Abstract
Purpose: Systemic administration of free chemotherapeutic drugs leads to severe toxic effects, and physiological characteristics of solid tumors restrain the drugs from reaching the hypoxic regions. E. coli Nissle 1917 (EcN) has been known to penetrate the barrier and proliferate in the interface between the viable and necrotic regions of tumors. This study aimed to fabricate a nanoscale minicell via genetic engineering of EcN for targeted delivery of chemotherapeutic drugs to the hypoxic regions of tumors for cancer therapy.
Methods: A large number of minicells were produced by knocking out the minCD gene and enhancing the minE expression in EcN. Then, a pH (low) insertion peptide (pHLIP) was displayed on the membrane surface through protein display technology to endow the cells with the ability to target the acidic microenvironments of tumors. The acidic-microenvironment targeting ability and therapeutic effect of the engineered minicells with chemotherapeutic drugs was thoroughly evaluated by using breast cancer cells and an orthotopic model of breast tumor.
Results: The EcN-derived minicells displaying pHLIP could be directly extracted from the fermentation broth and used for delivering chemotherapeutic drugs without any further modification. Targeting of doxorubicin (DOX)-loaded minicells to cancer cells via pHLIP resulted in rapid internalization and drug release in acidic media. Importantly, the pHLIP-mosaic minicells successfully invaded the necrotic and hypoxic regions of orthotopic breast cancers where free chemotherapeutic drugs could never get to because of vascular insufficiency and high interstitial fluid pressure. This invasion resulted in significant regression of an orthotopic breast tumor in a mouse model, while no seriously pathogenic effects were observed during the animal experiments.
Conclusions: This study provides a novel strategy for the fabrication of tumor-targeting carriers via genetic engineering based on biomaterials with the ability to penetrate hypoxic regions of tumors, high biocompatibility and low toxicity.
Keywords: E. coli Nissle 1917, minicell, low pH insert protein, hypoxic region, cancer therapy
Introduction
Severe toxicity of chemotherapy remains a thorny problem in cancer therapy. In addition, poorly vascularized microenvironments and high interstitial fluid pressures of solid tumors restrict the penetration of chemotherapeutic drugs into the centers of the injured tissues and greatly reduce the anticancer effects of these drugs 1-3. Targeted drug-delivery systems, such as liposomes 4, nanoparticles 5, and polymer micelles 6, are being explored to enhance the anticancer effects of chemotherapeutic drugs. By passive targeting (enhanced permeability and retention (EPR)) or active targeting (linking to tumor-targeting agents), the carriers successfully increase the accumulation of free chemotherapeutic drugs in tumors and lower the side effects of these drugs 7. However, most drug carriers accumulate in the perivascular regions after leakage from tumor vasculature, and the released drugs do not reach the hypoxic regions of the tumor because of the lack of arteries in the hypoxic regions and high interstitial fluid pressure 8-10.
Minicells, which are nanosized forms of bacteria, have been shown to be advantageous as ideal tumor-targeted drug-delivery systems to transport chemotherapeutic drugs into tumors 11-13. Knocking out minCD or overexpressing minE in bacteria can induce aberrant cell division and produce a large amount of minicells 14. These nanoscale cells contain the same cytoplasmic components as their parent bacteria except chromosomal DNA. The genome deficiency leads to a loss of the proliferation ability of minicells; however, minicells retain the other characteristics inherited from their parent bacteria 14, 15. In addition, some minicells have been used for targeted delivery of chemotherapeutic drugs into tumors and to inhibit cancer growth 11, 13. For example, minicells of Salmonella typhimurium (S. typhimurium) can be produced in large amounts using a genetically modified strain and can be loaded with diverse chemotherapeutic drugs without leakage 13. Drug-loaded minicells modified with antibodies to receptors on cancer cells can selectively enter tumors and release anticancer drugs to kill the cancer cells 11, 12. Importantly, paclitaxel-packaged minicells of S. typhimurium have achieved success in phase I clinical trials and did not have any serious toxic effects during observation 16.
In contrast to S. typhimurium and other bacteria that colonize in the necrosis, E. coli Nissle 1917 (EcN), with higher tumor-targeting ability, mainly proliferate in the interface between the necrotic and hypoxic regions of tumors 17-19, guaranteeing that EcN-loaded drugs can be released into hypoxic regions in cancer therapy. Additionally, the specific cell membrane of EcN can directly interact with the adaptive immune system and reduce inflammation 20, 21. Moreover, the serum-sensitive lipopolysaccharide (LPS) of the EcN membrane ensures the quick elimination of the strain from normal organs 22. Therefore, EcN-derived cells, with specific immune regulatory effects and the ability to target hypoxic regions, have potential applicability in the targeting of chemotherapeutic drugs into the deep centers of tumors to exert anticancer effects. Furthermore, the use of ligands against tumor-associated markers (HER2/EGFR) has been shown to have additive effects in enhancing the accumulation of minicells in solid tumors 13, 16. Acidosis is a typical feature of solid tumors 23. Using ligands against acidic microenvironments to mount drug vectors would significantly increase the targeting ability of drug vectors to most solid tumors and would enhance the anticancer effects of chemotherapeutic drugs. A low-pH insertion peptide (pHLIP) specifically targeting the acidic microenvironments of tumors has been employed to label RNA or nanoparticles to enhance the accumulation of these molecules in tumors and to enhance their therapeutic effects 24-27. Using genetic engineering to display the ligand on the membrane surfaces of minicells during fermentation will be much better than non-covalent bond formation between the antibody and antigen; what's more, this method does not require further modification.
Therefore, we fabricated a drug carrier based on EcN-derived minicells and pHLIP via genetic engineering for targeted delivery of chemotherapeutic drugs to the hypoxic regions of tumors. First, the scale production of minicells from EcN was conducted by deleting the minCD gene and enhancing the expression of the minE gene in the bacterial genome using Red/ET recombination technology. Next, the pHLIP was displayed on the membrane surfaces of the minicells using the Lpp-OmpA' protein display system for targeting vectors to solid tumors. In addition, bioluminescence with high signal-to-noise ratio was co-expressed with the pHLIP and used to monitor the in vivo distribution of the minicells. The minicells with the pHLIP could be directly extracted from the fermentation broth of the strain and used to load chemotherapeutic drugs. The results demonstrated that the pHLIP succeeded in enhancing the accumulation of DOX-loaded minicells in the necrotic and hypoxic regions of tumors. This resulted in significant inhibitory effects of drug-loaded minicells on the growth of orthotopic breast cancers without any notable toxicity.
Materials and methods
Minicell preparation from EcN using genetic engineering
Deletion of the minCD gene and overexpression of the minE in EcN were carried out using the Red/ET homologous recombination technique. The pSC101-BAD-gbaA-tet plasmid was electroporated into EcN cells and screened on a tetracycline (Tet)-containing lysogeny broth (LB) plate to obtain the positive strain (EcN harboring the pSC101-BAD-gbaA-tet plasmid) following the general protocol. The chloromycetin gene (Cm), with sequences homologous to the end of the minCD gene, and the ribosome binding site (RBS) were amplified from the pSUM plasmid using KTminCD1 and KTminCD2 as primers. The PCR products were purified and transferred to EcN cells harboring the pSC101-BAD-gbaA-tet plasmid by electroporation to conduct Red/ET homologous recombination 28. The derived strains were cultured on chloromycetin plates to obtain the EcN harboring a minCD deletion and overexpressing minE protein (ΔEcN). The positive ΔEcN strain was verified by sequencing the corresponding regions of minCD and minE in the genome using the primers IDCD1 and IDCD2. The gene sequence and the primers used in the study are listed in Supplementary Material (gene sequence and Table S1).
Construction of the plasmid with the protein display system and pHLIP
The gene of the Lpp-OmpA' protein with the nucleotide sequence of pHLIP was synthesized by GENEWIZ (Suzhou, China). The synthetic gene was fused with the Tet promoter by overlapping PCR to obtain the tet-Lpp-OmpA'-SSG-pHLIP-Histag DNA fragment using the primers Tet-R, Tet-F, pHLIP-R and pHLIP-F (Table S1). The derived DNA products and the pET28a plasmid were digested using the restriction endonucleases Nco I and Hind III, respectively. Then, the digested DNA fragments were ligated together to construct the pET28a-tet-Lpp-OmpA'-pHLIP-Histag plasmid. Next, the ligated products were transferred into E. coli GB2005 to obtain the positive strain and amplify the plasmid. Simultaneously, the LuxCDABE gene was amplified using the primers Luc1 and Luc2 and using the genome of Photorhabdus luminescens TTO1 as a template 29. The PCR products of the LuxCDABE gene cluster, the pET28a-tet-Lpp-OmpA'-pHLIP-Histag plasmid and the pET28a plasmid were digested using the restriction endonucleases Xho I and Not I, individually. Further, the digested DNA fragments were ligated by ligase to construct the plasmids pET28a-tet-Lpp-OmpA'-pHLIP-Histag-LuxCDABE (abbreviated as pET28a-pHLIP) and pET28a-LuxCDABE (named as pET28a-Lux). The plasmids were individually electroporated into BL21 and ΔEcN for protein expression. Additionally, the plasmid pUC18-GFP was transferred into ΔEcN to observe the budding process of the minicells.
Minicell characterization and purification
The ΔEcN cells harboring the pUC18-GFP plasmid were grown at 37 ℃ in LB medium overnight and transferred into fresh medium at a 2% (v/v) ratio. Next, when the OD600 value of the medium reached 1.1, the ΔEcN cells were collected and washed thrice with phosphate-buffered saline (PBS). The morphology of ΔEcN and the minicell budding process were captured using an LSM710 confocal laser scanning microscope (Carl Zeiss, Germany). Additionally, the bacteria were washed three times using sterile water and prepared by following the general protocol for morphological observation using an SU8000 field-emission scanning electron microscope (Hitachi, Japan).
Identification of pHLIP on the membranes of ΔEcN cells using Western blot and immunofluorescence
BL21 and ΔEcN cells harboring plasmids pET28a-pHLIP or pET28a-Lux were cultured in LB medium to mid-logarithmic phase. Then, 0.2 µg/mL Tet was added to the solution to induce the tet promoter and the expression of the Lpp-OmpA-pHLIP-Histag protein. Then, the bacteria were harvested and lysed using lysis buffer. The protein content of the supernatant was evaluated using SDS-PAGE and western blotting following a general protocol. Briefly, the proteins on the SDS-PAGE gel were transferred to a polyvinylidene fluoride (PVDF) membrane and incubated with anti-6×His rabbit polyclonal antibody (Sangon Biotech, Shanghai, China) at 1:1000 dilution at 4 °C overnight. Further, the PVDF membrane was washed three times using PBS supplemented with 0.5% (v/v) Tween 20 (TPBS) and further incubated with FITC-conjugated anti-rabbit antibody at a dilution of 1:100 for 1 h at room temperature. After washing thrice using TPBS, the FITC fluorescence on the membrane was captured using an imaging system 5200 Multi (Tanon, Shanghai, China). Furthermore, immunofluorescence was conducted to identify pHLIP on the cell membranes of the ΔEcN cells. The overnight culture of ΔEcN was transferred to fresh LB medium at a 2% ratio. Then, 1 mL of the strain was collected during the exponential growth phase by centrifugation and washed thrice using ice-cold PBS. The collected cells were resuspended in PBS containing 2% bovine serum albumin (BSA) and incubated for 30 min at room temperature. Then, the cells were collected, washed thrice using PBS, and incubated with anti-6×His rabbit polyclonal antibody (dilution 1:500) (Sangon Biotech, Shanghai, China) for 1 h on ice. Next, the ΔEcN cells was washed again with PBS and incubated with PBS containing 2% BSA and FITC-conjugated anti-rabbit antibody (Sangon Biotech, Shanghai, China) at a dilution of 1:50 for 1 h at room temperature. The immunofluorescence on the membrane surfaces of the ΔEcN cells was captured using a confocal laser scanning microscope.
Minicell extraction and drug packaging
The minicells were extracted and purified by gradient centrifugation in 20% sucrose solution. The ΔEcN cells harboring the pET28a-pHLIP or pET28a-Lux plasmids were cultured in 500 mL of LB medium at 37 ℃ overnight. The bacteria and minicells were harvested via centrifugation at 10000 ×g for 20 min. The pellets were resuspended in buffered saline gelatin (BSG) and vortexed for 10 min at maximum speed. The suspensions were first centrifuged for 10 min at 2000 ×g to remove most bacteria, and the supernatants were collected after centrifugation at 10000 ×g for 30 min. The obtained pellets were resuspended and cultured in LB solution containing 25 mg/L nalidixic acid and 50 μg/mL apramycin (Sangon Biotech, Shanghai, China) at 37 °C for 1 h to kill the residual bacteria. The resultant solutions were centrifuged at 10000 ×g for 30 min and washed using BSG. The obtained minicells were spread on an LB plate and cultured overnight to detect possible contamination. Finally, the minicell pellet was lyophilized in trehalose for long-term storage and reconstituted in PBS for in vitro and in vivo experiments.
DOX packaging and release assay
The minicells with pHLIP (minicellspHLIP) were incubated with 60 µg/mL of DOX (Sangon Biotech, Shanghai, China) for 0 h to 10 h or with different concentrations of the drug (50 µg/mL, 100 µg/mL, 150 µg/mL, 200 µg/mL, or 250 µg/mL) for 12 h at 37 ℃ in a shaking incubator (Eppendorf, Germany). The minicells were collected and washed three times. Next, the minicells were lysed by sonication to release the packaged DOX. The absorbance of the DOX in the supernatant was recorded at 498 nm using a Molecular Devices SpectraMax M5 (Molecular Devices Corporation, USA). Based on a standard curve, the concentrations of the packaged DOX in the minicells were calculated. The minicells harboring pHLIP and DOX (abbreviated as minicellspHLIP DOX) were visualized using confocal laser scanning microscopy. In addition, the minicellspHLIP DOX were incubated at 37 °C for 0 h to 16 h in a shaking incubator at a speed of 800 rpm. At specific time intervals, the minicells were collected, and the DOX contained in the cells was analyzed as in the above-mentioned packaging assay.
Targeted uptake of DOX and minicells by cancer cells
MCF-7 human breast cancer cells and 4T1 mouse breast cancer cells were purchased from the American Type Culture Collection (ATCC). Cells were cultured in RPMI 1640 (Gibco) medium supplemented with L-glutamine, 10% heat-inactivated fetal bovine serum (Gibco), 100 U/mL penicillin and 100 µg/mL streptomycin at 37 °C in a humidified incubator with a 5% CO2 atmosphere. The 4T1 and MCF-7 cells were seeded on glass coverslips overnight under cell culture conditions. Then, the media were replaced with pre-warmed media at pH 6.5 or 7.4 containing 109 DOX-packaged minicells (named minicells DOX) or minicellspHLIP DOX, and the cells were further cultured for 2 h. After treatment, the coverslips were washed with PBS three times, fixed in 4% paraformaldehyde for 15 min, and washed three times with PBS again. Next, the cells were stained with one drop of mounting medium containing DAPI Hoechst (0.5 µg/mL) to counter stain the nuclei. Images of cellular uptake and distribution of DOX in the cancer cells were captured using an inverted confocal laser scanning microscope. Furthermore, the 4T1 cells were seeded in a 96-well plate and cultured overnight. Then, the media was replaced with 100 µL of media at different pH values (pH 6.5 or 7.4) containing 109 minicells DOX and minicellspHLIP DOX per milliliter and incubated for 2 h. The cells were washed with PBS three times, and the bioluminescence of the cells was recorded and analyzed using an IVIS Spectrum system.
Flow cytometry analysis and immunofluorescence detection of minicellspHLIP DOX internalization by cancer cells
The 4T1 cells in the exponential growth phase were seeded in a six-well plate. After 24 h, the medium was replaced with fresh medium of different pH values (pH 7.4 or 6.5) containing PBS, minicells DOX (109/mL), minicellspHLIP DOX (109/mL) or 6.95 µg/mL DOX. After 2 h of incubation, the cells were harvested and resuspended in 500 µL of PBS. The fluorescence of DOX in the cells was recorded and analyzed by flow cytometry (CytoFLEX, Beckman Coulter, USA). Simultaneously, the cancer cells were seeded on a coverslip in a 12-well plate. After incubation overnight, the medium was replaced with minicells (109/mL) or minicellspHLIP (109/mL), and the cells were incubated for 2 h. Next, the cells were fixed in 4% paraformaldehyde and permeabilized using 0.1% Triton X-100. After washing thrice with PBS, 2% BSA was added into the wells to block non-specific antigens on the cancer cells. Then, the cells were incubated with anti-6×His rabbit polyclonal antibody (dilution 1:500) at 4 °C for 12 h. After washing thrice using PBS, the cells were further incubated with the fluorescein (FITC)-conjugated anti-rabbit antibody (Sangon Biotech, Shanghai, China) at a dilution of 1:50 for 1 h at room temperature. Next, fresh medium containing 3 µM DiIC183 was added to the wells, and the cells were incubated at 37 °C for 15 min. The excess DiIC183 was removed from the cells using warmed PBS, and a small drop of mounting media (glycerol antifade) containing DAPI was placed on the coverslip. Images of the distribution of minicellspHLIP in the cells was captured using an inverted confocal laser scanning microscope (LSM 710, Carl Zeiss, Germany).
Cytotoxicity of minicellspHLIP DOX against breast cancer cells
The 4T1 and MCF-7 breast cancer cells were seeded in a 96-well plate and cultured in a cell incubator overnight. Next, the culture media of the cells was replaced with fresh media of different pH values (pH 7.4 or 6.5) containing PBS, DOX, minicells, minicellspHLIP, minicellsDOX or minicellspHLIP DOX. The concentrations of DOX used were 0.5 µg/mL, 1.5 µg/mL, 3.0 µg/mL, 5.0 µg/mL, 7.0 µg/mL or 9.0 µg/mL. Given that the 109 minicellsDOX/minicellspHLIP DOX contain 6.95 µg of DOX, different numbers of minicells were added to the wells for the cytotoxicity assay. After 24 h of incubation, the media was removed and the cells were washed using PBS. One-hundred microliters of fresh media and 10 µL of cell counting kit-8 (Dojido, Japan) were added to each well of the 96-well plate and incubated for 1.5 h in a cell incubator. The absorbance of each well of the plate was recorded at 450 nm using a Molecular Devices SpectraMax M5.
In vivo distribution and antitumor effects of minicellspHLIP DOX
All animal experiments were performed following the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the experiments were approved by the Animals Ethics Committee of Hunan Normal University. Specific-pathogen-free (SPF) female BALB/C mice aged 6 to 8 weeks were purchased from the Hunan Tianqin biological company in Hunan Province, China. The animals were bred and maintained under SPF conditions for at least 3 days before use. The orthotopic breast tumors in the fourth mammary pads of the mice were established with 1 × 105 4T1 breast cancer cells in 30 µL of PBS. The tumor-targeting ability and antitumor effects of minicellspHLIP DOX were evaluated when the volumes of the 4T1 tumors grew to ~200 cm2. The tumor-bearing mice (each group had three mice) were intravenously (i.v.) injected with 100 µL of 1 × 1010 minicellsDOX or minicellspHLIP DOX. The bioluminescence signals from the mice were imaged at 0, 1, 2, 3 and 6 h using an IVIS Spectrum system to evaluate the distribution of minicellspHLIP DOX in the mice. Simultaneously, the mice with orthotopically implanted 4T1 breast tumors (each group contained eight mice) were i.v.-injected with 100 µL of PBS, 100 µL of 695 µg/mL DOX or 1 × 1010 of various minicells (minicells, minicellspHLIP, minicellsDOX and minicellspHLIP DOX) at 0, 7, 14 and 21 days. The 1 × 1010 minicellsDOX and minicellspHLIP DOX both contain 69.5 µg of DOX. Tumor volumes were recorded every three days using vernier calipers. At 3 h and day 25 after the first treatment, mice (n = 3) were sacrificed and the tumors were fixed in 4% paraformaldehyde overnight. Then, the tumor tissues were processed by hematoxylin and eosin (H&E) staining and immunohistochemistry. The sections were observed and recorded under an optical microscope (Olympus). Simultaneously, the healthy BALB/c mice were systemically administered 2 × 106 CFU (colony -forming units) of EcN to determine the in vivo distribution of the bacteria at 1 h and 3 h.
Toxicity
Toxicity of the engineered drug-vector in healthy BALB/c mice (n = 10) was evaluated by weekly i.v. administration of 1 × 1010 minicellspHLIP. Body weight was recorded every 2 days for 30 days. Finally, the mice were sacrificed by cervical dislocation, and the livers, spleens and kidneys were excised and fixed in 4% paraformaldehyde overnight and then stained for tissue analysis using a standard H&E procedure.
Statistical analysis
Statistical significance for all the experiments was determined by using Student's t-test. If the p-value was below 0.05, the differences between the experimental groups were considered significant.
Results
Construction of the minicellspHLIP for targeted delivery of chemotherapeutic drugs to hypoxic regions of solid tumors
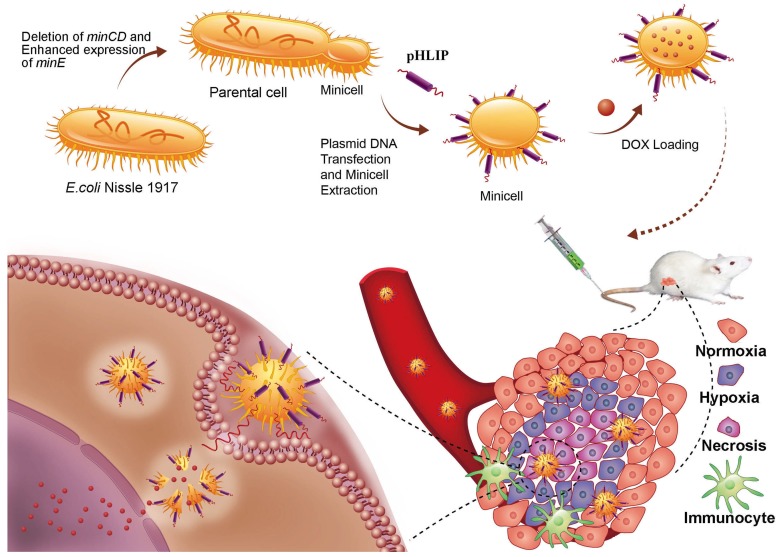
Deletion of the minCD gene and enhanced expression of the minE gene from the genome of EcN results in aberrant cell division and yields scores of minicells that lack chromosomal DNA (Figure 1). The genetically modified EcN cells were further transformed with the plasmid, which mainly included the Lpp-OmpA' protein display system, DNA fragments of pHLIP, and the LuxCDABE gene cluster. The Lpp-OmpA' protein display system, consisting of the first nine amino acids of the major E. coli lipoprotein and the transmembrane domain of OmpA (46-159), can efficiently anchor pHLIP to the membrane surfaces of the EcN-derived minicells 30. Simultaneously, LuxCDABE gene cluster was introduced to the minicells and used to trace their internalization and in vivo distribution 31. The engineered minicells displaying pHLIP can be unidirectionally loaded with chemotherapeutic drugs (DOX). The pHLIP could greatly increase the accumulation of the minicells in solid tumors. Taking advantage of the ability of EcN to penetrate into hypoxic regions, systemic administration of the engineered minicells could lead to invasion of the hypoxic regions of solid tumors by the minicells to release DOX and kill the cancer cells.
Figure 1.
Schematic illustration depicting the construction of the minicellspHLIP for targeted delivery of chemotherapeutic drugs into the hypoxic regions of solid tumors to kill cancer cells.
Display of pHLIP on the membrane surface of minicells
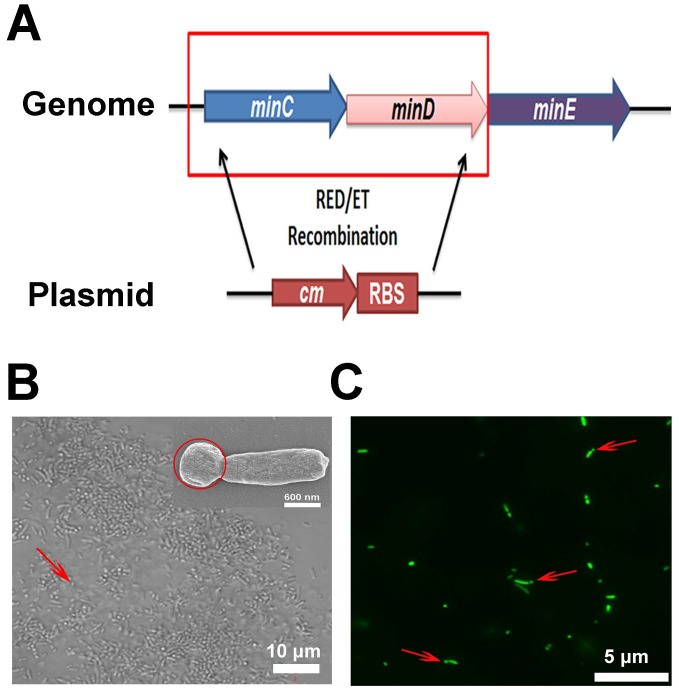
To obtain a large quantity of minicells from EcN, the minCD gene was deleted from the bacteria, while an RBS was introduced in front of the transcriptional start site of the minE gene using Red/ET homologous recombination technology (Figure 2A). The genome modification in EcN resulted in polar cell division and led to the production of a large number of achromosomal minicells (Figure 2B and C). The genetically modified strain was named ΔEcN. Using a differential interference contrast microscope (DIC), it was difficult to distinguish minicells from their parent EcN cells (Figure 2B). Scanning electron microscopy clearly showed the minicells budding from their parent cells (Figure 2B). Additionally, transformation of the cells with plasmids containing the green fluorescent protein (GFP) gene greatly favored the detection of minicells (red arrow) and the detection of the budding process of minicells from their parent cells (Figure 2C). The results indicated that deletion of minCD and overexpression of minE in EcN could induce disordered cell division in the bacteria, leading to the production of a large quantity of minicells from this strain.
Figure 2.
Construction of minicellspHLIP by gene knockout. (A) Modification of the EcN genome using Red/ET recombination technology (cm: chloromycetin; RBS: ribosomal binding site). (B) Morphology of ΔEcN-derived minicells (red arrow) as observed using optical microscopy by DIC and by field-emission scanning electron microscopy (red circle). (C) Observation of the budding of the minicell from the parent ΔEcN (red arrows) as seen using fluorescence microscopy.
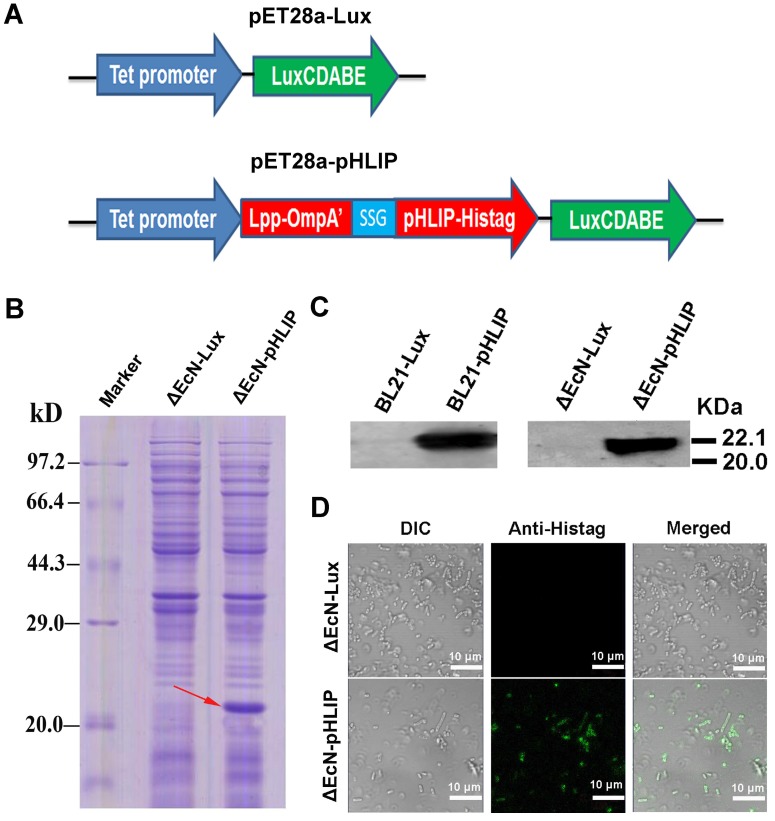
Minicells tagged with the antibody against the antigen on the receptors of cancer cells showed high tumor-targeting ability 16. Therefore, pHLIP, which has the ability to target the acidic microenvironments of tumors, was displayed on the EcN-derived minicells to enhance the accumulation of the minicells in tumors. The minicells with pHLIP and bioluminescence were constructed by electroporation of the pET28a-pHLIP plasmid into cells, while cells harboring the pET28a-Lux plasmid were used as the control group (Figure 3A). BL21, as the typical strain for protein expression, was used as a positive control in the experiment. The bacteria obtained from the transformation were named ΔEcN-pHLIP and ΔEcN-Lux. Similarly, the BL21 cells with the plasmids were individually designated BL21-pHLIP and BL21-Lux. The successful expression of the soluble Lpp-OmpA'-pHLIP-Histag protein in ΔEcN-pHLIP was confirmed by SDS-PAGE (red arrow) (Figure 3B) and western blotting (Figure 3C), while the control group (ΔEcN-Lux) did not show the presence of the indicated band (22 kDa). To exclude the possibility that the protein was only expressed in the cytoplasm, immunofluorescence was used to detect the presence of pHLIP on the membrane surface of ΔEcN. In contrast to the control group, the ΔEcN-pHLIP group clearly exhibited green fluorescence (Figure 3D). The intact membranes of the live strains would block the diffusion of the fluorescent antibody into the cells, which was confirmed by using the ΔEcN-Lux cells that did not express pHLIP (Figure 3D). The results demonstrated the successful display of pHLIP on the membrane surface of the ΔEcN-pHLIP strain. This result indicated that a potential tumor-targeting drug cargo was successfully constructed through genetic engineering and could be produced on a large scale by simple biological fermentation at very low cost. Considering the high biocompatibility of EcN and the tumor-targeting ability of pHLIP, the engineered minicells could be directly used to deliver chemotherapeutic drugs for cancer therapy without the need for the LPS purge.
Figure 3.
Plasmid construction and detection of pHLIP on the membrane surface of ΔEcN-pHLIP. (A) Diagram of the pET28a-LuxCDABE (pET28a-Lux) and pET28a-tet-Lpp-OmpA'-pHLIP-Histag-LuxCDABE (pET28a-pHLIP) plasmids. (B) Detection of pHLIP in the supernatant of ΔEcN containing the plasmid pET28a-pHLIP or pET28a-Lux using SDS-PAGE. (C) Detection of pHLIP in the supernatant of ΔEcN /BL21 with the plasmid pET28a-pHLIP or pET28a-Lux by western blotting. (D) Identification of pHLIP on the membrane surface of ΔEcN with the different plasmids (pET28a-pHLIP or pET28a-Lux) using immunofluorescence. These results are representative of 3 independent experiments.
Doxorubicin packaging and release assay
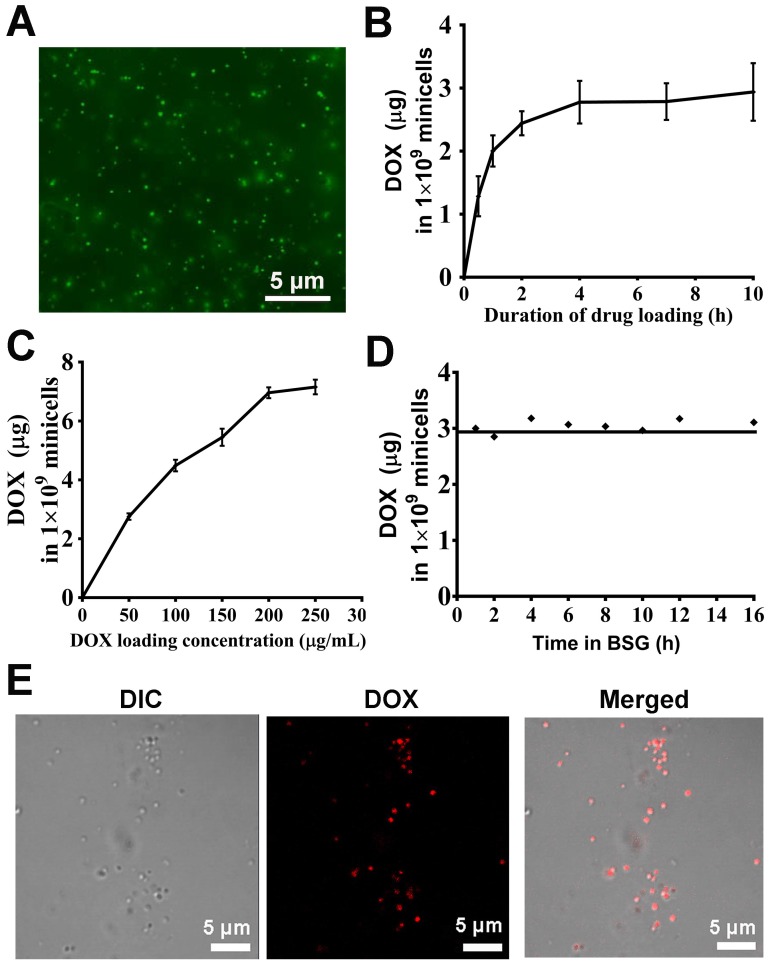
The anucleate minicellspHLIP produced by the ΔEcN strain can be extracted and purified directly from the fermentation broth. Gradient density centrifugation was first used to remove most of the bacteria, and the residual cells were further removed using the antibiotics nalidixic acid and apramycin. The antibiotics exert their cytotoxic effects against bacteria mainly by inhibiting the synthesis of DNA and ribonucleoproteins 32. Therefore, the membrane integrity and the components of the minicells would not be affected by the antibiotics. Fluorescence microscopy revealed that the morphologies of the purified minicells appear to be intact and uniform (Figure 4A). The drug-loading capacity of the ΔEcN-derived minicells was evaluated by incubation with various concentrations of DOX for different lengths of time. The minicellspHLIP were first incubated with 60 µg/mL of DOX for 0 h to 10 h. Figure 4B revealed that the packaged DOX in the minicells reached a maximum concentration (2.93 ± 0.45 µg per 1 × 109 minicellspHLIP) after 4 h of incubation, and the concentration did not increase any further over time, even after 10 h. What's more, the DOX content of the minicells also increased with the rise of drug concentrations. After incubation with 200 and 250 µg/mL of DOX, the packaged drugs in the minicellspHLIP reached concentrations of 6.95 ± 0.18 µg and 7.15 ± 0.24 µg per 109 minicellspHLIP, respectively (Figure 4C). This observation indicated that the amount of DOX in the minicellspHLIP almost reached a plateau after attaining a concentration of 200 µg/mL. Moreover, the leakage of DOX during transport is another key factor that should be carefully evaluated. Interestingly, the amounts of DOX packaged in the minicells appeared to be unchanged even after 16 h of incubation (Figure 4D). This indicated that DOX efflux did not occur after packaging into minicells under the experimental conditions. In other words, the minicells would not release the DOX until the minicells broke, which is beneficial for lowering the in vivo toxicity of chemotherapeutic drugs. Furthermore, confocal laser imaging clearly shows the minicellspHLIP loaded with DOX. The minicells were saturated with DOX against a clear background, indicating the membrane integrity of the cells after 12 h of incubation at 37 ℃ (Figure 4E). The unidirectionally-loaded DOX in the minicells guaranteed that there was no DOX released during blood circulation.
Figure 4.
DOX packaging and release assay. (A) Purified minicells visualized by fluorescence microscopy. (B) Drug-loading capacity of the minicellspHLIP after incubation with DOX for different lengths of time. (C) Drug-loading capacity of the minicellspHLIP incubated with different concentrations of DOX for 12 h. (D) DOX efflux from the minicellspHLIP DOX in buffered saline gelatin (BSG) from 0 h to 16 h. (E) Detection of minicellspHLIP DOX by confocal laser scanning microscopy. The results are expressed as the mean ± SD of 3 independent experiments.
MinicellspHLIP selectively target cancer cells in an acidic microenvironment
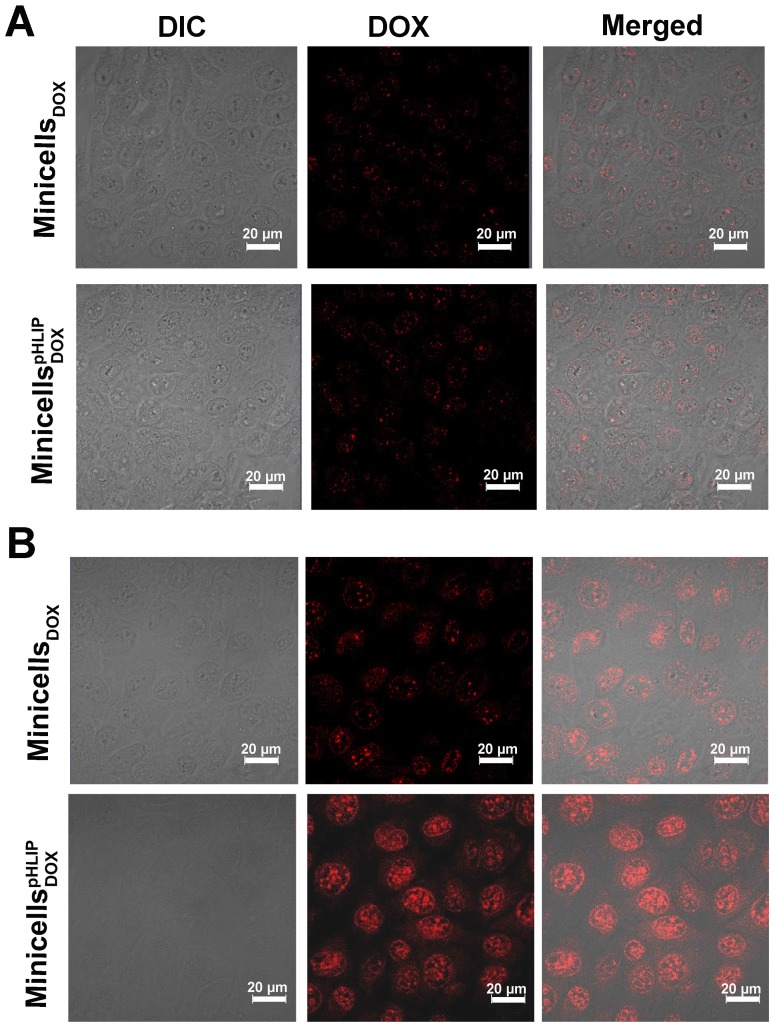
The pHLIP-mediated targeting of liposome or nanoparticles to acidic microenvironments of tumors has been well established 26, 27, 33. To determine whether pHLIP could enhance minicell internalization by cancer cells in acidic environments, the minicellspHLIP DOXand the minicellsDOX were each incubated with 4T1 breast cancer cells in media of different pH values (pH 6.5 or 7.4). The cellular uptake and intracellular distribution of DOX was visualized using confocal laser scanning microscopy. Figure 5A clearly shows that the 4T1 cancer cells display almost the same fluorescence intensity (red color) after 2 h of incubation with the minicellspHLIP DOX and minicellsDOX in the pH 7.4 medium, and the fluorescence signals were very weak. Because drug release only occurs when the minicellspHLIP DOX and minicells DOX are engulfed and digested by the cancer cells, the fluorescence intensity can be considered to be proportional to the number of minicells. This indicated that minicellspHLIP DOX could not selectively target cancer cells in the pH 7.4 medium. However, minicellspHLIP DOX-treated 4T1 cells showed much stronger fluorescence than the minicells DOX -treated group in the pH 6.5 medium (Figure 5B). Considering that the fluorescence intensity is proportional to the absorption of minicells, a much higher number of minicellspHLIP DOX was taken up by the cancer cells in the low-pH medium. However, the minicell DOX-treated group did not exhibit the selective targeting ability (Figure 5B). This indicated that it was due to pHLIP that the minicellspHLIP DOX could selectively enter 4T1 cancer cells in the acidic solution. A certain difference between the fluorescence intensities of cells incubated with minicells DOX and those incubated with minicellspHLIP DOX is also detectable in MCF-7 in the pH 7.4 medium; however, the difference is lower than that observed in the pH 6.5 solution (Figure S1). Figure 5 also demonstrated that the DOX (red) released from the minicells was mainly localized in the nucleus of the cancer cells. DOX prefers to enter the nucleus because the drug has a specific ability to stimulate peroxidation of the nuclear membrane. There, they exhibit cytotoxic effects through DNA interaction, lipid peroxidation and inhibition of topoisomerase II 34.
Figure 5.
Confocal imaging of minicells DOX and minicellspHLIP DOX internalization by 4T1 cancer cells in pH 7.4 (A) and pH 6.5 media (B). These results are representative of 3 independent experiments with similar results.
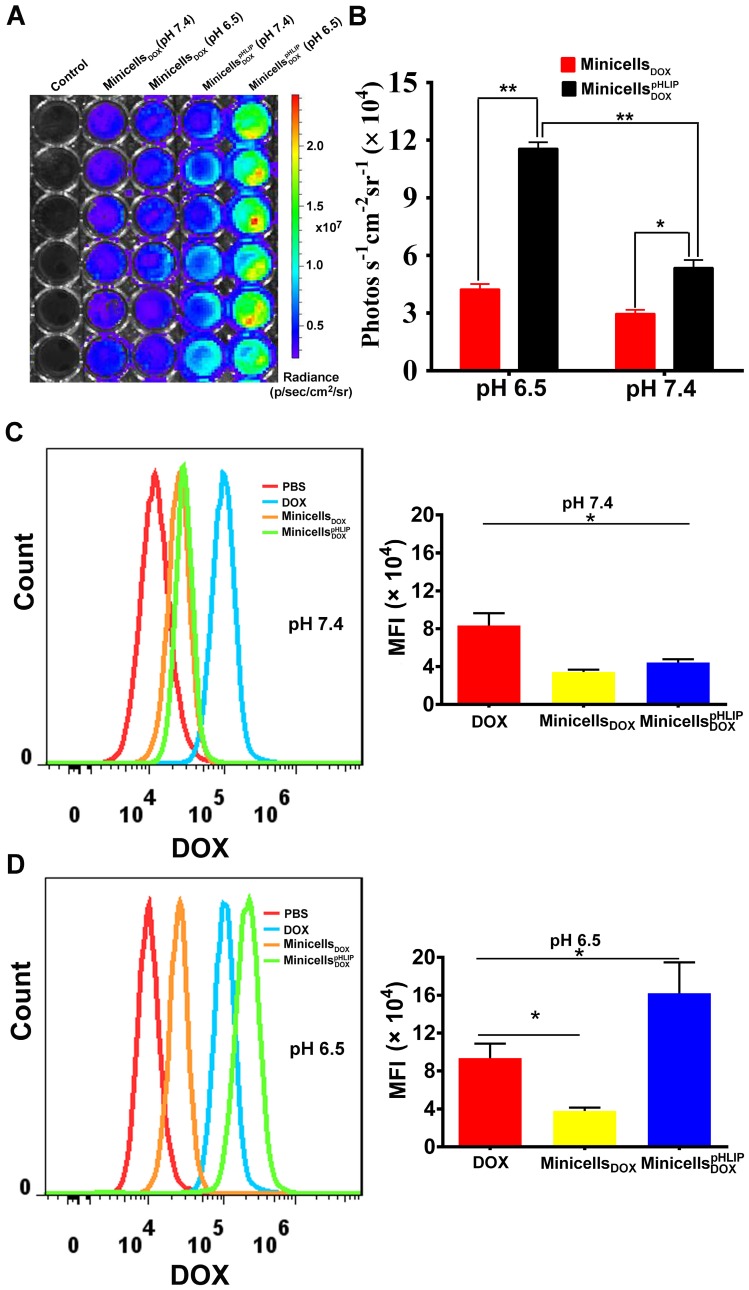
The ability of the minicellspHLIP DOX to target the acidic microenvironment of cancer cells was further evaluated using the IVIS Spectrum system. The minicells inherit the cytoplasmic components of the bacteria except the chromosomal DNA, indicating that the minicells possess the same bioluminescence as their parent cells. Indeed, the minicells displayed a strong bioluminescence signal (Figure S2), which greatly assisted in the quantification of minicells internalized by cancer cells in media of different pH values. As shown in Figure 6A, a significant difference in bioluminescence between the minicells DOX- and minicellspHLIP DOX-treated cells is recordable at pH 7.4 and at pH 6.5. However, the cancer cells in the minicellspHLIP DOX -treated group (11.56 ± 0.92 photos s-1 cm-2 sr-1) exhibited almost three times higher bioluminescence intensity than the cancer cells in the minicell DOX-treated group (4.05 ± 0.34 photos s-1 cm-2 sr-1) in the pH 6.5 medium (Figure 6B), which is in line with the findings from confocal laser scanning microscopy. To determine whether the DOX carried by minicellspHLIP DOX could successfully accumulate in cancer cells in solutions of different pH values, flow cytometry was used to analyze the fluorescence intensity of the chemotherapeutic drugs in the cells. After 2 h of incubation in the pH 7.4 solution, the fluorescence intensity of the minicellspHLIP DOX-treated group was almost the same as that of the minicellsDOX-treated group and two times lower than that of the DOX-treated group (Figure 6C). However, there was a sharp four-fold increase in the fluorescence intensity of the minicellspHLIP DOX-treated group in the pH 6.5 medium, while the fluorescence intensity of the DOX- and minicellsDOX-treated groups did not change significantly (Figure 6D). Moreover, the minicellspHLIP DOX-treated cells exhibited a 1.5-fold higher fluorescence intensity than the DOX-treated group in the low pH solutions (Figure 6D). The results further confirmed that low pH values can efficiently drive the entry of minicellspHLIP DOX into cancer cells, which is potentially useful for targeting acidic microenvironments of tumors in vivo for anticancer studies.
Figure 6.
Quantitative analysis of minicellspHLIP DOX internalization by 4T1 cells. Bioluminescence imaging (A) and signal quantification (B) of minicells DOX and minicellspHLIP DOX internalization by 4T1 cells after a 2 h co-incubation in the pH 6.5 and pH 7.4 media using the IVIS Spectrum system. Analysis of DOX uptake by 4T1 cells using flow cytometry after incubation with minicells DOX and minicellspHLIP DOX for 2 h in the pH 7.4 medium (C) and in the pH 6.5 medium(D); MFI stands for mean fluorescence intensity. The results are expressed as the mean ± SD of 3 independent experiments (*P < 0.05, **P < 0.01 in paired t-test).
Cellular localization of minicellspHLIP and selective cytotoxicity of minicellspHLIP DOX in vitro
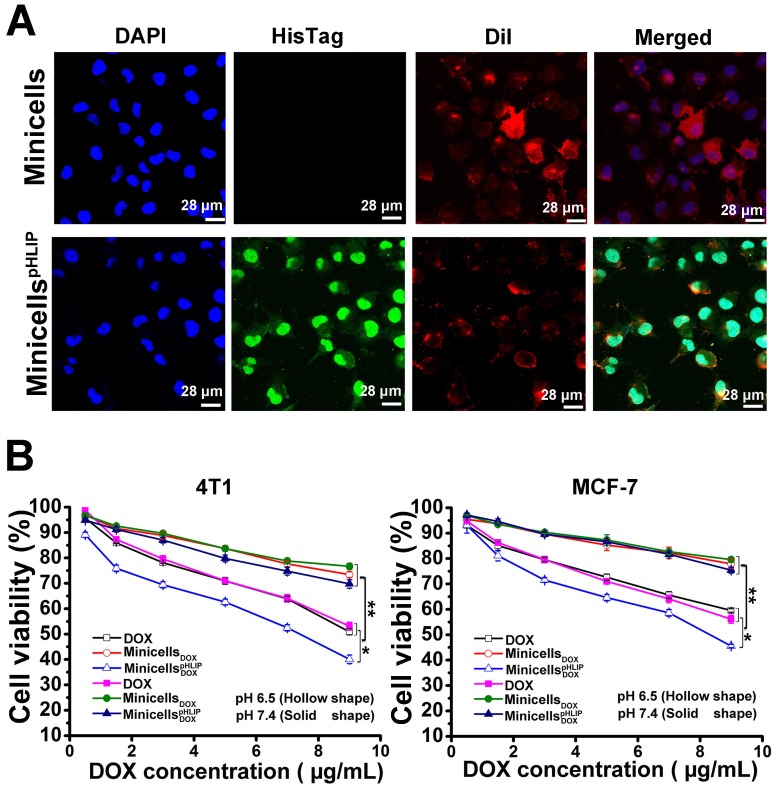
The IVIS Spectrum system detected the presence of minicells in the cancer cells by measuring the bioluminescence signal. The cellular localization of minicellspHLIP in the 4T1 cancer cells was identified by immunofluorescence. As shown in Figure 7A, the cells incubated with minicellspHLIP clearly exhibited green fluorescence (His-Tag antibody), while the minicell-treated group showed only red fluorescence (DiI for membrane staining) except for the DAPI fluorescence. This was observed because the green fluorescent protein-labeled antibody can indirectly bind to the six-His tag at the C-terminal of the pHLIP peptide, which is displayed on the cell membranes of minicellspHLIP. Although the green fluorescence was localized in the cytoplasm and nuclei of the cancer cells (Figure 7A), the nanocells have too large a volume to be able to penetrate the nuclear pore. Therefore, we can infer that the minicells localize in the cytoplasm, and the presence of green fluorescence in the nucleus should be due to the preference of the six-His-tag peptides from the degradation of minicellspHLIP in the cytoplasm to accumulate in the nucleus. DOX released from degradation of minicellspHLIP in the cytoplasm enter the nucleus to exhibit cytotoxic effects.
Figure 7.
Detection of minicellspHLIP in 4T1 cells and cytotoxicity of minicellspHLIP DOX and minicells DOX in solutions of different pH values. (A) Identification of minicellspHLIP DOX in 4T1 cells using immunofluorescence after incubation with 4T1 cells for 2 h in the pH 6.5 medium; minicells were used as the control group. (B) Inhibitory effects of minicells DOX and minicellspHLIP DOX on the growth of 4T1 and MCF-7 cells in the pH 6.5 (hollow shape) and pH 7.4 media (solid shape) after incubation for 24 h. The results are expressed as the mean ± SD of 3 independent experiments (*P < 0.05, **P < 0.01 in paired t-test).
Furthermore, mouse 4T1 breast cancer cells and human MCF-7 breast cancer cells were used to evaluate the in vitro cytotoxic effects of the engineered minicells. In agreement with the results obtained from the tumor-targeting assay in acidic microenvironments, the minicellspHLIP DOX and minicells DOX showed no significant differences in cytotoxicity to both the mouse and human breast cancer cells in neutral medium. The cell viability of the 4T1 and MCF-7 breast cancer cells treated with minicellspHLIP DOX or minicells DOX was more than 70% in the pH 7.4 medium (Figure 7B). However, in the acidic microenvironment (pH 6.5 medium), minicellspHLIP DOX exhibited higher cytotoxicity than minicells DOX against the 4T1 and MCF-7 cells. For example, the cell viability of the 4T1 cells was only 40.04% after 24 h of treatment with the minicellspHLIP DOXat the maximum concentration of DOX; the cell viability was much lower than that observed for the minicell DOX-treated group (73.42%). In addition, the cytotoxicity of minicellspHLIP DOX against the 4T1 cancer cells was also significantly higher than that of free DOX in the pH 6.5 medium. Additionally, the cell viability of MCF-7 cells in the minicellspHLIP DOX-treated group was only 45.56%, while that of the cells in the minicellsDOX-treated group reached 75.45% in the pH 6.5 medium (Figure 7B). In contrast, the two groups did not exhibit any significant difference in MCF-7 cell viability under the pH 7.4 condition. Moreover, the cell viability of the 4T1 and MCF-7 cancer cells in the DOX-treated group in the pH 6.5 medium and the pH 7.4 medium was not significantly different. Moreover, the minicellspHLIP DOX exhibited higher cytotoxic effects than DOX in the pH 6.5 medium. This demonstrated that the minicellspHLIP DOX not only selectively target cancer cells in acidic microenvironments but also kill the cells by releasing DOX.
MinicellspHLIP-mediated DOX delivery to orthotopic 4T1 breast cancer in vivo
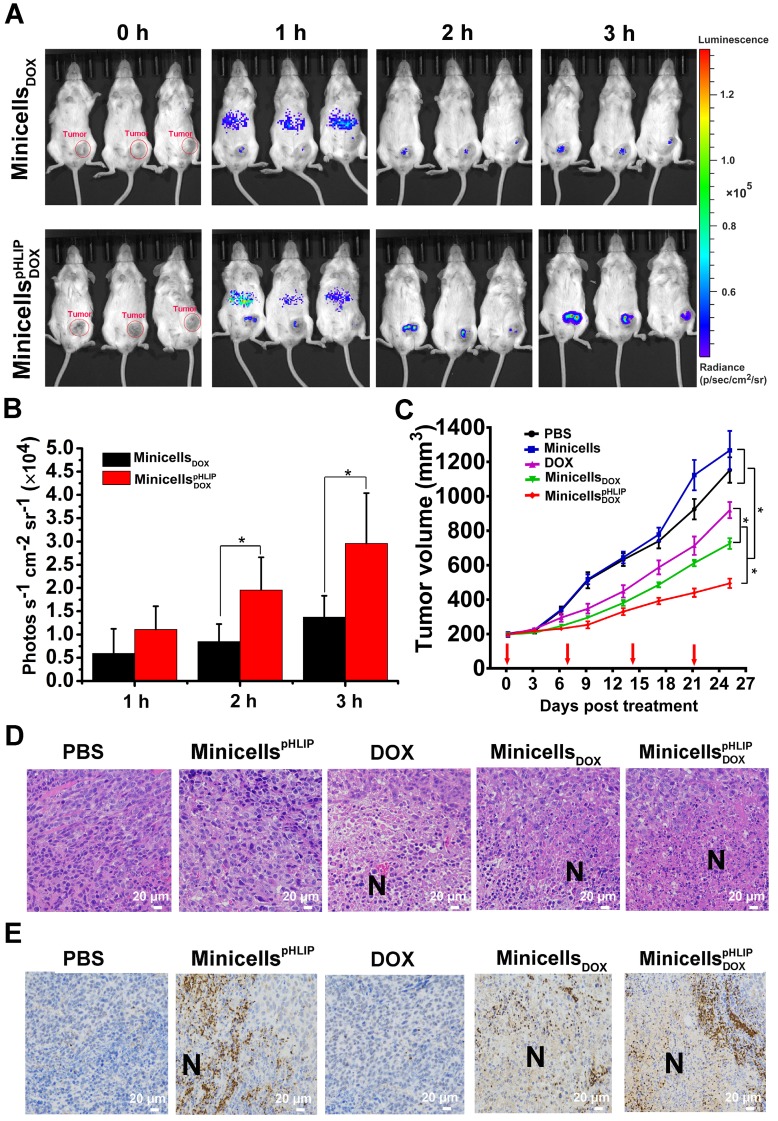
To verify the targeting ability of minicellspHLIP DOXto tumors in vivo, immunocompetent BALB/c mice with orthotopic 4T1 breast cancers were i.v. injected with minicellspHLIP DOX and minicells DOX. Owing to the bioluminescence of the minicells, the in vivo distribution of the minicells could be imaged using the IVIS Spectrum system at defined time points. As shown in Figure 8A, minicellspHLIP DOX and minicells DOX could rapidly enter tumors, but most minicells were retarded in the liver region of mice 1 h after i.v. administration. Over an extended period of time, the fluorescence intensity in the tumors clearly increased and peaked at 3 h after treatment; however, the fluorescence in the liver region rapidly vanished at 2 h (Figure 8A). Although there were no significant differences among the mice in the same group, the average fluorescence intensity of the minicellspHLIP DOX-treated group was two times higher than that of the minicell DOX-treated group in the experiments at 2 h and 3 h after treatment (Figure 8B). This finding indicated that the minicells with pHLIP have higher tumor-targeting abilities than the minicells without the peptide. The accumulation of minicellspHLIP DOX in cancers provides a solid platform on which to release DOX to kill cancer cells. The disappearance of minicells in the liver 1 h after treatment was most likely a result of clearance by the immune system. This phenomenon was also observed in the healthy BALB/c mice treated with live EcN. The bioluminescence signals peaked in the liver region at 1 h and quickly disappeared at 3 h after EcN treatment (Figure S3). Furthermore, the fluorescence disappeared in all the tested mice at 6 h after the minicells were i.v. injected (data not shown). This phenomenon probably occurred because the minicells were rapidly digested in tumors by various enzymes and engulfed by certain types of immune cells.
Figure 8.
Tumor-targeting ability, therapeutic effects and intra-tumor distribution of minicellspHLIP DOX. (A) 4T1-tumor-bearing BALB/c mice were i.v. administered with minicells DOX and minicellspHLIP DOX, and the bioluminescence signals from the mice were recorded using IVIS Spectrum system at defined time points. (B) Quantification of the bioluminescence signals from the IVIS Spectrum system after i.v. administration of minicells DOX and minicellspHLIP DOX at 1, 2 and 3 h. (C) BALB/c mice bearing 4T1 tumors were i.v. injected with PBS, minicellspHLIP, DOX, minicells DOX or minicellspHLIP DOX weekly (red arrows indicate the day of drug administration). The tumor volumes of the mice in each group (n = 8) were documented from the beginning of treatment. (D) H&E staining of tumor tissues. The tumors (each group has 3 mice) were separated at the end of drug treatment and fixed in 4% paraformaldehyde for H&E staining (400×). (E) Identification of the distribution of minicellspHLIP and minicellspHLIP DOX in tumor tissues by immunohistochemistry (400×). At 3 h after i.v. administration of PBS, minicellspHLIP, DOX, minicells DOX or minicellspHLIP DOX, the tumors were fixed in 4% paraformaldehyde for immunohistochemistry. Each group contains 3 mice. The images are representative of 3 similar results, and the results of the bar and line charts are expressed as the mean ± SD of 3 independent experiments (*P < 0.05 in paired t-test).
The therapeutic effects of minicellspHLIP DOX against cancers were evaluated using BALB/c mice with orthotopic 4T1 breast cancers. The tumor-bearing mice were treated by weekly i.v. administration of PBS, minicellspHLIP, DOX, minicells DOX or minicellspHLIP DOX for 25 days. As shown in Figure 8C, the minicellspHLIP DOX, minicells DOX and DOX showed significant inhibitory effects on the growth of 4T1 tumors compared with PBS- and minicells-treated groups. And, the minicellspHLIP DOX exhibited the best antitumor effects among the groups. At sacrifice, the average tumor volume of the minicellspHLIP DOX-treated group (494 mm3) corresponded to 42% of the PBS group and a 53% size reduction of the DOX group. Interestingly, the minicellsDOX (726 mm3) displayed higher anticancer effects than free DOX (919 mm3). This should be due to the EPR effects, by which the minicellsDOX can retain longer in the solid tumors than the free chemotherapeutic drug. The cumulative DOX in the solid tumors from the minicellsDOX group will be higher than the free DOX group, resulting in the significant difference of tumor regression. Moreover, the PBS group exhibited no significant differences than the minicells-treated group in terms of tumor regression. Previous studies have shown that live bacteria can restrain tumor growth by scrambling nutrients and stimulating immune response 18, 35. In this study, the minicells, which have no proliferation ability, could not inhibit the growth of 4T1 breast cancers without the help of DOX.
Histological examination of tumors was carried out at the end of the animal experiments. The tumor tissue revealed conspicuous necrotic extension in the minicellspHLIP DOX-treated groups while the DOX- and minicells DOX-treated groups showed a slight increase compared with the PBS- and minicells-treated groups (Figure 8D). Because of the lack of propagation, the minicells could not behave like their parent EcN cells, which could significantly extend the necrotic region. In this study, the amplification of necrosis in the tumors induced by the engineered minicells resulted from the cytotoxic effects of the DOX loaded in the minicells. Immunohistochemistry further revealed the distribution of the minicells in the tumor tissues. Figure 8E shows that the minicells with pHLIP appeared in the interfaces between necrotic and hypoxic regions and infiltrated into viable cancer cells. This distinguishing feature corresponds with the parent EcN that we described previously 18, 19. Generally, chemotherapeutic drugs cannot access the hypoxic regions because of the considerable amount of hindrance to interstitial transport in solid tumors 3, 36. In this study, the EcN-derived minicellspHLIP with DOX successfully invade the necrotic and hypoxic regions, similar to their parent EcN cells, guaranteeing that the loaded chemotherapeutic drugs could be released into the hypoxic regions of tumors.
The biosafety of the minicellspHLIP was preliminarily evaluated by measuring the weight loss of the mice and by pathological tissue analysis. The body weights of the BALB/c mice were recorded every 2 days during the treatment. The weekly i.v. administration of the minicellspHLIP did not affect the body weights of the mice or key organs compared with PBS (Figure S4A and B). The minicellspHLIP-treated mice behaved like the PBS-treated group, and no death occurred during the experiments. Also, no serious anaphylactic activity was found except for nose scratching during the first hour after minicellspHLIP administration. Additionally, histological analysis revealed that the key metabolic organ (liver), the vital immune organ (spleen) and the main organ of drug excretion (kidney) displayed no distinct difference between the minicellspHLIP-treated group and the PBS-treated group, as seen by the H&E staining assay (Figure S4C). The results demonstrated that the EcN-derived minicellspHLIP was not noticeably toxic to the mice.
Discussion
Because of physiological characteristics of solid tumors, most anticancer drugs, including free drugs and nanoparticles, cannot reach the hypoxic regions of solid tumors. In this study, we fabricated a tumor-targeting minicell via genetic engineering based on the probiotics EcN and pHLIP for targeted delivery of chemotherapeutic drugs to the hypoxic regions of solid tumors. Knocking out minCD and overexpression of minE in EcN resulted in the scalable production of minicells. Using the Lpp-OmpA' protein display system, pHLIP, with an ability to target acidic microenvironments, was successfully anchored to the membrane surfaces of the minicells. The minicellspHLIP could be produced at a very low cost via biological fermentation and used for drug delivery without any additional modification. Based on the acidic characteristics of solid tumors, pHLIP could efficiently enhance the internalization of minicells by cancer cells. Both the in vitro and in vivo experiments demonstrated that the DOX-loaded minicellspHLIP could selectively target the acidic microenvironments of tumors and invade the hypoxic regions of solid tumors to kill the cancer cells by releasing the drug; no serious pathogenic toxicity was observed during the animal experiments.
Our previous studies had revealed that the minicell parent strain, EcN, displayed high tumor-targeting ability and anticancer effects by expressing anticancer proteins 17, 18. However, the live bacterium has too great of an infection risk to be used in cancer therapy. In contrast, the EcN-derived minicells, which lack the ability to proliferate, inherit all the characteristics of their parent strain and can act as tumor-targeting drug vectors for cancer therapy. Therefore, we innovatively use a protein display system to exhibit the acid-targeting ligand pHLIP on the membrane surfaces of EcN-derived minicells to construct a tumor-targeting drug carrier for delivering drugs into the hypoxic regions of tumors. The peptide pHLIP, consisting of 37 amino acids, can form an inducible transmembrane α-helix under acidic conditions and can translocate nanoparticles into cells via a non-endocytic route 37. The typical acidity of solid tumors provides a low-pH environment for pHLIP to efficiently translocate exogenous substances into cancer cells. This response of pHLIP to acidic environments could greatly increase the accumulation of liposomes, RNA and other nanoparticles in solid tumors 25-27. In this study, minicells with pHLIP also displayed higher tumor-targeting abilities than the minicells without the peptide. Additionally, the minicellspHLIP successfully invaded the hypoxic regions in spite of the poorly vascularized microenvironment and high interstitial fluid pressure of solid tumors. This model provides an example of the potential application of bacteria-derived minicells and numerous ligands against tumor-associated markers, such as pHLIP, for tumor-targeted therapy. Moreover, the integration of the LuxCDABE gene cluster into the minicells makes it possible to detect the in vivo distribution of the nanocells after i.v. administration in real-time. This method is also very useful for recording the metabolism kinetics of the EcN-derived minicells and for evaluating the potential toxicity of the minicells.
To date, the S. typhimurium-derived minicells with an EGFR antibody has achieved great success in phase I clinical trials 16. In contrast to S. typhimurium and other bacteria, EcN displays high tumor-targeting ability and the specific ability to colonize in the interface between the necrotic and hypoxic regions 17, 18. Recent reports have focused on the application of the minicells derived from S. typhimurium to carry chemotherapeutic drugs for cancer therapy 13, 16. To construct a tumor-targeting minicell of S. typhimurium with low immunogenicity, the LPS must be removed to reduce inflammation: the tumor-targeting antibodies were mounted on the membrane surface through weak noncovalent interactions after minicell extraction 11, 13. However, the EcN-derived minicells with serum-sensitive characteristics and immune regulatory function ensures that they will be rapidly purged from normal organs and they induce low levels of inflammation 22, 38. Moreover, a protein display system was creatively used to securely anchor the tumor-targeting ligand to the membrane surfaces of minicells during the cell production. The results confirmed that the engineered minicells extracted from the fermentation broth could be directly used to deliver chemotherapeutic drugs, being free from the requirement of LPS purging, and that the minicells could successfully penetrate into the hypoxic regions of tumors by linking the minicells to tumor-targeting ligands. Considering the specific penetration ability of the minicells into the centers of tumors, the mechanisms underlying the activity and drug-loading capacity for other chemotherapeutic drugs will be further investigated in the near future.
In summary, we provide a novel strategy to prepare nanoscale biological carriers with tumor-targeting abilities, tumor penetrability, low toxicity and biodegradability at low cost using genetic engineering and biological fermentation. The minicells could be modified with most ligands of tumor-associated markers and formed tumor-targeting vectors. Alternatively, the strategy has the potential to be applied in other species, such as yeasts or microvesicles of human cells, to screen for ideal vectors for targeted delivery of chemotherapeutic drugs to hypoxic regions for cancer therapy.
Acknowledgments
This work was supported by the National Basic Research Program (973) of China (2012CB722301), the National High Technology Research and Development program (863) of China (2011AA10A203), the International Cooperation Project (0102011DFA32610), the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (20134486) and the National Natural Science Foundation of China (31770106, 81501538), the China Postdoctoral Science Foundation (2016M593035) and Jiangsu Planned Projects for Postdoctoral Research Funds (1501122c).
Abbreviations
- ATCC
American Type Culture Collection
- BSA
bovine serum albumin
- BSG
buffered saline gelatin
- Cm
chloromycetin
- DOX
doxorubicin
- EcN
E. coli Nissle 1917
- EGFR
epidermal growth factor receptor
- EPR
enhanced permeability and retention
- GFP
green fluorescent protein
- H&E
hematoxylin and eosin
- HER2
human epidermal growth factor receptor 2
- i.v.
intravenously
- LB
lysogeny broth
- LPS
lipopolysaccharide
- MFI
mean fluorescence intensity
- PBS
phosphate-buffered saline
- PVDF
polyvinylidene fluoride
- pHLIP
pH (low) insertion peptide
- RBS
ribosome binding site
- SPF
specific-pathogen-free
- Tet
tetracycline
- TPBS
PBS supplemented with 0.5% (v/v) Tween 20.
Supplementary Material
Supplementary methods, figures and tables.
References
- 1.Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307(5706):58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 2.Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat Rev Cancer. 2008;8(6):425–37. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heldin CH, Rubin K, Pietras K, Ostman A. High interstitial fluid pressure - an obstacle in cancer therapy. Nat Rev Cancer. 2004;4(10):806–13. doi: 10.1038/nrc1456. [DOI] [PubMed] [Google Scholar]
- 4.Drummond DC, Meyer O, Hong K, Kirpotin DB, Papahadjopoulos D. Optimizing liposomes for delivery of chemotherapeutic agents to solid tumors. Pharmacol Rev. 1999;51(4):691–743. [PubMed] [Google Scholar]
- 5.Yang E, Qian W, Cao Z, Wang L, Bozeman EN, Ward C. et al. Theranostic nanoparticles carrying doxorubicin attenuate targeting ligand specific antibody responses following systemic delivery. Theranostics. 2015;5(1):43–61. doi: 10.7150/thno.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng T, Liu J, Ren J, Huang F, Ou H, Ding Y. et al. Green Tea Catechin-Based Complex Micelles Combined with Doxorubicin to Overcome Cardiotoxicity and Multidrug Resistance. Theranostics. 2016;6(9):1277–92. doi: 10.7150/thno.15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muthu MS, Leong DT, Mei L, Feng SS. Nanotheranostics - application and further development of nanomedicine strategies for advanced theranostics. Theranostics. 2014;4(6):660–77. doi: 10.7150/thno.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.England CG, Gobin AM, Frieboes HB. Evaluation of uptake and distribution of gold nanoparticles in solid tumors. Eur Phys J Plus; 2015. p. 130. (11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chauhan VP, Jain RK. Strategies for advancing cancer nanomedicine. Nat Mater. 2013;12(11):958–62. doi: 10.1038/nmat3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishida T, Kiwada H. Alteration of tumor microenvironment for improved delivery and intratumor distribution of nanocarriers. Biol Pharm Bull. 2013;36(5):692–7. doi: 10.1248/bpb.b13-00121. [DOI] [PubMed] [Google Scholar]
- 11.MacDiarmid JA, Langova V, Bailey D, Pattison ST, Pattison SL, Christensen N. et al. Targeted Doxorubicin Delivery to Brain Tumors via Minicells: Proof of Principle Using Dogs with Spontaneously Occurring Tumors as a Model. PloS one. 2016;11(4):e0151832. doi: 10.1371/journal.pone.0151832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacDiarmid JA, Amaro-Mugridge NB, Madrid-Weiss J, Sedliarou I, Wetzel S, Kochar K. et al. Sequential treatment of drug-resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nat Biotechnol. 2009;27(7):643–51. doi: 10.1038/nbt.1547. [DOI] [PubMed] [Google Scholar]
- 13.MacDiarmid JA, Mugridge NB, Weiss JC, Phillips L, Burn AL, Paulin RP. et al. Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics. Cancer cell. 2007;11(5):431–45. doi: 10.1016/j.ccr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Yu XC, Margolin W. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J Bacteriol. 2000;182(21):6203–13. doi: 10.1128/jb.182.21.6203-6213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDiarmid JA, Madrid-Weiss J, Amaro-Mugridge NB, Phillips L, Brahmbhatt H. Bacterially-derived nanocells for tumor-targeted delivery of chemotherapeutics and cell cycle inhibitors. Cell cycle. 2007;6(17):2099–105. doi: 10.4161/cc.6.17.4648. [DOI] [PubMed] [Google Scholar]
- 16.Solomon BJ, Desai J, Rosenthal M, McArthur GA, Pattison ST, Pattison SL. et al. A First-Time-In-Human Phase I Clinical Trial of Bispecific Antibody-Targeted, Paclitaxel- Packaged Bacterial Minicells. PloS one. 2015;10(12):e0144559. doi: 10.1371/journal.pone.0144559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stritzker J, Weibel S, Hill PJ, Oelschlaeger TA, Goebel W, Szalay AA. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol. 2007;297(3):151–62. doi: 10.1016/j.ijmm.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Zhang Y, Xia L, Zhang X, Ding X, Yan F. et al. Escherichia coli Nissle 1917 targets and restrains mouse B16 melanoma and 4T1 breast tumors through expression of azurin protein. Appl Environ Microbiol. 2012;78(21):7603–10. doi: 10.1128/AEM.01390-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westphal K, Leschner S, Jablonska J, Loessner H, Weiss S. Containment of tumor-colonizing bacteria by host neutrophils. Cancer Res. 2008;68(8):2952–60. doi: 10.1158/0008-5472.CAN-07-2984. [DOI] [PubMed] [Google Scholar]
- 20.Sturm A, Rilling K, Baumgart DC, Gargas K, Abou-Ghazale T, Raupach B. et al. Escherichia coli Nissle 1917 distinctively modulates T-cell cycling and expansion via toll-like receptor 2 signaling. Infect Immun. 2005;73(3):1452–65. doi: 10.1128/IAI.73.3.1452-1465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arribas B, Rodriguez-Cabezas ME, Camuesco D, Comalada M, Bailon E, Utrilla P. et al. A probiotic strain of Escherichia coli, Nissle 1917, given orally exerts local and systemic anti-inflammatory effects in lipopolysaccharide-induced sepsis in mice. Br J Pharmacol. 2009;157(6):1024–33. doi: 10.1111/j.1476-5381.2009.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grozdanov L, Zahringer U, Blum-Oehler G, Brade L, Henne A, Knirel YA. et al. A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J Bacteriol. 2002;184(21):5912–25. doi: 10.1128/JB.184.21.5912-5925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11(9):671–7. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 24.Andreev OA, Engelman DM, Reshetnyak YK. Targeting diseased tissues by pHLIP insertion at low cell surface pH. Front Physiol. 2014;5:97. doi: 10.3389/fphys.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C. et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–10. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao L, Daniels J, Wijesinghe D, Andreev OA, Reshetnyak YK. pHLIP(R)-mediated delivery of PEGylated liposomes to cancer cells. J Control Release. 2013;167(3):228–37. doi: 10.1016/j.jconrel.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Dang M, Tian Y, Zhu Y, Liu W, Tian W. et al. Tumor Acidic Microenvironment Targeted Drug Delivery Based on pHLIP-Modified Mesoporous Organosilica Nanoparticles. ACS Appl Mater Interfaces. 2017;9(36):30543–52. doi: 10.1021/acsami.7b10840. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Sarov M, Rientjes J, Fu J, Hollak H, Kranz H. et al. An improved recombineering approach by adding RecA to lambda Red recombination. Molecular biotechnology. 2006;32(1):43–53. doi: 10.1385/mb:32:1:043. [DOI] [PubMed] [Google Scholar]
- 29.Bruckbauer ST, Kvitko BH, Karkhoff-Schweizer RR, Schweizer HP. Tn5/7-lux: a versatile tool for the identification and capture of promoters in gram-negative bacteria. BMC microbiology. 2015;15:17. doi: 10.1186/s12866-015-0354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daugherty PS, Olsen MJ, Iverson BL, Georgiou G. Development of an optimized expression system for the screening of antibody libraries displayed on the Escherichia coli surface. Protein Eng. 1999;12(7):613–21. doi: 10.1093/protein/12.7.613. [DOI] [PubMed] [Google Scholar]
- 31.Howe K, Karsi A, Germon P, Wills RW, Lawrence ML, Bailey RH. Development of stable reporter system cloning luxCDABE genes into chromosome of Salmonella enterica serotypes using Tn7 transposon. BMC microbiology. 2010;10:197. doi: 10.1186/1471-2180-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Netzker T, Schroeckh V, Gregory MA, Flak M, Krespach MK, Leadlay PF. et al. An Efficient Method To Generate Gene Deletion Mutants of the Rapamycin-Producing Bacterium Streptomyces iranensis HM 35. Appl Environ Microbiol. 2016;82(12):3481–92. doi: 10.1128/AEM.00371-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao L, Daniels J, Moshnikova A, Kuznetsov S, Ahmed A, Engelman DM. et al. pHLIP peptide targets nanogold particles to tumors. Proc Natl Acad Sci U S A. 2013;110(2):465–70. doi: 10.1073/pnas.1219665110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochemical pharmacology. 1999;57(7):727–41. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 35.Forbes NS, Munn LL, Fukumura D, Jain RK. Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Cancer Res. 2003;63(17):5188–93. [PubMed] [Google Scholar]
- 36.Ferretti S, Allegrini PR, Becquet MM, McSheehy PM. Tumor interstitial fluid pressure as an early-response marker for anticancer therapeutics. Neoplasia. 2009;11(9):874–81. doi: 10.1593/neo.09554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thevenin D, An M, Engelman DM. pHLIP-mediated translocation of membrane-impermeable molecules into cells. Chemistry & biology. 2009;16(7):754–62. doi: 10.1016/j.chembiol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guzy C, Paclik D, Schirbel A, Sonnenborn U, Wiedenmann B, Sturm A. The probiotic Escherichia coli strain Nissle 1917 induces gammadelta T cell apoptosis via caspase- and FasL-dependent pathways. International immunology. 2008;20(7):829–40. doi: 10.1093/intimm/dxn041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods, figures and tables.