regulation of cell surface protein expression, cholesterol metabolism, and carnitine transport, which are important for sperm maturation and motility.15,34,35,36 Pdzk1 prompts carnitine uptake by upregulating the activity of the carnitine transporter OCTN2.15,37
It has been reported that Pdzk1 is expressed in the epididymis and efferent ducts,14 which was confirmed by our findings. The cauda has a higher concentration of carnitine and the cauda sperm are more motile than that in the caput.38 However, we found that Pdzk1 was enriched in the caput fluid but not in the cauda fluid. The function of Pdzk1 in the cauda fluid may be compensated by another NHERF family member, NHERF1, which interacts with another carnitine transporter, OCTN1,15 since NHERF1 is enriched in the cauda fluid.
Pdzk1 was localized at the sperm tail, but had no expression in the testis. Pdzk1 may be secreted into the luminal fluid from the efferent ducts or epididymis and bound to the sperm surface. Since Pdzk1 has no typical signal sequence (Supplementary Figure 3 (83.5KB, tif) ), its secretion may be mediated by epididymosomes, specific exosomes in the epididymis.5 This manner of secretion was confirmed by the finding that Pdzk1 was present in urinary exosomes.39,40
Pdzk1 has no typical signal sequence. Pdzk1 protein sequence (NP_001139473) was analyzed by SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP) and no signal peptide was found. Pdzk1: PDZ domain containing 1.
The expression of Pdzk1 in human spermatozoa was analyzed and showed that Pdzk1 was decreased in asthenozoospermia. Mice without Pdzk1 are fertile but have increased serum cholesterol levels, which may affect sperm motility. However, this speculation requires further investigation because the sperm motility and sperm fertilizing ability in the knockout mice are unclear.41 The results obtained here indicate that Pdzk1 may be associated with sperm maturation and male infertility, but further research is needed to reveal the mechanisms behind this involvement.
CONCLUSION
Taking the obtained findings together, this work reveals the proteins differentially enriched between the caput and cauda fluid, shedding light on the distinct functions of the caput and cauda in sperm maturation. The epididymal luminal protein Pdzk1 newly identified here by iTRAQ has been associated with asthenozoospermia. This work provides a foundation for future mechanistic research on sperm maturation and male infertility.
AUTHOR CONTRIBUTIONS
AJL and GSW gathered the epididymal luminal proteins, performed proteomic analysis, and drafted the manuscript. PP and SGH collected human semen and performed western blotting analysis of Pdzk1 in human spermatozoa. YL and YM carried out the immunofluorescence analysis of Pdzk1. ZZD and HSW contributed to the western blotting analysis of Pdzk1 in mouse. FS conceived this study and helped to improve the manuscript. All authors have read and approved the final manuscript.
COMPETING INTERESTS
The authors declare that they have no competing interests.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China Grants (81430027 and 81671510 to FS; 81501309 to GSW) and the National Basic Research Program of China (2014CB943100 to FS).
Supplementary information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Lian J, Zhang X, Tian H, Liang N, Wang Y, et al. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol. 2009;7:13. doi: 10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navarro-Costa P, Goncalves J, Plancha CE. The AZFc region of the Y chromosome: at the crossroads between genetic diversity and male infertility. Hum Reprod Update. 2010;16:525–42. doi: 10.1093/humupd/dmq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoysman R. Epididymal causes of male infertility: pathogenesis and management. Int J Androl. 1982;5:120–34. [Google Scholar]
- 4.Schoysman R. Management of epididymal dysfunction: Correlation with basic physiology. In: Robaire B, Hinton BT, editors. The Epididymis: From Molecules to Clinical Practice. New York: Kluwer Academic/Plenum Publishers; 2002. pp. 473–82. [Google Scholar]
- 5.Cornwall GA. New insights into epididymal biology and function. Hum Reprod Update. 2009;15:213–27. doi: 10.1093/humupd/dmn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dacheux JL, Dacheux F. New insights into epididymal function in relation to sperm maturation. Reproduction. 2014;147:R27–42. doi: 10.1530/REP-13-0420. [DOI] [PubMed] [Google Scholar]
- 7.Dacheux JL, Belleannee C, Guyonnet B, Labas V, Teixeira-Gomes AP, et al. The contribution of proteomics to understanding epididymal maturation of mammalian spermatozoa. Syst Biol Reprod Med. 2012;58:197–210. doi: 10.3109/19396368.2012.663233. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan R, Saez F, Girouard J, Frenette G. Role of exosomes in sperm maturation during the transit along the male reproductive tract. Blood Cells Mol Dis. 2005;35:1–10. doi: 10.1016/j.bcmd.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Baker MA, Nixon B, Naumovski N, Aitken RJ. Proteomic insights into the maturation and capacitation of mammalian spermatozoa. Syst Biol Reprod Med. 2012;58:211–7. doi: 10.3109/19396368.2011.639844. [DOI] [PubMed] [Google Scholar]
- 10.Roberts KP, Ensrud KM, Hamilton DW. A comparative analysis of expression and processing of the rat epididymal fluid and sperm-bound forms of proteins D and E. Biol Reprod. 2002;67:525–33. doi: 10.1095/biolreprod67.2.525. [DOI] [PubMed] [Google Scholar]
- 11.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, et al. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–55. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, et al. QuickGO: a web-based tool for gene ontology searching. Bioinformatics. 2009;25:3045–6. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Zhang H, Wang L, Wang Y, Huang H, et al. Ca(2+)/calmodulin-dependent protein kinase IV promotes interplay of proteins in chromatoid body of male germ cells. Sci Rep. 2015;5:12126. doi: 10.1038/srep12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trepos-Pouplard M, Lardenois A, Staub C, Guitton N, Dorval-Coiffec I, et al. Proteome analysis and genome-wide regulatory motif prediction identify novel potentially sex-hormone regulated proteins in rat efferent ducts. Int J Androl. 2010;33:661–74. doi: 10.1111/j.1365-2605.2009.01006.x. [DOI] [PubMed] [Google Scholar]
- 15.Kato Y, Sai Y, Yoshida K, Watanabe C, Hirata T, et al. PDZK1 directly regulates the function of organic cation/carnitine transporter OCTN2. Mol Pharmacol. 2005;67:734–43. doi: 10.1124/mol.104.002212. [DOI] [PubMed] [Google Scholar]
- 16.Jelinsky SA, Turner TT, Bang HJ, Finger JN, Solarz MK, et al. The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol Reprod. 2007;76:561–70. doi: 10.1095/biolreprod.106.057323. [DOI] [PubMed] [Google Scholar]
- 17.Li JY, Wang HY, Liu J, Liu Q, Zhang JS, et al. Transcriptome analysis of a cDNA library from adult human epididymis. DNA Res. 2008;15:115–22. doi: 10.1093/dnares/dsn005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan H, Liu A, Zhang L, Zhou H, Wang Y, et al. Proteomic profiling of regionalized proteins in rat epididymis indicates consistency between specialized distribution and protein functions. J Proteome Res. 2006;5:299–307. doi: 10.1021/pr050324s. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Liu FJ, Jin SH, Shen XF, Wang YW. In-depth proteomic mapping of mouse (Mus musculus) epididymal constructive basis for sperm maturation. Proteome Sci. 2015;13:20. doi: 10.1186/s12953-015-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliva R, de Mateo S, Estanyol JM. Sperm cell proteomics. Proteomics. 2009;9:1004–17. doi: 10.1002/pmic.200800588. [DOI] [PubMed] [Google Scholar]
- 21.Brewis IA, Gadella BM. Sperm surface proteomics: from protein lists to biological function. Mol Hum Reprod. 2010;16:68–79. doi: 10.1093/molehr/gap077. [DOI] [PubMed] [Google Scholar]
- 22.Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci U S A. 2000;97:9390–5. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Liu F, Liu X, Liu J, Zhu P, et al. Mapping of the human testicular proteome and its relationship with that of the epididymis and spermatozoa. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004630. M110.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syntin P, Dacheux F, Druart X, Gatti JL, Okamura N, et al. Characterization and identification of proteins secreted in the various regions of the adult boar epididymis. Biol Reprod. 1996;55:956–74. doi: 10.1095/biolreprod55.5.956. [DOI] [PubMed] [Google Scholar]
- 25.Fouchecourt S, Metayer S, Locatelli A, Dacheux F, Dacheux JL. Stallion epididymal fluid proteome: qualitative and quantitative characterization; secretion and dynamic changes of major proteins. Biol Reprod. 2000;62:1790–803. doi: 10.1095/biolreprod62.6.1790. [DOI] [PubMed] [Google Scholar]
- 26.Belleannee C, Labas V, Teixeira-Gomes AP, Gatti JL, Dacheux JL, et al. Identification of luminal and secreted proteins in bull epididymis. J Proteomics. 2011;74:59–78. doi: 10.1016/j.jprot.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 27.Souza CE, Rego JP, Lobo CH, Oliveira JT, Nogueira FC, et al. Proteomic analysis of the reproductive tract fluids from tropically-adapted Santa Ines rams. J Proteomics. 2012;75:4436–56. doi: 10.1016/j.jprot.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Jin SH, Liu XX, Wang WJ, Liu FJ. Proteome profiling of the sperm maturation milieu in the rhesus monkey (Macaca mulatta) epididymis. Reprod Fertil Dev. 2016;28:732–41. doi: 10.1071/RD14322. [DOI] [PubMed] [Google Scholar]
- 29.Dacheux JL, Dacheux F, Labas V, Ecroyd H, Nixon B, et al. New proteins identified in epididymal fluid from the platypus (Ornithorhynchus anatinus) Reprod Fertil Dev. 2009;21:1002–7. doi: 10.1071/RD09091. [DOI] [PubMed] [Google Scholar]
- 30.Turner TT, Miller DW, Avery EA. Protein synthesis and secretion by the rat caput epididymidis in vivo: influence of the luminal microenvironment. Biol Reprod. 1995;52:1012–9. doi: 10.1095/biolreprod52.5.1012. [DOI] [PubMed] [Google Scholar]
- 31.Turner TT, Riley TA, Mruk DD, Cheng CY. Obstruction of the vas deferens alters protein secretion by the rat caput epididymidal epithelium in vivo. J Androl. 1999;20:289–97. [PubMed] [Google Scholar]
- 32.Liu X, Wang W, Liu F. New insight into the castrated mouse epididymis based on comparative proteomics. Reprod Fertil Dev. 2014;27:551–6. doi: 10.1071/RD13323. [DOI] [PubMed] [Google Scholar]
- 33.Turner TT, Riley TA, Vagnetti M, Flickinger CJ, Caldwell JA, et al. Postvasectomy alterations in protein synthesis and secretion in the rat caput epididymidis are not repaired after vasovasostomy. J Androl. 2000;21:276–90. [PubMed] [Google Scholar]
- 34.Kocher O, Yesilaltay A, Cirovic C, Pal R, Rigotti A, et al. Targeted disruption of the PDZK1 gene in mice causes tissue-specific depletion of the high density lipoprotein receptor scavenger receptor class B type I and altered lipoprotein metabolism. J Biol Chem. 2003;278:52820–5. doi: 10.1074/jbc.M310482200. [DOI] [PubMed] [Google Scholar]
- 35.Wang P, Wang JJ, Xiao Y, Murray JW, Novikoff PM, et al. Interaction with PDZK1 is required for expression of organic anion transporting protein 1A1 on the hepatocyte surface. J Biol Chem. 2005;280:30143–9. doi: 10.1074/jbc.M503969200. [DOI] [PubMed] [Google Scholar]
- 36.Mongioi L, Calogero AE, Vicari E, Condorelli RA, Russo GI, et al. The role of carnitine in male infertility. Andrology. 2016;4:800–7. doi: 10.1111/andr.12191. [DOI] [PubMed] [Google Scholar]
- 37.Gibb Z, Lambourne SR, Quadrelli J, Smith ND, Aitken RJ. L-carnitine and pyruvate are prosurvival factors during the storage of stallion spermatozoa at room temperature. Biol Reprod. 2015;93:104. doi: 10.1095/biolreprod.115.131326. [DOI] [PubMed] [Google Scholar]
- 38.Jeulin C, Lewin LM. Role of free L-carnitine and acetyl-L-carnitine in post-gonadal maturation of mammalian spermatozoa. Hum Reprod Update. 1996;2:87–102. doi: 10.1093/humupd/2.2.87. [DOI] [PubMed] [Google Scholar]
- 39.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–79. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huebner AR, Cheng L, Somparn P, Knepper MA, Fenton RA, et al. Deubiquitylation of protein cargo is not an essential step in exosome formation. Mol Cell Proteomics. 2016;15:1556–71. doi: 10.1074/mcp.M115.054965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kocher O, Pal R, Roberts M, Cirovic C, Gilchrist A. Targeted disruption of the PDZK1 gene by homologous recombination. Mol Cell Biol. 2003;23:1175–80. doi: 10.1128/MCB.23.4.1175-1180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blotting analysis of protamine 1 in the fluid of caput and cauda, and the cauda sperm. Gel is stained by Coomassie Brilliant Blue R-250.
The identifications by isobaric tag for relative and absolute quantitation
The differentially enriched proteins between the mouse caput and cauda luminal fluids
The unchanged proteins between the mouse caput and cauda luminal fluids
Complete scans of the different blots presented in Figure 2 and Figure 6 indicating the specificity of Pdzk1 antibody. (a) Uncropped picture presented in Figure 2a showing the expression of Pdzk1 in different organs of mouse. (b) Uncropped picture presented in Figure 6b showing the expression of Pdzk1 in human spermatozoa. Pdzk1: PDZ domain containing 1.
Pdzk1 has no typical signal sequence. Pdzk1 protein sequence (NP_001139473) was analyzed by SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP) and no signal peptide was found. Pdzk1: PDZ domain containing 1.


